Abstract
Cancer cells silence autosomal tumor suppressor genes by Knudson’s two-hit mechanism in which loss-of-function mutations and then loss of heterozygosity occur at the tumor suppressor gene loci. However, the identification of X-linked tumor suppressor genes has challenged the traditional theory of “two-hit inactivation” in tumor suppressor genes, introducing the novel concept that a single genetic hit can cause loss of tumor suppressor function. The mechanism through which these genes are silenced in human cancer is unclear, but elucidating the details will greatly enhance our understanding of the pathogenesis of human cancer. Here, we review the identification of X-linked tumor suppressor genes and discuss the potential mechanisms of their inactivation. In addition, we also discuss how the identification of X-linked tumor suppressor genes can potentially lead to new approaches to cancer therapy.
Keywords: X-linked genes, tumor suppressors, single-hit inactivation, X chromosome inactivation, cancer
Introduction
Cancer cells silence tumor suppressor genes by Knudson’s two-hit mechanism, [1] in which a loss-of-function mutation at the tumor suppressor locus is followed by a loss of heterozygosity (LOH), and therefore a loss of the wild-type tumor suppressor gene. Deletions, insertions, nonsense mutations, frame-shift mutations, missense mutations, or epigenetic alterations that inactivate functional activity of a protein are all observed in tumor suppressor genes. In most tumor suppressor genes, such as retinoblastoma (RB), both alleles of the gene must be inactivated for tumorigenesis [1]. In other tumor suppressor genes like TP53 and PTEN, a mutation at one allele may be sufficient to give rise to an altered cell phenotype, resulting in a lower level of tumor suppressor function during tumor development and progression [2, 3]. Males carry only one allele of the X-linked genes, and one allele in each female cell is inactivated as a method of dosage compensation between the sexes. These genes are therefore more susceptible to genetic damages promoting tumor development and progression [4–6] and a single genetic hit is sufficient to inactivate an X-linked tumor suppressor gene [7] as X chromosome inactivation acts as a functional LOH for X-linked tumor suppressor genes [8, 9].
Potential implication of X-linked tumor suppressor genes in human cancers
The possibility of tumor suppressor genes on the X chromosome was raised over two decades ago [7, 10]. The first piece of strong evidence for one such gene was provided by Klein et al. in 1991, using X-chromosome-transfer analysis [11]. Transfection of a normal Chinese hamster X chromosome to a nickel-transformed Chinese hamster cell line with an Xq deletion resulted in senescence of these previously immortal cells [11, 12]. Since then, numerous chromosomal regions including Xp11–22, Xq25–26, and Xq27–28 have been proposed as potential loci for tumor suppressor genes [13–24]. LOH and skewed X chromosome inactivation at these putative X-linked tumor suppressor loci have been frequently identified in breast, ovarian, and prostate cancers. Several studies have shown as high as 40% of ovarian cancer samples have LOH of X-linked genes [15, 16, 18, 19, 25–29]. In particular, LOH at Xp22.2–3 of the active X chromosome is frequently found in ovarian cancers with germline BRCA1 mutations [18]. LOH at Xp25–26 is significantly associated with TP53 LOH in ovarian cancer [28]. These results suggest that these loci may harbor tumor suppressor genes that functionally interact with other tumor suppressor genes [7, 9]. LOH at the active X chromosome may cause complete loss of tumor suppressor function of these X-linked genes, leaving individuals susceptible to cancer formation [7, 9]. In breast cancer, extensive LOH at the X chromosome has been identified [13, 14, 30, 31] and linked to higher tumor grade and lymph node metastasis [32, 36]. Interestingly, BRCA1 mutations have also been implicated in skewed X chromosome inactivation in breast cancer [33–35]. One report found a loss of X-linked gene expression in 22% of various cancers, including breast and ovarian cancers [36]. LOH at the X chromosome is also associated with sporadic colorectal carcinoma [37], renal-cell carcinoma [38, 39], melanoma [40] and neuroendocrine tumor [41–46].
Recent epidemiological studies have suggested a role for X-linked genes in susceptibility to human prostate cancer. Hereditary prostate cancer, X-linked (HPCX) region at Xq27–28 is a putative prostate cancer susceptibility locus [24, 47–50]. Xu et al. firstly identified the X-linked inheritance of familial prostate cancers at this locus, which may account for as much as 16% of hereditary prostate cancer cases [24]. Recent data clearly show that deletions within the HPCX locus are associated with sporadic prostate cancers, which raises the possibility that this locus may play a functional role as a tumor suppressor in prostate cancer [51]. Recently, a haplotype at Xq27.2 [24, 48] and common sequence variant at Xp11.22 [52] were shown to confer susceptibility to prostate cancer. While these two loci have been implicated in prostate cancer, the genes in these regions, including at the HPCX locus, have not been fully identified.
Identification of X-linked tumor suppressor genes
Although LOH analysis following Klein et al.’s paper [11] suggested the presence of X-linked tumor suppressor genes in human cancer, no specific genes were identified by such analysis. However, two studies published since 2007 have identified the first X-linked tumor suppressor genes [6, 53]. One study describes involvement of the X-linked gene, FAM123B (also call WTX) at Xq11.2 in Wilms tumor [53] and we have reported that FOXP3 at Xp11.23 is an X-linked tumor suppressor gene involved in both breast cancer and prostate cancer [5, 6], suggesting a single-hit inactivation of X-linked tumor suppressor genes in human cancer.
Recent whole genome-wide scan analyses provided substantial information regarding X-linked cancer-related genes and have identified a large number of somatic driver mutations in potential cancer-related genes [54–57]. These driver mutations have been proposed to contribute to the neoplastic process and are positively selected for during tumorigenesis. Interestingly, driver mutations are frequently found in numerous X-linked genes, which may be additional potential X-linked tumor suppressor genes. In breast cancer, a group of X-linked, cancer-related genes have been found, including FLNA, PFC, PRPS1, TARD8, MAGEE1, TAF and KLH4 [54–56]. In colorectal cancer, X-linked FLNA, TBX22, KIAA2022, IRS4, PCDH11X, GPR112 and F8 are proposed cancer-related genes. In melanoma, the suggested X-linked cancer-related genes include ZNF280C, IL3RA, PNMA3, NHS and FGD1. The role of these genes in tissue-specific carcinogenesis needs to be clarified and will be important in our understanding of the mechanism of single-hit inactivation of X-linked tumor suppressor genes during cancer initiation and progression. We describe below several confirmed and putative X-linked tumor suppressor genes (Table 1).
Table 1.
Putative X-linked tumor suppressor genes in human cancers
| Gene | Locus | X-linked in other mammals |
Function | Signaling pathway |
Involvement in cancer |
|---|---|---|---|---|---|
| FOXP3 | Xp11.23 | Mouse, Rat, Dog |
Transcriptional factor; Master regulator in the development and function of regulatory T cells; Tumor suppressor |
Nuclear transcritional regulation of the HER2, SKP2, c-Myc and p21 |
Down-regulated in breast cancer, prostate cancer and ovarain cancer tissues, but overexpression in pancreatic cancer tissues; Gene deletions and somatic mutations identified in breast cancer and prostate cancer |
| RBBP7 | Xp22.2 | Mouse, Rat, Dog |
Putative tumor suppressor involved in cell proliferation, differentiation and apoptosis |
Regulation of the Rb, BRCA1, WT1 and beta- catenin/TCF signalings |
Unknown |
| CD99 | Xp22.32 and Yp11.3 |
Dog, but not in Mouse, Rat |
cell-cell adhesion; Leukocyte migration, T- cell adhesion, ganglioside GM1 and transmembrane protein transport, and T-cell death and involved in in tumor malignancy and metastases |
Caspase- independent pathway and AKT-, ERK-, and JNK- dependent AP-1 activation signaling pathways |
Down-regulation in osteosarcoma, gallbladder carcinomas, nasopharyngeal carcinoma, gastric adenocarcinoma, and transitional cell carcinomas of urinary bladder, but up- regulated in desmoplastic small-cell tumours, synovial sarcomas, primitive neuroectodermal tumours and hepatocarcinomas; LOH at this locus and promoter methylation identified in gastric adenocarcinoma |
| FAM123B | Xq11.1 | Mouse, Rat, Dog |
Tumor suppressor for Wilms tumor |
Inhibition of the beta- catenin signaling and modulation of WT1 activity |
Gene deletion and somatic mutation identified in Wilms tumors |
| EDA2R | Xq12 | Mouse, Rat, but unknow in Dog |
Promoting apoptosis in tumor cells and epidermal morphogenesis during embryonic development |
Activation of the NF- kappaB and JNK pathways; downstream target of p53 |
Down-regulated in breast cancer and colorectal cancer; mutation and hypermethylation identified in colorectal cancer |
| RPS6KA6 | Xq21 | Mouse, Rat, Dog |
Inducing cell arrest and senescence and involved in invasion and chemotaxis |
Up-regulation of Rb, retinobl astoma- associated 46 kDa protein, p21, and claudin-2, but down- regulation of CXCR4 |
Down-regulated in colon carcinomas, colon adenomas, renal cell carcinomas, and endometrial cancers |
| ATRX | Xq21.1 | Mouse, Rat, Dog |
Nevol tumor suppressor involved in transcriptional regulation, nuclear architecture, and chromosome stability |
Regulation of chromatin remodeling, DNA methylation and gene transcription |
Down-regulation in acute myeloid leukemia, prostate cancer, breast cancer, esophageal squamous cell carcinoma; loss-of- function somatic mutations in pancreatic neuroendocrine tumor and paediatric glioblastoma |
| ELF4 | Xq26.1 | Mouse, Rat, Dog |
Transcriptional regulation, DNA damage response, and involved in natural killer (NK) cell development and function |
Transcriptional activation of the CSF2, IL3, IL8, and PRF1 and repression of MMP-9 and IL-8 |
LOH at this locus identified in ovarian and breast cancers; translocation in acute myelogenous leukemia; rearrangements of BCORL1-ELF4 in hepatocellular carcinomas |
| PHF6 | Xq26.3 | Mouse, Dog, but unknow in Rat |
Transcriptional regulation |
Regulation of homeobox transcription factor oncogenes TLX1 and TLX3 |
Mutation and deletion frequently found in T- cell acute lymphoblastic leukemia, adult acute myeloid leukemia, and hepatocellular carcinoma |
| LDOC1 | Xq27 | Mouse, Rat, Dog |
Inducing apoptosis in tumor cells |
Inhibition of the NF- kappaB signaling and the degradation of p53 |
Down-regulated in pancreatic and gastric cancer cell lines; deletions identified in prostate cancers, but high levels of LDOC1 correlate with poor prognosis in chronic lymphocytic leukemia |
| RPL10 | Xq28 | Mouse, Rat, Dog |
Putative tumor suppressor for hormonally dependent cancers |
Repression of the c-Jun and AP-1- mediated transcriptional activation |
LOH and microsatellite instability at this locus frequently occurred in ovarian cancer; down- regulated in prostate cancer and multiple endocrine neoplasia type 1 |
| DKC1 | Xq28 |
Mouse, Rat, Dog |
Multiple functions in telomerase, ageing and cancerr |
RNA processing and modification; p53- and p27- denpendent translational control in ribosome biogenesis |
Unknown |
Family with sequence similarity 123B (FAM123B) / Wilms Tumor on the X (WTX)
Wilms tumor is cancer of the kidneys that typically occurs in children, rarely in adults [58–60]. Wilms tumor 1 (WT1) at chromosome 11p13 is a zinc finger transcription factor gene and is the most completely characterized gene in the field of Wilms tumor genetics, but just 5~10% of cases are caused by inactivation of the WT1 tumor-suppressor gene [60]. Recently, a genome-wide scan for DNA copy-number changes in 51 primary tumor specimens found small overlapping deletions at Xq11.1 in approximately 30% (15/51) of tumors [53]. The deletions were associated with an uncharacterized gene (FAM123B) that the researchers named WTX [53]. Of interest, the FAM123B deletions were all heterozygous in female Wilms tumors and targeted to the active X chromosome, leading to gene inactivation by a single-hit mechanism [53]. Similarly, heterozygous intragenic truncating mutations were found in 7.3% (6/82) of Wilms tumors examined. Only the mutant copy was expressed, indicating placement on the active X chromosome. In addition, the tumors with FAM123B deletions or mutation did not carry mutations of WT1, suggesting FAM123B genomic alterations may occur independently of WT1 mutations in Wilms tumor.
