SUMMARY
Mdm2 E3 ubiquitin ligase-mediated p53 degradation is generally accepted as the major mechanism for p53 regulation; nevertheless, the in vivo significance of this function has not been unequivocally established. Here, we have generated an Mdm2Y487A knock-in mouse; Mdm2Y487A mutation inactivates Mdm2 E3 ligase function without affecting its ability to bind its homologue MdmX. Unexpectedly, Mdm2Y487A/Y487A mice were viable and developed normally into adulthood. While disruption of Mdm2 E3 ligase function resulted in p53 accumulation, p53 transcriptional activity remained low; however, exposure to sub-lethal stress resulted in hyperactive p53 and p53-dependent mortality in Mdm2Y487A/Y487A mice. These findings reveal a potentially dispensable nature for Mdm2 E3 ligase function in p53 regulation, providing insight that may affect how this pathway is targeted therapeutically.
INTRODUCTION
Classically, p53 has been characterized as a tumor suppressor, functioning as a transcription factor for a number of genes important in the regulation of cell cycle arrest, apoptosis, and senescence (Levine, 1997). In more recent years, a multitude of research has emerged, demonstrating p53 to also be critical in meiosis, reproduction, and metabolism, further broadening the role of p53 (Levine et al., 2011; Lu et al., 2010; Vousden and Ryan, 2009).
The oncoprotein Mdm2 is generally accepted to function as the primary negative regulator of p53 (Kruse and Gu, 2009; Wade et al., 2010). The RING finger domain of Mdm2 confers its E3 ubiquitin ligase activity, and through this function Mdm2 controls p53 stability, targeting it for export from the nucleus and subsequent degradation by the proteasome (Haupt et al., 1997; Honda et al., 1997; Kubbutat et al., 1997). In addition to regulating p53 protein levels, Mdm2 also regulates p53 activity by binding via its N-terminus directly to the p53 transactivation domain (Momand et al., 1992; Oliner et al., 1993). In vivo, deletion of Mdm2 results in high levels of p53 protein and transcriptional activity, and early embryonic lethality in Mdm2−/− mice; this phenotype is rescued by concomitant deletion of Trp53, further demonstrating the essential role of Mdm2 in p53 regulation (Jones et al., 1995; Montes de Oca Luna et al., 1995).
MdmX, an Mdm2 homologue, is similarly capable of binding at its N-terminus to the p53 transactivation domain to control p53 activity, however, MdmX does not harbor intrinsic E3 ubiquitin ligase activity (Jackson and Berberich, 2000; Poyurovsky et al., 2007; Shvarts et al., 1996). Similar to Mdm2−/− mice, p53-dependent early embryonic lethality is also observed in Mdmx−/− mice (Parant et al., 2001). While the early embryonic lethality observed in both Mdm2 and Mdmx KO mouse models suggests that individually Mdm2 and MdmX play unique and critical roles in the regulation of p53 activity; the embryonic lethality observed in two recently developed Mdmx knock-in mouse models that disrupt Mdm2:MdmX heterodimerization suggests that the ability of these two proteins to interact at their respective C-terminal RING finger domains and function collaboratively is essential for p53 regulation (Huang et al., 2011; Pant et al., 2011).
Mdm2:MdmX heterodimerization is also disrupted by mutation of the Mdm2 RING finger domain, as observed in the Mdm2C462A knock-in mouse model (Clegg et al., 2012; Itahana et al., 2007). This mutation substitutes cysteine 462 of murine Mdm2 to alanine, thus disrupting Mdm2:MdmX interaction and ablating Mdm2 E3 ligase activity, but maintaining Mdm2:p53 interaction. Homozygous Mdm2C462A mutation results in early embryonic lethality due to excessive p53 activation, suggesting that Mdm2:p53 binding alone is insufficient for controlling p53 activity, and calling into question the extent to which Mdm2:p53 interaction can regulate p53 activity in vivo (Itahana et al., 2007). Because both Mdm2 E3 ligase activity and Mdm2:MdmX heterodimerization are disrupted by the Mdm2C462A mutation, the independent effect of loss of each of these RING finger domain functions cannot be deduced from this model. Therefore, the in vivo role of Mdm2 E3 ubiquitin ligase activity, believed to be a critical Mdm2 function in p53 regulation, is yet to be unequivocally addressed. We carried out the current study to address this question.
RESULTS
Mdm2Y487A/Y487A Mice Are Viable and Phenotypically Normal
In vitro, the extreme C-terminal 5 amino acids of Mdm2 are necessary for E3 ubiquitin ligase activity; mutations within this region disrupt Mdm2 E3 ligase activity, without affecting its ability to homo-oligomerize with Mdm2 or hetero-oligomerize with MdmX (Poyurovsky et al., 2007; Uldrijan et al., 2007). Specifically, retention of an aromatic amino acid at the residue 489 of human Mdm2 is essential for Mdm2 E3 ligase activity, and Tyr-to-Ala mutation (Y489A) ablates Mdm2 E3 ligase activity without affecting its interaction with MdmX (Uldrijan et al., 2007). In vitro characterization of the mouse Mdm2Y487A recapitulated the published study of the corresponding human Mdm2Y489A; expression of both the human and mouse mutant Mdm2 in U2OS cells correlated with accumulation of ectopically expressed and endogenous p53 (Figure S1A–D). Furthermore, co-overexpression of MdmX with Mdm2Y487A resulted in p53 levels similar to that observed in the presence of WT Mdm2, in agreement with observations made in the human Mdm2Y489A, and indicative of a potential role for MdmX in improving, and at times rescuing, Mdm2 E3 ligase activity (Okamoto et al., 2009; Poyurovsky et al., 2007; Uldrijan et al., 2007). Titration of MdmX demonstrated that coexpression of MdmX with mutant Mdm2 at a 1:1 ratio reduced p53 stabilization but when MdmX was coexpressed with Mdm2 at a 1:5 or 1:10 ratio, similar to ratios previously observed in human cells (Wang et al., 2009; Wang et al., 2007), a greatly diminished effect on p53 stability was noted, pointing to the importance of the relative levels of Mdm2 and MdmX in their function (Figure S1E–F).
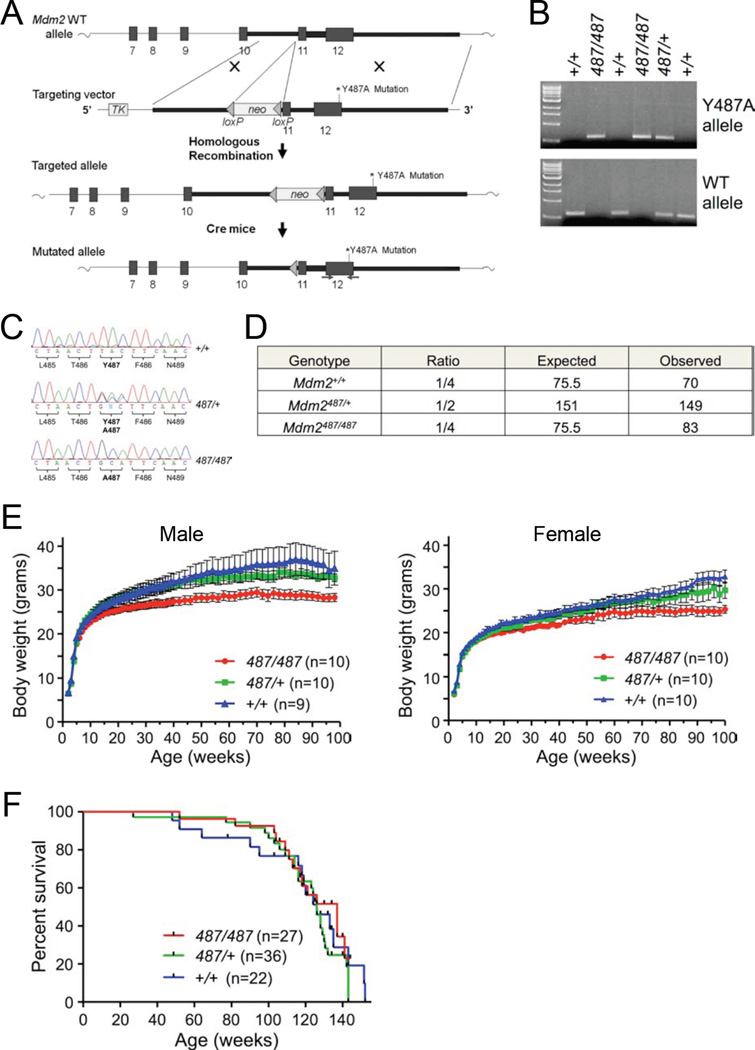
To assess if the Mdm2Y487A mutant maintains this phenotype in vivo, site directed mutagenesis was used to introduce a Tyr 487-to-Ala substitution in mouse genomic Mdm2 (Figure 1A). Correct substitution of TAC to GCA in exon 12 of Mdm2 was confirmed via DNA sequencing. The targeting vector containing the Mdm2 Y487A substitution was injected into 129/Sv embryonic stem (ES) cells, and the ES cells were then microinjected into C57BL/6 blastocysts to produce chimeras. Following mating of chimeric male mice with C57BL/6 females, PCR (Figure 1B) and sequencing (Figure 1C) of mouse genomic DNA was used to confirm germline transmission of the Mdm2Y487A mutation. The neomycin cassette was removed from the neo-Mdm2Y487A/+ mice by mating against an EIIa-Cre transgenic mouse, which targets expression of Cre recombinase early in mouse embryogenesis, providing fairly ubiquitous excision of the marker from the targeted allele in the mouse. Further intercrossing of Cre-treated mice was performed to outbreed the Cre-transgene, followed by five generations of backcrossing to C57BL/6 to generate mutant mice with a 97% C57BL/6 genetic background.
