Abstract
Streptococcus mutans is a Gram-positive oral bacterium that is a primary etiological agent associated with human dental caries. In the oral cavity, S. mutans adheres to immobilized salivary agglutinin (SAG) contained within the salivary pellicle on the tooth surface. Binding to SAG is mediated by cell surface P1, a multifunctional adhesin that is also capable of interacting with extracellular matrix proteins. This may be of particular importance outside of the oral cavity as S. mutans has been associated with infective endocarditis and detected in atherosclerotic plaque. Despite the biomedical importance of P1, its binding mechanisms are not completely understood. In this work, we use atomic force microscopy-based single-molecule and single-cell force spectroscopy to quantify the nanoscale forces driving P1-mediated adhesion. Single-molecule experiments show that full-length P1, as well as fragments containing only the P1 globular head or C-terminal region, binds to SAG with relatively weak forces (~50 pN). In contrast, single-cell analyses reveal that adhesion of a single S. mutans cell to SAG is mediated by strong (~500 pN) and long-range (up to 6000 nm) forces. This is likely due to the binding of multiple P1 adhesins to self-associated gp340 glycoproteins. Such a cooperative, long-range character of the S. mutans–SAG interaction would therefore dramatically increase the strength and duration of cell adhesion. We also demonstrate, at single-molecule and single-cell levels, the interaction of P1 with fibronectin and collagen, as well as with hydrophobic, but not hydrophilic, substrates. The binding mechanism (strong forces, cooperativity, broad specificity) of P1 provides a molecular basis for its multifunctional adhesion properties. Our methodology represents a valuable approach to probe the binding forces of bacterial adhesins and offers a tractable methodology to assess anti-adhesion therapy.
Keywords: Streptococcus mutans, AFM, single-cell force spectroscopy, single-molecule force spectroscopy, bacterial adhesion, salivary agglutinin, P1, antigen I/II
Streptococcus mutans is an acidogenic Gram-positive oral bacterium that is a primary disease-causing agent associated with tooth decay.1 In the oral cavity, S. mutans colonization depends on sucrose-dependent as well as sucrose-independent mechanisms. Sucrose-independent adhesion of S. mutans cells to tooth surfaces involves cell surface proteins such as the cell wall-anchored adhesin P1 (also referred to as Ag I/II, PAc, SpaP or antigen B).2–6 P1 is a multifunctional adhesin that contributes to S. mutans’ ability to adhere to tooth pellicles and cause tooth decay.7–13 In addition to its etiologic association with human dental caries, S. mutans has also been linked to cases of bacterial endocarditis and has been detected in atherosclerotic plaque.14–16 Some strains of S. mutans have been reported to invade human coronary endothelial cells.17,18 In the oral environment, P1 interacts primarily with the glycoprotein salivary agglutinin (SAG) complex predominantly composed of the scavenger receptor glycoprotein 340 (gp340/DMBT1), contained within the salivary pellicle on tooth surfaces.2,4,5,19–23 P1 has also been shown to bind to extracellular matrix proteins such as collagen (Coll)24–27 and fibronectin (Fn),26,28,29 and is involved in cell–cell adhesion as well.30 The ability of P1 to promote bacterial adherence and to affect colonization, cariogenicity and biofilm formation has made it of interest as a therapeutic target.20,31–34
The primary amino acid sequence of P1 consists of a signal sequence, an N-terminal region, alanine-rich repeats (A1–3), an intervening segment containing a variable (V) region, proline-rich repeats (P1–3), C-terminal region consisting of three domains (C1–3,) and a wall-spanning region.35,36 A structural model of P1 derived from crystal structures demonstrates that the A-repeats form a long α-helix that intimately intertwines into a left-handed supercoiled structure with the helical polyproline P-repeats to form an unusually long and narrow stalk.37 A β-rich “globular head” domain, predicted to contain a carbohydrate-binding trench, intervenes the A- and P-repeats at the tip of the extended stalk. The C-terminal region is also globular and comprised of three structurally related β-sandwich domains stabilized by covalent isopeptide bonds.38,39 Within the context of the complete fully folded molecule, this region resides at the opposite end of the stalk. In addition to its unusual tertiary structure, our recent work has demonstrated that a complex ultrastructure of P1 also exists on the cell surface such that a scaffold of covalently attached P1 interacts with more loosely associated adhesive P1 fragments, including the C-terminal fragment previously identified as Antigen II,40 to form the functional adhesive layer.
Various techniques have been used to locate binding domains within both P1 and SAG. An earlier study using a sandwich-type assay, identified the N-terminal segment of P1 (PAc) as a saliva-binding region.41 A subsequent study employing Scatchard analysis suggested the presence of two binding sites within P1 (Ag I/II).20 More recently, surface plasmon resonance (SPR) was used to demonstrate that the apical head region (A3VP1) and the C-terminal fragment (C1–3) each are capable of independent and noncompetitive adherence to immobilized SAG.37 The same group specifically identified the scavenger rich cysteine repeat (SRCR) domains of gp340 as the binding sites within SAG to which A3VP1 and C-terminal P1 fragments adhere.22 It was further shown that calcium-dependent adherence of P1 to SAG stems from calcium-induced conformational changes in the SRCR domains of gp340 that increase their thermal stability.22 Though the aforementioned studies shed light on the adhesion of P1 to the SAG glycoprotein complex, the binding forces involved in the interaction of P1 to SAG as well as to other extracellular matrix proteins have yet to be quantified and the molecular mechanisms fully elucidated.
Atomic force microscopy (AFM) offers unprecedented opportunities for exploring the forces involved in microbial cell adhesion and biofilm formation. Single-molecule force spectroscopy (SMFS) with biospecific AFM tips allows researchers to gain insight into the localization, binding strength and elasticity of individual adhesins, either on purified systems or on live cells. In the pathogenesis context, examples of thoroughly investigated adhesins are the mycobacterial heparin-binding hemagglutinin adhesin (HBHA),42 the agglutinin-like sequence (Als) proteins from Candida albicans,43 and fibronectin binding proteins (FnBPs) from Staphylococcus aureus.44 In addition, single-cell force spectroscopy (SCFS), in which AFM cantilevers are functionalized with microbial cells, makes it possible to quantify adhesive interactions on a single-cell basis. New protocols have been developed for the reliable SCFS analysis of microbes. One approach is FluidFM, where hollow AFM probes with nanoscale apertures are used for manipulating and probing individual cells.45 Another assay for reliable single microbial cell manipulation uses colloidal probes coated with bioinspired polydopamine wet adhesive.46 Currently, an exciting challenge is to combine SMFS and SCFS to gain insight into the binding mechanisms of adhesins. In one such study, Herman et al.47 probed the binding strength of the SdrG adhesin from Staphylococcus epidermidis, revealing that it binds to the blood plasma protein fibrinogen with a strength equivalent to that of a covalent bond, thus much larger than the force of any adhesin investigated so far. Here, we used SMFS and SCFS techniques to unravel the nanoscale adhesion forces between S. mutans P1 and target receptors (SAG, Fn and Coll) as well as to hydrophobic and hydrophilic substrates.
RESULTS AND DISCUSSION
Strength of Single P1–SAG Bonds
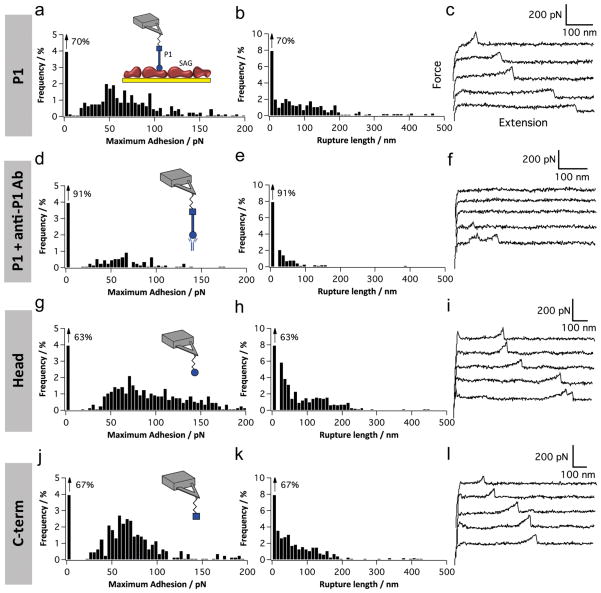
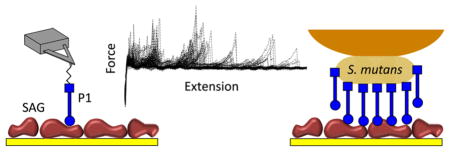
SMFS was used to understand how P1 adhesins adhere to salivary components and extracellular matrix molecules at the molecular level. We measured the forces between AFM tips functionalized with complete P1 molecules or P1 fragments (globular head, C-terminal region) and SAG proteins randomly immobilized on solid substrates (Figure 1a, inset). To ensure single-adhesin detection, the tip was functionalized with a PEG-benzaldehyde linker using a well-established protocol.48 The adhesion force and rupture length histograms, as well as representative retraction force curves obtained at a pulling speed of 1000 nm s−1 for the P1–SAG interaction, are shown in Figure 1a–c. Considerable (~30%) binding events were observed with adhesion forces in the range of 30–200 pN. The most probable force was 57 ± 30 pN (mean values and SD from five independent experiments), a value in the range of the forces reported for other adhesins at similar loading rate (i.e., the rate at which the force is applied to the complex).49 To test the specificity of binding, we blocked the P1-tip using an anti-P1 antibody (mAb 1-6F)50 directed against the head region (Figure 1d–f).51 We observed a substantial decrease in binding frequency (down to ~9%) and in rupture length (<100 nm). The same result was obtained when using an irrelevant BSA-functionalized tip or a surface without SAG proteins confirming the specificity of the measured P1–SAG forces.
