Abstract
The oral pathogen Streptococcus mutans expresses a surface protein, P1, which interacts with the salivary pellicle on the tooth surface or with fluid-phase saliva, resulting in bacterial adhesion or aggregation, respectively. P1 is a target of protective immunity. Its N-terminal region has been associated with adhesion and aggregation functions and contains epitopes recognized by efficacious antibodies. In this study, we used Bacillus subtilis, a gram-positive expression host, to produce a recombinant N-terminal polypeptide of P1 (P139–512) derived from the S. mutans strain UA159. Purified P139–512 reacted with an anti-full-length P1 antiserum as well as one raised against intact S. mutans cells, indicating preserved antigenicity. Immunization of mice with soluble and heat-denatured P139–512 induced antibodies that reacted specifically with native P1 on the surface of S. mutans cells. The anti-P139–512 antiserum was as effective at blocking saliva-mediated aggregation of S. mutans cells and better at blocking bacterial adhesion to saliva-coated plastic surfaces compared with the anti-full-length P1 antiserum. In addition, adsorption of the anti-P1 antiserum with P139–512 eliminated its ability to block the adhesion of S. mutans cells to abiotic surfaces. The present results indicate that P139–512, expressed and purified from a recombinant B. subtilis strain, maintains important immunological features of the native protein and represents an additional tool for the development of anticaries vaccines.
Keywords: Streptococcus mutans, P1 protein, recombinant proteins, Bacillus subtilis, antibody responses, saliva-binding region
Introduction
Streptococcus mutans is a major etiologic agent contributing to the causation of dental caries, one of the most common human infectious diseases (Russell, 2000). Bacterial adherence to the tooth surface involves both sucrose-dependent and -independent mechanisms in which hydrogen bonds and hydrophobic interactions are formed between the bacterial cells and the salivary components present within dental pellicles (Gibbons, 1984; Gibbons et al., 1986). Streptococcus mutans also interacts with other oral micro-organisms including primary colonizing commensal bacteria such as Streptococcus gordonii (Jenkinson & Lamont, 2005; Nobbs et al., 2009). The complex multifactorial adherence process involves the Mr ~185 kDa, major surface antigen P1, originally identified as Antigen I/II (Russell & Lehner, 1978), and also referred to as PAc (Koga et al., 1990).
P1 family proteins are expressed by almost all oral streptococci and orthologs have now also been identified in Streptococcus pyogenes and Streptococcus agalactiae (Zhang et al., 2006). P1 is also required for saliva-mediated bacterial aggregation, possibly a nonimmune host defense mechanism, which is abolished in S. mutans isogenic mutants in which the spaP (pac) gene has been disrupted (Lee et al., 1989; Koga et al., 1990). The cariogenicity of S. mutans is diminished in the absence of P1 in a gnotobiotic rat model (Crowley et al., 1999). P1 has been reported as a promising target for protective immunity in naturally sensitized humans and has been studied as a vaccine candidate in active immunization studies in primates and rodents (Lehner et al., 1981; Michalek et al., 2001b; Smith & Mattos-Graner, 2008; Shivakumar et al., 2009). Passive immunization with certain, but not all, monoclonal antibodies against P1 has also been reported to confer protection against S. mutans colonization in macaques (Lehner et al., 1985) and human subjects (Ma et al., 1987).
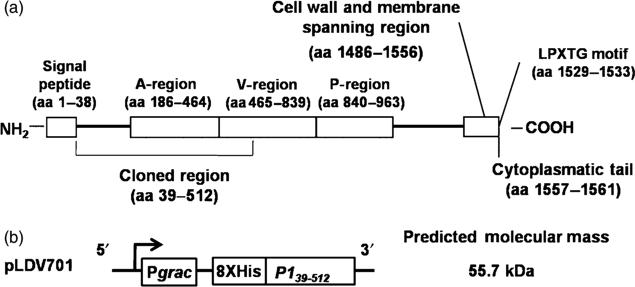
The primary sequence of S. mutans P1 shows several distinct features (see Fig. 1a). The N-terminal portion encompasses the signal peptide (amino acid residues 1–38) and an alanine-rich region (A-region, amino acid residues 186–464). The A-region shows a seven-residue periodicity that adopts an extended alpha-helix structure (Larson et al., 2010). The central region has a proline-rich segment (P-region, amino acid residues 840–963); between the A- and P-regions lies a segment commonly referred to as the variable or the V-region because this is where most of the sequence differences between S. mutans Antigen I/II family members are clustered (Brady et al., 1991). The A- and P-regions of P1 interact, and these and segments flanking them contribute to the formation of complex discontinuous eptiopes (Rhodin et al., 2004; Seifert et al., 2004; van Dolleweerd et al., 2004; McArthur et al., 2007). At the C-terminal end, P1 displays cell wall and membrane-spanning regions, an LPXTG motif and a cytoplasmatic tail characteristic of cell surface-localized sortase substrates (Fischetti et al., 1990; Ton-That et al., 2004).
Fig. 1.
Schematic representation of the Streptococcus mutans P1 protein (a) and the recombinant gene encoding the truncated P139–512 polypeptide (b) cloned in Bacillus subtilis pLDV701 vector. The predicted molecular mass of the recombinant P139–512 protein is indicated on the right of (b).
The A-region located within amino-terminal end of P1 has been reported to contribute to the interaction of S. mutans with salivary components, including salivary agglutinin (SAG), now known to represent the lung scavenger receptor cysteine-rich protein, gp340 (Prakobphol et al., 2000). Because of its interactions with salivary constituents, the A-region has sometimes been referred as the saliva-binding region (SBR) (Nakai et al., 1993; Hajishengallis et al., 1995, 1998). This segment of P1 competitively inhibits both adherence of S. mutans to immobilized SAG and aggregation in the presence of fluid-phase SAG (Crowley et al., 1993); however, different determinants have been implicated in these two processes (Brady et al., 1992). The A-region is also antigenic and encompasses both B-cell and T-cell epitopes (Takahashi et al., 1991; Kelly et al., 1995; Senpuku et al., 1996). Antibodies against this segment of P1 have been reported to block the aggregation of S. mutans and to reduce the development of dental caries (Takahashi et al., 1991; Munro et al., 1993; Huang et al., 2001; Michalek et al., 2001a; Tsuha et al., 2004).
Previous studies have reported the development of anti-caries vaccine approaches based on a recombinant P1 N-terminal region expressed in Escherichia coli. Usually, the recombinant S. mutans protein has been genetically fused to high-molecular-weight protein carriers and/or secreted into the periplasm of the host bacterial strains in order to reduce proteolytic degradation or simplify the purification process (Crowley et al., 1993; Okahashi et al., 1993; Hajishengallis et al., 1995; Toida et al., 1997; Matsumoto-Nakano et al., 2008). Because hybrid proteins expressed in heterologous bacterial hosts can potentially lose important native-like immunological determinants, the generation of recombinant nonfused P1-derived peptides in an alternative expression system may result in an improved vaccine antigen and may contribute toward a better understanding of the functional and immunological features of the S. mutans protein.
Bacillus subtilis is a gram-positive spore-forming soil bacterium with a long history of industrial and technological uses, such as the production of proteases and fermented foods. Spores are also used as probiotics for different animal species and as growth promoters and biological control agents for several cultivated plants (Harwood, 1992; Paulitz & Bélanger, 2001; Ferreira et al., 2005; Paccez et al., 2006, 2007). The present knowledge of its physiology and genetics, as well as the availability of gene cloning and expression tools, make B. subtilis an important alternative to E. coli as a host for the expression of heterologous proteins with preserved biological activity devoid of lipopolysaccharide contamination that may affect immunological studies (Meima et al., 2004; Westers et al., 2004).