FAM123B at Xq11.1 is close to the centromere and encodes an 1135 amino acid protein with no conserved functional domains except for a predicted nuclear localization signal. Functional analyses in cultured Xenophus and zebra fish cells have provided a possible mechanism for the tumor suppressor activity of FAM123B in Wilms tumor by demonstrating that FAM123B promotes β-catenin ubiquitination and degradation, antagonizing Wnt/β -catenin signaling [61]. A recent study determined that FAM123B shuttles between the cytoplasm and the nucleus, where it is present in a distinct subnuclear compartment implicated in transcription and RNA processing [62]. Moreover, the C-terminus of FAM123B binds to the transcription factor WT1 and modulates its transcriptional activity [62]. Thus, FAM123B may play a role in the transcriptional regulation of genes determining cellular differentiation [61, 62].
Rivera et al. have determined that approximately 20% of Wilms tumor cases are caused by inactivation of FAM123B, independent of mutations in WT1 [53]. This observation has been confirmed by other recent studies. Ruteshouser et al. identified 18.4% (23/125) of Wilms tumors carrying 16 deletions and 8 mutations of FAM123B; of these, 20% had accompanying mutations in WT1 and/or CTNNB1 (coding for β –catenin) vs. 17.5% with mutations in FAM123B alone [63]. Perotti et al. identified Wilms tumors with FAM123B deletions in 11% (5/45) of male samples and LOH in 18% (9/50) of female samples. In the latter group, only two cases had a deletion in the active X chromosome, and sequence analyses detected an inactivating somatic mutation in just one tumor [64]. However, just 7% of Wilms tumor cases had functionally null genes, suggesting that previously reported estimates on the proportion of Wilms tumors due to FAM123B alterations should be reconsidered [64]. Wegert et al. examined mutations of FAM123B in a large set of 429 Wilms tumors [65]. They found that 11.5% of tumor samples lacked expression of FAM123B mRNA. Gene deletion was identified in 17% of Wilms tumor cases, equally distributed between males (deletion) and females (LOH), but point mutations were found in just 2% of tumors. These genetic alterations appear to be independent of WT1 mutations. However, they did not find a significant correlation between FAM123B deletion status or expression level and clinical parameters, suggesting that FAM123B alteration is not an essential and early mutation necessary to drive tumorigenesis, but rather a later event that may affect only a fraction of cells with unclear clinical relevance [65]. Overall, these data show that FAM123B has been somatically inactivated in 11~29% of cases of Wilms tumor [53, 63–65]. Recently, Fukuzawa et al. indicated that the expression levels of the FAM123B were not reduced in females bearing this gene deletion, suggesting mosaic deletion or deletions occurring on the inactive X chromosome [66] Although they found somatic missense mutations with one allele in 2 patients, the wild-type allele is still expressed. Perotti et al. also suggested that gene deletions can occur randomly on both the active and inactive X chromosome [64, 67]. More recently, Jenkins et al. identified germline mutations of FAM123B that caused an X-linked sclerosing bone dysplasia, osteopathia striata congenita with cranial sclerosis [68]. This condition is lethal in males and causes increased bone density and craniofacial malformations in females [68]. However, patients do not have any predisposition to Wilms tumor [68, 69]. These data suggest that Wilms tumor may require the acquisition of a specific order of mutations for tumorigenesis, in which somatic mutation of FAM123B occurs later in tumor development [65, 69], while germline mutation of this gene does not predispose individuals to Wilms tumor [69].
Forkhead box P3 (FOXP3)
FOXP3 at Xp11.23 is a member of the forkhead-box/winged-helix transcription factor family. It was identified during positional cloning of Scurfin, a gene causing X-linked autoimmune diseases in mice and humans [70–73]. Scurfin was later renamed FOXP3 and found to be the master regulator of development and function of regulatory T cells [74]. However, FOXP3 is expressed not only in lymphocytes but also in epithelial tissues of the breast, lung, and prostate [75]. The human FOXP3 gene has alterative splicing variants, including Δ2, Δ3, Δ7, Δ3–4, Δ3/8 and Δ8. Normal cells express the splice variants Δ2 and Δ7 [76–80], while cancer cells predominantly express splice variants Δ3, Δ3–4, Δ3/8 and Δ8 [6, 81–83].
Our lab previously observed that mice with a spontaneous mutation of FoxP3 developed mammary tumors at a high rate. Nuclear FOXP3 protein is expressed in normal human breast epithelial cells but is lost in 80% of human breast cancers [6]. The significance of FOXP3 as an X-linked tumor suppressor gene in humans is supported by the prevalence of somatic mutations (36%), gene deletions (13%) and lack of nuclear FOXP3 in the majority of breast cancer samples examined [6]. Interestingly, an overwhelming majority of the mutations are heterozygous and the deletion of the gene is heterozygous in all cases. These data reaffirm the notion that a single-hit is sufficient to inactivate X-linked genes. Furthermore, mammary carcinomas exhibited FOXP3 inactivation while over-expressing two critical oncogenes, HER2/ErbB2 [6] and SKP2 [84], and have low expression of the tumor suppressor genes p21 [85] and LATS1/2 [86] in both mice and humans. Functional analysis demonstrated that FOXP3 inhibits breast tumor growth by directly repressing the transcriptional activity of oncogenes HER2 and SKP2 while inducing transcription of tumor suppressor genes p21 and LATS1/2 [6, 84–86]. These data identified FOXP3 as an X-linked tumor suppressor in breast cancer in both mice and humans.
Recent epidemiological studies have indicated two loci at Xp11.22 [52] and Xq27–28 [24, 48] associated with susceptibility to prostate cancer, but the genes in these regions have not been identified. FOXP3 resides near the Xp11.22 region and has significant linkage disequilibrium (LD) between these two loci, raising the possibility that the FOXP3 locus may contribute to X-linked prostate cancer susceptibility and may, in fact, act as a tumor suppressor in prostate cancer. Consequently, we analyzed FOXP3 expression in a large panel of human prostate cancer samples. As expected, nuclear FOXP3 is present in normal human prostate epithelial cells but is lost in approximately 70% of human prostate cancers. FOXP3 is frequently inactivated in prostate cancer samples by deletion (14%) and somatic mutation (25%). An inverse correlation was observed between FOXP3 and c-MYC expression in human primary prostate cancers [5]. Functional analysis showed that FOXP3-mediated transcriptional repression of c-MYC is necessary to control c-MYC levels in normal prostate epithelial cells, contributing to the widespread overexpression of c-MYC in prostate cancer [5]. Wild-type (WT) FOXP3 exhibits strong growth inhibition of prostate cancer cell lines [5]. Importantly, prostate-specific ablation of FoxP3 in the mouse caused early onset of prostate hyperplasia and prostatic intraepithelial neoplasia (PIN) [5]. These data indicate that FOXP3 is also an X-linked tumor suppressor gene in prostate cancer.
However, FOXP3 expression in human cancers, as reported in recent studies, is subject to debate. While Merlo et al. reported a 2-fold higher rate of FOXP3 expression in breast cancer samples [87] than that in our report [6], the former did not discriminate cytoplasmic from nuclear forms and the latter only considered nuclear FOXP3 protein. The majority of mutant FOXP3 is located in the cytoplasm, whereas WT FOXP3 is a nuclear protein [5, 6, 84, 85]. Reported mutants of FOXP3 in prostate cancer cells abrogate normal FOXP3 function by disrupting its translocation into nuclei [5]. Ladoire et al. recently revealed that while FOXP3 is expressed in 57% of breast cancer samples, expression is limited to the cytoplasm in HER2 mutant-positive breast cancer samples [88, 89]. Karanikas et al. broadly identified FOXP3 expression in 25 tumor cell lines of different tissue origins including lung cancer, colon cancer, breast cancer, melanoma, erythroid leukemia, and acute T cell leukemia, with both FOXP3 mRNA and protein detected in all tumor cell lines [90]. Immunohistochemical staining of these cell lines showed predominantly cytoplasmic expression of FOXP3 in melanoma (GERL), colon (HCA 2.6) and breast cancer (MCF7) cell lines, and both cytoplasmic and nuclear expression in lung cancer (GILI) and T lymphoblastic leukemia (JURKAT) cell lines. Similarly, cytoplasmic staining of FOXP3 was detected in pancreatic cancers [91]. Currently, there is debate regarding the clinical significance of FOXP3 expression in human cancers. FOXP3 expression was correlated with better disease outcome and survival in patients with HER2+-breast cancer [92] and colorectal cancer [93], but the converse was true in patients with breast cancer [87], non-small cell lung cancer, urinary bladder cancer [94], hepatocellular carcinoma, [95] and tongue squamous cell carcinoma [96]. However, most of them did not divide FOXP3 expression by cellular localization, which is essential for FOXP3 tumor suppressor function. The cytoplasmic localization is associated with the loss of tumor inhibition [97], thus nuclear FOXP3 may be a useful marker for predicting disease outcome in human cancers.
Ebert et al. identified the FOXP3 mRNA variant lacking Δ3–4 as widely expressed in human melanoma cell lines but absent in T regulatory cells. This alternative splicing event introduces a translation frame-shift that is predicted to encode a novel protein in tumor cells and therefore may contribute to the malignant progression of cells [81]. Likewise, recent studies revealed that specific splice variants of FOXP3, such as deletion of Δ3, Δ3–4, Δ3/8 and Δ8, are preferably expressed in breast and ovarian cancers, and malignant melanomas and malignant T cells of Sezary syndrome [6, 81–83, 92]. Inactivation of FOXP3 function by disruption of intracellular localization or alternative splicing events might help to reconcile reported differences in FOXP3 expression in human cancer samples [4, 5].