Figure 1.
Single-molecule force spectroscopy of the P1–SAG interaction. (a–l) Adhesion force histogram (a, d, g, and j), rupture length histogram (b, e, h, and k), and typical force curves (c, f, i, and l) obtained by recording force curves (n = 1024 curves for each condition) in PBS buffer with 1 mM Ca2+ between solid substrates functionalized with SAG, and AFM tips functionalized with full-length P1 (a–c), full-length P1 blocked with a solution of monoclonal antibodies directed against the P1 head (mAb 1-6F) (d–f), P1 head region (g–i), or P1 C-terminal region (j–l). The schematic representations emphasize the oriented, rod-like structure of P1 and its main structural regions, i.e., globular head (circle), stalk (bar), and C-terminal region (square). For each condition, similar data were obtained using at least 5 different tips and 5 different substrates.
Pulling on P1–SAG bonds gave rise to rupture lengths up to 200 nm, indicating that the mechanically stretched molecules were elongated. While P1 has a stiff, rod-like structure,37 SAG is a large glycoprotein complex that is expected to readily elongate under force. P1 is known to bind glycoprotein 340 (gp340, DMBT-1),5,52,53 a Mr ~ 340-kDa glycoprotein that contains 14 SRCR (scavenger receptor cysteine rich) domains of about 100–110 amino acids each.5,20 Considering that each amino acid contributes 0.36 nm to the contour length of a fully extended polypeptide chain, and that gp340 has 2413 amino acids, full extension of the protein would give a length of ~868 nm. This is much larger than our measured rupture lengths, meaning that most gp340 domains were not unfolded, in line with the presence of disulfide bonds within the SRCR domains.2
Recent studies revealed that the globular head and C-terminal regions of P1 are capable of independent adherence to SAG.37,38 Figure 1g–l shows that the force data obtained for each of these domains were similar to those obtained for full-length P1. This reiterates that both domains contribute to the P1–SAG interaction. The similar force profiles obtained for these two separate fragments suggest they share similar binding properties. In summary our results show (i) that the force of single P1–SAG bonds is relatively weak, (ii) that pulling on these bonds does not lead to the unfolding of gp340 SRCR domains, and (iii) that both the globular head and C-terminal regions bind SAG with the same forces as the full-length molecule.
Binding of P1 to Fibronectin and Collagen
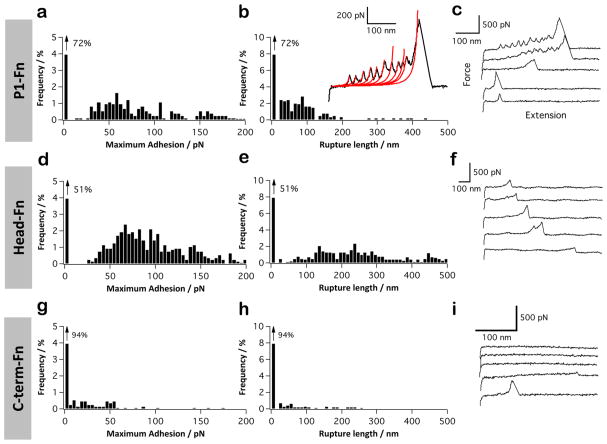
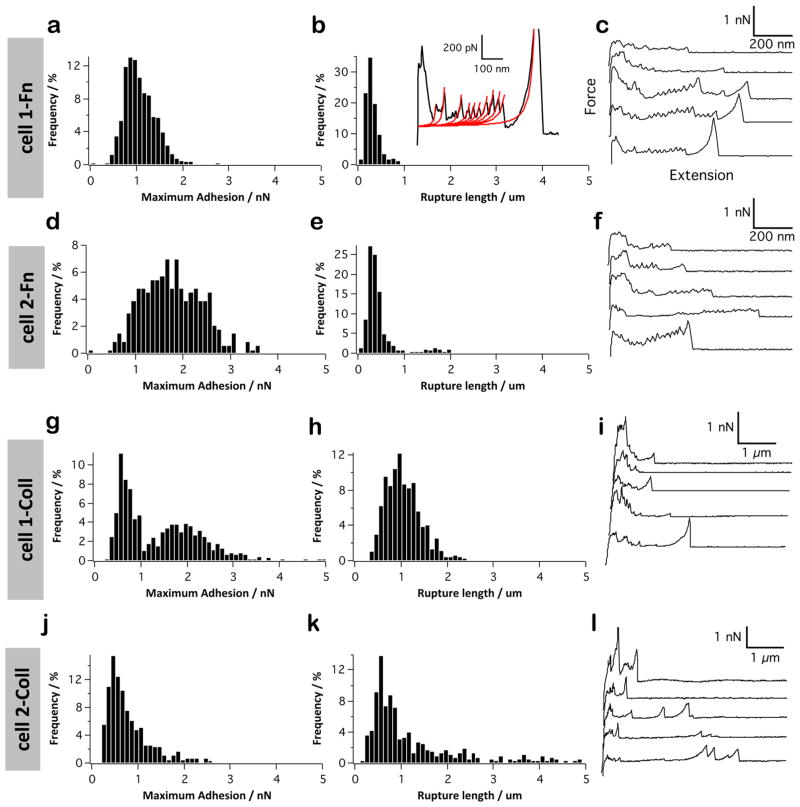
P1 has been reported to mediate adhesion to extracellular matrix proteins,25,54–56 but the mechanisms involved are not fully characterized. We quantified the binding force of P1 with both Fn and Coll, using a constant pulling speed of 1000 nm s−1. Figure 2a–c shows the histograms of adhesion forces and rupture lengths, with representative force curves, recorded between P1-tips and Fn-coated substrates. The binding frequency, ~28%, mean adhesion force, 46 ± 27 pN, and rupture lengths, 25–200 nm, were similar to those measured for the P1–SAG interaction. We note that our adhesion forces and rupture lengths are smaller than those reported by Soell et al. (~250 pN, 50–750 nm).26 However, the surface chemistry used in the earlier study was different and less defined; i.e., P1 molecules were adsorbed onto silicon nitride tips, thus favoring the probing of multiple and loosely immobilized adhesins. In the current study, in a few cases (<1%) multiple force peaks of 100–300 pN were observed, suggesting that multiple protein domains were unfolded one after the other (Figure 2b, inset). Force peaks were well described by the worm-like-chain model (WLC), the change in contour length between consecutive peaks being ΔLc = 25.0 ± 0.6 nm (n = 50 from 15 curves). This value is consistent with the 28–31 nm expected for the ~90 amino acids of single Fn repeats.57,58 The mean unfolding forces, 116 ± 34 and 261 ± 66 pN, are consistent with earlier observations by Oberhauser et al., where they showed a hierarchical unfolding of FnIII domains.57 When the globular head fragment of P1 was used (Figure 2d–f) rather than the full-length molecule, more binding events were observed (49% vs 28%). By contrast, binding events were significantly reduced when we used the C-terminal domain (Figure 2g–i). Although we cannot exclude the involvement of other regions, our data strongly suggest that the globular head is the primary Fn-binding domain. The higher binding probability observed for the head fragment, compared to full P1, could be due to the better exposure of the Fn-binding region on the AFM tip.
Figure 2.
P1 binds fibronectin via the head region. (a–i) Adhesion force histogram (a, d, and g), rupture length histogram (b, e, and h), and typical force curves (c, f, and i) obtained by recording force curves (n = 1024 curves for each condition) in PBS buffer between solid substrates functionalized with fibronectin (Fn), and AFM tips functionalized with full-length P1 (a–c), P1 globular head (d–f), or C-terminal region (g–i). The red lines in panel (b) shows that for the Fn interaction, sawtooth force profiles were observed and well-described by the worm-like-chain model, using a persistence length lp of 0.4 nm: F(x) = kBT/lp [0.25(1−x/Lc) −2 + x/Lc −0.25], where Lc is the contour length of the molecule, kB is the Boltzmann constant, and T the absolute temperature. For each condition, similar data were obtained using at least 3 different tips and 3 different substrates.