Figure 1. Generation of Mdm2Y487A Knock-in Mice.
(A) Schematic representation of the Mdm2 targeting vector. Solid boxes represent regions of vector homology to the target locus; thick numbered boxes represent the last six Mdm2 exons. Triangles indicate the presence and orientation of loxP sites. Site directed mutagenesis was used to introduce the Tyr-to-Ala mutation at the 487 amino acid residue.
(B) PCR amplification of genomic DNA isolated from toe snips of mouse pups.
(C) Sequencing analysis of genomic DNA isolated from offspring of Mdm2Y487A/+ heterozygous intercrosses. DNA was PCR amplified (primer location indicated with arrows in A) and sequenced.
(D) Observed and expected birth ratios from Mdm2Y487A/+ intercrosses; total of 302 offspring observed.
(E) Mdm2+/+, Mdm2Y487A/+, and Mdm2Y487A/Y487A mice were weighed and observed weekly. Data represent means +/− SEM.
(F) Mdm2+/+, Mdm2Y487A/+, and Mdm2Y487A/Y487A male and female mice were observed weekly for activity and viability. Kaplan-Meier survival curve is shown.
See also Figure S1.
Mice heterozygous for Mdm2Y487A were viable, and appeared phenotypically normal and fertile. Surprisingly, upon intercrossing Mdm2Y487A/+ mice, Mdm2Y487A/Y487A mice were observed at normal Mendelian ratios at birth, indicating that no embryonic lethality is associated with homozygosity of this mutation (Figure 1D). To determine if the mutation had effects later in development, a cohort of male and female mice was assembled and monitored weekly for weight, activity, and overt signs of disease. Both male and female homozygous mutant mice appeared to be somewhat smaller than their heterozygous and wild type (WT) counterparts (Figure 1E). Mdm2Y487A/Y487A mice did not demonstrate any notable differences in disease occurrence, spontaneous tumor formation, or mortality compared to WT mice (Figure 1F).
Endogenous Mdm2Y487A Does Not Degrade p53 but Maintains Normal Binding Dynamics
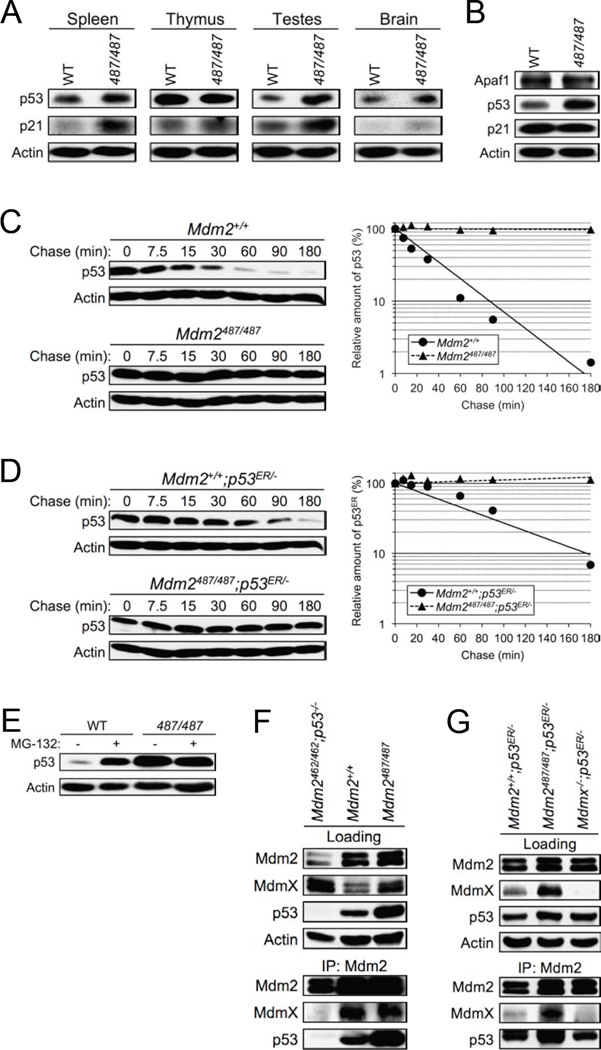
The viability and apparent normality of Mdm2Y487A/Y487A mice was surprising; thus, we examined endogenous Mdm2Y487A to determine its E3 ligase function and its ability to interact with MdmX. In some tissue types, such as the spleen and testes, and in mouse embryonic fibroblasts (MEFs), increased levels of p53 were observed in the Mdm2Y487A/Y487A mutant compared to the WT, suggestive of disrupted Mdm2 E3 ligase activity for p53 degradation; this increase was more dramatic in MEFs compared to tissues (Figure 2A–B). While somewhat higher p21 levels were observed in some Mdm2Y487A/Y487A tissues, potentially indicative of higher p53 activity, in Mdm2Y487A/Y487A MEFs, elevated levels of p53 did not lead to increased levels of the p53 targets p21 and Apaf1, suggesting that p53 transcriptional activity is not increased in Mdm2Y487A/Y487A MEFs.
Figure 2. Mdm2Y487A Is Unable to Degrade p53, but Maintains Normal Binding Dynamics.
(A) Lysates from the spleen, thymus, testes, and brain of 8-week old Mdm2+/+ (WT) and Mdm2Y487A/Y487A (487/487) mice were analyzed by western blotting for p53 and p21 levels. Actin was used as a loading control.
(B) Early passage Mdm2+/+ (WT) and Mdm2Y487A/Y487A (487/487) MEF cell lysates were analyzed by western blotting for Apaf1, p53 and p21 levels. Actin was used as a loading control.
(C) Early passage Mdm2+/+ and Mdm2Y487A/Y487A MEFs were treated with 100 µg/mL cycloheximide and harvested using SDS lysis buffer at the time points indicated. The levels of p53 and actin were analyzed by western blotting. The amount of p53 was quantified by densiometry, normalized to the level of actin, and plotted.
(D) Levels of p53ER were measured and analyzed as in C.
(E) Mdm2+/+ (WT) and Mdm2Y487A/Y487A (487/487) MEFs were treated with 25 µM MG-132 for 5 hr as indicated. Lysates were collected and the level of p53 was analyzed by western blotting. Actin was used as a loading control.
(F) Mdm2+/+ and Mdm2Y487A/Y487A MEFs were immunoprecipitated and immunoblotted with the indicated antibodies. Loading represents 1% of total cell lysate utilized for IP. Mdm2C462A/C462A;p53−/− MEFs were utilized as a negative control for Mdm2:MdmX binding.
(G) Mdm2+/+;p53ER/− and Mdm2Y487A/Y487A;p53ER/− MEFs were immunoprecipitated and immunoblotted with the indicated antibodies as in F, MdmX−/−;p53ER/− MEFs were utilized as a negative control for Mdm2:MdmX binding.
See also Figure S2.
To more directly analyze the E3 ligase activity of Mdm2Y487A for p53, a half-life assay using the protein synthesis inhibitor cycloheximide was performed in Mdm2+/+ and Mdm2Y487A/Y487A MEFs. In the presence of WT Mdm2, p53 has a half-life of approximately 30 minutes; however, in the presence of only Mdm2Y487A, no degradation of p53 was observed (Figure 2C). Further, following treatment with the proteasome inhibitor MG-132, p53 was significantly accumulated in WT MEFs, whereas in Mdm2Y487A/Y487A MEFs no further accumulation of p53 was observed (Figure 2E). The increased levels of p53 under unstressed conditions and the lack of p53 accumulation by MG-132 treatment in Mdm2Y487A/Y487A MEFs suggest that loss of Mdm2 E3 ligase activity significantly disrupts proteasome-mediated degradation of p53, and supports the role of Mdm2 as the primary E3 ubiquitin ligase for p53 degradation in vivo. In agreement with previous in vitro studies showing that Mdm2Y489A does not degrade p53 (Poyurovsky et al., 2007; Uldrijan et al., 2007), this data demonstrates that endogenous Mdm2Y487A has similarly lost this function.
While Mdm2Y487A/Y487A mice are viable in the presence of p53, in order to directly compare this mutant to the embryonic lethal Mdm2C462A/C462A mutant (Itahana et al., 2007), an inducible p53 background, p53ERTAM (p53ER hereafter), was utilized. In the p53ER background, endogenous p53 is substituted with a p53 fusion protein, providing the ability for p53 to be turned “on” and “off” in the presence and absence of 4-hydroxytamoxifen (4-OHT), respectively (Christophorou et al., 2005). Similar to MEFs harboring the Mdm2C462A RING domain mutant, half-life assay of Mdm2Y487A/Y487A;p53ER/− MEFs demonstrated no degradation of p53ER (Figure 2D). Furthermore, as demonstrated by immunoprecipitation of Mdm2 in MEFs, Mdm2Y487A mutant protein maintains the ability to bind to MdmX, p53, and p53ER (Figure 2F–G).