FOXP3 has been demonstrated to be a master regulator controlling transcriptional activity of SKP2, HER-2/ErbB2 and p21 in breast cancer and c-MYC in prostate cancer [5, 6, 84, 85]. The context of FOXP3 regulation during tumor development remains largely unexplored. A recent study demonstrated that FOXP3 up-regulation in human breast and colon carcinoma cells requires p53 function. DNA-damaging agents can induce FOXP3 expression in p53-positive carcinoma cells, but not in cells lacking p53 function. However, knockdown of FOXP3 can blunt this p53-mediated growth inhibition [93]. These results indicate that FOXP3 expression is directly regulated by p53 during DNA damage responses. FOXP3 may therefore be a key determinant of cell fate in p53-dependent DNA damage responses. In addition, Zhang et al. found that FOXP3 is expressed weakly or not at all in ovarian cancer cells. Up-regulation of FOXP3 led to decreased expression of Ki-67 and cyclin-dependent kinases, resulting in inhibition of cell proliferation. Up-regulation of FOXP3 also caused reduced cell migration and invasion, possibly due to reduced expression of matrix metalloproteinase-2 and urokinase-type plasminogen activator [83]. The authors further proposed that FOXP3 inhibits the activation of mammalian target of rapamycin (mTOR) and nuclear factor (NF)-κB signaling. Therefore, FOXP3 might play divergent functional roles in different tissues and tumor stages.
Ribosomal protein L10 (RPL10)
This gene is located at Xq28 and encodes a ribosomal protein that is a component of the 60S subunit and most likely involved in the later steps of the 60S ribosomal subunit assembly [94, 95]. RPL10 was initially identified as a candidate tumor suppressor gene in Wilms tumor [96], but later studies refuted this claim [97]. RPL10 can repress transcriptional activity of the proto-oncogene c-Jun in vitro, but these activities have not been verified in vivo [97–103]. c-Jun works with c-Fos to form the transcription factor AP-1, which facilitates signaling through the estrogen and androgen receptors [104–108]. Inhibition of AP-1 activity presumably suppresses development and growth of sex hormone-regulated tumor cells. RPL10 may therefore function as a tumor suppressor in hormonally-dependent cancers through its inhibition of c-Jun and AP-1 activity. Furthermore, it may play a role in multiple endocrine neoplasia type 1 (MEN1), which is caused by inactivation of the tumor suppressor MEN1. RPL10 was 8-fold down-regulated in a neuroendocrine tumor cell line, BON1, transfected with MEN1 [109]. RPL10 is expressed in islets, tumors, and exocrine cells of the pancreas in human MEN1 carriers and Men1 heterozygous mice [110], suggesting that RPL10 may play a role in MEN1-related pancreatic tumorigenesis. It has been reported that LOH (18~30%) and microsatellite instability (12~50%) at Xq28 frequently occur in ovarian cancer and are significantly associated with susceptibility to ovarian cancer [111]. In prostate cancer, decreased RPL10 expression may be associated with early development of prostate cancer, but high levels of RPL10 at later stages of tumor development may facilitate progression into a more aggressive phenotype [112]. It is therefore premature to draw any conclusions regarding the functional role of RPL10 in human cancer.
Retinoblastoma binding protein 7 (RBBP7, also known as BRAP46)
This gene is located at Xp22.2 and codes for a ubiquitously-expressed nuclear protein that binds to Rb protein, which regulates cell proliferation [113]. WT1 also signals through RBBP7 to inhibit growth [116]. Ectopic expression of RBBP7 in several tumor cell lines suppresses cell growth and proliferation in vitro and in vivo [117], suggesting that RBBP7 may be a tumor suppressor in some cancers [115]. In human breast cancer, RBBP7 can activate c-Jun NH2-terminal kinase-dependent apoptosis and suppress tumorigenicity and estrogen-stimulated progression of breast cancer in vitro and in vivo [118–121]. The suppressive activity of RBBP7 may be mediated by up-regulation of GSK-3β, which attenuates β-catenin/TCF signaling [122]. However, other studies have suggested that RBBP7 induces epithelial-mesenchymal transition in mammary epithelial cells [123]. High expression of RBBP7 may be associated with development/progression of human breast cancer [124], and RBBP7 protein can inhibit BRCA1 transactivation activity and may therefore attenuate DNA damage response [114, 115]. The levels of RBBP7 serum, mRNA and protein were significantly increased in non-small cell lung cancer and elevated serum level was highly correlated with distant metastasis, suggesting RBBP7 is involved in cancer metastasis through regulation of E-cadherin [125, 126]. Recent studies indicate that RBBP7 is specifically expressed in brain tumors [127] and can directly convert mitotic germ cells into specific neuronal cells by several histone remodeling and modifying [128]. However, the functional role of RBBP7 during tumor development requires further experiments to be explained.
Ectodysplasin A2 receptor (EDA2R)
EDA2R is located at Xq12 and is highly expressed during embryonic development, with a particular role in epidermal morphogenesis [129]. Mutations in this gene manifest clinically as loss of hair, sweat glands, and teeth [129, 130]. Mice lacking EDA2R are indistinguishable from their WT littermates, but myodegeneration induced by EDA-A2 (the ligand of EDA2R) can be prevented by EDA2R deficiency [131]. EDA2R activates the NF-κB and JNK pathways [132, 133] while inducing apoptosis through a caspase 8-dependent mechanism [134]. Studies suggest that EDA2R is a potential downstream effector of p53-induced apoptosis in cancer cells [136, 137] and may therefore be a potential tumor suppressor to carcinogenesis., It is down-regulated in breast and colorectal cancers [135], and mutations and promoter hypermethylation of EDA2R have been identified in colorectal cancer cells [136, 137].
PHD finger protein 6 (PHF6)
PHF6 at Xq26.3 is a member of the plant homeodomain (PHD)-like finger family. It encodes a protein with two PHD-type zinc finger domains, indicating a potential role in transcriptional regulation localizing proteins to the nucleolus [138]. Mutations affecting the coding region of this gene, splicing of the gene transcript, or gene deletions have been associated with Borjeson-Forssman-Lehmann syndrome (BFLS), characterized by mental retardation, epilepsy, hypogonadism, hypometabolism, obesity, swelling of facial subcutaneous tissue, narrow palpebral fissures, and large ears [138, 139]. Van Vlierberghe et al. identified inactivating mutations and deletions in the X-linked PHF6 in 16% of pediatric and 38% of adult primary T cell acute lymphoblastic leukemia (T-ALL) samples [140]. Notably, PHF6 mutations are almost exclusively found in T-ALL samples from male subjects. Mutational loss of PHF6 function is associated with leukemias driven by aberrant expression of the homeobox transcription factor oncogenes TLX1 and TLX3 [140]. Additionally, recent studies support the involvement of this gene in adult acute myeloid leukemia [141, 142], T-cell lymphoblastic leukemia [142–144] and hepatocellular carcinoma [142]. Consistent with these data, Phf6 alterations have been identified in murine models of lymphoma [145]. These results suggested that PHF6 may be a new X-linked tumor suppressor in leukemias.
E74-like factor 4 (ELF4, also known as MEF)
ELF4 is located on chromosome Xq26.1 and its protein is a transcriptional activator that binds and activates CSF2, IL3, IL8, and PRF1 [146, 147]. The encoded protein is involved in natural killer (NK) cell development and function, innate immunity, and induction of cell cycle arrest in naive CD8+ cells [146, 147]. Early studies identified LOH at the ELF4 locus in both ovarian and breast cancers [14, 28]. A later study reported that ELF4 suppress the transcription of MMP-9 and IL-8 in non-small-cell lung cancer cells, suggesting that ELF4 may be a candidate tumor suppressor gene [148]. Although ELF4 inhibits tumor growth in vitro, Elf4-deficient mice do not spontaneously develop tumors [148, 149].
Chromosomal rearrangements resulting in the fusion transcript BCORL1-ELF4 have been identified in hepatocellular carcinomas [150]. ELF4 is also targeted by the t(X;21)(q26;q22) in acute myelogenous leukemia. Lacorazza et al. suggested that ELF4 regulates the proliferation of primitive hematopoietic progenitor cells at steady state by controlling their entry into the cell cycle, thus affecting the decision of stem/primitive progenitor cells to divide or remain quiescent [151]. However, recent studies have also indicated that ELF4 may be a candidate oncogene [152, 153]. ELF4 functions as a tumor promoting factor in ovarian cancer [152], activating MDM2 expression and blocking oncogene-induced p16 activation [153]. However, recent functional analysis showed that ELF4 contributes to the persistence of γH2AX DNA damage foci and promotes the DNA damage response, leading to the induction of apoptosis [154]. These contrary roles of ELF4 need to be clarified by further studies.
Leucine zipper, down-regulated in cancer 1 (LDOC1)
LDOC1 is located at Xq27 and encodes a nuclear protein down-regulated in some cancer cell lines [155]. It is thought to play a role in inhibiting the NF-κB pathway and promoting apoptosis of cancer cells [156, 157]. Interestingly, LDOC1 is located in the HPCX region, which is a major locus for hereditary prostate cancer, and deletions here are also associated with sporadic prostate cancers [24, 48, 51, 158];, no prostate susceptibility genes have been identified thus far. LDOC1 inhibits the degradation of p53 [159] and has therefore may be a potential X-linked tumor suppressor gene. However, high levels of LDOC1 correlate with poor prognosis in untreated chronic lymphocytic leukemia patients, suggesting that LDOC1 functions as an oncogene [160]. Further study is therefore needed.
Dyskeratosis congenita 1 (DKC1)
This gene is located at Xq28 and encodes the protein dyskerin, which is involved in rRNA processing and modification [161, 162]. Mutations in this gene lead to X-linked dyskeratosis congenita, characterized by reticulate skin pigmentation, mucosal leukoplakia, nail dystrophy, and progressive bone marrow failure [161, 162]. DKC1 has been proposed to function in telomerase activity, ageing and cancer [163]. However, recent functional analyses indicate that DKC1 may be a putative tumor suppressor by promoting p53- and p27-dependent translational control of ribosome biogenesis, suggesting a role of dyskerin in the inactivation of p53 in human tumors [164–166]. Therefore, DKC1 is a promising candidate X-linked tumor suppressor gene.