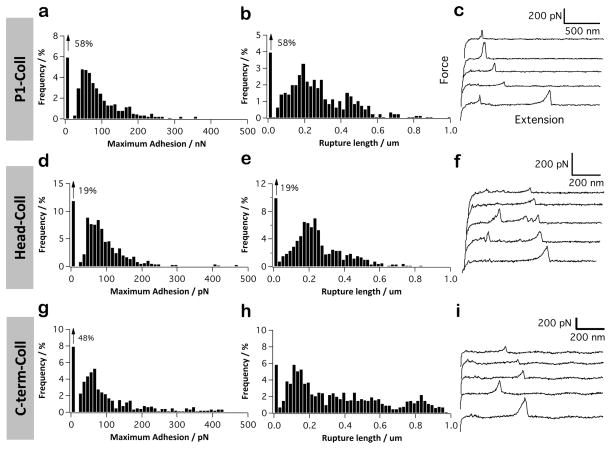
We next probed the binding of P1 to collagen. Figure 3a–c shows that the force data for the P1–Coll interaction were different from those of the P1-Fn interaction in that they showed larger adhesion frequency (~42%), mean adhesion force (50–200 pN) and rupture distances (up to 600 nm). The larger adhesion forces could indicate that single P1–Coll bonds are stronger and/or that single P1 are able to bind to multiple sites on the Coll molecule. The longer extensions reflect the long, fibrillar structure of collagen. Similar to Fn, more binding events were observed with the globular head fragment than with the full-length protein (Figure 3d–f), suggesting it is the primary Coll-binding domain. However, substantial binding was also observed with the C-terminal region (Figure 3g–i), supporting the notion that both the globular head and the C-terminal regions are Coll-binding domains.
Figure 3.
P1 binds collagen via both the head and C-terminal regions. (a–i) Adhesion force histogram (a, d, and g), rupture length histogram (b, e, and h), and typical force curves (c, f, and i) obtained by recording force curves (n = 1024 curves for each condition) in PBS with 1 mM Ca2+ buffer between solid substrates functionalized with collagen (Coll), and AFM tips functionalized with full-length P1 (a–c), P1 globular head (d–f), or C-terminal region (g–i). For each condition, similar data were obtained using at least 3 different tips and 3 different substrates.
P1 Mediates Hydrophobic Interactions
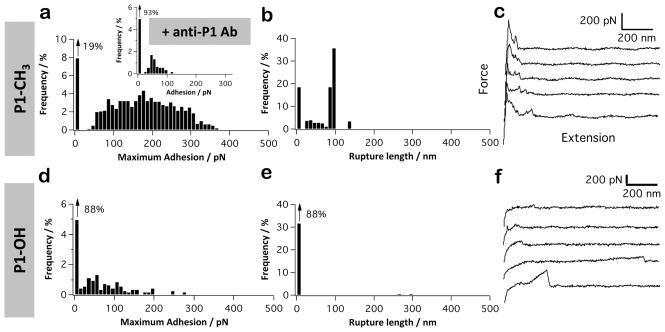
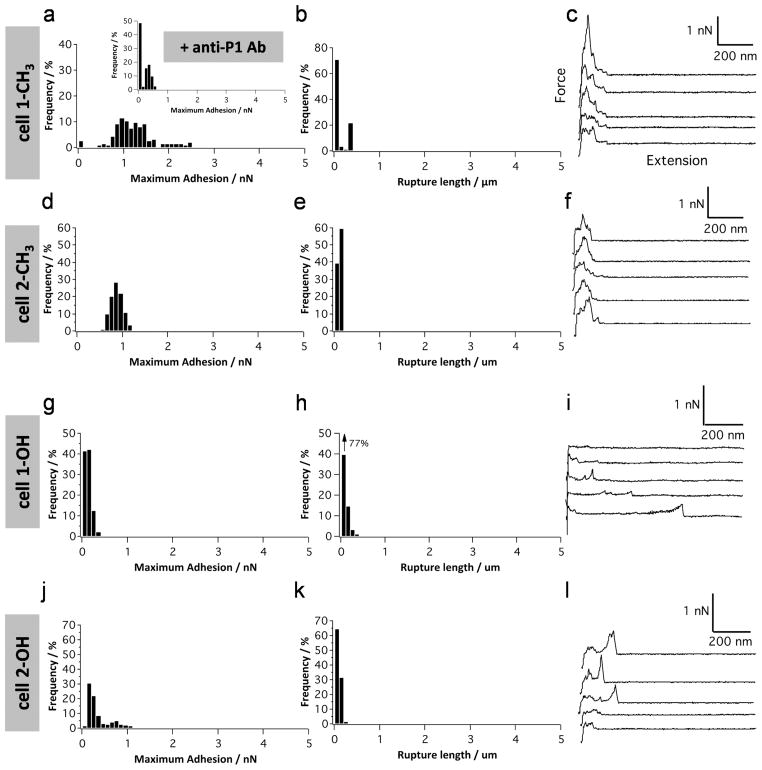
Hydrophobic interactions represent an important driving force for the adhesion of microbial pathogens.59 Hydrophobic components can be covalently bound to the cell wall, like cell wall proteins, or intercalated into the outer membrane, like fimbriae. In the oral cavity context, Streptococcus sanguis60 and Streptococcus salivarius61 have been shown to attach to saliva-coated hydroxylapatite via hydrophobic interactions. Cell surface proteins of S. mutans, including P1, are known to contribute to S. mutans hydrophobicity;62–64 therefore, we measured the forces between single adhesins and hydrophobic, methyl-terminated substrates at a pulling speed of 1000 nm s−1. As can be seen in Figure 4a–c, force signatures differed from those associated with Fn and Coll. Most force curves (81%) showed force peaks of 50–350 pN magnitude and 20–100 nm rupture length. We attribute these to hydrophobic interactions with P1 for the following reasons. First, rupture lengths were much shorter than for protein-coated substrates, a finding consistent with the notion that hydrophobic forces are short-range (<10 nm) in nature. Presumably, the 20–100 nm ruptures correspond to the extension of one or two P1 molecules, not surprising since P1 has been shown to self-associate. Second, adhesive events were substantially diminished upon blocking the P1-tip with antibodies directed against the apical head of P1 (mAb 1-6F), suggesting that this region contributes to hydrophobic binding (Figure 4a, inset). Third, a substantial reduction of adhesion frequency was noted when measuring forces on hydrophilic hydroxyl-terminated substrates (Figure 4d–f). Hence, our single-molecule data provide direct evidence that P1 mediates strong nonspecific hydrophobic interactions, in line with the notion that P1 is rich in hydrophobic amino acids that are prominently exposed on the surface of the crystal structures of the protein.37,38,65 This property is relevant as hydrophobic forces may represent a clinically important mechanism for S. mutans to colonize the tooth environment. Patients with high dental caries scores demonstrated a higher prevalence of hydrophobic bacteria than low caries patients.66 In addition, compounds that diminish the surface hydrophobicity of S. mutans are now being reported to inhibit bacterial adherence in vitro67–69 and cariogenicity in an animal model.67
Figure 4.
P1 displays strong hydrophobic properties. (a–f) Adhesion force histogram (a and d), rupture length histogram (b and e), and typical force curves (c and f) obtained by recording force curves (n = 1024 curves for each condition) in PBS buffer with 1 mM Ca2+ between AFM tips functionalized with full-length P1 and methyl-terminated substrates (a–c) or hydroxyl-terminated substrates (d–f). The inset in (a) shows the adhesion force histogram obtained between P1 and a methyl-terminated substrate following blocking with a solution of monoclonal antibodies directed against the P1 head (mAb 1-6F). For each condition, similar data were obtained using at least 3 different tips and 3 different substrates.