Mdm2 has previously been identified as the E3 ligase responsible for MdmX degradation (Kawai et al., 2003; Linke et al., 2008; Pan and Chen, 2003), and as expected, in the presence of Mdm2Y487A, stabilization of MdmX is observed (Figure 2F–G). Cycloheximide mediated half-life assay indicated that the degradation of MdmX is indeed attenuated in the presence of Mdm2Y487A (Figure S2A–B). Moreover, while total protein levels of p53, Mdm2, and MdmX are increased in Mdm2Y487A/Y487A MEFs, the subcellular distribution of each of these proteins remained similar to that observed in WT MEFs (Figure S2C–F). Of note, although Mdm2 and MdmX levels are increased in the presence of Mdm2Y487A, this increase does not appear to alter the Mdm2:MdmX ratio sufficiently to result in efficient degradation of p53 (Figure 2C–E).
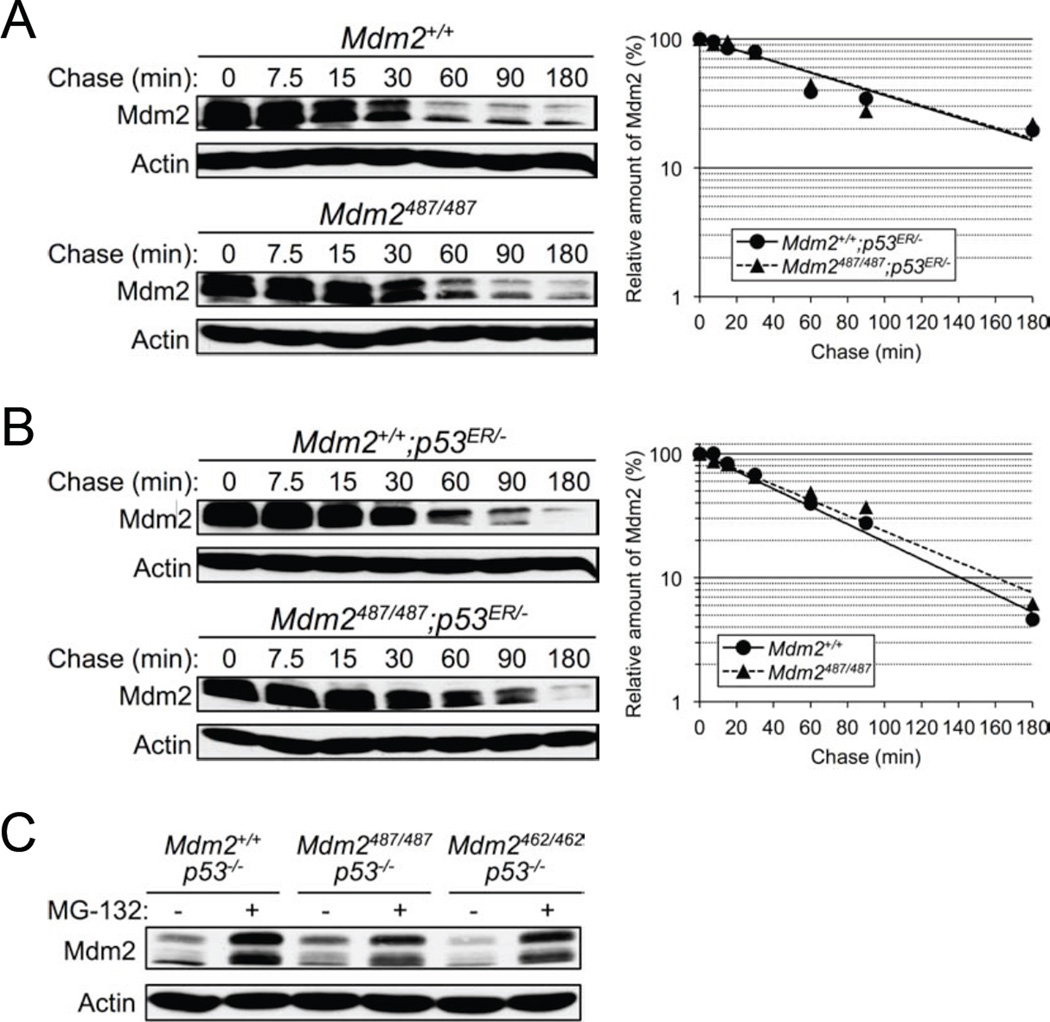
Y487A Mutation Does Not Affect Mdm2 Degradation
Regulation of Mdm2 stability is believed to be mediated primarily through autoubiquitination (Fang et al., 2000; Honda and Yasuda, 2000; Stommel and Wahl, 2004); however, the advent of the Mdm2C462A knock-in mouse provided strong evidence suggesting that Mdm2 does not in fact autoubiquitinate in vivo, but that instead an unknown E3(s) is responsible for Mdm2 degradation (Itahana et al., 2007). Nonetheless, the Mdm2C462A model has a caveat in that it disrupts Mdm2 RING structure, along with Mdm2 E3 ligase function and Mdm2:MdmX binding, and the possibility of this structural change adversely affecting Mdm2 stability cannot be ruled out. With the Mdm2Y487A model we wanted to further explore the mechanism of Mdm2 degradation in vivo. Despite disrupting Mdm2-mediated degradation of p53 and MdmX (Figure 2C–D and Figure S2A–B), and higher levels of Mdm2 in Mdm2Y487A/Y487A MEFs (Figure 2F–G), Mdm2 was degraded at a similar rate in Mdm2Y487A/Y487A MEFs as in WT MEFs (Figure 3A). Similar to previous observations made in Mdm2C462A/C462A;p53ER/− MEFs (Itahana et al., 2007), a normal rate of Mdm2 degradation was observed in Mdm2Y487A/Y487A;p53ER/− MEFs (Figure 3B). To further investigate the mechanism of Mdm2 degradation, while avoiding the confounding effects of p53 transcriptional activation of Mdm2, a p53-null genetic background was utilized. MG-132 treatment resulted in similar stabilization of Mdm2 in Mdm2+/+;p53−/−, Mdm2Y487A/Y487A;p53−/−, and Mdm2C462A/C462A;p53−/− MEFs, indicating that Mdm2 degradation is mediated by proteasomal proteolysis, and further suggesting the role of an E3 ligase other than Mdm2 itself in Mdm2 ubiquitination and degradation (Figure 3C).
Figure 3. Y487A Mutation Does Not Affect Mdm2 Degradation.
(A) Early passage Mdm2+/+ and Mdm2Y487A/Y487A MEFs were treated with 100 µg/mL cycloheximide and harvested using SDS lysis buffer at the time points indicated. The levels of Mdm2 and actin were analyzed by western blotting. The amount of Mdm2 was quantified by densiometry, normalized to the level of actin, and plotted.
(B) The degradation of Mdm2 in Mdm2+/+;p53ER/− and Mdm2Y487A/Y487A;p53ER/− MEFs was analyzed as in A.
(C) Mdm2+/+;p53−/−, Mdm2Y487A/Y487A;p53−/−, and Mdm2C462A/C462A;p53−/− MEFs were treated with 25 µM MG-132 for 8 hr as indicated. Lysates were collected and the level of Mdm2 was analyzed by western blotting. Actin was used as a loading control.
See also Figure S3.
While both Mdm2Y487A and Mdm2C462A disrupt p53 degradation and maintain Mdm2 degradation, the location of the Y487 residue, C-terminal of the Mdm2 RING finger domain, should not disrupt zinc coordination, thus proper folding and the structure of the RING domain should remain intact (Figure S3) (Linke et al., 2008). The observation of a second, mechanistically unique, E3 inactive Mdm2 mutant capable of maintaining normal degradation provides additional evidence against the necessity of autoubiquitination for in vivo degradation of Mdm2.