The aim of this study was to evaluate the immunological properties of a P1-derived polypeptide spanning the amino-terminus and encompassing the whole A-region and a short segment C-terminal, expressed and purified from a B. subtilis recombinant strain, and to determine its possible utility as a vaccine antigen.
Material and methods
Bacterial strains and growth conditions
All the bacterial strains and plasmids used in this work are listed in Table 1. Escherichia coli and B. subtilis strains were routinely cultivated in Luria–Bertani broth at 37 1C. Antibiotics were added to the growth media as necessary according to the strain and plasmids used. Competent E. coli cells were prepared using the CaCl2-mediated transformation protocol (Sambrook & Russell, 2001), while natural B. subtilis competent cells were generated using the two-step transformation method (Cutting et al., 1990). Streptococcus mutans strains were cultivated in brain–heart infusion broth for 16 h at 37 °C in 5% CO2; PC3370 and PC3370C strains were grown with tetracycline (10 μg mL−1) and kanamycin (500 mg mL−1), respectively.
Table 1.
Bacterial strains and plasmids used in this work
| Main characteristics | References | |
|---|---|---|
| Strains | ||
| S. mutans | ||
| UA 159 | Kmr | Adjic et al. (2002) |
| PC3370 | S. mutans NG8 spaP::tetr | Crowley et al. (1999) |
| PC3370C | S. mutans NG8 spaP::tetr; complemented with pMAD (pDL289+spaP) | Brady et al. (1998) |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15Δ(lacZYA-argF) U169 deoR, recA1 endA1 hsdR17 (rk - mk+ phoA supE44λ- thi-1 gyrA96 relA1 | Invitrogen™ |
| CG14 | M15(pREP4) carrying pCG14 | Brady et al. (1998) |
| B. subtilis | ||
| WW02 | leuA8 metB5 trpC2 hsrdRM1 amyE::neo | Wehrl et al. (2000) |
| LDV700 | WW02 carrying pHT08 [empty vector - IPTG-inducible promoter (Pgrac)] | This work |
| LDV701 | WW02 carrying pLDV701 (P139-512 under the control of Pgrac) | This work |
| Plasmids | ||
| pGEM-T-Easy™ | Ampr; operon lac;T-end | Promega™ |
| pCG14 | Derived from pQE-30 with a cloned spaP gene coding for 39 to 1561 amino acids of the P1 protein | Brady et al. (1992) |
| pGP1N | Ampr; pGEM-T-Easy™ with cloned P139-152 | This work |
| pHT08 | Ampr; Cmr; IPTG-inducible PgraC promoter; His-tag | Nguyen et al. (2007) |
| pLDV701 | Derived from pHT08 with cloned P139-152 | This work |
Plasmid constructions
A B. subtilis strain expressing a polypeptide corresponding to the S. mutans to aa 39–512 (P139–512) of the P1 protein (see Fig. 1a) was obtained after cloning the coding sequence into the plasmid pHT08 under control of the Pgrac promoter (Nguyen et al., 2007). The spaP region encoding the amino-terminus of P1 through the A-region and without a signal peptide was amplified by PCR as a 1.2-kb fragment using S. mutans UA159 chromosomal DNA as the template and primers FWsbr (5′-AAAggATCCATggATgAAACgACCACTAC) and RVsbr (5′-CgCgACgTCATTTggCTCAAgATCATA gAC) with restriction sites for BamHI and AatII (underlined sequences), respectively. The amplified fragment was cloned into the pGEM-T-Easy™ vector, resulting in the recombinant pGP1N vector. The pGP1N was digested with BamHI and AatII and the released fragment was subcloned into pHT08 to generate the recombinant plasmid named pLDV701. The correct nucleotide sequence of the cloned insert was confirmed by automated nucleotide sequencing.
Expression of the recombinant P139-512 by genetically modified B. subtilis
The plasmids pLDV701 and pHT08 were introduced into naturally competent B. subtilis WW02. One clone transformed with pLDV701 and another transformed with pHT08 was selected and the corresponding strains were named LDV701 and LDV700, respectively. Induction of protein expression and preparation of whole-cell protein extracts of genetically modified B. subtilis strains were performed as described previously (Paccez et al., 2006; Nguyen et al., 2007).
Purification of the recombinant P139-512 expressed by B. subtilis strain LDV701
Bacillus subtilis cells were suspended in buffer A (0.1 M Tris HCl, 0.5 M NaCl, pH 7.5) and lysozyme (800 μmg mL−1) and incubated for 30 min to 37 1C. Sodium dodecyl sulfate (SDS) (0.01%) and phenylmethylsulfonyl fluoride (PMSF) (0.01 M) were added to the suspension and the cells were disrupted by sonication. The soluble supernatant was collected by centrifugation and subjected to affinity chromatography using a nickel–resin column in an AKTA FPLC device (Amersham Biosciences). Bound proteins were eluted with buffer B (0.1 M Tris HCl, 0.5 M NaCl, pH 7.5) containing final imidazole concentrations ranging from 0.05 to 1 M. The recombinant S. mutans P1 protein was purified from the previously described E. coli CG14 strain (Brady et al., 1998). Expression of the recombinant protein was achieved following incubation of the E. coli CG14 strain in a medium containing 0.1 mM of IPTG for 4 h. The cell pellets were suspended in 0.1 M Tris HCl, 0.2 M NaCl and 4 M urea (pH 8.0) containing lysozyme (100 μg mL−1) and incubated for 30 min to 37 °C. SDS (0.01%) and PMSF (0.01 M) were added to the suspension before sonic disruption of the cells. The soluble supernatant was applied in a nickel-affinity chromatography column and eluted under the same conditions as those described for the purification of the P139–512. The protein concentrations of the eluted fractions were determined using the BCA protein quantification assay (Pierce™), with bovine serum albumin as a standard.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blots
SDS-PAGE was performed following standard procedures using a Mini Protean II vertical electrophoresis unit (Mini-protean, BioRad). Western blots were carried out following the incubation of nitrocellulose membranes with P1-specific (1 : 3000) or anti-P139–512-specific (1 : 2000) sera. The anti-P1 serum was raised in mice immunized with the recombinant P1 protein generated in E. coli while the anti-P139–512 serum was collected from mice immunized with the protein purified from B. subtilis LDV701 strain. Reactive bands were detected using a chemiluminescent kit (Super Signal™, Pierce), as described by the manufacturer.