Binding Mechanism of S. mutans: Cooperativity, Broad Specificity
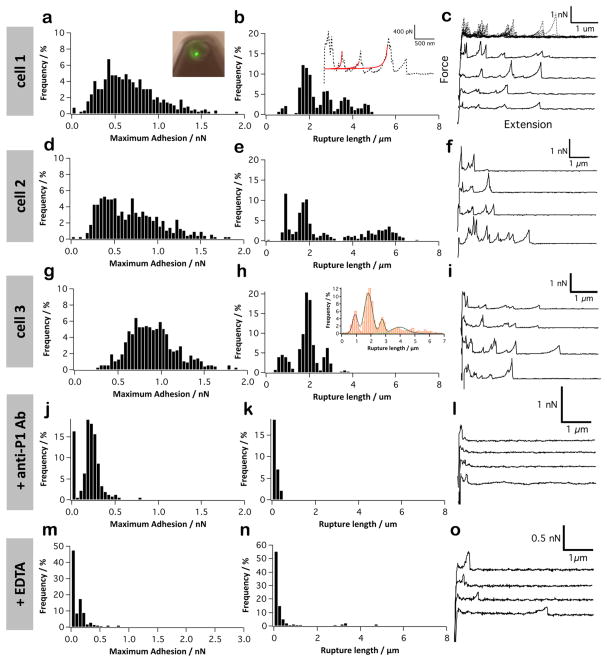
Next, SCFS was used to measure the binding forces between whole cells and SAG. Individual S. mutans bacteria were attached to AFM cantilevers without altering their viability (Figure 5a, inset). Force–distance curves were then collected between the cellular probes and biotic and abiotic substrates. For both SAG and Coll interactions, we used a pulling speed of 5000 nm s−1, rather than 1000 nm s−1 as in SMFS, because we observed that at lower pulling speeds the cells either died or detached from the cantilever. Figure 5 shows that the force data for the interaction between S. mutans cells and SAG surfaces were notably different from those of single P1–SAG bonds. As shown in Figure 5a–i, the magnitude (100–1500 pN) and range (500–6000 nm) of the adhesion force profiles were substantially larger than those observed with single-P1 molecules, implying that the S. mutans–SAG interaction involves multiple bonds. These interaction forces were inhibited by anti-P1 antibodies directed against the apical head (mAb 1-6F) (Figure 5j–l) and upon addition of EDTA (Figure 5m–o), supporting the notion that P1 binding to SAG is specific and calcium-dependent.19,22,38 We conclude that the measured forces therefore originate from cell surface-localized P1 molecules. These results reinforce that antibodies of appropriate specificity would be of value in immunotherapeutic approaches to disrupt S. mutans adherence to immobilized SAG. Secretory IgA and serum IgG present in gingival crevicular fluid are both associated with active protection in immunized and naturally sensitized animals or human subjects.31–35 P1 has shown promise not only as a vaccine component for active immunization, but also as a target of passively administered monoclonal antibodies, which appear to work by influencing the conformation of the antigen on the cell surface.51
Figure 5.
Single-cell force spectroscopy of the S. mutans-SAG interaction. (a–i) Adhesion force histogram (a, d, and g), rupture length histogram (b, e, and h), and typical force curves (c, f, and i) obtained by recording force curves (n = 300 curves for each condition) in PBS buffer with 1 mM Ca2+ between three different S. mutans bacteria and solid substrates functionalized with SAG. (j–l) Control experiment in which the bacterial cell surface was blocked with a solution of monoclonal antibodies directed against the P1 head. (m–o) Control experiment in which the bacterial cell surface was treated with 10 mM EDTA. The fluorescence image in (a) shows a single living bacterium (in green) immobilized on a colloidal cantilever (see text for details). The inset in (b) shows that most curves with large adhesion force peaks were well-described by the WLC model (red lines). The inset in (h) is the rupture length histogram obtained by merging the data sets of the three cells. A total of 8 different cells from 5 independent cultures were probed.
Comparison of the mean adhesion forces of single P1 (50 pN) and single whole cells (500 pN) suggests that the cell–SAG interaction involves about 10 adhesins. Note that this is a rough estimate that should be considered with caution. For technical reasons, we used different pulling speeds, thus loading rates, for probing P1–SAG (1000 nm s−1) and cell–SAG (5000 nm s−1) interactions. This difference in loading rate, although not very large, will influence the measured adhesion forces. Importantly, this straightforward calculation assumes parallel loading of all bonds and linearly additive adhesion forces, which may not be strictly correct for single-live cell experiments.70 When multiple bonds are exposed to increasing external force, they establish a cooperative coupled system. Each bond is exposed to an individual force equal to the external force divided by the number of bonds. As the probability distribution for rupture forces is widely spread, it is likely that, in a multiple binding complex, at least one of many individual bonds ruptures at a low force. As a result, the force acting on each of the remaining attached bonds increases and the process of unbinding accelerates.71 Also, active rebinding may increase the lifetime of multivalent attachments. For instance, ligand dissociation from a surface might be strongly influenced by receptor clustering. Hence, it is not straightforward to link the rupture of multiple bonds to the behavior of single bonds.
Nevertheless, a remarkable feature, not observed with single-molecule analyses of P1, was the presence of repeated force patterns in the curves. Superimposition of multiple curves indeed revealed the presence of four main adhesion force peaks of 500–800 pN magnitude (most prominent in cell 1, Figure 5c) and equally spaced by ~900 nm (multiple peak Gaussian fits of the rupture distribution yielded the following peaks: 0.90 ± 0.1, 1.8 ± 0.2, 2.7 ± 0.1, 3.9 ± 0.5 μm, see inset of Figure 5h). Our finding that the strength and spacing of the main adhesion peaks did not vary much, even from one cell to another, suggests that these events result from the rupture of laterally associated P1–gp340 complexes rather than from the synchronous breakage of independent bonds. Therefore, we postulate that bacterial adhesion to SAG is enhanced by cooperative binding of multiple P1–gp340 complexes. Such a cooperative model, reminiscent of the binding mechanism of cadherins,72 is strongly supported by the fact that both the adhesin and its gp340 binding partner contain more than one binding site. Larson and colleagues showed that P1 has two SAG adherence regions adhering in a noncompetitive manner, suggesting that multivalency plays an important role in adhesion.38 The presence of repeated P1 binding peptides in the multiple SRCR domains of gp340 has been suggested to promote multivalent interaction with bacterial cells.21 Our AFM data provide direct evidence supporting this speculation. In the same vein, compared to single SRCR domains, three tandem SRCR domains displayed a cooperative increase in their bacterial adherence.22
It is tempting to speculate on the molecular origin of the ~900 nm repeating distances in the force profiles. This value is very close to the length expected for a fully extended gp340, ~868 nm, leading us to believe that the four force peaks correspond to the sequential unfolding of four gp340 proteins. This is in contrast to our single molecule experiments in which the force was insufficient to unfold gp340. That multiple proteins are being stretched is not surprising as gp340 has the ability to self-associate and form higher order complexes, as large as 5000 kDa.22 Hence, pulling on one protein would lead to the stretching of self-associated gp340 aggregates. The speculated occurrence of such protein unfolding events is supported by the fact that force peaks were well described with the WLC model (Figure 5b inset). In summary, while single P1–SAG bonds are too weak to unfold gp340 proteins, we found that the interaction of S. mutans bacteria with SAG involves cooperative, multivalent bonds between multiple P1 adhesins and self-associated gp340 proteins. When subjected to force, the cell–SAG bond shows strong, long-range forces reflecting the unfolding of multiple gp340 proteins. In the host environment, we expect that the multivalent and long-range character of the P1–SAG bonds would dramatically increase the strength and duration of the bacterial–substrate interaction, thus helping bacteria to resist physiological shear stress and remain attached.
We also probed the interaction of whole S. mutans cells with Fn and Coll (Figure 6). Compared to the SAG interaction, forces higher than 1000 pN were much frequently observed, and the rupture length shorter. Comparison with SMFS data (Figures 2 and 3) suggests that Fn and Coll interactions involve ~15 and ~30 P1 molecules, respectively. Note that for reasons explained above these rough estimates should be considered with caution. Unfolding patterns were frequently observed for Fn, with an average unfolding force of 265 ± 78 pN (~12% of the force curves, inset in Figure 6b). Fitting the peaks with the WLC model yielded a change in contour length between consecutive peaks of ΔLc = 26.7 ± 2 nm (n = 120 from 20 force curves). These values, similar to those observed by SMFS, are consistent with the unraveling of FnIII domains from different molecular constructs.57,58
Figure 6.
Interaction forces between S. mutans and fibronectin/collagen. Adhesion force histogram (a, d, g, and j), rupture length histogram (b, e, h, and k), and typical force curves (c, f, i, and l) obtained by recording force curves (n = 300 curves for each condition) in PBS buffer with 1 mM Ca2+ between two different S. mutans bacteria and solid substrates functionalized with either Fn (a–f) or Coll (g–l). The inset in (b) demonstrates that some Fn curves showed sawtooth patterns well-described by the WLC model (red lines).
Lastly, to confirm the involvement of hydrophobic interactions in P1-mediated adhesion, we measured the forces between S. mutans and a hydrophobic surface (Figure 7a–f). All force curves showed force peaks of 500–1500 pN magnitude and short (<100 nm) rupture length. In the light of the SMFS data (Figure 4), we attribute these forces to the hydrophobic interaction of ~10 P1 molecules. Lower adhesion forces were observed upon blocking with an anti-P1 antibody (mAb 1-6F) (Figure 7a, inset), strongly suggesting the involvement of P1 in hydrophobic interactions. As in SMFS, a pronounced reduction of adhesion frequency was noted on a hydrophilic surface (Figure 7g–l).
Figure 7.
S. mutans adhesion involves hydrophobic interactions. Adhesion force histogram (a, d, g, and j), rupture length histogram (b, e, h, and k), and typical force curves (c, f, i, and l) obtained by recording force curves (n = 300 curves for each condition) in PBS buffer with 1 mM Ca2+ between two different S. mutans bacteria and methyl-terminated substrates (a–f) or hydroxyl-terminated substrates (g–l). The inset in (a) shows the adhesion force histogram obtained on a methyl-terminated substrate following blocking with a solution of monoclonal antibodies directed against the P1 head (mAb 1-6F).