Mdm2:MdmX Heterodimerization Is Essential for p53 Suppression
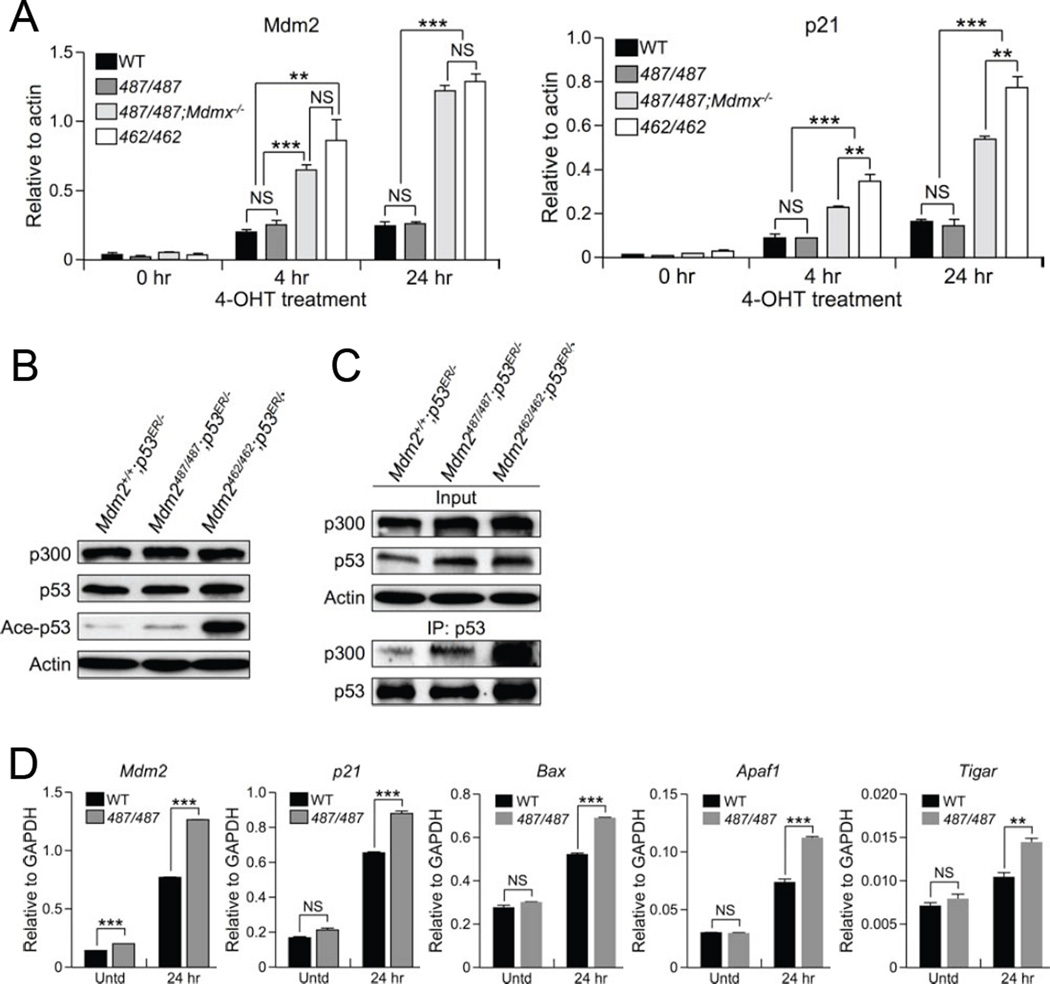
In light of the embryonic lethality observed in MdmxΔRING (Pant et al., 2011), MdmxC462A (Huang et al., 2011), and Mdm2C462A (Itahana et al., 2007) homozygous mutants, all of which disrupt Mdm2:MdmX heterodimerization, we sought to use the Mdm2Y487A model in conjunction with the Mdm2C462A model for a comparative study of the effect of Mdm2:MdmX heterodimerization on p53 activity, independent of Mdm2 E3 ligase function. To this end, qRT-PCR was performed to compare the relative mRNA levels of p53 target genes in MEFs. Upon activation of p53 with 4-OHT, Mdm2Y487A/Y487A;MdmX+/+;p53ER/−(487/487) MEFs maintained similarly low levels of mRNA in a number of p53 target genes, including Mdm2, p21, Apaf1, and Bax, as compared to Mdm2+/+;MdmX+/+;p53ER/− (WT) MEFs (Figure 4A and Figure S4A). Disruption of Mdm2:MdmX heterodimerization via concomitant deletion of MdmX, as in Mdm2Y487A/Y487A;Mdmx−/−;p53ER/− (487/487;Mdmx−/−), or via Mdm2 RING finger mutation, as in Mdm2C462A/C462A;Mdmx+/+;p53ER/− (462/462), was correlated with higher mRNA levels of p53 and p53 target genes, indicative of higher p53 activity in the absence of Mdm2:MdmX interaction (Figure 4A and Figure S4A). The E3 inactive status of the Mdm2Y487A mutant in vivo suggests that the function of Mdm2:MdmX interaction goes beyond E3 ligase activity; following activation with 4-OHT, p53ER levels are similarly reduced in both the presence and absence of MdmX (Figure S4B), indicating that under unstressed conditions the degradation of p53 by Mdm2 does not require MdmX binding. Together, this suggests that the ability of Mdm2 to heterodimerize with MdmX, but not its E3 ligase activity, is essential for suppression of p53 activity.
Figure 4. Mdm2:MdmX Heterodimerization Is Essential for p53 Suppression.
(A) mRNA levels of Mdm2 (left) and p21 (right) in Mdm2+/+;Mdmx+/+;p53ER/− (WT), Mdm2Y487A/Y487A;Mdmx+/+;p53ER/− (487/487), Mdm2Y487A/Y487A;Mdmx−/−;p53ER/− (487/487;Mdmx−/−), and Mdm2C462A/C462A ;Mdmx+/+ ;p53ER/− (462/462) MEFs after 4-OHT treatment were measured by qRT-PCR. All samples were analyzed in triplicate. Data are means +/− SD normalized to Actin.
(B) Mdm2+/+;p53ER/−, Mdm2Y487A/Y487A;p53ER/−, and Mdm2C462A/C462A;p53ER/− MEFs were incubated with 4-OHT for 4 hr, harvested and analyzed by western blotting. Actin was used as a loading control.
(C) Mdm2+/+;p53ER/−, Mdm2Y487A/Y487A;p53ER/−, and Mdm2C462A/C462A;p53ER/− MEF lysates were cross-linked and then immunoprecipitated and immunoblotted as indicated. Cells were incubated with 4-OHT for 4 hr prior to cross-linking.
(D) Expression of p53 target genes, Mdm2, p21, Bax, Apaf1, and Tigar in Mdm2+/+ (WT) and Mdm2Y487A/Y487A (487/487) MEFs was measured by qRT-PCR after 25 J/m2 UV treatment as indicated. Untreated (Untd) MEFs were used as a control. All samples were analyzed in triplicate minimally. Data are means +/− SD normalized to GAPDH.
See also Figure S4.
To elucidate the mechanism by which the Mdm2:MdmX heterodimer suppresses p53 activity we assessed p53 acetylation, an important post-translational modification for the enhancement of p53 activity (Gu and Roeder, 1997). While total p53 levels were comparable between genotypes, a higher level of acetylated p53 was observed in Mdm2C462A/C462A;p53ER/− MEFs lacking Mdm2:MdmX heterodimerization compared to either Mdm2Y487A/Y487A;p53ER/− or Mdm2+/+;p53ER/− MEFs, both of which retain intact Mdm2:MdmX heterodimer formation (Figure 4B). Additionally, loss of heterodimerization correlated with increased binding between p53 and the histone acetyltransferase, p300 (Figure 4C). This data suggests that Mdm2:MdmX heterodimerization functions to control p53 activity, at least in part, through inhibiting p53:p300 interaction and p53 acetylation.
Taking into account the outward normalcy of Mdm2Y487A/Y487A mice, despite lacking Mdm2 E3 ligase activity, we wanted to further explore if and when the E3 ligase function of Mdm2 is necessary. As a transcriptional target of p53, Mdm2 transcription is stimulated following p53 activation, presumably to degrade p53 and return it to pre-stress levels, and effectively forming a feedback loop between Mdm2 and p53 (Barak et al., 1993; Juven et al., 1993). We sought to test whether the absence of Mdm2 E3 ligase activity, an essential component of the Mdm2-p53 negative feedback loop, affects the p53-mediated stress response. Prior to stress, WT and Mdm2Y487A/Y487A MEFs exhibited relatively low levels of mRNA in a number of well-characterized p53 target genes, including Mdm2, the cell cycle regulator p21, and the pro-apoptotic genes Bax, Apaf1, and Tigar, as determined by qRT-PCR (Figure 4D). However, following UV irradiation (IR), Mdm2Y487A/Y487A MEFs demonstrated significantly higher mRNA levels of the p53 target genes assessed compared to WT MEFs, indicative of higher levels of p53 activity in the absence of Mdm2 E3 ligase function (Figure 4D).
To determine if the increased p53 activity observed with UV activation of the p53 pathway was a more universal response, qRT-PCR was utilized to analyze the mRNA levels of the above-mentioned p53 target genes following additional forms of DNA damage, including 5 Gy γ IR, and 1 µM doxorubicin, as well as ribosomal stress, as induced by a low dose (5 nM) of actinomycin D. Regardless of stimuli, Mdm2Y487A/Y487A MEFs were observed to have increased mRNA levels compared to WT MEFs in all of the p53 target genes analyzed, although the magnitude of activation varied between treatments (Figure S4D). Together these data suggest that although the E3 inactive Mdm2Y487A can suppress p53 in the absence of stress, it cannot efficiently suppress stress-stimulated hyper-p53 activity.