Ribosomal protein S6 kinase, 90kDa, polypeptide 6 (RPS6KA6, also known as RSK4)
This gene is located at Xq21 and encodes a member of the ribosomal S6 kinase family, a group of serine-threonine protein kinases regulated by growth factors [167]. Exogenous expression of RPS6KA6 resulted in decreased cell proliferation, increased accumulation of cells in G(0)-G(1) phase, and increased expression of tumor suppressor genes such as Rb, RbAp46, and p21 [168, 169]. In vitro over-expression of RPS6KA6 also induced cell arrest and senescence in normal fibroblasts and malignant colon cancer cell lines. Interestingly, RPS6KA6 mRNA levels in these cell lines were increased both in replicative- and stress-induced senescence. Cells expressing E1A or RB short interfering RNA were resistant to RPS6KA6-mediated senescence [170]. Overexpression of RPS6KA6 also led to reduced colony formation in soft agar and suppressed invasive and migratory activities of MDA-MB-231 cells both in vitro and in vivo. These effects may have been due to up-regulation of claudin-2 and down-regulation of CXCR4, both of which play roles in invasion and chemotaxis [168]. One study determined that RPS6KA6 was down-regulated in 90% of colon carcinomas, 86% of colon adenomas, and 80% of renal cell carcinomas examined [170]. These results suggest that RPS6KA6 may act as an important tumor suppressor gene in breast cancer by inhibiting invasion and migration of cancer cells and in colon and renal cell carcinomas by modulating induction of senescence and controlling cell proliferation. Although no functional mutation has been identified in human cancers, a recent study showed that RSK4 is expressed in normal uterine tissue but is absent or reduced by hypermethylation in both endometrial cancer cell lines and primary tumors [171].
CD99 molecule (CD99)
This gene is found in the pseudoautosomal regions of the X and Y chromosomes (Xp22.32 and Yp11.3, respectively) and escapes X chromosome inactivation [172]. CD99 is a cell surface glycoprotein involved in leukocyte migration, T cell adhesion, ganglioside and protein transport, and apoptosis of T cells [174]. This gene encodes two different proteins produced by alternative splicing, with the short form harboring a deletion in the cytoplasmic domain [173].. While the longer isoform inhibited migration and metastasis in tumors, the shorter form actually increased motility and MMP-9 expression of human breast cancer cells through the AP-1 activated AKT, ERK, and JNK signaling pathways [175, 176]. CD99 appears to induce tumors and bone metastases in human Ewing sarcoma cell lines in vitro and in vivo [177], suggesting that CD99 may actually be an oncogene in human Ewing sarcoma cells. In two studies, 78–100% of pancreatic neuroendocrine tumors stained positive for CD99 [178, 179]. However, a recent study indicated that down-regulation of CD99 protein (62/100 in tumors but 34/35 in normal tissues) by gene promoter hypermethylation is a critical event in the transitional cell carcinomas of urinary bladder, especially in advanced stages [180], implicating a tumor suppressor function.
Alpha thalassemia/mental retardation syndrome X-linked (ATRX)
ATRX is located at Xq21.1 and its protein contains an ATPase/helicase domain, belonging to the SWI/SNF family of chromatin remodeling proteins. This gene is a major epigenetic regulator of transcription, nuclear architecture, and chromosome stability in mammalian cells [181, 182]. Mutations of this gene affect chromatin remodeling, DNA methylation, and transcriptional regulation. Such mutations are associated with an X-linked mental retardation syndrome, often complicated by alpha-thalassaemia and myelodysplastic syndrome [183–187]. Interestingly, skewed X chromosome inactivation was found in patients with alpha-thalassaemia syndrome and abnormal imprinted X chromosome inactivation was also identified in ATRX-deficient mice [182].
Recent evidence indicates that ATRX is abnormally regulated in several types of cancers, including acute myeloid leukemia, prostate cancer, breast cancer, esophageal squamous cell carcinoma, and pancreatic neuroendocrine tumors [46, 188–192]. In 132 patients with de novo acute myeloid leukemia, low expression levels of this gene correlated with an adverse karyotype and poor prognosis [192]. The low miRNA levels of this gene in gene expression profiling experiments were also observed in primary prostate cancers [189], irradiated breast cancers, [191] and esophageal squamous cell carcinomas [188]. Importantly, recent studies revealed that frameshift and nonsense somatic mutations of ATRX are frequently observed in pancreatic neuroendocrine tumors and loss of DAXX or ATRX function in 43–45% of tumors with high levels of chromosome instability [46, 190, 193, 194]. More recently, somatic mutations in the H3.3-ATRX-DAXX chromatin remodelling pathway were also identified in 44% of pediatric glioblastoma [195]. Therefore, ATRX is a novel candidate X-linked tumor suppressor gene.
Mechanism of inactivation of X-linked tumor suppressor genes
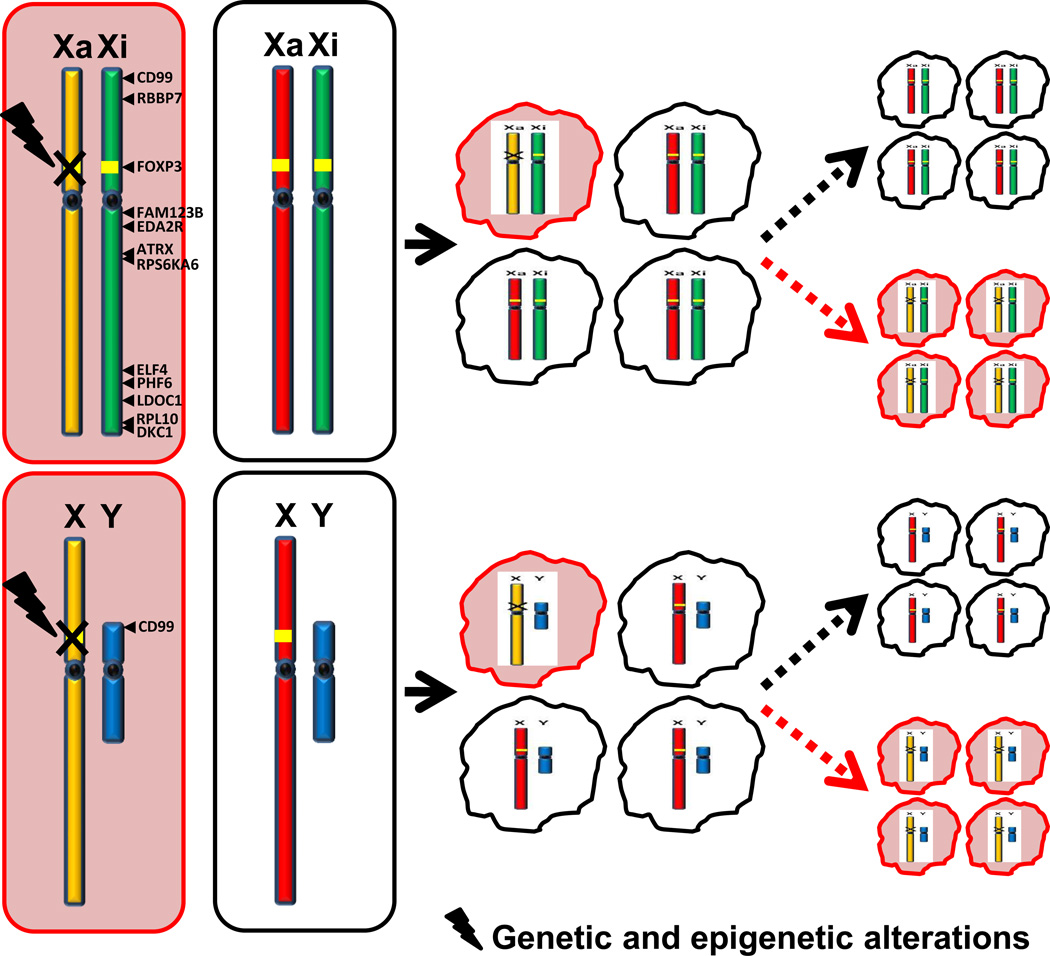
Identification of X-linked tumor suppressor genes has furthered our understanding of the pathogenesis of human cancer. However, the silencing mechanism of these genes in human cancer is still unclear. Since females have two X chromosomes while males have only one, the inactivation mechanism of these genes in human cancer should be different between male and females (Fig. 1). This mechanism provides a novel genetic paradigm in the pathogenesis of human cancer.
Fig. 1.
Mechanism of inactivation of X-linked tumor suppressor gene. X-linked tumor suppressor genes in male cancer may be inactivated by multiple genetic or epigenetic events such as gene deletion, mutation, DNA methylation and post-transcriptional modification, etc., and in female cancer may also be implicated in LOH, bi-allelic methylation and skewed X-inactivation, etc. Cells with those genetic or epigenetic alterations will undergo a negative or positive selection during tumorigenesis. The pink elliptic boxes or irregular circles indicate cells with genetic or epigenetic damage in an active allele. The black dotted arrows indicate a negative selection while the red dotted arrows indicate a positive selection. The black solid arrows indicate a location of the putative tumor suppressor genes. Xa: active X-chromosome; Xi: inactive X-chromosome.
Inactivation of X-linked tumor suppressor genes in females
X chromosome inactivation is a mechanism of gene dosage compensation that has evolved in mammals. One X chromosome in each female cell is randomly inactivated and remains inactive throughout the lifetime of the cell and its descendants. Females are thus a mosaic of two cell populations, expressing either paternal or maternal X-linked alleles. In females, an X-linked tumor suppressor gene is more susceptible to additional genetic damage since one of the alleles is already silenced by X-chromosome inactivation. Here we propose that X-linked tumor suppressor genes in female cancer may be inactivated by multiple events such as LOH, mutation, bi-allelic methylation and skewed X-inactivation, etc (Fig. 1).
Gene deletion and mutation
The role of heterozygous tumor suppressors in cancer pathogenesis has been demonstrated recently with the identification of two X-linked tumor suppressor genes, FOXP3 [6] and WTX [53], which are inactivated by a single genetic hit [4]. Female mice with a FoxP3-heterozygous mutation develop spontaneous breast cancer at a higher rate than WT mice [6]. The majority of the mutations and all deletions of FOXP3 are heterozygous in human breast cancer [6]. Likewise, all mutations and deletions of WTX identified are heterozygous in Wilms tumor [53]. Rivera et al. used DNA sequencing to identify inactivating mutations in the WTX allele on the active X chromosome in female Wilms tumors. FISH analysis confirmed the deletion of WTX on the active X chromosome in several of these tumors. WT WTX allele was only found on the inactive X chromosome. [53]. These data suggest that X-linked tumor suppressor genes may be inactivated by a single gene deletion or mutation targeting the active allele.
It should be noted that while X-linked tumor suppressor genes are subjected to X inactivation, some X-linked genes can escape X chromosome inactivation such as RBBP7, CD99, PHF6 and LDOC1 [209]. It has been suggested that escape from X inactivation may be regulated by long non-coding RNA (lncRNA) within the escaping chromosomal domains [210]. Many of the genes which escape inactivation are present in particular regions of the X chromosome and silenced and escape regions have been shown to have distinct chromatin marks [211], but the precise mechanism of inactivation of such X-linked tumor suppressor genes in cancer is unknown.