CONCLUSIONS
Recent SCFS experiments have revealed that microbial cell adhesion molecules display a variety of mechanical responses that are important for cell adhesion. Example of such mechanical properties include ligand recognition by specific adhesins,47 unfolding of multifunctional adhesins,73 nanospring properties of Gram-positive pili,74 extension of Gram-negative pili,75 macromolecular bonds in bacterial–yeast adhesion,76 and lipid tethers in microbe–host interactions.75 By combining SMFS and SCFS, we have shown here that the interaction of S. mutans bacteria with SAG is mediated by strong, long-range forces between multiple P1 adhesins and several gp340 glycoproteins, thus favoring a model in which cooperative binding greatly enhances bacterial adhesion.
Characterizing how P1 mediates S. mutans adhesion to salivary components and extracellular matrix molecules is important to a comprehensive understanding of the organism’s ability to cause pathology. In the past years, AFM has been a valuable method in analyzing the interaction forces of S. mutans.26,28,77–79 However, the complete binding mechanisms of P1 are not fully described. An important finding of the current work is that the mechanical response of P1–SAG bonds is dramatically different in single-molecule and single-cell systems. We showed that the force of single P1–SAG bonds is weak (50 pN), involves both the globular head and C-terminal region of P1, and does not trigger the unfolding of the SRCR domains of gp340 proteins. In contrast, the interaction of whole S. mutans bacteria with SAG is mediated by strong, long-range forces (~500 pN) between multiple (up to 10) P1 adhesins and multiple (up to 4) gp340 glycoproteins. These results suggest that the weak unit binding force of P1 is amplified by the lateral association of multiple P1–gp340 complexes. The cooperative and long-range bonds between S. mutans may therefore increase the strength and duration of the bacterial–host interaction. We also found that S. mutans binds to Fn and Coll with forces stronger but shorter than those of the S. mutans–SAG interaction. Our finding of strong binding of P1 to these extracellular matrix (ECM) proteins may also be of particular biological relevance outside of the oral cavity as S. mutans has been associated with infective endocarditis as well as detected in atherosclerotic plaque.14–16 The exact roles of ECM-binding proteins to the etiology of heart disease remain to be dissected; however, S. mutans strains that have been isolated from cardiac lesions15,16 or that can invade human coronary artery endothelial cells in vivo17,18 are known to produce P1 and/or another collagen binding protein, Cnm. Lastly, P1 mediates strong, short-range interactions of S. mutans with a hydrophobic surface, implying that hydrophobic forces may contribute to the adhesion of S. mutans. In addition to adhesion to salivary constituents such as SAG, bacterial hydrophobicity has also been reported to correlate with adhesion of oral species, including S. mutans, to dental biomaterials.80 The observed composite binding mechanisms of P1 (strong forces, cooperativity, unfolding patterns, broad specificity) helps to explain its wide-ranging properties as a multifunctional adhesin. There are several issues to address in future research. In the present study, force measurements were performed at room temperature. As the temperature in the oral cavity is higher, it would be interesting to assess to what extent an increase in temperature would impact the observed force behaviors. Also, varying the loading rate in force experiments should made it possible to determine the off-rate of specific P1 bonds.
METHODS
Bacterial Strains and Growth Conditions
Cells from S. mutans wild-type strain NG881 were grown in Tryptic Soy Casein (TSC) broth (Bio-Rad, France) by inoculating a single colony of bacteria into 10 mL of TSC and incubating for 18–24 h at 37 °C. Cells were harvested by centrifugation at 5000g for 10 min, washed twice with PBS and resuspended in 5 mL of PBS. The original suspension was further diluted in PBS with 1 mM CaCl2 to obtain an optical density of 0.01 at 540 nm. The cell suspension was gently vortexed to release cells from aggregates that form in standing cultures before use.
Anti-P1 Monoclonal Antibodies
Monoclonal antibody 1-6F50 directed against the globular head of P151 was purified from murine ascites fluid using Protein A affinity chromatography as described previously.82
Preparation of Biotic and Abiotic Surfaces
SAG, Fn and Coll proteins were covalently immobilized, in a random orientation, onto self-assembled monolayers (SAMs) of carboxyl-terminated alkanethiols. Glass coverslips coated with a thin (~30 nm) gold layer were immersed overnight in ethanol solutions containing 1 mM 16-mercaptohexadecanoic acid (Sigma-Aldrich, Belgium) and 11-mercapto-1-undecanol (Sigma-Aldrich) in a 1:9 ratio and then rinsed with ethanol and dried. A brief (~30 s) sonication was applied to remove loosely bound alkanethiol aggregates. The resulting monolayers were further immersed for 30 min in a solution containing 20 mg/mL N-hydroxysuccinimide (NHS) (Sigma-Aldrich) and 50 mg/mL 1-ethyl-3-(3-(dimethylamino)propyl)-carbodiimide (EDC) (Sigma-Aldrich) and subsequently rinsed with water. The activated surfaces were then incubated with either 100 ng/mL SAG,83 0.2 mg/mL Fn (from human plasma, Sigma-Aldrich), or 1 mg/mL collagen (Type 1 from rat tail, Sigma-Aldrich), in PBS for at least 2 h, followed by rinsing with the same buffer.
For preparing hydrophobic and hydrophilic substrates, gold-coated glass coverslips were immersed overnight in a solution of 1 mM of either 1-dodecanethiol (Sigma-Aldrich) or 11-mercapto-1-undecanol (Sigma-Aldrich), then rinsed with ethanol and dried under N2. All surfaces were freshly prepared and used the same day.
Functionalization of AFM Tips with Full-Length P1 and P1 Fragments
For SMFS, AFM tips were functionalized with either the full-length P1 adhesin, or globular head (A3VP1) or C-terminal fragments via a 6 nm-long polyethylene glycol (PEG) linker. MSCT cantilevers (Olympus) were first washed with chloroform and ethanol, placed in an UV-ozone cleaner for 15 min, and immersed overnight in 5.6 M ethanolamine hydrochloride (in DMSO) to generate amino groups on the tip surface. The cantilevers were then reacted with PEG linkers carrying benzaldehyde groups on their free-tangling end48 and further immersed in 1% citric acid solution for 10 min. After a rinsing step with Milli-Q water (ELGA LabWater), the cantilevers were incubated in a 100–200-μL droplet of 0.2 mg/mL full-length P1 or P1 fragments in PBS containing 10 mM NaCNBH3. After 1 h of incubation, the unreacted aldehyde groups were passivated with 5 μL of a 1 M ethanolamine hydrochloride solution (pH 9.5) for 10 min. Finally, the cantilevers were washed with PBS and stored in the same buffer with 1% NaN3 until use (within 7 days).
Single-Molecule Force Spectroscopy
For single-molecule experiments, force–distance curves were recorded at room temperature (20 °C) in PBS buffer (pH 7.4) with 1 mM CaCl2 using a Nanoscope VIII Multimode AFM (Bruker Corporation, Santa Barbara, CA) with the above-mentioned functionalized tips and substrates. Protein-coated substrates (SAG, Fn, or Coll) were gently rinsed with five baths of PBS, and attached to a steel sample puck (Bruker Corporation) using a double-sided adhesive and mounted onto the AFM liquid cell. The spring constants of the cantilevers were typically in the range of 0.01–0.04 N/m, as determined by thermal noise method.84 Force mapping was performed by collecting a 32 × 32 array of force–distance curves on a 1 × 1 μm2 area of the protein surfaces. All force curves were recorded at ~100 ms contact time, with a maximum applied force of 250 pN and 1000 nm s−1 approach and retraction speeds.
Cell Probe Preparation
Cell probes for SCFS were prepared using a protocol that combines colloidal probe cantilevers and bioinspired polydopamine glue.46 With the use of a Nano-scope VIII Multimode AFM, triangular shaped tipless cantilevers (NPO10, Microlevers, Veeco Metrology Group) were brought into contact with a thin (~2 μL spread onto a 12 mm diameter glass coverslip) layer of UV-curable glue (NOA 63, Norland Edmund Optics), and subsequently onto a silica microsphere (~6.1 mm diameter, Bangs Laboratories). After ~1 min of contact, the colloidal probe was cured for 12–15 min under a UV-lamp. The cantilever was then immersed for 1–4 h in 4 mg/mL of dopamine hydrochloride (99%, Sigma-Aldrich, Belgium) in 10 mM Tris buffer (pH 8.5), rinsed and gently mounted onto the AFM tip holder. To attach a single bacterium onto the colloidal probe, 50 μL of the bacterial suspension (~107 cells/mL) was incubated on a glass-bottomed Petri dish, which also contains the functionalized surfaces. To the bacterial suspension was added 2 μL of a 1:1 Syto 9 (green fluorescent nucleic acid stain)/Propidium iodide [red-fluorescent nuclear and chromosome counterstain, Live–dead Baclight viability kit (Invitrogen, kit L7012)] mixture at 1.5 mM to confirm the viability of the bacteria. The Petri dish was then flooded with PBS buffer (pH 7.4) with 1 mM CaCl2. Before attaching a bacterium, the spring constant of the cell probe, which was typically in the range of 0.05–0.1 N/m, was determined using the thermal noise method. The colloidal probe was then mounted into the AFM and brought into contact with an isolated bacterium. Proper attachment and positioning of single bacterium on the colloidal probe was achieved using a Bioscope Catalyst (Bruker Corporation, Santa Barbara, CA) equipped with a Zeiss Axio Observer Z1 and a Hamamatsu camera C10600. After confirming proper attachment of the bacterial cell by fluorescence imaging, the bacterial cell probe was positioned over the functionalized surface of interest without dewetting.