Mouse immunizations and serum collection
BALB/c mice were supplied by the Isogenic Mouse Breeding Facility of the Department of Parasitology, Biomedical Sciences Institute (ICB), University of São Paulo (USP). All animal handling procedures were in accordance with the principles of the Brazilian code for the use of laboratory animals. Groups of five female mice, 6–8 weeks of age, were immunized subcutaneously with five doses of 10 μg of purified P1 or P139–512 proteins at 2-week intervals. The first dose was administered with complete Freund's adjuvant and the subsequent doses were administered with incomplete Freund's adjuvant. The recombinant P139–512 was administered as a soluble protein or after heat denaturation (boiling for 10 min) (P139–512d). Antisera against whole S. mutans PC3370 (anti-SmuΔP1) or PC3370C (anti-Smu) were raised by intravenous immunization of five female mice with five doses at 2-week intervals of 100 μL containing 7 × 109 CFU in phosphate-buffered saline (PBS) of whole cells harvested from cultures grown to an OD600nm of 2. Serum samples were collected 2 weeks after the last inoculation, pooled and stored at −20 °C for subsequent analysis. Samples were tested for reactivity with full-length P1 and P139–512 antigens by an enzyme-linked immunosorbant assay (ELISA). Adsorption of anti-P1 serum samples with purified P139–512 or P139–512d was carried out in ELISA plates previously treated with 200 ng per well of purified recombinant protein as the solid phase-bound antigen. Aliquots of anti-P1 serum diluted 1 : 105 were applied to the plate wells and kept for 2 h at 37 °C. The serum samples were removed and checked for residual reactivity to the purified P139–512 in Western blots. The procedure was repeated until no reaction with the P139–512 was detected.
ELISA
The determination of specific antibody levels in serum samples was performed by ELISA and serial end-point titration in 96-well MaxiSorp (Nunc) plates coated with the recombinant truncated or full-length P1 proteins according to previously described procedures (Luiz et al., 2008). Briefly, each purified protein tested (100 ng per well) diluted in PBS was added to the plates and kept for 16 h at 4 °C. Plates were washed twice with PBS–Tween 0.05% and then blocked with PBS–Tween – 5% milk for 2 h at 37 °C. One hundred microliters of twofold serial dilutions of the primary antibody beginning at 1/25 were added to the wells and incubated for 2 h at 37 °C. After a second washing step, peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Sigma) (1/3000) was added and plates were incubated for 2 h at 37 °C. Following a final wash, the plates were developed with O-phenylenediamine (0.4 mg mL−1; Sigma) substrate solution containing 12% H2O2 and the reactions were stopped after addition of 2 M H2SO4. A492nm was measured on a microplate reader (LabSystem). All samples were assayed in duplicate. An absorbance value of preimmune sera was used as a reference blank. Dilution curves were drawn for each sample and end-point titers were calculated as the reciprocal values of the last dilution with an optical density of 0.05.
Immunofluorescence analysis
Streptococcus mutans PC3370 and PC3370C strains were spread on glass slides, heat fixed and evaluated for reactivity with the anti-P139–512 antiserum, followed by fluorescein isothiocyanate-conjugated goat anti-mouse IgG (Invitrogen™, Carlsbad, CA) as described by Homonylo-McGavin & Lee (1996). The anti-P1 antiserum and preimmune sera served as positive and negative controls. The stained cells were viewed using a Nikon TE300 inverted epifluorescence microscope (495 nm excitation, 525 nm emission).
Inhibition of saliva-mediated aggregation of S. mutans
Assays were carried out with clarified saliva, as described previously (Rundegren & Arnold, 1987). Aggregation and inhibition of aggregation assays in the presence of the test sera were performed according to a previously described method (Brady et al., 1992). All tests were repeated independently at least three times.
Inhibition of S.mutans adhesion to immobilized saliva
Adhesion of S. mutans to immobilized saliva, and adherence inhibition mediated by different serum samples, were based on a method described previously (Jakubovics et al., 2005). As a positive control reaction for inhibition of P1-mediated bacterial adhesion, we used 3 mM EDTA added to the bacterial cell suspension to interrupt the calcium-dependent interaction between P1 and gp340, as described previously (Crowley et al., 1993). Nonimmune serum was used as a negative control and the S. mutans strain PC3370 devoid of P1 was also used as a negative control. The numbers of bound cells were calculated from a standard curve relating A600nm to the cell number of S. mutans. All tests were independently repeated at least three times.
Statistical analyses
Results were analyzed in graphpad prism 5 and were expressed as mean ± SD. One-way anova was used to compare the inhibition of adherence by anti-P139–512 sera. Statistically significant differences were considered with P values below 0.05.
Results
Expression of S. mutans P139-512 by genetically modified B. subtilis strains
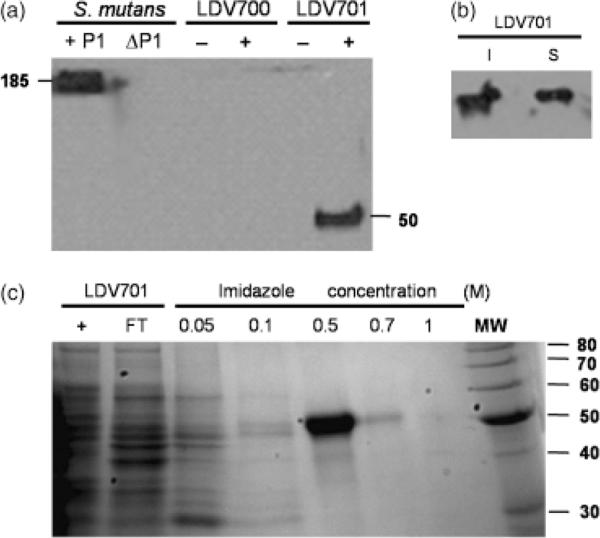
The pLDV701 vector encodes the truncated recombinant S. mutans P1 polypeptide with an N-terminal histidine tag under control of the IPTG-inducible Pgrac promoter (Fig. 1b). The encoded protein has a predicted molecular mass of 55.7 kDa. A single band, with an apparent molecular mass of Mr~50 kDa, reactive with an anti-P1 antiserum was detected in immunoblots of whole-cell extracts of the B. subtilis LDV701 strain following incubation with IPTG to induce protein expression. The same anti-P1-specific polyclonal serum detected only the Mr 185 kDa P1 protein in extracts of the S. mutans PC3370C-positive control strain. No cross-reacting bands were observed in extracts of B. subtilis transformed with pHT08 or the LDV701 strain cultivated in the absence of the inducer (Fig. 2a). The recombinant polypeptide accumulated in the cytoplasm of the B. subtilis LDV701 strain and approximately half of the protein was present in a soluble form (Fig. 2b and data not shown). Cellular extracts containing soluble proteins of B. subtilis strain LDV701 were loaded on a column of nickel-containing resin and the recombinant protein was eluted at 0.5 M imidazole concentration (Fig. 2c). A final protein yield of 15 mg L−1 of culture was achieved following incubation at 37 °C for 4 h in the presence of IPTG. No significant improvements in the recovery yields of the soluble protein were achieved following incubation of the B. subtilis at different growth temperatures, agitation levels or varying the inducer concentrations and induction periods in the presence of IPTG (data not shown).
Fig. 2.
Detection and purification of the recombinant P139–512 protein expressed by Bacillus subtilis cells. (a) Immunodetection of P1 or P139–512 expressed by Streptococcus mutans PC3370C (1P1) or B. subtilis LDV701 (LDV701) strains, respectively. Negative controls include S. mutans PC3370 (ΔP1) and the B. subtilis LDV700 strain (LDV700). − and + denote cells induced or not with IPTG, respectively. Estimated molecular weights (kDa) of the reactive protein bands are indicated. (b) Immunodetection of recombinant P139–512 in the soluble (S) or insoluble (I) fractions of the B. subtilis LDV701 following induction with IPTG. (c) Purification of recombinant P139–512 expressed in B. subtilis. Imidazole concentrations used to elute the protein from the nickel-containing resin are indicated. Proteins were separated in 12.5% polyacrylamide gels and stained with Comassie Brilliant Blue. MW, molecular weight markers indicated in kilodaltons (kDa) at the right side of the figure.