Bi-allelic epigenetic inactivation
In a two-hit mechanism for inactivation of autosomal tumor suppressor genes, methylation of one allele may serve as the second hit in cases with a functional mutation or LOH of the other allele. In females, one of the alleles in X-linked genes is silenced by X chromosome inactivation under a complex mechanism involving a long noncoding RNA, XIST (X inactivation specific transcript) [10, 212, 213], as well as XIST is expressed at high levels on the inactive X chromosome and is not expressed on the active X-chromosome [212–214]. DNA/histone methylation and histone hypoacetylation [196, 197] also contribute to X chromosome inactivation. Methylation of most X-linked genes occurs on the inactive X chromosome. Our analysis using pyrosequencing technology revealed that the CpG island motif of FOXP3 was approximately 50% methylated in normal breast epithelial cells (unpublished data), which is consistent with X-inactivation. Although somatic genetic defects at the FOXP3 locus have been identified in 36% of human breast cancers, it is still unexplained why there is a lack of FOXP3 in the majority of breast cancers [6]. It is therefore of interest whether the transcriptional activity of FOXP3 is down-regulated by the epigenetic changes in breast cancers such as bi-allelic methylation. Moreover, evidence suggests that BRCA1 is associated with XIST regulation on the active X chromosome, and XIST can be abnormally expressed on an active X-linked allele in breast cancer cells, silencing the gene [30, 198]. In addition, recent studies indicate that X-linked EDA2R is also down-regulated in breast cancer by promoter hypermethylation [135, 137]. Thus, bi-allelic epigenetic inactivation may be more frequent in X-linked genes than autosomal genes and is a potential mechanism for inactivation of X-linked tumor suppressor genes during cancer development. This mechanism needs to be validated by further study.
Skewed X-inactivation
X-inactivation is random and therefore expression of the paternal and maternal alleles of a gene should be roughly 50% for each. Skewed X-inactivation can occur naturally, such as in the peripheral blood cells of older females [202] and in a very small percentage (2.7~4.5%) of the general female population [35, 202, 203]. In females carrying mutations in critical X-linked genes, selective X-inactivation is observed due to negative selection of cells expressing the mutant allele [204]. This selection allows for phenotypic suppression of X-linked dominant disorders in females by selecting for expression of the WT allele in relevant tissues [204]. However, in female carriers of a mutant allele, if skewed X-inactivation selects against the WT allele, the cells will undergo a positive selection for the mutant allele (Fig. 1) [6, 204]. Consequently, it is hypothesized that skewed X-inactivation in combination with a single genetic hit can inactivate an X-linked tumor suppressor gene [4, 204]. Previous studies [8, 35, 205, 206] have reported an increased frequency of skewed X-inactivation in DNA from peripheral blood cells of patients with ovarian and breast cancer and BRCA1 mutation carriers; BRCA1 is involved the regulation of many X-linked genes [34, 207] and may contribute to the maintenance of the inactive X chromosome [33, 208]. Skewed X-inactivation, through either chance or selection, in females who are heterozygous carriers of an X-linked disorder can lead to the clinical manifestation of a nominally recessive disease. In cancer, a gain-of-function or loss-of-function mutation in an X-linked oncogene or tumor suppressor, respectively, may give a proliferative advantage to cells carrying this mutation on the active X chromosome, causing not only skewed X-inactivation but also an increased risk of cancer [35, 205]. Additionally, if X-linked gene inactivation affects a cluster of neighboring genes, as has been suggested, the active allele of an X-linked tumor suppressor gene can be silenced during the inactivation of a nearby mutated gene [9]. However, these hypotheses have not been tested experimentally and the prevalence of skewed X-inactivation in human cancer cells is still unknown.
Inactivation of X-linked tumor suppressor genes in males
FOXP3 deletion has been identified in 23 (14%) of 165 prostate cancer samples [5]. Among them, 5 of the 23 cases showed an increase in X chromosome number. Interestingly, the deletion was complete in all X chromosomes [5], suggesting that X chromosome duplications in cancer tissues likely occurred after deletion of FOXP3. Our sequencing analyses identified single base-pair changes in 5 (25%) of 20 samples (4 missense and one intronic) leading to the functional inactivation of FOXP3 [5]. Although FOXP3 is frequently inactivated in prostate cancer by deletion and somatic mutation, approximately 70% of prostate cancer samples exhibited a loss of nuclear FOXP3 expression that was not fully explained by the two somatic alterations [5]. Thus, inactivation of FOXP3 in prostate cancer may also be caused by additional mechanisms such as DNA methylation, histone hypoacetylation and gene regulation, etc (Fig. 1). To date, there is no more information regarding inactivation of X-linked tumor suppressors in male-specific cancers.
Conclusion
The identification of X-linked tumor suppressor genes has challenged the traditional theory of “two-hit inactivation” of tumor suppressors, suggesting a tumorigenic mechanism involving a single genetic hit. In contrast to bi-allelic inactivation of autosomal tumor-suppressor genes, single-hit damage is sufficient to functionally inactivate X-linked tumor suppressor genes in human cancers [4]. In males, the single-hit events can be caused by genetic or epigenetic alterations of somatic or germline origin (Fig. 1). In females, the potential single-hit events include those affecting males as well as skewed X-inactivation and positive selection (Fig. 1). A functional single-hit event in a X-linked tumor suppressor gene on the active X chromosome will result in gene silencing in female cells, unless these cells are subjected to negative selection, inhibiting tumor formation. Otherwise, cells lacking tumor suppressor function are likely to undergo positive selection, with skewed X-inactivation and an increased risk of developing cancer [35, 205]. However, the mechanism of skewed X inactivation in X-linked tumor suppressor genes is still unknown. Between 25–35% of human X-linked genes either are not inactivated or subject to varying inactivation [215]. If an X-linked tumor suppressor gene is located in either of these clusters, the inactivation of this gene in females should behave like autosomal tumor suppressor genes and follow Knudson’s two-hit mechanism. While there are numerous putative X-linked tumor suppressor genes currently being researched, only two have been verified by repeated study, FOXP3 and WTX. Neither is located in the regions of the X chromosome that partially or completely escape inactivation and, therefore, are susceptible to a novel single-hit genetic activation.
Future perspectives
The identification of X-linked tumor suppressor genes has significant clinical potential in the treatment of human cancer. One of the biggest challenges in cancer therapy is restoration of tumor suppressor function following inactivation, since it is difficult to repair what is genetically broken [4]. Deletions and/or mutations identified in known X-linked tumor suppressor genes FOXP3 and WTX are often heterozygous in female cancer cells [6, 53]. While the inactive X chromosome is sometimes lost during cancer development [30, 34], in cancers retaining the inactive chromosome there is the possibility of reactivating the silenced WT allele of these genes for cancer therapy. For example, FOXP3 expression is lost in the majority of breast cancers and up-regulation of WT FOXP3 inhibits cell growth and proliferation in breast cancer cells [6]. If FOXP3 is indeed mutated exclusively on the active X chromosome, then the inactive X chromosome contains a WT allele that may serve as an important target for treatment of breast cancer. Development of such a therapy requires further study into the mechanisms inactivating X-linked tumor suppressor genes in cancer as well as strategies to selectively activate the inactive WT allele cancer cells. A recent study observed that anisomycin treatment induced FOXP3 expression in both mouse and human breast cancer cell lines [216]. Such induction resulted in increased apoptosis of cancer cells and reduced growth of established mouse mammary tumors [216]. This observation raises the intriguing possibility of restoring FOXP3 function through drug treatment.
DNA methylation, XIST, and histone hypoacetylation work in concert to maintain inactivation of X-linked genes [10, 212–214], and epigenetic changes and XIST deregulation observed during cancer development may reduce the steps needed to reactivate X-linked tumor suppressor genes [44]. However, restoration of gene expression in the inactive X chromosome may bring about unexpected or undesired results due to loss of dosage compensation of various important X-linked genes. We therefore need to develop a specific therapeutic strategy for the targeted reactivation of only the X-linked tumor suppressor gene of interest. Furthermore, some X-linked tumor suppressor genes have multiple functions at different stages of development or in different tissues and cell types. For instance, FOXP3 is both an X-linked tumor suppressor in breast and prostate tissue [5, 6] and a master regulator of regulatory T cell development and function [74]. Reactivation of FOXP3 expression from the inactive X chromosomes may therefore affect immune responses. Thus, the specific therapeutic strategy should be localized to tumor tissues and the toxicity and side effects of any drug must be also considered.
A recent study has evaluated the clinical efficacy of a monoclonal antibody Fv-FOXP3 protein in therapies for breast, ovarian, and colon cancer [217]. The Fv-FOXP3 is a fusion protein produced by Pichia pastoris. Treatment with Fv-FOXP3 resulted in dose-dependent cell death of cancer cells in vitro, with increased production of the p17-activated fragment of caspase-3 leading to apoptosis of cancer cells. In vivo, Fv-FOXP3 treatment led to a significant reduction in tumor burden in a syngeneic mouse model of metastatic colon cancer. Therefore, the identification of FOXP3 as an X-linked tumor suppressor gene has already been used in clinical research to develop a novel potential treatment of human cancers.
Numerous studies have cited other X-linked genes with putative tumor suppressor functions. However, many of these genes have conflicting functions that give them a dual characterization of tumor suppressor gene and oncogene in different cell types and tumor stages. Their exact, context-specific functional roles in cancer development remain to be fully clarified. Future studies will need to explore how to best identify true X-linked tumor suppressor genes and specifically reactivate their expression, as well as further elucidate the differences between X-linked and autosomal tumor suppressor genes.
Executive summary.
Introduction: The existence of X-linked tumor suppressors has long been suspected, particularly in breast, ovarian, and prostate cancers.
Potential implication of X-linked tumor suppressor genes in human cancers: These genes represent a novel mechanism of tumorigenesis, since X-inactivation in females and the presence of only one X chromosome in males allows for inactivation of an X-linked gene by a single-hit mechanism.
Identification of X-linked tumor suppressor genes: Two X-linked tumor suppressor genes have been verified by repeated study: FOXP3 in breast and prostate cancer and FAM123B/WTX in Wilms tumor. Over half a dozen putative X-linked tumor suppressor genes have been identified, but further study is needed to determine their true function in tumorigenesis.
Mechanism of inactivation of X-linked tumor suppressor genes: A single genetic hit, in the form of a mutation, deletion, or aberrant methylation, can lead to loss of tumor suppressor function in males; in females, this may occur if combined with skewed X-inactivation.
Future perspectives: A female cell that has undergone a loss-of-function genetic alteration of an X-linked tumor suppressor gene may still possess a wild-type allele of the gene on the inactive X chromosome. Activation of this gene and restoration of tumor suppressor function represents a novel potential therapy for female cancers.
References
- 1.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine AJ, Finlay CA, Hinds PW. P53 is a tumor suppressor gene. Cell. 2004;116(2 Suppl):S67–S69. doi: 10.1016/s0092-8674(04)00036-4. 61 p following S69. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Gao J, Lei Q, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4(3):209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 4. Liu Y, Wang L, Zheng P. X-linked tumor suppressors: perplexing inheritance, a unique therapeutic opportunity. Trends Genet. 2010;26(6):260–265. doi: 10.1016/j.tig.2010.03.004. *Another recent review of X-linked tumor suppressor genes.
- 5.Wang L, Liu R, Li W, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009;16(4):336–346. doi: 10.1016/j.ccr.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zuo T, Wang L, Morrison C, et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007;129(7):1275–1286. doi: 10.1016/j.cell.2007.04.034. **This paper verified that FOXP3 is an X-linked tumor suppressor gene.