Single-Cell Force Spectroscopy
SCFS measurements were performed at room temperature (20 °C) in PBS buffer (pH 7.4) with 1 mM CaCl2 using a Bioscope Catalyst AFM. With the inverted optical microscope, the bacterial probe was engaged onto the protein surface of interest. Force curves were recorded on 3–5 different spots. For both SAG and Coll surfaces, 250 pN of contact force, ~100 ms delay and 5000 nm s−1 approach and retraction speeds were used. On Fn surfaces, 1000 nm s−1 approach and retraction speeds were applied. For each condition, 3–10 bacterial cells from independent cultures were probed. The same procedure was used to perform SMFS and SCFS data extraction, calculation and reporting.74
Acknowledgments
Work at the Université Catholique de Louvain was supported by the National Fund for Scientific Research (FNRS), the Université Catholique de Louvain (Fondation Louvain-Prix De Merre), the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Y.F.D. is a Research Director of the FNRS. Work at the University of Florida was supported by National Institute of Health/National Institute of Dental and Craniofacial Research Grants R01DE08007 and R01DE21789 to L.J.B.
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
REFERENCES AND NOTES
- 1.Hamada S, Slade HD. Biology, Immunology, and Cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bikker FJ, Ligtenberg AJ, Nazmi K, Veerman EC, van’t Hof W, Bolscher JG, Poustka A, Nieuw Amerongen AV, Mollenhauer J. Identification of the Bacteria-Binding Peptide Domain on Salivary Agglutinin (gp-340/DMBT1), a Member of the Scavenger Receptor Cysteine-rich Superfamily. J Biol Chem. 2002;277:32109–32115. doi: 10.1074/jbc.M203788200. [DOI] [PubMed] [Google Scholar]
- 3.Brady LJ, Piacentini DA, Crowley PJ, Oyston PC, Bleiweis AS. Differentiation of Salivary Agglutinin-mediated Adherence and Aggregation of Mutans Streptococci by Use of Monoclonal Antibodies against the Major Surface Adhesin P1. Infect Immun. 1992;60:1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oho T, Yu H, Yamashita Y, Koga T. Binding of Salivary Glycoprotein-secretory Immunoglobulin A Complex to the Surface Protein Antigen of Streptococcus mutans. Infect Immun. 1998;66:115–121. doi: 10.1128/iai.66.1.115-121.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prakobphol A, Xu F, Hoang VM, Larsson T, Bergstrom J, Johansson I, Frangsmyr L, Holmskov U, Leffer H, Nilsson C, et al. Salivary Agglutinin, which Binds Streptococcus mutans and Helicobacter pylori, is the Lung Scavenger Receptor Cysteine-rich Protein gp-340. J Biol Chem. 2000;275:39860–39866. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- 6.Ray CA, Gfell LE, Buller TL, Gregory RL. Interactions of Streptococcus mutans Fimbria-associated Surface Proteins with Salivary Components. Clin Diagn Lab Immunol. 1999;6:400–404. doi: 10.1128/cdli.6.3.400-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuramitsu HK. Streptococcus mutans: Molecular Genetic Analysis. American Society for Microbiology; Washington, DC: 2000. [Google Scholar]
- 8.Crowley PJ, Brady LJ, Michalek SM, Bleiweis AS. Virulence of a spaP Mutant of Streptococcus mutans in a Gnotobiotic Rat Model. Infect Immun. 1999;67:1201–1206. doi: 10.1128/iai.67.3.1201-1206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cvitkovitch DG, Li YH, Ellen RP. Quorum Sensing and Biofilm Formation in Streptococcal Infections. J Clin Invest. 2003;112:1626–1632. doi: 10.1172/JCI20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolenbrander PE. Oral Microbial Communities: Biofilms, Interactions, and Genetic Systems. Annu Rev Microbiol. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 11.Levesque CM, Voronejskaia E, Huang YC, Mair RW, Ellen RP, Cvitkovitch DG. Involvement of Sortase Anchoring of Cell Wall Proteins in Biofilm Formation by Streptococcus mutans. Infect Immun. 2005;73:3773–3777. doi: 10.1128/IAI.73.6.3773-3777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell TJ. The Pathogenesis of Streptococcal Infections: From Tooth Decay to Meningitis. Nat Rev Microbiol. 2003;1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- 13.Shemesh M, Tam A, Steinberg D. Expression of Biofilm-Associated Genes of Streptococcus mutans in Response to Glucose and Sucrose. J Med Microbiol. 2007;56:1528–1535. doi: 10.1099/jmm.0.47146-0. [DOI] [PubMed] [Google Scholar]
- 14.Gendron R, Grenier D, Maheu-Robert L. The Oral Cavity as a Reservoir of Bacterial Pathogens for Focal Infections. Microbes Infect. 2000;2:897–906. doi: 10.1016/s1286-4579(00)00391-9. [DOI] [PubMed] [Google Scholar]
- 15.Nakano K, Inaba H, Nomura R, Nemoto H, Takeda M, Yoshioka H, Matsue H, Takahashi T, Taniguchi K, Amano A, et al. Detection of Cariogenic Streptococcus mutans in Extirpated Heart Valve and Atheromatous Plaque Specimens. J Clin Microbiol. 2006;44:3313–3317. doi: 10.1128/JCM.00377-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nomura R, Nakano K, Nemoto H, Fujita K, Inagaki S, Takahashi T, Taniguchi K, Takeda M, Yoshioka H, Amano A, et al. Isolation and Characterization of Streptococcus mutans in Heart Valve and Dental Plaque Specimens from a Patient with Infective Endocarditis. J Med Microbiol. 2006;55:1135–1140. doi: 10.1099/jmm.0.46609-0. [DOI] [PubMed] [Google Scholar]
- 17.Abranches J, Miller JH, Martinez AR, Simpson-Haidaris PJ, Burne RA, Lemos JA. The Collagen-binding Protein Cnm is Required for Streptococcus mutans Adherence to and Intracellular Invasion of Human Coronary Artery Endothelial Cells. Infect Immun. 2011;79:2277–2284. doi: 10.1128/IAI.00767-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abranches J, Zeng L, Belanger M, Rodrigues PH, Simpson-Haidaris PJ, Akin D, Dunn WA, Jr, Progulske-Fox A, Burne RA. Invasion of Human Coronary Artery Endothelial Cells by Streptococcus mutans OMZ175. Oral Microbiol Immunol. 2009;24:141–145. doi: 10.1111/j.1399-302X.2008.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ericson T, Rundegren J. Characterization of a Salivary Agglutinin Reacting with a Serotype c Strain of Streptococcus mutans. Eur J Biochem. 1983;133:255–261. doi: 10.1111/j.1432-1033.1983.tb07456.x. [DOI] [PubMed] [Google Scholar]
- 20.Hajishengallis G, Koga T, Russell MW. Affinity and Specificity of the Interactions between Streptococcus mutans Antigen I/II and Salivary Components. J Dent Res. 1994;73:1493–1502. doi: 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- 21.Bikker FJ, Ligtenberg AJ, End C, Renner M, Blaich S, Lyer S, Wittig R, van’t Hof W, Veerman EC, Nazmi K, et al. Bacteria Binding by DMBT1/SAG/gp-340 is Confined to the VEVLXXXXW Motif in its Scavenger Receptor Cysteine-rich Domains. J Biol Chem. 2004;279:47699–47703. doi: 10.1074/jbc.M406095200. [DOI] [PubMed] [Google Scholar]
- 22.Purushotham S, Deivanayagam C. The Calcium-induced Conformation and Glycosylation of Scavenger-rich Cysteine Repeat (SRCR) Domains of Glycoprotein 340 Influence the High Affinity Interaction with Antigen I/II Homologs. J Biol Chem. 2014;289:21877–21887. doi: 10.1074/jbc.M114.565507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell MW, Mansson-Rahemtulla B. Interaction between Surface Protein Antigens of Streptococcus mutans and Human Salivary Components. Oral Microbiol Immunol. 1989;4:106–111. doi: 10.1111/j.1399-302x.1989.tb00107.x. [DOI] [PubMed] [Google Scholar]