Antibodies raised in mice immunized with P139-512 expressed in B. subtilis recognize the native and recombinant S. mutans P1 protein
Two groups of female BALB/c mice were immunized subcutaneously with five doses of the recombinant P139–512 protein: one group with the soluble protein purified from the B. subtilis LDV701 strain and another group immunized with the same protein boiled for 10 min (P139–512d). Antibodies raised in mice immunized with untreated or heat-denatured P139–512, as well as serum raised in mice immunized with purified P1 expressed in E. coli, were tested in immunoblots and showed no cross reaction with unspecific B. subtilis or S. mutans proteins (data not shown and Fig. 3). As expected, both sera reacted with P139–512 in whole-cell extracts of IPTG-treated B. subtilis LDV701 (Fig. 3). A major protein band of approximately 85 kDa was specifically detected in the whole-cell extract of S. mutans PC3370C by sera raised in mice against P139–512 or P139–512d. This probably represents a stable degradation product of the native streptococcal P1 protein (Fig. 3). That the 185 kDa P1 protein was not well recognized by the anti-P139–512 antisera suggests that epitopes contained within this truncated polypetide may be masked in the context of the full-length molecule, a property that has been observed previously when monoclonal antibody reactivity against full-length P1 and derivatives has been evaluated by Western blot (McArthur et al., 2007).
Fig. 3.
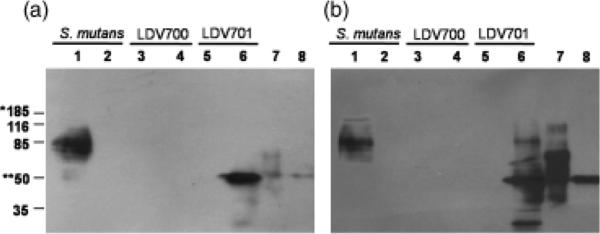
Antigen specificity of antibodies raised in mice immunized with P139–512. (a) Western blots developed with serum samples collected from mice immunized with P139–512 or (b) P139–512d. Samples: 1,Streptococcus mutans PC3370C strain; 2, S. mutans PC3370 strain; 3, Bacillus subtilis LDV700 strain following incubation without IPTG; 4, B. subtilis LDV700 strain following incubation with IPTG; 5, B. subtilis LDV701 strain following incubation without IPTG; 6, B. subtilis LDV701 strain following incubation with IPTG; 7, recombinant P1 protein; 8, P139–512 purified protein. Proteins were sorted in 12.5% polyacrylamide gels. The molecular weights of the reactive protein bands are indicated. (*) and (**) indicate the molecular weights of P1 and P139–512 proteins, respectively.
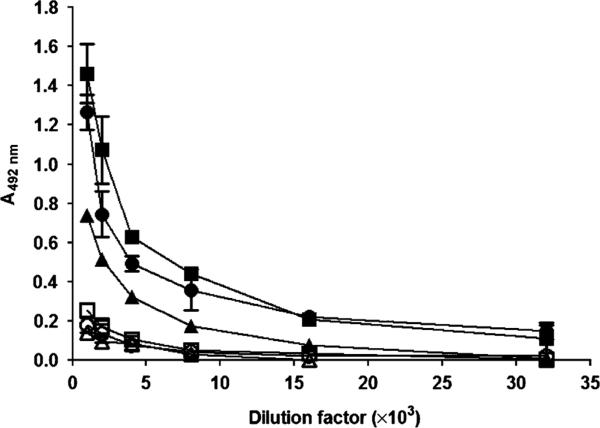
The antiserum raised in mice immunized with P1, P139–512 or P139–512d was titrated in ELISA plates using purified full-length P1, P139–512 or P139–512d, as the solid phase-bound antigens. Not surprisingly, the highest measured titer was observed with the anti-P1 antiserum against the homologous full-length P1 protein (109) because additional epitopes would be contained within this molecule that are not present in the P139–512 derivative (Table 2). As shown in Table 2 and Fig. 4, ELISAs carried out with the anti-P1 antiserum against P139–512 demonstrated IgG titers of 104, while lower values (103) were obtained on plates coated with P139–512d. This suggests that at least some antigenic epitopes were affected by heat denaturation. When ELISA tests were carried out with antisera raised in mice immunized with P139–512 or P139–512d, the anti-P1 IgG titers were similar (7.0 × 103 and 8.4 × 103, respectively), while higher values were recorded for reactivity against P139–512 in comparison with P139–512d (Table 2). Taken together, these results suggest that antibodies raised in mice immunized with P139–512 or P139–512d produced in B. subtilis recognize epitopes found in the S. mutans P1 protein and that the immunogenicity and antigenicity of P139–512 is diminished by heat denaturation. Additionally, the lower titer of anti-P139–512 and anti-P139–512d sera measured against full-length P1 compared with P139–512 again suggests that epitopes contained within the truncated construct may be masked in the intact protein.
Table 2.
Reactivity of antibodies raised in mice immunized with recombinant proteins or whole Streptococcus mutans cells with recombinant P1, P139-512 or P139-512d
| Plates coated with |
|||
|---|---|---|---|
| Antiserum | P1 | P139-512 | P139-512d |
| Anti-P1 | 1.65 × 109 | 1.3 × 104 | 6.3 × 103 |
| Anti-P1 39-512 | 7.0 × 103 | 7.6 × 104 | 6.8 × 103 |
| Anti-P1 39-512d | 8.4 × 103 | 1.1 × 104 | 5.1 × 103 |
| Anti-Smu* | ND | 1.7 × 105 | 7.0 × 104 |
| Anti-SmuΔP1† | ND | 2.0 × 102 | 1.4 × 102 |
Antisera raised in mice immunized with the whole cell of the S. mutans PC3370C strain.
Antisera raised in mice immunized with the whole cell of the S. mutans PC3370 (P1-knockout) strain.
ND, not determined.
Fig. 4.
IgG titration in antisera raised in mice immunized with full-length P1, expressed in Escherichia coli, and with P139–512 or P139–512d, produced in recombinant Bacillus subtilis. Serum dilutions were reacted in ELISA plates coated with P139–512 (closed symbols) or heat-denatured P139–512d (open symbols). Symbols: (circles) anti-P1; (squares) anti-P139–512; and (triangles) anti-P139–512d.