- 7. Spatz A, Borg C, Feunteun J. X-chromosome genetics and human cancer. Nat Rev Cancer. 2004;4(8):617–629. doi: 10.1038/nrc1413. *An excellent review of X chromosome abnormalities in cancer.
- 8.Buller RE, Sood AK, Lallas T, Buekers T, Skilling JS. Association between nonrandom X-chromosome inactivation and BRCA1 mutation in germline DNA of patients with ovarian cancer. J Natl Cancer Inst. 1999;91(4):339–346. doi: 10.1093/jnci/91.4.339. [DOI] [PubMed] [Google Scholar]
- 9.Liao DJ, Du QQ, Yu BW, Grignon D, Sarkar FH. Novel perspective: focusing on the X chromosome in reproductive cancers. Cancer Invest. 2003;21(4):641–658. doi: 10.1081/cnv-120022385. [DOI] [PubMed] [Google Scholar]
- 10.Pageau GJ, Hall LL, Ganesan S, Livingston DM, Lawrence JB. The disappearing Barr body in breast and ovarian cancers. Nat Rev Cancer. 2007;7(8):628–633. doi: 10.1038/nrc2172. [DOI] [PubMed] [Google Scholar]
- 11. Klein CB, Conway K, Wang XW, et al. Senescence of nickel-transformed cells by an X chromosome: possible epigenetic control. Science. 1991;251(4995):796–799. doi: 10.1126/science.1990442. *The earliest paper implying the existence of X-linked tumor suppressor genes.
- 12.Wang XW, Lin X, Klein CB, Bhamra RK, Lee YW, Costa M. A conserved region in human and Chinese hamster X chromosomes can induce cellular senescence of nickel-transformed Chinese hamster cell lines. Carcinogenesis. 1992;13(4):555–561. doi: 10.1093/carcin/13.4.555. [DOI] [PubMed] [Google Scholar]
- 13.Loupart ML, Adams S, Armour JA, Walker R, Brammar W, Varley J. Loss of heterozygosity on the X chromosome in human breast cancer. Genes Chromosomes Cancer. 1995;13(4):229–238. doi: 10.1002/gcc.2870130402. [DOI] [PubMed] [Google Scholar]
- 14.Choi C, Kim MH, Juhng SW. Loss of heterozygosity on chromosome XP22.2-p22.13 and Xq26.1-q27.1 in human breast carcinomas. J Korean Med Sci. 1998;13(3):311–316. doi: 10.3346/jkms.1998.13.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang-Feng TL, Li S, Han H, Schwartz PE. Frequent loss of heterozygosity on chromosomes Xp and 13q in human ovarian cancer. Int J Cancer. 1992;52(4):575–580. doi: 10.1002/ijc.2910520414. [DOI] [PubMed] [Google Scholar]
- 16.Yang-Feng TL, Han H, Chen KC, et al. Allelic loss in ovarian cancer. Int J Cancer. 1993;54(4):546–551. doi: 10.1002/ijc.2910540405. [DOI] [PubMed] [Google Scholar]
- 17.Feder M, Liu Z, Apostolou S, Greenberg RE, Testa JR. Loss of chromosomes 1 and X in a renal oncocytoma: implications for a possible pseudoautosomal tumor suppressor locus. Cancer Genet Cytogenet. 2000;123(1):71–72. doi: 10.1016/s0165-4608(00)00304-6. [DOI] [PubMed] [Google Scholar]
- 18.Buekers TE, Lallas TA, Buller RE. Xp22.2-3 loss of heterozygosity is associated with germline BRCA1 mutation in ovarian cancer. Gynecol Oncol. 2000;76(3):418–422. doi: 10.1006/gyno.1999.5713. [DOI] [PubMed] [Google Scholar]
- 19.Edelson MI, Lau CC, Colitti CV, et al. A one centimorgan deletion unit on chromosome Xq12 is commonly lost in borderline and invasive epithelial ovarian tumors. Oncogene. 1998;16(2):197–202. doi: 10.1038/sj.onc.1201479. [DOI] [PubMed] [Google Scholar]
- 20.Fujino T, Risinger JI, Collins NK, et al. Allelotype of endometrial carcinoma. Cancer Res. 1994;54(16):4294–4298. [PubMed] [Google Scholar]
- 21.Mitra AB, Murty VV, Li RG, Pratap M, Luthra UK, Chaganti RS. Allelotype analysis of cervical carcinoma. Cancer Res. 1994;54(16):4481–4487. [PubMed] [Google Scholar]
- 22.Thrash-Bingham CA, Salazar H, Greenberg RE, Tartof KD. Loss of heterozygosity studies indicate that chromosome arm 1p harbors a tumor supressor gene for renal oncocytomas. Genes Chromosomes Cancer. 1996;16(1):64–67. doi: 10.1002/(SICI)1098-2264(199605)16:1<64::AID-GCC9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 23.Rohrbach H, Haas CJ, Baretton GB, et al. Microsatellite instability and loss of heterozygosity in prostatic carcinomas: comparison of primary tumors, and of corresponding recurrences after androgen-deprivation therapy and lymph-node metastases. Prostate. 1999;40(1):20–27. doi: 10.1002/(sici)1097-0045(19990615)40:1<20::aid-pros3>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 24. Xu J, Meyers D, Freije D, et al. Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat Genet. 1998;20(2):175–179. doi: 10.1038/2477. **This paper identified a prostate cancer susceptibility locus on the X-chromosome.
- 25.Cheng PC, Gosewehr JA, Kim TM, et al. Potential role of the inactivated X chromosome in ovarian epithelial tumor development. J Natl Cancer Inst. 1996;88(8):510–518. doi: 10.1093/jnci/88.8.510. [DOI] [PubMed] [Google Scholar]
- 26.Osborne RJ, Leech V. Polymerase chain reaction allelotyping of human ovarian cancer. Br J Cancer. 1994;69(3):429–438. doi: 10.1038/bjc.1994.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dodson MK, Hartmann LC, Cliby WA, et al. Comparison of loss of heterozygosity patterns in invasive low-grade and high-grade epithelial ovarian carcinomas. Cancer Res. 1993;53(19):4456–4460. [PubMed] [Google Scholar]
- 28.Choi C, Cho S, Horikawa I, et al. Loss of heterozygosity at chromosome segment Xq25-26.1 in advanced human ovarian carcinomas. Genes Chromosomes Cancer. 1997;20(3):234–242. [PubMed] [Google Scholar]
- 29.Chenevix-Trench G, Kerr J, Hurst T, et al. Analysis of loss of heterozygosity and KRAS2 mutations in ovarian neoplasms: clinicopathological correlations. Genes Chromosomes Cancer. 1997;18(2):75–83. doi: 10.1002/(sici)1098-2264(199702)18:2<75::aid-gcc1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 30.Sirchia SM, Ramoscelli L, Grati FR, et al. Loss of the inactive X chromosome and replication of the active X in BRCA1-defective and wild-type breast cancer cells. Cancer Res. 2005;65(6):2139–2146. doi: 10.1158/0008-5472.CAN-04-3465. [DOI] [PubMed] [Google Scholar]
- 31.Wang N, Cedrone E, Skuse GR, Insel R, Dry J. Two identical active X chromosomes in human mammary carcinoma cells. Cancer Genet Cytogenet. 1990;46(2):271–280. doi: 10.1016/0165-4608(90)90112-n. [DOI] [PubMed] [Google Scholar]
- 32.Piao Z, Malkhosyan SR. Frequent loss Xq25 on the inactive X chromosome in primary breast carcinomas is associated with tumor grade and axillary lymph node metastasis. Genes Chromosomes Cancer. 2002;33(3):262–269. doi: 10.1002/gcc.10024. [DOI] [PubMed] [Google Scholar]
- 33.Ganesan S, Silver DP, Greenberg RA, et al. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111(3):393–405. doi: 10.1016/s0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- 34. Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–132. doi: 10.1016/j.ccr.2006.01.013. *An excellent analysis of abberant X chromosomes in cancers.
- 35.Kristiansen M, Knudsen GP, Maguire P, et al. High incidence of skewed X chromosome inactivation in young patients with familial non-BRCA1/BRCA2 breast cancer. J Med Genet. 2005;42(11):877–880. doi: 10.1136/jmg.2005.032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knuutila S, Aalto Y, Autio K, et al. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155(3):683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bottarelli L, Azzoni C, Necchi F, et al. Sex chromosome alterations associate with tumor progression in sporadic colorectal carcinomas. Clin Cancer Res. 2007;13(15 Pt 1):4365–4370. doi: 10.1158/1078-0432.CCR-06-2736. [DOI] [PubMed] [Google Scholar]
- 38.Jiang F, Richter J, Schraml P, et al. Chromosomal imbalances in papillary renal cell carcinoma: genetic differences between histological subtypes. Am J Pathol. 1998;153(5):1467–1473. doi: 10.1016/S0002-9440(10)65734-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathur M, Das S, Samuels HH. PSF-TFE3 oncoprotein in papillary renal cell carcinoma inactivates TFE3 and p53 through cytoplasmic sequestration. Oncogene. 2003;22(32):5031–5044. doi: 10.1038/sj.onc.1206643. [DOI] [PubMed] [Google Scholar]
- 40.Indsto JO, Nassif NT, Kefford RF, Mann GJ. Frequent loss of heterozygosity targeting the inactive X chromosome in melanoma. Clin Cancer Res. 2003;9(17):6476–6482. [PubMed] [Google Scholar]
- 41.Pizzi S, D'adda T, Azzoni C, et al. Malignancy-associated allelic losses on the X-chromosome in foregut but not in midgut endocrine tumours. J Pathol. 2002;196(4):401–407. doi: 10.1002/path.1075. [DOI] [PubMed] [Google Scholar]
- 42.Chen YJ, Vortmeyer A, Zhuang Z, Gibril F, Jensen RT. X-chromosome loss of heterozygosity frequently occurs in gastrinomas and is correlated with aggressive tumor growth. Cancer. 2004;100(7):1379–1387. doi: 10.1002/cncr.20104. [DOI] [PubMed] [Google Scholar]
- 43.D'adda T, Bottarelli L, Azzoni C, et al. Malignancy-associated X chromosome allelic losses in foregut endocrine neoplasms: further evidence from lung tumors. Mod Pathol. 2005;18(6):795–805. doi: 10.1038/modpathol.3800353. [DOI] [PubMed] [Google Scholar]
- 44.D'adda T, Candidus S, Denk H, Bordi C, Hofler H. Gastric neuroendocrine neoplasms: tumour clonality and malignancy-associated large X-chromosomal deletions. J Pathol. 1999;189(3):394–401. doi: 10.1002/(SICI)1096-9896(199911)189:3<394::AID-PATH444>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 45.Azzoni C, Bottarelli L, Pizzi S, D'adda T, Rindi G, Bordi C. Xq25 and Xq26 identify the common minimal deletion region in malignant gastroenteropancreatic endocrine carcinomas. Virchows Arch. 2006;448(2):119–126. doi: 10.1007/s00428-005-0058-4. [DOI] [PubMed] [Google Scholar]
- 46.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters MA, Jarvik GP, Janer M, et al. Genetic linkage analysis of prostate cancer families to Xq27-28. Hum Hered. 2001;51(1–2):107–113. doi: 10.1159/000022965. [DOI] [PubMed] [Google Scholar]
- 48.Yaspan BL, Mcreynolds KM, Elmore JB, Breyer JP, Bradley KM, Smith JR. A haplotype at chromosome Xq27.2 confers susceptibility to prostate cancer. Hum Genet. 2008;123(4):379–386. doi: 10.1007/s00439-008-0486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lange EM, Chen H, Brierley K, et al. Linkage analysis of 153 prostate cancer families over a 30-cM region containing the putative susceptibility locus HPCX. Clin Cancer Res. 1999;5(12):4013–4020. [PubMed] [Google Scholar]
- 50.Stephan DA, Howell GR, Teslovich TM, et al. Physical and transcript map of the hereditary prostate cancer region at xq27. Genomics. 2002;79(1):41–50. doi: 10.1006/geno.2001.6681. [DOI] [PubMed] [Google Scholar]
- 51.Kibel AS, Faith DA, Bova GS, Isaacs WB. Xq27-28 deletions in prostate carcinoma. Genes Chromosomes Cancer. 2003;37(4):381–388. doi: 10.1002/gcc.10230. [DOI] [PubMed] [Google Scholar]
- 52.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nature Genetics. 2008;40(3):281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rivera MN, Kim WJ, Wells J, et al. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science. 2007;315(5812):642–645. doi: 10.1126/science.1137509. **This paper verified the existence of the first identified X-linked tumor suppressor gene, WTX.