- 24.Love RM, McMillan MD, Jenkinson HF. Invasion of Dentinal Tubules by Oral Streptococci is Associated with Collagen Recognition Mediated by the Antigen I/II Family of Polypeptides. Infect Immun. 1997;65:5157–5164. doi: 10.1128/iai.65.12.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen FC, Assev S, van der Mei HC, Busscher HJ, Scheie AA. Functional Variation of the Antigen I/II Surface Protein in Streptococcus mutans and Streptococcus intermedius. Infect Immun. 2002;70:249–256. doi: 10.1128/IAI.70.1.249-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soell M, Hemmerle J, Hannig M, Haikel Y, Sano H, Selimovic D. Molecular Force Probe Measurement of Antigen I/II-Matrix Protein Interactions. Eur J Oral Sci. 2010;118:590–595. doi: 10.1111/j.1600-0722.2010.00785.x. [DOI] [PubMed] [Google Scholar]
- 27.Switalski LM, Butcher WG, Caufield PC, Lantz MS. Collagen Mediates Adhesion of Streptococcus mutans to Human Dentin. Infect Immun. 1993;61:4119–4125. doi: 10.1128/iai.61.10.4119-4125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busscher HJ, van de Belt-Gritter B, Dijkstra RJ, Norde W, van der Mei HC. Streptococcus mutans and Streptococcus intermedius Adhesion to Fibronectin Films Are Oppositely Influenced by Ionic Strength. Langmuir. 2008;24:10968–10973. doi: 10.1021/la8016968. [DOI] [PubMed] [Google Scholar]
- 29.Kelemen L, Rizk S, Debreczeny M, Ogier J, Szalontai B. Streptococcal Antigen I/II Binds to Extracellular Proteins through Intermolecular Beta-sheets. FEBS Lett. 2004;566:190–194. doi: 10.1016/j.febslet.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 30.Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus Adherence and Colonization. Microbiol Mol Biol Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kt S, Kmk M, NB, Jimson S, RS Dental Caries Vaccine—A Possible Option? J Clin Diagn Res. 2013;7:1250–1253. doi: 10.7860/JCDR/2013/5246.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Q, Katz J, Zhang P, Ashtekar AR, Gaddis DE, Fan M, Michalek SM. Contribution of a Streptococcus mutans Antigen Expressed by a Salmonella Vector Vaccine in Dendritic Cell Activation. Infect Immun. 2011;79:3792–3800. doi: 10.1128/IAI.05338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S. Dental Caries and Vaccination Strategy Against the Major Cariogenic Pathogen, Streptococcus mutans. Curr Pharm Biotechnol. 2014;14:960–966. doi: 10.2174/1389201014666131226144339. [DOI] [PubMed] [Google Scholar]
- 34.Zhao W, Minderman H, Russell MW. Identification and Characterization of Intestinal Antigen-presenting Cells Involved in Uptake and Processing of a Nontoxic Recombinant Chimeric Mucosal Immunogen Based on Cholera Toxin Using Imaging Flow Cytometry. Clin Vaccine Immunol. 2014;21:74–84. doi: 10.1128/CVI.00452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, Jenkinson HF. The Changing Faces of Streptococcus Antigen I/II Polypeptide Family Adhesins. Mol Microbiol. 2010;77:276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heim KP, Crowley PJ, Brady LJ. An Intramolecular Interaction Involving the N terminus of a Streptococcal Adhesin Affects Its Conformation and Adhesive Function. J Biol Chem. 2013;288:13762–13774. doi: 10.1074/jbc.M113.459974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larson MR, Rajashankar KR, Patel MH, Robinette RA, Crowley PJ, Michalek S, Brady LJ, Deivanayagam C. Elongated Fibrillar Structure of a Streptococcal Adhesin Assembled by the High-Affinity Association of Alpha- and PPII-helices. Proc Natl Acad Sci US A. 2010;107:5983–5988. doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larson MR, Rajashankar KR, Crowley PJ, Kelly C, Mitchell TJ, Brady LJ, Deivanayagam C. Crystal Structure of the C-terminal Region of Streptococcus mutans Antigen I/II and Characterization of Salivary Agglutinin Adherence Domains. J Biol Chem. 2011;286:21657–21666. doi: 10.1074/jbc.M111.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nylander A, Forsgren N, Persson K. Structure of the C-terminal Domain of the Surface Antigen SpaP from the Caries Pathogen Streptococcus mutans. Acta Crystallogr Sect F. 2011;67:23–26. doi: 10.1107/S174430911004443X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly C, Evans P, Bergmeier L, Lee SF, Progulske-Fox A, Harris AC, Aitken A, Bleiweis AS, Lehner T. Sequence Analysis of the Cloned Streptococcal Cell Surface Antigen I/II. FEBS Lett. 1989;258:127–132. doi: 10.1016/0014-5793(89)81632-1. [DOI] [PubMed] [Google Scholar]
- 41.Nakai M, Okahashi N, Ohta H, Koga T. Saliva-Binding Region of Streptococcus mutans Surface Protein Antigen. Infect Immun. 1993;61:4344–4349. doi: 10.1128/iai.61.10.4344-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupres V, Menozzi FD, Locht C, Clare BH, Abbott NL, Cuenot S, Bompard C, Raze D, Dufrêne YF. Nanoscale Mapping and Functional Analysis of Individual Adhesins on Living Bacteria. Nat Methods. 2005;2:515–520. doi: 10.1038/nmeth769. [DOI] [PubMed] [Google Scholar]
- 43.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. Force-Induced Formation and Propagation of Adhesion Nanodomains in Living Fungal Cells. Proc Natl Acad Sci US A. 2010;107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lower SK, Lamlertthon S, Casillas-Ituarte NN, Lins RD, Yongsunthon R, Taylor ES, DiBartola AC, Edmonson C, McIntyre LM, Reller LB, et al. Polymorphisms in Fibronectin Binding Protein A of Staphylococcus aureus Are Associated with Infection of Cardiovascular Devices. Proc Natl Acad Sci US A. 2011;108:8372–18377. doi: 10.1073/pnas.1109071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guillaume-Gentil O, Potthoff E, Ossola D, Franz CM, Zambelli T, Vorholt JA. Force-Controlled Manipulation of Single Cells: From AFM to FluidFM. Trends Biotechnol. 2014;32:381–388. doi: 10.1016/j.tibtech.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Beaussart A, El-Kirat-Chatel S, Sullan RMA, Alsteens D, Herman P, Derclaye S, Dufrêne YF. Quantifying the Forces Guiding Microbial Cell Adhesion Using Single-Cell Force Spectroscopy. Nat Protoc. 2014;9:1049–1055. doi: 10.1038/nprot.2014.066. [DOI] [PubMed] [Google Scholar]
- 47.Herman P, El-Kirat-Chatel S, Beaussart A, Geoghegan JA, Foster TJ, Dufrêne YF. The Binding Force of the Staphylococcal Adhesin SdrG Is Remarkably Strong. Mol Microbiol. 2014;93:356–368. doi: 10.1111/mmi.12663. [DOI] [PubMed] [Google Scholar]
- 48.Ebner A, Wildling L, Kamruzzahan AS, Rankl C, Wruss J, Hahn CD, Holzl M, Zhu R, Kienberger F, Blaas D, et al. A New, Simple Method for Linking of Antibodies to Atomic Force Microscopy Tips. Bioconjugate Chem. 2007;18:1176–1184. doi: 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
- 49.Dufrêne YF. Atomic Force Microscopy in Microbiology: New Structural and Functional Insights into the Microbial Cell Surface. mBio. 2014;5:e01363–14. doi: 10.1128/mBio.01363-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ayakawa GY, Boushell LW, Crowley PJ, Erdos GW, McArthur WP, Bleiweis AS. Isolation and Characterization of Monoclonal Antibodies Specific for Antigen P1, a Major Surface Protein of Mutans Streptococci. Infect Immun. 1987;55:2759–2767. doi: 10.1128/iai.55.11.2759-2767.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinette RA, Heim KP, Oli MW, Crowley PJ, McArthur WP, Brady LJ. Alterations in Immunodominance of Streptococcus mutans AgI/II: Lessons Learned from Immunomodulatory Antibodies. Vaccine. 2014;32:375–382. doi: 10.1016/j.vaccine.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jakubovics NS, Stromberg N, van Dolleweerd CJ, Kelly CG, Jenkinson HF. Differential Binding Specificities of Oral Streptococcal Antigen I/II Family Adhesins for Human or Bacterial Ligands. Mol Microbiol. 2005;55:1591–1605. doi: 10.1111/j.1365-2958.2005.04495.x. [DOI] [PubMed] [Google Scholar]