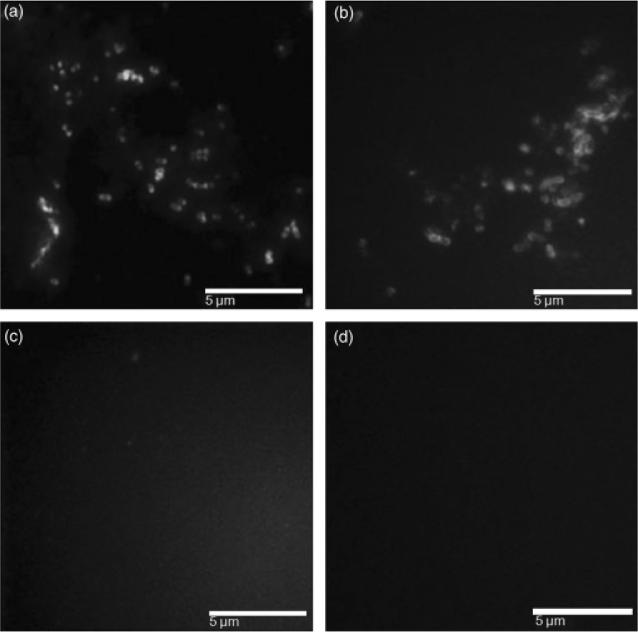
To further evaluate whether epitopes present in the native S. mutans P1 protein are conserved in the recombinant P139–512, antisera raised in mice against the S. mutans PC3370 strain lacking the spaP gene encoding P1, or against PC3370C, the deletion mutant complemented with a plasmid encoding P1, were reacted with P139–512 and P139–512d polypetides in ELISA (Table 2). Antibodies generated against S. mutans PC3370C, but not those raised against PC3370, reacted with both of these antigens, further indicating that epitopes reflective of cell surface-localized streptococcal P1 are maintained in the recombinant P139–512 expressed in B. subtilis. As additional evidence of the specifi-city of the anti-P139–512 antiserum, S. mutans PC3370C and PC3370 were reacted with serum samples harvested from mice immunized with P139–512 or P139–512d. As shown in Fig. 5, PC3370C, but not PC3370, was labeled with both serum samples. These results indicate that the recombinant P139–512 polypeptide preserved surface-exposed epitopes, even after the heat treatment.
Fig. 5.
Immunofluorescence labeling of Streptococcus mutans cells using antisera from mice immunized with P139–512 or P139–512d. Samples: (a) S. mutans PC3370C strain reacted with the anti-P139–512 antiserum; (b) S. mutans PC3370C strain reacted with anti-P139–512 d antiserum; (c) S. mutans PC3370 (ΔP1) strain reacted with anti-P139–512 antiserum; (d) S. mutans PC3370 (ΔP1) strain reacted with anti-P139–512d serum. Labeled S. mutans cells were revealed with fluorescein isothiocyanate -conjugated goat anti-mouse IgG.
Streptococcus mutans saliva-mediated aggregation and adherence is inhibited by antisera raised in mice immunized with recombinant P139-512 protein
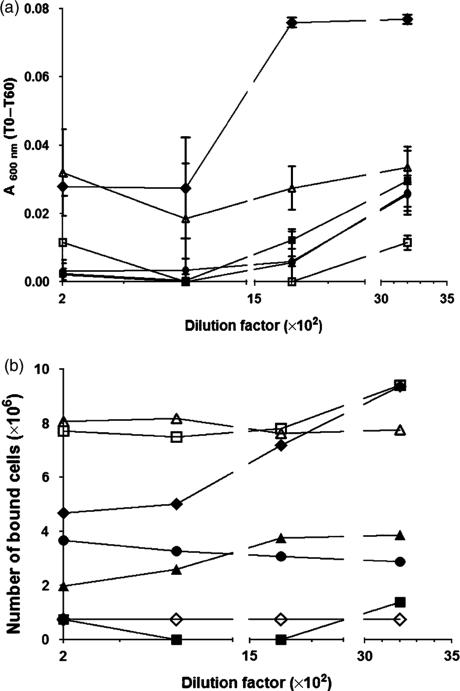
To evaluate the functional characteristics of antibodies raised in mice immunized with the recombinant P139–512 protein, we tested serum samples both for inhibition of saliva-mediated bacterial aggregation and for inhibition of S. mutans adherence to an abiotic surface. As demonstrated in Fig. 6a, anti-P139–512 and anti-P139–512d serum samples inhibited saliva-mediated aggregation of S. mutans cells at levels comparable to that from mice immunized with the full-length P1 protein (Table 2). The nonimmune mouse control serum did not show any effect on saliva-mediated S. mutans aggregation. The antiaggregation effect of the anti-P1 antiserum was partially, but not completely eliminated by adsorption with P139–512d, suggesting that antibodies against heat-sensitive epitopes contained within this poly-peptide may contribute to aggregation inhibition. In contrast, adsorption with P139–512 did not decrease the ability of the anti-P1 antiserum to inhibit aggregation and the inhibitory effect was even slightly improved (Fig. 6a). This suggests that the relative levels and balance of specific antibodies within the polyclonal anti-P1 reagent are a factor contributing to its biological function.
Fig. 6.
Functional characterization of antibodies raised in mice immunized with P139–512 produced in Bacillus subtilis. (a) Inhibition of saliva-mediated aggregation of Streptococcus mutans PC3370C cells. Samples containing bacterial cells and the tested serum were incubated for 5 min at 37 °C and the OD600nm was measured (T0). After an additional 60-min incubation period at 37 1C, the OD (T60) was measured once more. The values are expressed as the difference between T0 and T60 for each tested serum dilution. Symbols: (◆) nonimmune serum; (●) anti-P1 serum; (■) anti-P139–512 serum; (▲) anti-P139–512d serum; (□) anti-P1 serum adsorbed with P139–512; and (△) anti-P1 serum adsorbed with P139–512d. (b) Inhibition of S. mutans adherence to immobilized saliva in microplate wells. Streptococcus mutans PC3370C cells were mixed with diluted tested serum samples, incubated for 30 min to 37 °C, and transferred to microplate wells coated with clarified saliva and incubated for an additional 2 h. Bound cells were revealed by staining with 0.5% crystal violet and determination of absorbance of the solubilized cell-bound stain at 600 nm. Symbols are the same as those shown in (a) and (◇) cells incubated in the presence of 3 mM EDTA. All tested sera were diluted in order to reach a final titer of 104. Data represent the average of three independent experiments. Values are expressed as mean ± SD.
Experiments testing the inhibition of S. mutans adhesion to saliva-coated microplate wells showed that anti-P1, anti-P139–512d and anti-P139–512 antisera all significantly inhibited the binding of S. mutans PC3370C to the immobilized salivary constituents (Fig. 6b). The anti-P139–512 reagent was most effective in this regard. The interaction of P1 with gp340 is calcium-dependent and, as expected, S. mutans cells incubated in the presence of EDTA did not adhere to the saliva-coated microplate wells. A low level of background inhibition was observed with the nonimmune mouse serum that was eliminated at the higher dilutions. No significant aggregation or adherence was observed with the S. mutans PC3370 P1-deficient negative control strain (data not shown). In contrast to the results observed in the aggregation inhibition assays, the anti-P1 serum adsorbed with P139–512 or P139–512d completely lost its ability to inhibit the adhesion of PC3370C to immobilized salivary constituents (Fig. 6b). It has long been recognized that bacterial aggregation and adherence represent distinct properties that can be distinguished by anti-P1 antibodies of differing specificities (Brady et al., 1992). Importantly, the loss of adherence-inhibiting antibodies by adsorption with the purified B. subtilis P139–512 truncated polypeptide indicates that relevant functional epitopes are contained within this construction.
Discussion
The P1 adhesin of S. mutans represents an important target of protective immunity and represents a candidate antigen for anticaries vaccines (Shivakumar et al., 2009). In this study, we expressed and purified a P1-derivatived polypep-tide using a heterologous prokaryotic protein expression system based on a genetically modified B. subtilis strains. Our results demonstrate that the truncated P139–512 poly- peptide produced by Bacillus is recognized by antibodies generated in mice immunized with either S. mutans whole cells or with the purified full-length P1 protein. Conversely, immunization of mice with the B. subtilis-derived recombinant P139–512 induced serum IgG antibodies that recognize the full-length molecule as well as the native protein expressed on S. mutans cells. Finally, the anti-P139–512 antibodies were shown to be functionally active as demonstrated by an ability to interfere both with aggregation and adhesion to saliva-coated surfaces of S. mutans.