Mdm2 E3 Ligase Function Is Necessary for Recovery from DNA Damage
Classically, Mdm2 E3 ligase mediated ubiquitination is thought to be critical in the downregulation of p53 after its stabilization in response to stress, and thus we wanted to explore the effect of loss of this function on p53 stability and activity following activation of the p53 stress response pathway. Under untreated conditions, despite significant differences in total p53 levels, Mdm2Y487A/Y487A MEFs demonstrated low p53 activity as indicated by levels of p53 phosphorylation (Figure 5A and Figure S5A–B), p21 protein expression (Figure 2B and Figure 5A), and mRNA expression of number of p53 target genes (Figure 4D, and Figure S4D). Following γ IR, UV IR, or doxorubicin induced DNA damage, WT MEFs demonstrated initial stabilization of total p53 and phosphorylated p53, followed by a return towards low levels; in Mdm2Y487A/Y487A MEFs, sustained high levels were observed (Figure 5A–C and Figure S5A–B). The continued increase of total p53 protein in the mutant MEFs after DNA damage is likely due to the combined effect of increased p53 transcription (Raman et al., 2000; Webster and Perkins, 1999) (Figure S5C), increased p53 translation (Candeias et al., 2008; Gajjar et al., 2012; Takagi et al., 2005), and the inability of the Mdm2Y487A mutant to degrade p53. The effect of increased levels of p53 and the corresponding increase in p53 activity in Mdm2Y487A/Y487A MEFs following exposure to DNA damaging reagents is further reflected in increased cell cycle arrest (Figure S5D) and decreased cell survival (Figure S5E). Additionally, we observed that growth of WT and Mdm2Y487A/Y487A MEFs was similar when cultured at 3% O2, which better emulates physiological levels of oxygen, but at 20% O2, the oxidative stress of standard cell culture conditions was sufficient to reduce cell growth in otherwise untreated, unstressed Mdm2Y487A/Y487A MEFs (Figure S5F). Furthermore, 3% O2 cell culturing conditions attenuate the p53 response to 5 Gy X-ray IR in WT MEFs (Achison and Hupp, 2003; Parrinello et al., 2003; Wang et al., 2006).
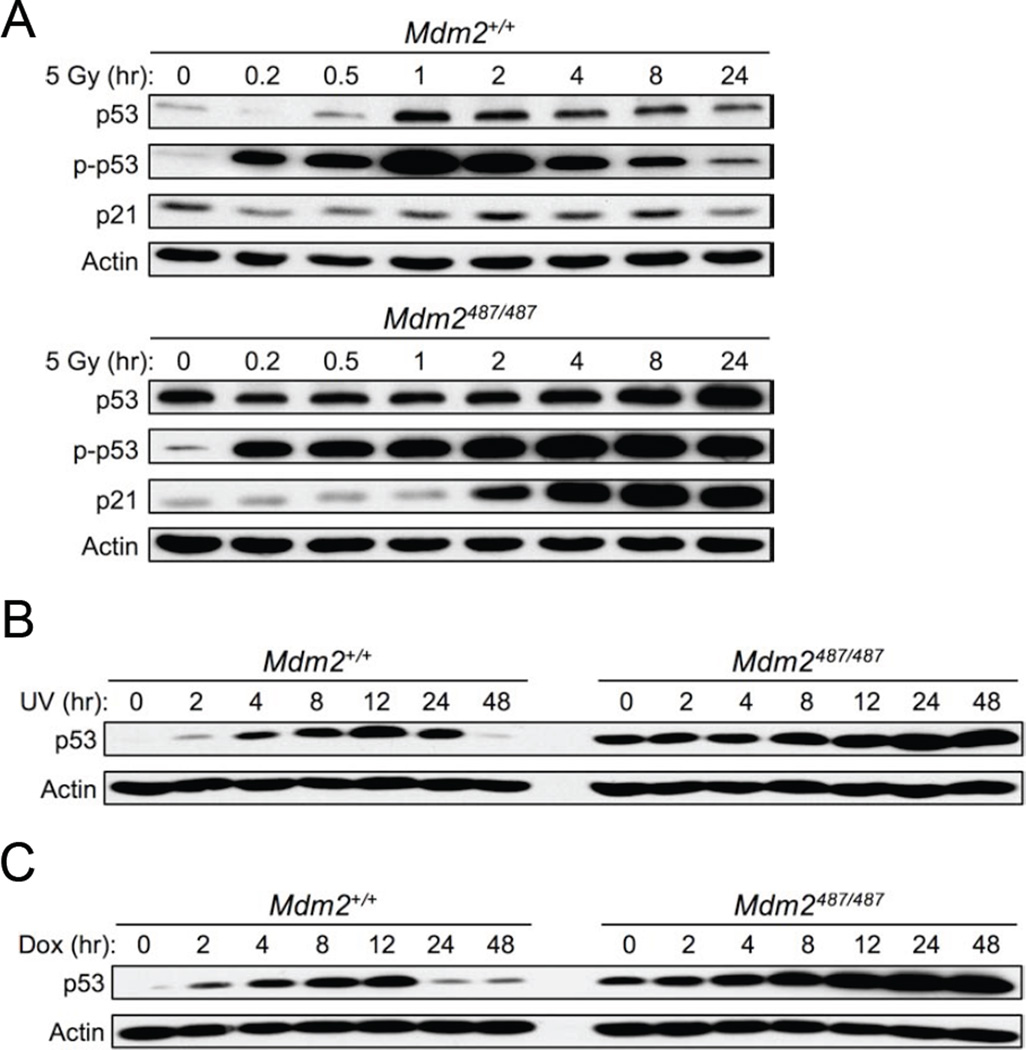
Figure 5. Mdm2-Mediated Degradation of p53 Is Necessary to Regulate p53 Protein and Activity Levels After Stress.
(A) Mdm2+/+ (top) and Mdm2Y487A/Y487A (bottom) MEFs were treated with 5 Gy X-ray IR, cell lysates were collected at the indicated time points and analyzed by western blotting with antibodies to p53, S-18 phosphorylated p53 (p-p53), and p21. Actin was used as a loading control.
(B and C) DNA damage induced by 25 J/m2 UV (B) or 1 µM doxorubicin (C) was utilized to activate the p53 pathway in Mdm2+/+ and Mdm2Y487A/Y487A MEFs, cell lysates were collected at the indicated time points and analyzed by western blotting. In (C) drugs were removed after 4 hr of treatment.
See also Figure S5.
In addition to DNA damage, in response to non-DNA damaging reagents, such as low levels of actinomycin D, a similar pattern of p53 stabilization was observed (Figure S5G). Together, these data indicate that the Mdm2 E3 ligase function is critical for controlling the magnitude of p53 activation in response to stress, and restoring p53 to basal levels after stress.
Mdm2Y487A/Y487A Mice Demonstrate Increased Radiosensitivity
The enhanced p53 stabilization and activity in Mdm2Y487A/Y487A MEFs following stress, and the apparent inability of cells to recover following activation of the p53-mediated stress response, pointed to the importance of Mdm2 E3 ligase function in regulating p53 following exposure to stressors. To test if the aberrant activation of p53 observed in Mdm2Y487A/Y487A MEFs would be replicated in vivo, WT and Mdm2Y847A/Y487A mice were treated with a sublethal dose (5 Gy) of γ IR. As expected, this dose of IR did not cause any immediate disease or affect mortality in WT mice; however, in Mdm2Y847A/Y487A mice 5 Gy γ IR resulted in significant weight loss and 100% lethality by 22 days post treatment (Figure 6A and Figure S6A). To determine whether the observed phenotype is p53 dependent, Mdm2Y847A/Y487A mice with concomitant deletion of either one or both copies of p53 were similarly treated with 5 Gy γ IR. In contrast to Mdm2Y487A/Y487 mice with intact p53, both Mdm2Y847A/Y487A;p53+/− and Mdm2Y847A/Y487A;p53−/− mice were resistant to 5 Gy γ IR treatment and demonstrated no significant changes in body weight or viability (Figure 6B and Figure S6B), indicating that the lethality observed in the Mdm2Y847A/Y487A mice was p53 dependent. While deletion of p53 can serve a protective function against the short term consequences associated with exposure to IR, including apoptosis and hematopoietic failure, we observed increased survival in Mdm2Y487A/Y487A;p53+/− mice compared to Mdm2Y487A/Y487A;p53−/− mice; this observation supports the idea that both too much p53, as in Mdm2Y487A/Y487A;p53+/+ mice, as well as too little or no p53, as in Mdm2Y487A/Y487A;p53−/− mice, can be deleterious (Donehower et al., 1992; Mendrysa et al., 2003).