- 53.Loimaranta V, Jakubovics NS, Hytonen J, Finne J, Jenkinson HF, Stromberg N. Fluid- or Surface-Phase Human Salivary Scavenger Protein gp340 Exposes Different Bacterial Recognition Properties. Infect Immun. 2005;73:2245–2252. doi: 10.1128/IAI.73.4.2245-2252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beg AM, Jones MN, Miller-Torbert T, Holt RG. Binding of Streptococcus mutans to Extracellular Matrix Molecules and Fibrinogen. Biochem Biophys Res Commun. 2002;298:75–79. doi: 10.1016/s0006-291x(02)02390-2. [DOI] [PubMed] [Google Scholar]
- 55.Handley PS, Harty DW, Wyatt JE, Brown CR, Doran JP, Gibbs AC. A Comparison of the Adhesion, Coaggregation and Cell-surface Hydrophobicity Properties of Fibrillar and Fimbriate Strains of Streptococcus salivarius. J Gen Microbiol. 1987;133:3207–3217. doi: 10.1099/00221287-133-11-3207. [DOI] [PubMed] [Google Scholar]
- 56.Jenkinson HF, Demuth DR. Structure, Function and Immunogenicity of Streptococcal Antigen I/II Polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 57.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The Mechanical Hierarchies of Fibronectin Observed with Single-Molecule AFM. J Mol Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 58.Rief M, Gautel M, Schemmel A, Gaub HE. The Mechanical Stability of Immunoglobulin and Fibronectin III Domains in the Muscle Protein Titin Measured by Atomic Force Microscopy. Biophys J. 1998;75:3008–3014. doi: 10.1016/S0006-3495(98)77741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doyle RJ. Contribution of the Hydrophobic Effect to Microbial Infection. Microbes Infect. 2000;2:391–400. doi: 10.1016/s1286-4579(00)00328-2. [DOI] [PubMed] [Google Scholar]
- 60.Nesbitt WE, Doyle RJ, Taylor KG. Hydrophobic Interactions and the Adherence of Streptococcus sanguis to Hydroxylapatite. Infect Immun. 1982;38:637–644. doi: 10.1128/iai.38.2.637-644.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hogg SD, Manning JE. The Hydrophobicity of ‘Viridans’ Streptococci Isolated From the Human Mouth. J Appl Bacteriol. 1987;63:311–318. doi: 10.1111/j.1365-2672.1987.tb02708.x. [DOI] [PubMed] [Google Scholar]
- 62.Guo L, Wu T, Hu W, He X, Sharma S, Webster P, Gimzewski JK, Zhou X, Lux R, Shi W. Phenotypic Characterization of the Foldase Homologue PrsA in Streptococcus mutans. Mol Oral Microbiol. 2013;28:154–165. doi: 10.1111/omi.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SF, Progulske-Fox A, Erdos GW, Piacentini DA, Ayakawa GY, Crowley PJ, Bleiweis AS. Construction and Characterization of Isogenic Mutants of Streptococcus mutans Deficient in Major Surface Protein Antigen P1 (I/II) Infect Immun. 1989;57:3306–3313. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamura H, Yamada A, Kato H. Molecular Characterization of the Dextran-Binding Lectin B Gene dblB of Streptococcus criceti in Streptococcus mutans Strain GS-5 with Mutations in Both gbpC and spaP Genes. Genes Genet Syst. 2014;89:41–50. doi: 10.1266/ggs.89.41. [DOI] [PubMed] [Google Scholar]
- 65.McBride BC, Morris EJ, Ganeshkumar N. Relationship of Streptococcal Cell Surface Proteins to Hydrophobicity and Adherence. American Society for Microbiology; Washington, DC: 1985. [Google Scholar]
- 66.Lucena PH, Biondi L, de Torres R. Hydrophobicity Test in Mutans Streptococci. Acta Odontol Latinoam. 2010;23:210–215. [PubMed] [Google Scholar]
- 67.Matsumoto-Nakano M, Nagayama K, Kitagori H, Fujita K, Inagaki S, Takashima Y, Tamesada M, Kawabata S, Ooshima T. Inhibitory Effects of Oenothera biennis (Evening Primrose) Seed Extract on Streptococcus mutans and S. mutans-induced Dental Caries in Rats. Caries Res. 2011;45:56–63. doi: 10.1159/000323376. [DOI] [PubMed] [Google Scholar]
- 68.Yamanaka A, Kimizuka R, Kato T, Okuda K. Inhibitory Effects of Cranberry Juice on Attachment of Oral Streptococci and Biofilm Formation. Oral Microbiol Immun. 2004;19:150–154. doi: 10.1111/j.0902-0055.2004.00130.x. [DOI] [PubMed] [Google Scholar]
- 69.Yamanaka-Okada A, Sato E, Kouchi T, Kimizuka R, Kato T, Okuda K. Inhibitory Effect of Cranberry Polyphenol on Cariogenic Bacteria. Bull Tokyo Dent Coll. 2008;49:107–112. doi: 10.2209/tdcpublication.49.107. [DOI] [PubMed] [Google Scholar]
- 70.Helenius J, Heisenberg CP, Gaub HE, Muller DJ. Single-Cell Force Spectroscopy. J Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 71.Williams PM. Analytical Descriptions of Dynamic Force Spectroscopy: Behaviour of Multiple Connections. Anal Chim Acta. 2003;479:107–115. [Google Scholar]
- 72.Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, Drenckhahn D. Cadherin Interaction Probed by Atomic Force Microscopy. Proc Natl Acad Sci US A. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.El-Kirat-Chatel S, Beaussart A, Boyd CD, O’Toole GA, Dufrene YF. Single-Cell and Single-Molecule Analysis Deciphers the Localization, Adhesion, and Mechanics of the Biofilm Adhesin LapA. ACS Chem Biol. 2014;9:485–494. doi: 10.1021/cb400794e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sullan RM, Beaussart A, Tripathi P, Derclaye S, El-Kirat-Chatel S, Li JK, Schneider YJ, Vanderleyden J, Lebeer S, Dufrêne YF. Single-Cell Force Spectroscopy of Pili-Mediated Adhesion. Nanoscale. 2014;6:1134–1143. doi: 10.1039/c3nr05462d. [DOI] [PubMed] [Google Scholar]
- 75.Beaussart A, Baker AE, Kuchma SL, El-Kirat-Chatel S, O’Toole GA, Dufrêne YF. Nanoscale Adhesion Forces of Pseudomonas aeruginosa Type IV Pili. ACS Nano. 2014;8:10723–10733. doi: 10.1021/nn5044383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beaussart A, Herman P, El-Kirat-Chatel S, Lipke PN, Kucharíková S, Van Dijck P, Dufrêne YF. Single-Cell Force Spectroscopy of the Medically-Important Staphylococcus epidermidis-Candida albicans interaction. Nanoscale. 2013;5:10894–10900. doi: 10.1039/c3nr03272h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Busscher HJ, van de Belt-Gritter B, Dijkstra RJ, Norde W, Petersen FC, Scheie AA, van der Mei HC. Intermolecular Forces and Enthalpies in the Adhesion of Streptococcus mutans and an Antigen I/II-deficient Mutant to Laminin Films. J Bacteriol. 2007;189:2988–2995. doi: 10.1128/JB.01731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van der Mei HC, Rustema-Abbing M, de Vries J, Busscher HJ. Bond Strengthening in Oral Bacterial Adhesion to Salivary Conditioning Films. Appl Environ Microb. 2008;74:5511–5515. doi: 10.1128/AEM.01119-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu CP, van de Belt-Gritter B, Dijkstra RJ, Norde W, van der Mei HC, Busscher HJ. Interaction Forces between Salivary Proteins and Streptococcus mutans with and without Antigen I/II. Langmuir. 2007;23:9423–9428. doi: 10.1021/la701477w. [DOI] [PubMed] [Google Scholar]
- 80.Grivet M, Morrier JJ, Benay G, Barsotti O. Effect of Hydrophobicity on in Vitro Streptococcal Adhesion to Dental Alloys. J Mater Sci Mater Med. 2000;11:637–642. doi: 10.1023/a:1008913915399. [DOI] [PubMed] [Google Scholar]
- 81.Knox KW, Wicken AJ. Effect of Growth Conditions on the Antigenic Components of Streptococcus mutans and Lactobacilli. Adv Exp Med Biol. 1978;107:629–637. doi: 10.1007/978-1-4684-3369-2_71. [DOI] [PubMed] [Google Scholar]
- 82.Rhodin NR, Van Tilburg ML, Oli MW, McArthur WP, Brady LJ. Further Characterization of Immunomodulation by a Monoclonal Antibody against Streptococcus mutans Antigen P1. Infect Immun. 2004;72:13–21. doi: 10.1128/IAI.72.1.13-21.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oli MW, McArthur WP, Brady LJ. A Whole Cell BIAcore Assay to Evaluate P1-Mediated Adherence of Streptococcus mutans to Human Salivary Agglutinin and Inhibition by Specific Antibodies. J Microbiol Methods. 2006;65:503–511. doi: 10.1016/j.mimet.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 84.Hutter JL, Bechhoefer J. Calibration of Atomic-Force Microscope Tips. Rev Sci Instrum. 1993;64:3342–3342. [Google Scholar]