Low cost and high yields are the two major reasons for the widespread acceptance of E. coli strains as bacterial hosts for the expression of heterologous proteins both by academic laboratories and by industry. Nonetheless, problems associated with reduced expression, generation of insoluble proteins and contamination with endotoxin, due to the presence of outer membrane fragments, may constrain the use of E. coli as a host for purification of heterologous proteins. Bacillus subtilis strains show a good capacity to express recombinant proteins with preserved biological activity and do not have lipopolysaccharides, representing a promising alternative for the expression and purification of recombinant proteins (Harwood, 1992; Terpe, 2006; Schumann, 2007). Indeed, some proteins derived from gram-positive bacteria, such as streptokinase, have a greater stability when produced in B. subtilis (Wong et al., 1994). Similarly, the human interleukin-3 has been successfully expressed and purified from B. subtilis at recovery yields above 100 mg L−1 (Westers et al., 2005). Our initial attempts to produce S. mutans P139–512 in E. coli failed due to low expression levels and the insolubility of the recombinant protein. In contrast, the same protein was expressed in B. subtilis at reasonable levels (15 mg L−1) in a soluble state. Practical application of recombinant proteins generated in prokaryotic expression systems requires good expression levels and conservation of structural and functional features of the native protein. The recombinant P139–512 produced in B. subtilis was recognized by antisera generated against purified full-length recombinant P1 from E. coli as well as whole S. mutans cells. Several studies have shown that the P1 amino-terminal region containing the alanine-rich region is antigenic, as evidenced by reactivity of sera from patients naturally infected with S. mutans (Takahashi et al., 1991; Okahashi et al., 1993; Senpuku et al., 1996, 1998). Our data indicate that P139–512 preserves both antigenic and immunogenic determinants of the native S. mutans P1 protein. In the current work, with boiled and untreated polypeptides, we found that both heat-stable and heat-denaturable epitopes of P1, including those exposed at the cell surface, are contained within the recombinant P139–152. Interestingly, boiling the recombinant protein did not completely eliminate P139–512 antigenicity or immunogenicity, suggesting that both linear as well as heat-labile structural features play important roles in the immunological properties of P1.
The interaction of S. mutans with salivary components is a multifunctional process that determines whether the tooth surface will be colonized or not. The P1 protein interacts with salivary constituents adsorbed to the tooth surface leading to adherence (Lee et al., 1988, 1989; Koga et al., 1990). P1 also interacts with soluble fluid-phase components promoting aggregation and, presumably, elimination of S. mutans cells from the oral cavity (Ericson & Rundegren, 1983; Demuth et al., 1988; Koga et al., 1990). These dual roles of S. mutans P1 are contributed to by amino acid residues within P139–512 (Crowley et al., 1993; Nakai et al., 1993; Hajishengallis et al., 1998; Matsumoto-Nakano et al., 2008). Aggregation and adherence of S. mutans are inhibited by different anti-P1 monoclonal antibodies (Brady et al., 1992), indicating that the composite specificity of the host immune response would represent a key feature in determining the neutralization activity of elicited antibodies to a specific vaccine immunogen. In our current study, antibodies generated in mice immunized with the recombinant P139–512 or P139–512d protein were effective in blocking both bacterial aggregation and the adherence of S. mutans to abiotic surfaces. These results indicate that relevant immunological determinants required both for adhesion and for aggregation of the native S. mutans protein are maintained in the recombinant protein expressed in B. subtilis cells even after the denaturation process.
Consistent with previous findings that P1 determinants mediating bacterial aggregation and adherence can be differentiated with certain monoclonal antibodies, experiments with the anti-P1 antiserum adsorbed with, and hence depleted of antibodies against P139–512 or P139–512d, demonstrated a difference in the relative effect on aggregation inhibition compared with adherence inhibition. The ability of the anti-P1 antiserum to inhibit aggregation was impaired more by adsorption with P139–512d compared with P139–512, suggesting that antibodies against linear epitopes may be more relevant for this biological activity. That immunization of mice with P139–512 induced the formation of antibodies better able to inhibit bacterial adherence to salivary components compared with antibodies elicited against P139–512d or full-length P1 is informative. First, this suggests that antibodies against heat-labile epitopes may contribute to adherence inhibition. Second, the higher degree of adherence inhibition induced by P139–512 compared with the intact molecule suggests an effect of truncation on immunodominance and that important determinants may be better exposed in the truncated poly-peptide. It has been shown previously that certain anti-P1 monoclonal antibodies can alter the adhesin's immunogeni-city by increasing epitope exposure. Furthermore, these results were replicated by immunization with truncated variants of the protein (Robinette et al., 2009). The current results reiterate that less may in fact be more when it comes to induction of an optimal and efficacious response.
In conclusion, we have shown that a recombinant P139–512 polypeptide expressed and purified from B. subtilis cells preserves the relevant antigenic and immunogenic determinants of the native S. mutans adhesin. Because recombinant purified antigens represent an important approach for the development of vaccines against dental caries and systemic damage caused by S. mutans (Hajishengallis et al., 1995; Toida et al., 1997; Zhang et al., 2002), the ability to utilize B. subtilis as a promising new expression system represents an additional tool for the generation of recombinant antigens with application in acellular-based vaccines.
Acknowledgements
This work was supported by grants from Fundação de Amparoà Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional do Desenvolvimento Científico e Tecnológico (CNPq) and by the National Institute for Dental and Craniofacial Research R01 DE13882. We are grateful to Dr Wolfgang Schumann for helpful discussions. We gratefully acknowledge the skillful technical support of Loren C. da Silva, Natalie M. de Souza and Aline Teixeira.