- 54.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 55.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 56.Shah SP, Morin RD, Khattra J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 57. Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. **An in-depth review of somatic mutations in human cancers.
- 58.Miller RW, Young JL, Jr, Novakovic B. Childhood cancer. Cancer. 1995;75(1 Suppl):395–405. doi: 10.1002/1097-0142(19950101)75:1+<395::aid-cncr2820751321>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 59.Varan A. Wilms' tumor in children: an overview. Nephron Clin Pract. 2008;108(2):c83–c90. doi: 10.1159/000113012. [DOI] [PubMed] [Google Scholar]
- 60.Rivera MN, Haber DA. Wilms' tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer. 2005;5(9):699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- 61.Major MB, Camp ND, Berndt JD, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316(5827):1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 62.Rivera MN, Kim WJ, Wells J, et al. The tumor suppressor WTX shuttles to the nucleus and modulates WT1 activity. Proc Natl Acad Sci U S A. 2009;106(20):8338–8343. doi: 10.1073/pnas.0811349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruteshouser EC, Robinson SM, Huff V. Wilms tumor genetics: mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer. 2008;47(6):461–470. doi: 10.1002/gcc.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perotti D, Gamba B, Sardella M, et al. Functional inactivation of the WTX gene is not a frequent event in Wilms' tumors. Oncogene. 2008;27(33):4625–4632. doi: 10.1038/onc.2008.93. [DOI] [PubMed] [Google Scholar]
- 65.Wegert J, Wittmann S, Leuschner I, Geissinger E, Graf N, Gessler M. WTX inactivation is a frequent, but late event in Wilms tumors without apparent clinical impact. Genes Chromosomes Cancer. 2009;48(12):1102–1111. doi: 10.1002/gcc.20712. [DOI] [PubMed] [Google Scholar]
- 66.Fukuzawa R, Anaka MR, Weeks RJ, Morison IM, Reeve AE. Canonical WNT signalling determines lineage specificity in Wilms tumour. Oncogene. 2009;28(8):1063–1075. doi: 10.1038/onc.2008.455. [DOI] [PubMed] [Google Scholar]
- 67. Perotti D, Radice P. Is WTX a suitable target for cancer therapy? Pediatr Blood Cancer. 2011;56(4):682. doi: 10.1002/pbc.22947. *A recent study examining the therapeutic potention of WTX.
- 68.Jenkins ZA, Van Kogelenberg M, Morgan T, et al. Germline mutations in WTX cause a sclerosing skeletal dysplasia but do not predispose to tumorigenesis. Nat Genet. 2009;41(1):95–100. doi: 10.1038/ng.270. [DOI] [PubMed] [Google Scholar]
- 69.Fukuzawa R, Holman SK, Chow CW, Savarirayan R, Reeve AE, Robertson SP. WTX mutations can occur both early and late in the pathogenesis of Wilms tumour. J Med Genet. 2010;47(11):791–794. doi: 10.1136/jmg.2010.080663. [DOI] [PubMed] [Google Scholar]
- 70.Chatila TA, Blaeser F, Ho N, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106(12):R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brunkow ME, Jeffery EW, Hjerrild KA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 72.Bennett CL, Christie J, Ramsdell F, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27(1):20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 73.Wildin RS, Ramsdell F, Peake J, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27(1):18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 74.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science (New York, N.Y. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 75.Chen GY, Chen C, Wang L, Chang X, Zheng P, Liu Y. Cutting edge: Broad expression of the FoxP3 locus in epithelial cells: a caution against early interpretation of fatal inflammatory diseases following in vivo depletion of FoxP3-expressing cells. J Immunol. 2008;180(8):5163–5166. doi: 10.4049/jimmunol.180.8.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Allan SE, Passerini L, Bacchetta R, et al. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115(11):3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaur G, Goodall JC, Jarvis LB, Hill Gaston JS. Characterisation of Foxp3 splice variants in human CD4+ and CD8+ T cells--identification of Foxp3Delta7 in human regulatory T cells. Mol Immunol. 2010;48(1–3):321–332. doi: 10.1016/j.molimm.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 78.Scotto L, Naiyer AJ, Galluzzo S, et al. Overlap between molecular markers expressed by naturally occurring CD4+CD25+ regulatory T cells and antigen specific CD4+CD25+ and CD8+CD28- T suppressor cells. Hum Immunol. 2004;65(11):1297–1306. doi: 10.1016/j.humimm.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 79.Smith EL, Finney HM, Nesbitt AM, Ramsdell F, Robinson MK. Splice variants of human FOXP3 are functional inhibitors of human CD4+ T-cell activation. Immunology. 2006;119(2):203–211. doi: 10.1111/j.1365-2567.2006.02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yagi H, Nomura T, Nakamura K, et al. Crucial role of FOXP3 in the development and function of human CD25+CD4+ regulatory T cells. Int Immunol. 2004;16(11):1643–1656. doi: 10.1093/intimm/dxh165. [DOI] [PubMed] [Google Scholar]
- 81.Ebert LM, Tan BS, Browning J, et al. The regulatory T cell-associated transcription factor FoxP3 is expressed by tumor cells. Cancer Res. 2008;68(8):3001–3009. doi: 10.1158/0008-5472.CAN-07-5664. [DOI] [PubMed] [Google Scholar]
- 82.Krejsgaard T, Gjerdrum LM, Ralfkiaer E, et al. Malignant Tregs express low molecular splice forms of FOXP3 in Sezary syndrome. Leukemia. 2008;22(12):2230–2239. doi: 10.1038/leu.2008.224. [DOI] [PubMed] [Google Scholar]
- 83.Zhang HY, Sun H. Up-regulation of Foxp3 inhibits cell proliferation, migration and invasion in epithelial ovarian cancer. Cancer Lett. 2010;287(1):91–97. doi: 10.1016/j.canlet.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 84.Zuo T, Liu R, Zhang H, et al. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest. 2007;117(12):3765–3773. doi: 10.1172/JCI32538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu R, Wang L, Chen G, et al. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/histone deacetylase 4 association to the locus. Cancer Res. 2009;69(6):2252–2259. doi: 10.1158/0008-5472.CAN-08-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li W, Wang L, Katoh H, Liu R, Zheng P, Liu Y. Identification of a tumor suppressor relay between the FOXP3 and the Hippo pathways in breast and prostate cancers. Cancer Res. 2011;71(6):2162–2171. doi: 10.1158/0008-5472.CAN-10-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Merlo A, Casalini P, Carcangiu ML, et al. FOXP3 expression and overall survival in breast cancer. J Clin Oncol. 2009;27(11):1746–1752. doi: 10.1200/JCO.2008.17.9036. [DOI] [PubMed] [Google Scholar]
- 88.Ladoire S, Arnould L, Mignot G, et al. Presence of Foxp3 expression in tumor cells predicts better survival in HER2-overexpressing breast cancer patients treated with neoadjuvant chemotherapy. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-010-0831-1. [DOI] [PubMed] [Google Scholar]
- 89.Martin F, Ladoire S, Mignot G, Apetoh L, Ghiringhelli F. Human FOXP3 and cancer. Oncogene. 2010;29(29):4121–4129. doi: 10.1038/onc.2010.174. [DOI] [PubMed] [Google Scholar]
- 90.Karanikas V, Speletas M, Zamanakou M, et al. Foxp3 expression in human cancer cells. J Transl Med. 2008;6:19. doi: 10.1186/1479-5876-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hinz S, Pagerols-Raluy L, Oberg HH, et al. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res. 2007;67(17):8344–8350. doi: 10.1158/0008-5472.CAN-06-3304. [DOI] [PubMed] [Google Scholar]
- 92.Katoh H, Zheng P, Liu Y. Signalling through FOXP3 as an X-linked tumor suppressor. Int J Biochem Cell Biol. 2010;42(11):1784–1787. doi: 10.1016/j.biocel.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jung DJ, Jin DH, Hong SW, et al. Foxp3 expression in p53-dependent DNA damage responses. J Biol Chem. 2010;285(11):7995–8002. doi: 10.1074/jbc.M109.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan YL, Diaz JJ, Denoroy L, Madjar JJ, Wool IG. The primary structure of rat ribosomal protein L10: relationship to a Jun-binding protein and to a putative Wilms' tumor suppressor. Biochem Biophys Res Commun. 1996;225(3):952–956. doi: 10.1006/bbrc.1996.1277. [DOI] [PubMed] [Google Scholar]
- 95.Nguyen YH, Mills AA, Stanbridge EJ. Assembly of the QM protein onto the 60S ribosomal subunit occurs in the cytoplasm. J Cell Biochem. 1998;68(2):281–285. [PubMed] [Google Scholar]
- 96.Dowdy SF, Lai KM, Weissman BE, Matsui Y, Hogan BL, Stanbridge EJ. The isolation and characterization of a novel cDNA demonstrating an altered mRNA level in nontumorigenic Wilms' microcell hybrid cells. Nucleic Acids Res. 1991;19(20):5763–5769. doi: 10.1093/nar/19.20.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Van Den Ouweland AM, Verdijk M, Mannens MM, Van Oost BA. The QM gene is X-linked and therefore not involved in suppression of tumorigenesis in Wilms' tumor. Hum Genet. 1992;90(1–2):144–146. doi: 10.1007/BF00210759. [DOI] [PubMed] [Google Scholar]