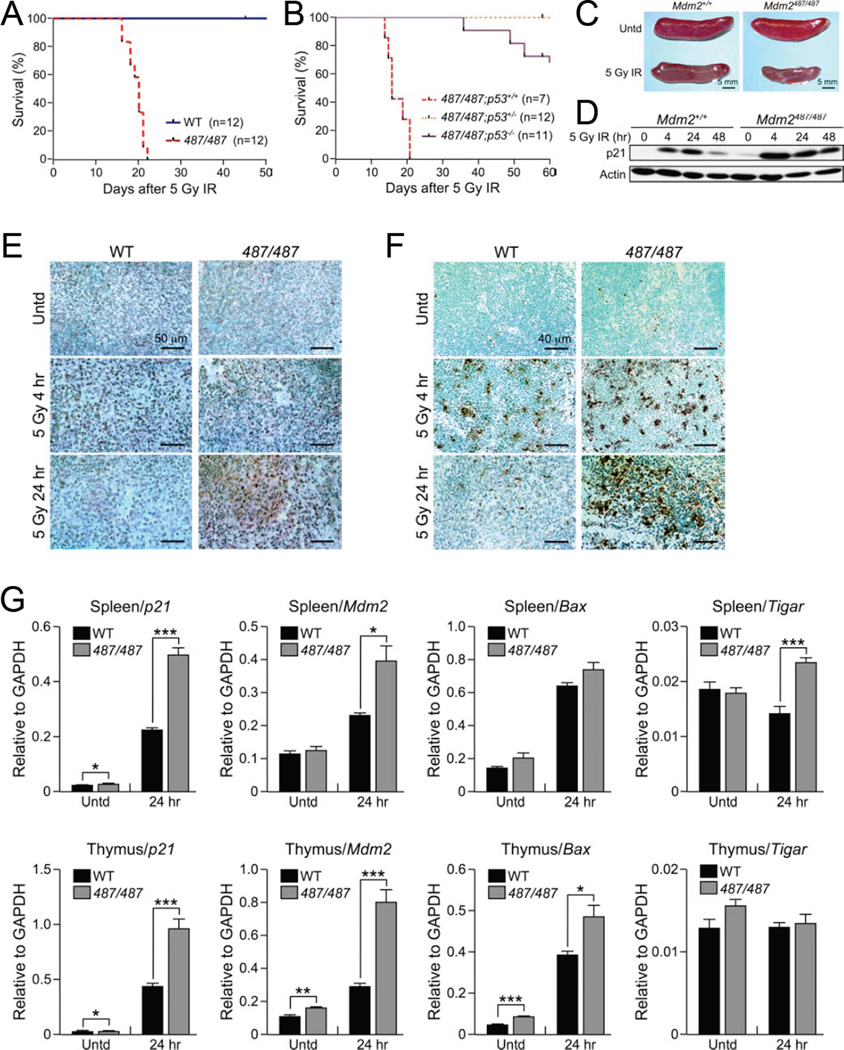
Figure 6. Mdm2Y487A/Y487A Mice Demonstrate Increased Radiosensitivity.
(A) Kaplan-Meier survival curve for Mdm2+/+ (WT) and Mdm2Y487A/Y487A (487/487) female mice treated with 5 Gy γ-IR. Mice were between 5.5 and 8.5 weeks old at the time of treatment.
(B) Kaplan-Meier survival curve for Mdm2Y487A/Y487A;p53+/+ (487/487;p53+/+), Mdm2Y487A/Y487A;p53+/− (487/487;p53+/−), and Mdm2Y487A/Y487A;p53−/− (487/487;p53−/−) male mice treated with 5 Gy γ-IR. Mice were between 6.5 and 7.5 weeks old at time of treatment.
(C) Representative gross histology of Mdm2+/+ and Mdm2Y487A/Y487A spleens of 6.5 week old male mice either untreated or treated with 5 Gy γ-IR as indicated; spleens were collected 14 days after treatment.
(D) Fourteen-week old Mdm2+/+ and Mdm2Y487A/Y487A female mice were treated with 5 Gy γ-IR. Splenic tissue harvested at the indicated time points was analyzed by western blotting for p21 levels. Actin was used as a loading control.
(E) Immunohistochemistry staining for p53 in splenic tissue from Mdm2+/+ (WT) and Mdm2Y487A/Y487A (487/487) mice left untreated or after 5 Gy γ IR as indicated.
(F) Splenic tissue from Mdm2+/+ (WT) and Mdm2Y487A/Y487A (487/487) mice left untreated or after 5 Gy γ IR as indicated, and TUNEL staining was was used to assay for apoptosis.
(G) mRNA levels of the p53 target genes p21, Mdm2, Bax , and Tigar in the spleen and thymus of Mdm2+/+ (WT) and Mdm2Y487A/Y487A (487/487) female mice either untreated or treated with 5 Gy γ IR as indicated was measured by qRT-PCR. Mice were between 11 and 18 weeks old at time of treatment. All samples were analyzed in triplicate. Data are means +/− SD normalized to GAPDH. n=3 for each genotype in each treatment.
See also Figure S6.
Gross examination revealed a significantly smaller spleen, and significantly lower levels of red blood cells and white blood cells in Mdm2Y487A/Y487A mice compared to the WT (Figure 6C and Figure S6C–D). The time of death following treatment, together with decreased splenic size and blood counts, is suggestive of hematopoietic failure as the ultimate cause of death in the irradiated mutant animals (Gudkov and Komarova, 2003; Mendrysa et al., 2003; Wang et al., 2011).
Histological examination indicated that both WT and Mdm2Y487A/Y487A mice have similarly low levels of p53 in the spleen prior to IR, and increased levels of p53 by 4 hr after treatment with 5 Gy γ IR (Figure 6E). In the presence of WT Mdm2, p53 levels in the spleen returned to near untreated levels by 24 hr after 5 Gy γ IR, while in the Mdm2Y487A/Y487A spleen, dense areas with high p53 remain (Figure 6E). Corresponding to observations in the spleen, p53 levels were relatively low and similar in the testes, intestine, and liver of untreated WT and Mdm2Y487A/Y487A mice (Figure S6E–H). Following 5 Gy γ IR, however, p53 levels were higher in Mdm2Y487A/Y487A mice compared to WT mice at 4 hr and 24 hr post treatment in the intestine (Figure S6G), and at 24 hr in the testes (Figure S6F); the liver is generally considered to be radioresistant and did not demonstrate any changes in observed p53 (Figure S6H).
To assess the effect of the increase in p53 protein on p53 activity, qRT-PCR of several p53 target genes, including p21, Mdm2, Bax, and Tigar, in splenic and thymic tissue from WT and Mdm2Y487A/Y487A mice was performed. Following 5 Gy γ IR, p53 activity is significantly higher in Mdm2Y487A/Y487A mice compared to the WT, as indicated by higher mRNA expression of p53 target genes, providing evidence that the higher p53 protein levels in Mdm2Y487A/Y487A mice correlate with increased p53 activity (Figure 6G). The dynamics of the p53 stress response in the spleen and thymus were further examined by western blot; in response to 5 Gy γ IR treatment, p21 was increased and maintained at a higher level in the Mdm2Y487A/Y487A mutant compared to the WT, further indicating that the accumulation of p53 protein correlates with increased p53 activity (Figure 6D and Figure S6I). To infer the biological consequences of increased p53 levels and activity in the mouse, TUNEL staining was utilized to assess the level of apoptosis; in the spleen, corresponding to increased p53 levels and activity, increased levels of apoptosis were observed in Mdm2Y487A/Y487A mice following IR (Figure 6F).
DISCUSSION
Mdm2 has long been considered the gatekeeper of both p53 stability and activity, and as such, it is critically important to have a thorough understanding of Mdm2-mediated p53 regulation in vivo. It has long been accepted that Mdm2 E3 ligase activity is an essential mechanism for Mdm2-mediated p53 regulation; with this in mind, the most unanticipated finding of the current study is that fully intact Mdm2 E3 ligase activity towards p53 is dispensable in vivo, as demonstrated by the viability, normal development, and lifespan of Mdm2Y487A/Y487A mice. In vivo, the loss of Mdm2 E3 ligase activity resulted in increased stabilization of p53, but loss of E3 activity alone, without disruption of Mdm2:MdmX heterodimerization, was not sufficient to significantly increase p53 activity. Following activation of the p53 pathway, Mdm2 E3 ligase function becomes essential in controlling p53 abundance and activity.
A number of recent knock-in mouse models, including MdmxΔRING, MdmxC462A, and Mdm2C462A, have implicated Mdm2:MdmX heterodimerization as an essential component of p53 regulation, in large part due to the embryonic lethality observed when this interaction is disrupted (Huang et al., 2011; Itahana et al., 2007; Pant et al., 2011). In this regard, the Mdm2Y487A model provided vital complementary data, further pointing towards Mdm2:MdmX heterodimerization as the key to understanding p53 regulation. Uniquely, the Mdm2Y487A model allows for analysis of RING finger binding dynamics independent of Mdm2 E3 ubiquitin ligase activity. In contrast with previous in vitro data, the study of the Mdm2Y487A knock-in mouse model suggests that Mdm2 E3 ligase activity is significantly disrupted even in the presence of Mdm2:MdmX heterodimerization (Okamoto et al., 2009; Poyurovsky et al., 2007; Uldrijan et al., 2007). The apparent discrepancy between Mdm2Y487A in vitro, which demonstrated near normal levels of p53 stabilization in the presence of MdmX, and in vivo, which demonstrated near, if not complete disruption of p53 degradation, serves as an additional example as to the importance of examining proteins at physiologically relevant levels.