References
- Adjic D, McLaughlin R, Savic G, et al. Genome sequence of Streptococcus mutans UA 159, a cariogenic dental pathogen. P Natl Acad Sci USA. 2002;99:14439–14494. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Crowley PJ, Ma JCK, Kell C, Lee SF, Lehner T, Bleiweis AS. Restriction fragment length polymorphisms and sequence variation within the spaP gene of Streptococcus mutans serotype c isolates. Infect Immun. 1991;59:1803–1810. doi: 10.1128/iai.59.5.1803-1810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Piacentini DA, Crowley PJ, Oyston PC, Bleiweis AS. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesion P1. Infect Immun. 1992;60:1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Cvitkovitch DG, Geric CM, Addison MN, Joyce JP, Crowley PJ, Bleiweis AS. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect Immun. 1998;66:4274–4282. doi: 10.1128/iai.66.9.4274-4282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PJ, Brady LJ, Piacentini DA, Bleiweis AS. Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect Immun. 1993;61:1547–1552. doi: 10.1128/iai.61.4.1547-1552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PJ, Brady LJ, Michalek SM, Bleiweis AS. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect Immun. 1999;67:1201–1206. doi: 10.1128/iai.67.3.1201-1206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- Demuth DR, Davis CA, Corner AM, Lamont RJ, Leboy PS, Malamud D. Saliva mediated aggregation of Enterococcus faecalis transformed with a Streptococus sanguis gene encoding the SSP-5 surface antigen. Infect Immun. 1988;57:1470–1475. doi: 10.1128/iai.57.5.1470-1475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson T, Rundegren J. Characterization of a salivary agglutinin reacting with a serotype c strain of Streptococcus mutans. Eur J Biochem. 1983;133:255–261. doi: 10.1111/j.1432-1033.1983.tb07456.x. [DOI] [PubMed] [Google Scholar]
- Ferreira LCS, Ferreira RCC, Schumann W. Bacillus subtilis as a tool for vaccine development: from antigen factories to delivery vectors. An Acad Bras Cienc. 2005;77:113–124. doi: 10.1590/s0001-37652005000100009. [DOI] [PubMed] [Google Scholar]
- Gibbons R. Adherent interactions which may affect microbial ecology in the mouth. J Dent Res. 1984;63:378–385. doi: 10.1177/00220345840630030401. [DOI] [PubMed] [Google Scholar]
- Gibbons R, Cohen L, Hay D. Strains of Streptococcus mutans and Streptococcus sobrinus attach to different pellicle receptors. Infect Immun. 1986;52:555–561. doi: 10.1128/iai.52.2.555-561.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Hollingshead SK, Koga T, Russell MW. Mucosal immunization with a bacterial protein antigen genetically coupled to cholera toxin A2/B subunits. J Immunol. 1995;154:4322–4332. [PubMed] [Google Scholar]
- Hajishengallis G, Russell MW, Michalek SM. Comparison of an adherence domain and a structural region of Streptococcus mutans antigen I/II in protective immunity against dental caries in rats after intranasal immunization. Infect Immun. 1998;66:1740–1743. doi: 10.1128/iai.66.4.1740-1743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR. Bacillus subtilis and its relatives: molecular biological and industrial workhorses. Trends Biotechnol. 1992;10:247–256. doi: 10.1016/0167-7799(92)90233-l. [DOI] [PubMed] [Google Scholar]
- Homonylo-McGavin M, Lee SF. Role of the C terminus in antigen P1 surface localization in Streptococcus mutans and two related cocci. J Bacteriol. 1996;178:801–807. doi: 10.1128/jb.178.3.801-807.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Hajishengallis G, Michalek SM. Induction of protective immunity against Streptococcus mutans colonization after mucosal immunization with attenuated S. enterica serovar typhimurium expressing an S. mutans adhesion under control of in vivo-inducible nirB promoter. Infect Immun. 2001;69:2154–2161. doi: 10.1128/IAI.69.4.2154-2161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N, Kerrigan S, Nobbs A, Strömberg N, Dolleweerd C, Cox D, Kelly C, Jenkinson H. Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect Immun. 2005;73:6629–6638. doi: 10.1128/IAI.73.10.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson H, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Kelly CG, Todryk S, Kendal HL, Munro G, Lehner T. T-cell, adhesion, and B-cell epitopes of the cell surface Streptococcus mutans protein antigen I/II. Infect Immun. 1995;63:3649–3658. doi: 10.1128/iai.63.9.3649-3658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Okahashi N, Takahashi I, Kanamoto T, Asakawa H, Iwaki M. Surface hydrophobicity, adherence, and aggregation of cell surface protein antigen mutants of Streptococcus mutans serotype c. Infect Immun. 1990;58:289–296. doi: 10.1128/iai.58.2.289-296.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MR, Rajashankar KR, Patel MH, Robinette RA, Crowley PJ, Michalek S, Brady LJ, Deivanayagam C. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of {alpha} - and PPII-helices. Proc Natl Acad Sci. 2010 doi: 10.1073/pnas.0912293107. DOI: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Progulske-Fox A, Bleiweis AS. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect Immun. 1988;56:2114–2119. doi: 10.1128/iai.56.8.2114-2119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Progulske-Fox A, Erdos GW, Piacentini D, Ayakawa GY, Crowley PJ, Bleiweis AS. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II). Infect Immun. 1989;57:3306–3313. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner T, Russell MW, Caldwell J, Smith R. Immunization with purified protein antigens from Streptococcus mutans against dental caries in Rhesus monkeys. Infect Immun. 1981;34:407–415. doi: 10.1128/iai.34.2.407-415.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner T, Caldwell J, Smith T. Local passive immunization by monoclonal antibodies against streptococcal antigen I/II in the prevention of dental caries. Infect Immun. 1985;50:796–799. doi: 10.1128/iai.50.3.796-799.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luiz WB, Cavalcante RCM, Paccez JD, Souza RD, Sbrogio-Almeida ME, Ferreira RRC, Ferreira LCS. Boosting systemic and secreted antibody responses in mice orally immunized with recombinant Bacillus subtilis strains following parenteral priming with a DNA vaccine encoding the enterotoxigenic Escherichia coli (ETEC) CFA/I fimbriae B subunit. Vaccine. 2008;26:3998–4005. doi: 10.1016/j.vaccine.2008.05.030. [DOI] [PubMed] [Google Scholar]
- Ma JKC, Smith R, Lehner T. Use of monoclonal antibodies in local passive immunization to prevent colonization of human teeth by Streptococcus mutans. Infect Immun. 1987;55:1274–1278. doi: 10.1128/iai.55.5.1274-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Nakano M, Tsuji M, Amano A, Ooshima T. Molecular interactions of alanine-rich and proline-rich regions of cell surface protein antigen c in Streptococcus mutans. Oral Microbiol Immun. 2008;23:265–270. doi: 10.1111/j.1399-302X.2007.00421.x. [DOI] [PubMed] [Google Scholar]
- McArthur W, Rhodin N, Seifert T, Oli M, Robinette R, Demuth D, Brady LJ. Characterization of epitopes recognized by anti-Streptococcus mutans P1 monoclonal antibodies. Immunol Med Microbiol. 2007;50:342–353. doi: 10.1111/j.1574-695X.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- Meima R, Van Dijl JM, Bron S. Expression systems in Bacillus. In: Baneyx F, editor. Protein Expression Technologies. Horizon Bioscience; Norfolk, UK: 2004. pp. 199–252. [Google Scholar]
- Michalek SM, Huang Y, Hajishengallis G. Induction of protective immunity against Streptococcus mutans colonization after mucosal immunization with attenuated Salmonella enterica serovar typhimurium expressing an S. mutans adhesion under the control of in vivo-inducible nirB promoter. Infect Immun. 2001a;69:4344–4349. doi: 10.1128/IAI.69.4.2154-2161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek SM, Katz J, Childers NK. A vaccine against dental caries: an overview. Biodrugs. 2001b;15:501–508. doi: 10.2165/00063030-200115080-00002. [DOI] [PubMed] [Google Scholar]
- Munro G, Evans P, Todryk S, Buckett P, Kelly CG, Lehner T. A protein fragment of Streptococcal cell surface antigen I/II which prevents adhesion of Streptococcus mutans. Infect Immun. 1993;61:4590–4598. doi: 10.1128/iai.61.11.4590-4598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M, Okahashi N, Ohta H, Koga T. Saliva-binding region of Streptococcus mutans surface protein antigen. Infect Immun. 1993;61:4344–4349. doi: 10.1128/iai.61.10.4344-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HD, Phan TTP, Schumann W. Expression vectors for the rapid purification of recombinant proteins in Bacillus subtilis. Curr Microbiol. 2007;55:89–93. doi: 10.1007/s00284-006-0419-5. [DOI] [PubMed] [Google Scholar]
- Nobbs A, Lamont RJ, Jenkinson H. Streptococcus adherence and colonization. Microbiol Mol Biol R. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahashi N, Takahashi I, Nakai M, Senpuku H, Nisizawa T, Koga T. Identification of antigenic epitopes in an alanine-rich repeating region of surface protein antigen of Streptococcus mutans. Infect Immun. 1993;61:1301–1306. doi: 10.1128/iai.61.4.1301-1306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccez JD, Luiz WB, Sbrogio-Almeida ME, Ferreira RCC, Schumann W, Ferreira LCS. Stable episomal expression system under control of a stress inducible promoter enhances the immunogenicity of Bacillus subtilis as a vector for antigen delivery. Vaccine. 2006;24:2935–2943. doi: 10.1016/j.vaccine.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Paccez JD, Nguyen HD, Luiz WB, Ferreira RCC, Sbrogio-Almeida ME, Schuman W, Ferreira LCS. Evaluation of different promoter sequences and antigen sorting signals on the immunogenicity of Bacillus subtilis vaccine vehicles. Vaccine. 2007;25:4671–4680. doi: 10.1016/j.vaccine.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Pancholi V, Fischetti VA. Identification of an endogenous membrane anchor-cleaving enzyme for group A streptococcal M protein. Its implication for the attachment of surface proteins in gram-positive bacteria. J Exp Med. 1989;170:2119–2133. doi: 10.1084/jem.170.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulitz TC, Bélanger RR. Biological control in greenhouse systems. Annu Rev Phytopathol. 2001;39:103–133. doi: 10.1146/annurev.phyto.39.1.103. [DOI] [PubMed] [Google Scholar]
- Prakobphol A, Xu F, Hoang VM, et al. Salivary agglutinin, which binds Streptococcus mutans P1 epitope recognized by immunomodulatory monoclonal antibody 6-11A. Infect Immun. 2000;72:4680–4688. doi: 10.1128/IAI.72.8.4680-4688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodin NR, Cutalo JM, Tomer TN, McArthur WP, Brady LJ. Characterization of the Streptococcus mutans P1 epitope recognize by immunomodulatory monoclonal antibody 6-11A. Infect Immun. 2004;72:4680–4688. doi: 10.1128/IAI.72.8.4680-4688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette RA, Oli MW, McArthur WP, Brady LJ. Beneficial immunomodulation by Streptococcus mutans anti-P1 monoclonal antibodies is Fc independent and correlates with increased exposure of a relevant target epitope. J Immunol. 2009;183:4628–4638. doi: 10.4049/jimmunol.0803300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundegren JL, Arnold RR. Bacteria-agglutinating characteristics of secretory IgA and a salivary agglutinin. Adv Exp Med Biol. 1987;216:1005–1013. [PubMed] [Google Scholar]
- Russell R. Pathogenesis of oral streptococci. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rool JI, editors. Gram-Positive Pathogens. American Society for Microbiology; Washington, DC.: 2000. pp. 272–279. [Google Scholar]
- Russell RRB, Lehner T. Characterization of antigens extracted from cells and culture fluids of Streptococcus mutans serotype c. Arch Oral Biol. 1978;23:7–15. doi: 10.1016/0003-9969(78)90047-x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Schumann W. Production of recombinant proteins in Bacillus subtilis. Adv Appl Microbiol. 2007;62:137–189. doi: 10.1016/S0065-2164(07)62006-1. [DOI] [PubMed] [Google Scholar]
- Seifert T, Bleiweis AS, Brady LJ. Contribution of the alanine-rich region of S. mutans P1 to antigenicity, surface expression, and interaction with the proline-rich repeat domain. Infect Immun. 2004;72:4699–4706. doi: 10.1128/IAI.72.8.4699-4706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senpuku H, Iizima T, Yamaguchi Y, Nagata S, Ueno Y, Saito M, Hanada N, Nisizawa T. Immunogenicity of peptides coupled with multiple T-cell epitopes of a surface protein antigen of Streptococcus mutans. Immunology. 1996;88:275–283. doi: 10.1111/j.1365-2567.1996.tb00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senpuku H, Yanagi K, Nisizawa T. Identification of Streptococcus mutans PAc peptide motif binding with human MHC class II molecules (DRB1 0802, 1101, 1401 and 1405). Immunology. 1998;95:322–330. doi: 10.1046/j.1365-2567.1998.00583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar KM, Vidya SK, Chandu GN. Dental caries vaccine. Indian J Dent Res. 2009;20:99–106. doi: 10.4103/0970-9290.49066. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Mattos-Graner RO. Secretory immunity following mutans streptococcal infection or immunization. Curr Top Microbiol. 2008;319:131–156. doi: 10.1007/978-3-540-73900-5_6. [DOI] [PubMed] [Google Scholar]
- Takahashi I, Okahashi N, Matsushita K, Tokuda M, Kanamoto T, Munekata E, Russell MW, Koga T. Immunogenicity and protective effect against oral colonization by Streptococcus mutans of synthetic peptides of a streptococcal surface protein antigen. J Immunol. 1991;146:332–336. [PubMed] [Google Scholar]
- Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biot. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- Toida N, Hajishengallis G, Wu H, Russell MW. Oral immunization with saliva-binding region of Streptococcus mutans Ag I/II genetically coupled to the cholera toxin B subunit elicits T-helper-cell responses in gut-associated lymphoid tissues. Infect Immun. 1997;65:909–915. doi: 10.1128/iai.65.3.909-915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Tsuha Y, Hanada N, Asano T, Abeis T, Yamaguchi S, Salam M, Nakao R, Takeuchi H. Role of peptide antigen for induction of inhibitory antibodies to S. mutans in the human oral cavity. Clin Exp Immunol. 2004;137:393–401. doi: 10.1111/j.1365-2249.2004.02548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dolleweerd CJ, Kelly CG, Chargelegue D, Ma J. Peptide mapping of a novel discontinuous epitope of the major surface adhesin from Streptococcus mutans. J Biol Chem. 2004;279:22198–22203. doi: 10.1074/jbc.M400820200. [DOI] [PubMed] [Google Scholar]
- Wehrl W, Niederweis M, Schumann W. The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation. J Bacteriol. 2000;182:3870–3873. doi: 10.1128/jb.182.13.3870-3873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westers H, Darmon E, Zanen G, Veening JW, Kuipers OP, Bron S, Quax WJ, Van Dijl JM. The Bacillus secretion stress response is an indicator for alpha-amylase production levels. Lett Appl Microbiol. 2004;39:65–73. doi: 10.1111/j.1472-765X.2004.01539.x. [DOI] [PubMed] [Google Scholar]
- Westers L, Dijkstra DS, Westers H, Van Dijl JM, Quax WJ. Secretion of functional human interleukin-3 from Bacillus subtilis. J Biotechnol. 2005;123:211–214. doi: 10.1016/j.jbiotec.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Wong SL, Ye R, Nathoo S. Engineering and production of streptokinase in a Bacillus subtilis expression–secretion system. Appl Environ Microb. 1994;60:517–523. doi: 10.1128/aem.60.2.517-523.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Jespersgaard C, Lamberty-Mallory L, Katz J, Huang Y, Hajishengallis G, Michalek SM. Enhanced immunogenicity of a genetic chimeric protein consisting of two virulence antigens of Streptococcus mutans and protection against infection. Infect Immun. 2002;70:6779–6787. doi: 10.1128/IAI.70.12.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Green NM, Sitkiewicz I, LeFebvre RB, Musser JM. Identification and characterization of an antigen I/II family protein produced by group A Streptococcus. Infect Immun. 2006;74:4200–4213. doi: 10.1128/IAI.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]