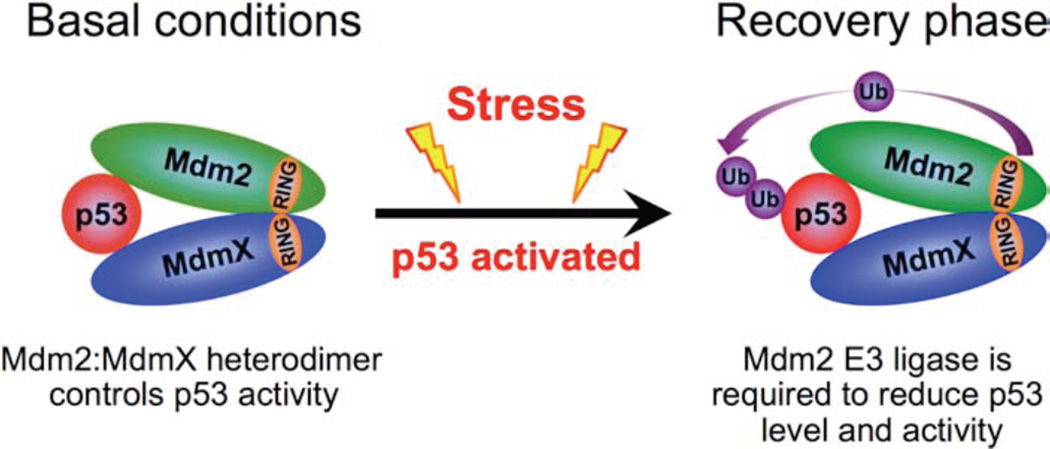
While correlated with the regulation of p53 stability and activity, the mechanism through which the Mdm2:MdmX heterodimer functions in these roles remains inadequately understood. In concert with existing mouse models we believe our data supports the following model: Under unstressed conditions the Mdm2:MdmX heterodimer is able to suppress p53 activity through binding, conceivably by inhibiting p300-mediated p53 acetylation, without Mdm2 mediated p53 degradation. Following stress and the subsequent post-translational modifications to p53, Mdm2, and MdmX, p53 levels and activity are increased. Once p53 has been stabilized and activated, the Mdm2:MdmX heterodimer is no longer sufficient to control p53, and Mdm2 E3 ubiquitin ligase activity becomes critical for recovery from the p53-mediated stress response (Figure 7).
Figure 7. Mdm2:MdmX Heterodimer Regulation of p53.
Model depicting Mdm2:MdmX heterodimer regulation of p53 before and after activation of the p53 stress response pathway.
Although MdmX is generally cytoplasmic, while Mdm2 and p53 are primarily localized to the nucleus, the Mdm2Y487A mouse further implicates the interaction between Mdm2 and MdmX to be critical in p53 regulation. One explanation for the apparent necessity for Mdm2:MdmX interaction is the role of Mdm2 in translocation of MdmX to the nucleus, as MdmX does not carry its own nuclear localization signal or nuclear export signal (Li et al., 2002; Marine and Jochemsen, 2005). Alternatively, the fraction of Mdm2 and p53 localized to the cytoplasm may allow for Mdm2:MdmX heterodimer formation and function; previous data has implicated cytoplasmic MdmX in p53 regulation (Jimenez et al., 1999; Moll et al., 1995; Ohtsubo et al., 2009). Regardless, further investigation to characterize the mechanism illuminating how Mdm2 and MdmX work together to inhibit p53, despite differences in subcellular localization, remains an important yet unresolved issue in the field.
In addition to implications on the role of Mdm2 E3 ligase activity in the regulation of p53 stability, the Mdm2Y487A mouse model has also provided additional insight into Mdm2 degradation. While the E3 ligase responsible for Mdm2 degradation in vivo is yet to be identified, this model provides further evidence that under physiological conditions autoubiquitination is not the primary mechanism for Mdm2 degradation. Additional research into the regulation of Mdm2 stability, and identification of E3 ubiquitin ligases with specificity for Mdm2 are important ventures to be explored.
Due to the high rate of p53 mutation, deletion, and misregulation in many cancers, in theory, the p53 pathway is an attractive drug target; however, effective therapeutics utilizing this pathway are not yet found in the clinic (Danovi et al., 2004; Hainaut et al., 1997; Momand et al., 1998; Wade et al., 2013). As brought to light by our study, the basic understanding of p53 regulation in vivo is continuing to evolve. The key findings from this study have important clinical implications, as a number of drugs targeting the Mdm2-p53 pathway, including Nutlin-3a, were developed to target Mdm2:p53 interaction and/or Mdm2-mediated ubiquitination of p53 (Vassilev et al., 2004; Wade et al., 2013). Although many factors in the tumor microenviroment are known to activate the p53 pathway, it remains to be determined if, as observed in response to DNA damage, disruption of Mdm2 E3 ligase activity will result in p53 hyper-activation in response to the tumor microenvironment itself (Bieging et al., 2014). Along with a number of recent knock-in mouse models, the Mdm2Y487A mouse points to the significance of Mdm2:MdmX heterodimerization in both p53 stability and activity, and thus disruption of this interaction may be a potent therapeutic target. Focused study of the interaction between Mdm2 and p53 at physiological levels has provided additional insight into both p53 and Mdm2 regulation, which will allow for more effective application of therapeutics targeting this pathway, as well as additional targets for the development of effective drugs.
EXPERIMENTAL PROCEDURES
Generation of Mdm2Y487A Knock-in Mice
Murine 129/Sv genomic DNA containing the last six exons (7–12) of Mdm2 was a gift from Dr. Guillermina Lozano (University of Texas, M.D. Anderson Cancer Center). The targeting vector was constructed in a PGK neo vector previously described (Itahana et al., 2007). A Tyr-to-Ala substitution (TAC to GCA) was introduced in codon 487 using site-directed mutagenesis. The final construct was sequenced using overlapping primers. Electroporation of 129/Sv-derived AB2.2 ES cells was performed by the UNC Animal Models Core Facility. DNA isolated from G418-resistant ES colonies were confirmed for the presence of the Y487A substitution by PCR amplification of genomic DNA using primers flanking exon 12, followed by sequencing. Two positive clones were injected into C57BL/6 blastocysts, and the blastocysts were transferred into pseudopregnant female recipients. The resulting chimeric males were mated with C57BL/6 females; germline transmission was confirmed by PCR analysis and DNA sequencing. Following germline transmission of the Mdm2Y487A mutation, Mdm2Y487A/+ mice were mated with Ella-Cre transgenic mice (Jackson Laboratories, stock number 003724) to excise the neomycin selection marker. The resulting mice were backcrossed to C57BL/6 for five generations. Mice were bred and maintained strictly under protocols 10–045 and 13–044 approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill Animal Care Facility.
Mouse Breeding, Genotyping and Experiments
PCR was utilized to identify the Mdm2Y487A mutation using primers for the WT (m305wF 5’-TTATTGAAGGACTATTGGAAGTGTACCTCA-3’; mY487A–W-R-4 5’-GTGAGCAGGTCAGCTAGTTGAAGTA-3’) and mutant (m305wF; mY487A–M-R-1 5’-TTGTGAGCAGGTCAGCTAGTTGAATGC-3’) alleles. The Mdmx−/− mice were a gift from Guillermina Lozano (University of Texas, M.D. Anderson Cancer Center). The p53−/− mice were purchased from The Jackson Laboratory (stock number 002101). PCR primers for WT (p53-X7, 5′-GGATGGTGGTATACTCAGAGCC-3′; p53-X6, 5′-ACAGCGTGGTGGTACCTTAT-3′) and mutant (p53-X7; NEO18.5, 5′-TCCTCGTGCTTTACGGTATC-3′) p53 alleles were described by The Jackson Laboratory. p53ERTAM knock-in mice were a gift from Gerard Evan (University of Cambridge, UK) and genotyping of the p53ERTAM alleles was performed as described previously (Christophorou et al., 2005).
At 5.5 to 8.5 weeks of age, mice were treated with 5 Gy γ IR (Mark I Model 68-1 Cesium-137 Irradiator) at a dose rate of 1.89 Gy/min. For survival studies following IR, mice were monitored and weighed every other day, and moribund mice were humanely euthanized. Mouse tissue was fixed in formalin for histopathology and snap frozen in liquid nitrogen for protein extraction.
Cell Culture
Primary MEFs were cultured in a 37°C incubator with 5% CO2, and 3% O2 in DMEM supplied with 10% FBS and penicillin (100 IU/ml)/streptomycin (100 µg/ml). For activation of p53ERTAM, 100 nM 4-OHT (Sigma) dissolved in 100% ethanol was added to the culture medium. MEFs were treated as indicated with 25 J/m2 UV, 5 nM actinomycin D (Act D), 1 µM doxorubicin (Dox), or 5 Gy γ IR.
Protein Analysis
Mouse monoclonal Mdm2 (2A10, Calbiochem), MdmX (MdmX-82, Sigma), p53 (NCL-505, Leica), phospho-p53 (Ser15, Cell Signaling), acetyl-p53 (Lys320, Cell Signaling), actin (MAB1501, Chemicon International), and goat polyclonal p21 (C-19; Santa Cruz) antibodies were purchased commercially. Rabbit polyclonal antibody to p21 was a gift from Yue Xiong (UNC-Chapel Hill). Mouse monoclonal antibody to MdmX (7A8) was a gift from Jiandong Chen (University of South Florida). MEFs were lysed in 0.1% NP-40 buffer for immunoprecipitation (IP) and 0.5% NP-40 buffer for straight western blotting. Tissue was lysed in tissue lysis buffer (0.2% Triton X-100, 0.3% NP-40). Procedures and conditions for IP and immunoblotting were described previously (Itahana et al., 2003). Protein cross-linking was performed prior to p53 IP as described previously (Clegg et al., 2012; Itahana et al., 2007). The half-life of p53 and Mdm2 was measured by treating cells with cycloheximide (100 µg/ml) for the indicated length of time, followed by lysis in 2% SDS lysis buffer. The protein level was analyzed by western blotting, and the intensity of the bands in the linear range of exposure was quantified by densitometry. Subcellular fractionation was performed using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific) according to manufacturer’s instructions.
Quantitative Real-time PCR
Total RNA was isolated using an RNeasy Kit (Qiagen) and cDNA was synthesized using SuperScript III (Invitrogen). qRT-PCR was performed with SYBR green master mix (Applied Biosystems) using the7900HT Fast Real-Time PCR System (Applied Biosystems) according to manufacturer instructions. Data was collected and exported with SDS 2.2.2 software. Relative expression was calculated using GAPDH or Actin as an internal control as indicated. Primers used were as follows: Mdm2, 5’CCA ACCATCGACTTCCAGCAGCATT3’ and 5’GAT TGGCTGTCTGCACACTGGG3’; p21, 5’CCTGGTGATGTCCGACCTG3’ and 5’CCATGAGCG CATCGCAATC3’; Apaf1, 5’CGGTGAAGGTGTGGAATGTCATTACCG3’ and 5’GGATTTCTCC ATTGTCATCTCCAGTTGC3’; Bax, 5’GGACAGCAATATGGAGCTGCAGAGG3’ and 5’GGAGG AAGTCCAGTGTCCAGCC3’; TIGAR, 5’CGATCTCACGAGGACTAAGCAGACC3’ and 5’GCCA AAGAGCTTTCCAAACCGCTGC3’; GAPDH, 5’AGGTCGGTGTGAACGGATTTG3’ and 5’TGTA GACCATGTAGTTGAGGTCA3’; Actin, 5’CCACAGCTGAGAGGGAAATCGTGC 3’ and 5’CCAG AGCAGTAATCTCCTTCTGCATCC3’, p53, 5’ATGGAGGAGCCGCAGTCAGATC3’ and 5’CCATTGTTCAATATCGTCCGGG3’.
Immunohistochemistry and TUNEL staining
Tissue from Mdm2+/+ and Mdm2Y487A/Y487A mice was harvested and fixed in 10% formalin for 24 hr. Five micrometer sections were stained by immunohistochemistry for p53 (NCL-p53-CM5p, Leica Biosystems) and DAB staining. TUNEL staining for apoptosis was performed using the Apoptag Peroxidase In Situ Apoptosis Detection Kit (Millipore) according to manufacturer’s instructions. Stained tissue was analyzed using an Olympus IX-81 microscope fitted with a SPOT camera and software.
Statistical Analysis
Statistical analysis was carried out using GraphPad Prism 4 Software (GraphPad Software, San Diego, CA). For lifespan analysis and IR experiments a Kaplan-Meier survival curve was carried out. A two-tailed unpaired t-test analysis was utilized to compare mRNA levels in qRT-PCR experiments.
Supplementary Material
Highlights.
-
-
Mdm2Y487A loses E3 ligase activity for p53 degradation but retains MdmX binding.
-
-
Robust Mdm2 E3 ligase function is dispensable for embryogenesis and development.
-
-
In vivo, Mdm2Y487A mutation results in increased p53 stability but not activity.
-
-
Mdm2Y487A/Y487A mice demonstrate p53-dependent hyper-radiosensitivity.
SIGNIFICANCE.
Classically characterized as an overarching tumor suppressor, p53 is frequently mutated, deleted, and misregulated in human cancers. Inhibition of Mdm2 E3 ubiquitin ligase-mediated degradation of p53 is considered to be an essential mechanism for p53 activation and serves as the underpinning of many cancer treatments aimed at reactivation of p53. As observed in our Mdm2Y487A knock-in mouse, inactivation of Mdm2 E3 ligase function without obliterating Mdm2:MdmX interaction does not efficiently activate p53; however, disruption of Mdm2 E3 ligase function sensitizes mice to irradiation induced DNA damage and lethality. This study provides insight into the Mdm2-p53 regulatory loop and sheds light on the current problems surrounding the development of effective cancer therapeutics targeting the p53 pathway.
ACKNOWLEDGEMENTS
We thank Dale Cowley from UNC Animal Models Core Facility for developing the Mdm2 knock-in mice, and Everardo Macias, Yizhou He, Yong Liu, Hengming Ke, and Patrick Leslie for their helpful advice and technical assistance. We are in debt to Jiandong Chen, Yue Xiong, Guillermina Lozano, Gerard Evan, Aart Jochemsen, Fabiola Moretti, Stephen Jones, Moshe Oren, and Zhi-Min Yuan for their generosity in sharing reagents. This research was supported by grants from the National Institutes of Health (CA127770 and CA167637) to Y.Z. and UNC Genetics and Molecular Biology Training Grant to L.T. This research was also supported by grants from the Leukemia and Lymphoma Society, American Cancer Society, and NSFC from China to Y.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Achison M, Hupp TR. Hypoxia attenuates the p53 response to cellular damage. Oncogene. 2003;22:3431–3440. doi: 10.1038/sj.onc.1206434. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, Bourougaa K, Calvo F, Fahraeus R. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098–1105. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, Evan GI. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37:718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- Clegg HV, Itahana Y, Itahana K, Ramalingam S, Zhang Y. Mdm2 RING mutation enhances p53 transcriptional activity and p53-p300 interaction. PLoS One. 2012;7:e38212. doi: 10.1371/journal.pone.0038212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- Gajjar M, Candeias MM, Malbert-Colas L, Mazars A, Fujita J, Olivares-Illana V, Fahraeus R. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell. 2012;21:25–35. doi: 10.1016/j.ccr.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer. 2003;3:117–129. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Soussi T, Shomer B, Hollstein M, Greenblatt M, Hovig E, Harris CC, Montesano R. Database of p53 gene somatic mutations in human tumors and cell lines: updated compilation and future prospects. Nucleic Acids Res. 1997;25:151–157. doi: 10.1093/nar/25.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, Zuo Y, Kawai H, Shadfan M, Ganapathy S, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci U S A. 2011;108:12001–12006. doi: 10.1073/pnas.1102309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol. 2000;20:1001–1007. doi: 10.1128/mcb.20.3.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez GS, Khan SH, Stommel JM, Wahl GM. p53 regulation by post-translational modification and nuclear retention in response to diverse stresses. Oncogene. 1999;18:7656–7665. doi: 10.1038/sj.onc.1203013. [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Juven T, Barak Y, Zauberman A, George DL, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278:45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol. 2011;12:259–265. doi: 10.1038/nrm3086. [DOI] [PubMed] [Google Scholar]
- Li C, Chen L, Chen J. DNA damage induces MDMX nuclear translocation by p53-dependent and -independent mechanisms. Mol Cell Biol. 2002;22:7562–7571. doi: 10.1128/MCB.22.21.7562-7571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Chapo J, Roig I, Abrams JM. Meiotic recombination provokes functional activation of the p53 regulatory network. Science. 2010;328:1278–1281. doi: 10.1126/science.1185640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Jochemsen AG. Mdmx as an essential regulator of p53 activity. Biochem Biophys Res Commun. 2005;331:750–760. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, McElwee MK, Michalowski J, O'Leary KA, Young KM, Perry ME. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23:462–472. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, LaQuaglia M, Benard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci U S A. 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo C, Shiokawa D, Kodama M, Gaiddon C, Nakagama H, Jochemsen AG, Taya Y, Okamoto K. Cytoplasmic tethering is involved in synergistic inhibition of p53 by Mdmx and Mdm2. Cancer Sci. 2009;100:1291–1299. doi: 10.1111/j.1349-7006.2009.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583:2710–2714. doi: 10.1016/j.febslet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Xiong S, Iwakuma T, Quintas-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci U S A. 2011;108:11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Biju MP, Wang MH, Haase VH, Dong Z. Cytoprotective effects of hypoxia against cisplatin-induced tubular cell apoptosis: involvement of mitochondrial inhibition and p53 suppression. Journal of the American Society of Nephrology : JASN. 2006;17:1875–1885. doi: 10.1681/ASN.2005121371. [DOI] [PubMed] [Google Scholar]
- Wang YV, Leblanc M, Fox N, Mao JH, Tinkum KL, Krummel K, Engle D, Piwnica-Worms D, Piwnica-Worms H, Balmain A, et al. Fine-tuning p53 activity through C-terminal modification significantly contributes to HSC homeostasis and mouse radiosensitivity. Genes Dev. 2011;25:1426–1438. doi: 10.1101/gad.2024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YV, Wade M, Wahl GM. Guarding the guardian: Mdmx plays important roles in setting p53 basal activity and determining biological responses in vivo. Cell Cycle. 2009;8:3443–3444. doi: 10.4161/cc.8.21.9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YV, Wade M, Wong E, Li YC, Rodewald LW, Wahl GM. Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc Natl Acad Sci U S A. 2007;104:12365–12370. doi: 10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster GA, Perkins ND. Transcriptional cross talk between NF-kappaB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.