- 98.Monteclaro FS, Vogt PK. A Jun-binding protein related to a putative tumor suppressor. Proc Natl Acad Sci U S A. 1993;90(14):6726–6730. doi: 10.1073/pnas.90.14.6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Imafuku I, Masaki T, Waragai M, et al. Presenilin 1 suppresses the function of c-Jun homodimers via interaction with QM/Jif-1. J Cell Biol. 1999;147(1):121–134. doi: 10.1083/jcb.147.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaneko K, Kobayashi H, Onodera O, Miyatake T, Tsuji S. Genomic organization of a cDNA (QM) demonstrating an altered mRNA level in nontumorigenic Wilms' microcell hybrid cells and its localization to Xq28. Hum Mol Genet. 1992;1(7):529–533. doi: 10.1093/hmg/1.7.529. [DOI] [PubMed] [Google Scholar]
- 101.Oh HS, Kwon H, Sun SK, Yang CH. QM, a putative tumor suppressor, regulates proto-oncogene c-yes. J Biol Chem. 2002;277(39):36489–36498. doi: 10.1074/jbc.M201859200. [DOI] [PubMed] [Google Scholar]
- 102.Lillico SG, Mottram JC, Murphy NB, Welburn SC. Characterisation of the QM gene of Trypanosoma brucei. FEMS Microbiol Lett. 2002;211(2):123–128. doi: 10.1111/j.1574-6968.2002.tb11213.x. [DOI] [PubMed] [Google Scholar]
- 103.Stanbridge E, Farmer A, Mills A, et al. Molecular characterization of QM, a novel gene with properties consistent with tumor suppressor function. Cold Spring Harb Symp Quant Biol. 1994;59:573–576. doi: 10.1101/sqb.1994.059.01.064. [DOI] [PubMed] [Google Scholar]
- 104.Kushner PJ, Agard DA, Greene GL, et al. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74(5):311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 105.Prall OW, Rogan EM, Sutherland RL. Estrogen regulation of cell cycle progression in breast cancer cells. J Steroid Biochem Mol Biol. 1998;65(1–6):169–174. doi: 10.1016/s0960-0760(98)00021-1. [DOI] [PubMed] [Google Scholar]
- 106.Bamberger AM, Milde-Langosch K, Rossing E, Goemann C, Loning T. Expression pattern of the AP-1 family in endometrial cancer: correlations with cell cycle regulators. J Cancer Res Clin Oncol. 2001;127(9):545–550. doi: 10.1007/s004320100255. [DOI] [PubMed] [Google Scholar]
- 107.Horninger W, Reissigl A, Rogatsch H, et al. Prostate cancer screening in Tyrol, Austria: experience and results. Eur Urol. 1999;35(5–6):523–538. doi: 10.1159/000019893. [DOI] [PubMed] [Google Scholar]
- 108.Fronsdal K, Engedal N, Slagsvold T, Saatcioglu F. CREB binding protein is a coactivator for the androgen receptor and mediates cross-talk with AP-1. J Biol Chem. 1998;273(48):31853–31859. doi: 10.1074/jbc.273.48.31853. [DOI] [PubMed] [Google Scholar]
- 109.Stalberg P, Grimfjard P, Santesson M, et al. Transfection of the multiple endocrine neoplasia type 1 gene to a human endocrine pancreatic tumor cell line inhibits cell growth and affects expression of JunD, delta-like protein 1/preadipocyte factor-1, proliferating cell nuclear antigen, and QM/Jif-1. J Clin Endocrinol Metab. 2004;89(5):2326–2337. doi: 10.1210/jc.2003-031228. [DOI] [PubMed] [Google Scholar]
- 110.Lejonklou MH, Edfeldt K, Johansson TA, Stalberg P, Skogseid B. Neurogenin 3 and neurogenic differentiation 1 are retained in the cytoplasm of multiple endocrine neoplasia type 1 islet and pancreatic endocrine tumor cells. Pancreas. 2009;38(3):259–266. doi: 10.1097/MPA.0b013e3181930818. [DOI] [PubMed] [Google Scholar]
- 111.Shen XJ, Ali-Fehmi R, Weng CR, Sarkar FH, Grignon D, Liao DJ. Loss of heterozygosity and microsatellite instability at the Xq28 and the A/G heterozygosity of the QM gene are associated with ovarian cancer. Cancer Biol Ther. 2006;5(5):523–528. doi: 10.4161/cbt.5.5.2610. [DOI] [PubMed] [Google Scholar]
- 112.Altinok G, Powell IJ, Che M, et al. Reduction of QM protein expression correlates with tumor grade in prostatic adenocarcinoma. Prostate Cancer Prostatic Dis. 2006;9(1):77–82. doi: 10.1038/sj.pcan.4500848. [DOI] [PubMed] [Google Scholar]
- 113.Huang S, Lee WH, Lee EY. A cellular protein that competes with SV40 T antigen for binding to the retinoblastoma gene product. Nature. 1991;350(6314):160–162. doi: 10.1038/350160a0. [DOI] [PubMed] [Google Scholar]
- 114.Yarden RI, Brody LC. BRCA1 interacts with components of the histone deacetylase complex. Proc Natl Acad Sci U S A. 1999;96(9):4983–4988. doi: 10.1073/pnas.96.9.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen GC, Guan LS, Yu JH, Li GC, Choi Kim HR, Wang ZY. Rb-associated protein 46 (RbAp46) inhibits transcriptional transactivation mediated by BRCA1. Biochem Biophys Res Commun. 2001;284(2):507–514. doi: 10.1006/bbrc.2001.5003. [DOI] [PubMed] [Google Scholar]
- 116.Guan LS, Rauchman M, Wang ZY. Induction of Rb-associated protein (RbAp46) by Wilms' tumor suppressor WT1 mediates growth inhibition. J Biol Chem. 1998;273(42):27047–27050. doi: 10.1074/jbc.273.42.27047. [DOI] [PubMed] [Google Scholar]
- 117.Guan LS, Li GC, Chen CC, Liu LQ, Wang ZY. Rb-associated protein 46 (RbAp46) suppresses the tumorigenicity of adenovirus-transformed human embryonic kidney 293 cells. Int J Cancer. 2001;93(3):333–338. doi: 10.1002/ijc.1338. [DOI] [PubMed] [Google Scholar]
- 118.Li GC, Guan LS, Wang ZY. Overexpression of RbAp46 facilitates stress-induced apoptosis and suppresses tumorigenicity of neoplastigenic breast epithelial cells. Int J Cancer. 2003;105(6):762–768. doi: 10.1002/ijc.11148. [DOI] [PubMed] [Google Scholar]
- 119.Zhang TF, Yu SQ, Deuel TF, Wang ZY. Constitutive expression of Rb associated protein 46 (RbAp46) reverts transformed phenotypes of breast cancer cells. Anticancer Res. 2003;23(5A):3735–3740. [PubMed] [Google Scholar]
- 120.Zhang TF, Yu SQ, Loggie BW, Wang ZY. Inducible expression of RbAp46 activates c-Jun NH2-terminal kinase-dependent apoptosis and suppresses progressive growth of tumor xenografts in nude mice. Anticancer Res. 2003;23(6C):4621–4627. [PubMed] [Google Scholar]
- 121.Zhang TF, Yu SQ, Wang ZY. RbAp46 inhibits estrogen-stimulated progression of neoplastigenic breast epithelial cells. Anticancer Res. 2007;27(5A):3205–3209. [PubMed] [Google Scholar]
- 122.Li GC, Wang ZY. Retinoblastoma suppressor associated protein 46 (RbAp46) attenuates the beta-catenin/TCF signaling through up-regulation of GSK-3beta expression. Anticancer Res. 2006;26(6B):4511–4518. [PubMed] [Google Scholar]
- 123.Li GC, Wang ZY. Constitutive expression of RbAp46 induces epithelial-mesenchymal transition in mammary epithelial cells. Anticancer Res. 2006;26(5A):3555–3560. [PubMed] [Google Scholar]
- 124.Thakur A, Rahman KW, Wu J, et al. Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Mol Cancer Res. 2007;5(2):171–181. doi: 10.1158/1541-7786.MCR-06-0071. [DOI] [PubMed] [Google Scholar]
- 125.Wang CL, Wang CI, Liao PC, et al. Discovery of retinoblastoma-associated binding protein 46 as a novel prognostic marker for distant metastasis in nonsmall cell lung cancer by combined analysis of cancer cell secretome and pleural effusion proteome. J Proteome Res. 2009;8(10):4428–4440. doi: 10.1021/pr900160h. [DOI] [PubMed] [Google Scholar]
- 126.Fu J, Qin L, He T, et al. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21(2):275–289. doi: 10.1038/cr.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Crea F, Hurt EM, Farrar WL. Clinical significance of Polycomb gene expression in brain tumors. Mol Cancer. 2010;9:265. doi: 10.1186/1476-4598-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331(6015):304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yan M, Wang LC, Hymowitz SG, et al. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science. 2000;290(5491):523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 130.Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Newton K, French DM, Yan M, Frantz GD, Dixit VM. Myodegeneration in EDA-A2 transgenic mice is prevented by XEDAR deficiency. Mol Cell Biol. 2004;24(4):1608–1613. doi: 10.1128/MCB.24.4.1608-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sinha SK, Zachariah S, Quinones HI, Shindo M, Chaudhary PM. Role of TRAF3 and −6 in the activation of the NF-kappa B and JNK pathways by X-linked ectodermal dysplasia receptor. J Biol Chem. 2002;277(47):44953–44961. doi: 10.1074/jbc.M207923200. [DOI] [PubMed] [Google Scholar]
- 133.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424(6950):793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 134.Sinha SK, Chaudhary PM. Induction of apoptosis by X-linked ectodermal dysplasia receptor via a caspase 8-dependent mechanism. J Biol Chem. 2004;279(40):41873–41881. doi: 10.1074/jbc.M407363200. [DOI] [PubMed] [Google Scholar]
- 135.Punj V, Matta H, Chaudhary PM. X-linked ectodermal dysplasia receptor is downregulated in breast cancer via promoter methylation. Clin Cancer Res. 2010;16(4):1140–1148. doi: 10.1158/1078-0432.CCR-09-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tanikawa C, Ri C, Kumar V, Nakamura Y, Matsuda K. Crosstalk of EDA-A2/XEDAR in the p53 signaling pathway. Mol Cancer Res. 2010;8(6):855–863. doi: 10.1158/1541-7786.MCR-09-0484. [DOI] [PubMed] [Google Scholar]
- 137.Tanikawa C, Furukawa Y, Yoshida N, Arakawa H, Nakamura Y, Matsuda K. XEDAR as a putative colorectal tumor suppressor that mediates p53-regulated anoikis pathway. Oncogene. 2009;28(34):3081–3092. doi: 10.1038/onc.2009.154. [DOI] [PubMed] [Google Scholar]
- 138.Lower KM, Turner G, Kerr BA, et al. Mutations in PHF6 are associated with Borjeson-Forssman-Lehmann syndrome. Nat Genet. 2002;32(4):661–665. doi: 10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- 139.Berland S, Alme K, Brendehaug A, Houge G, Hovland R. PHF6 Deletions May Cause Borjeson-Forssman-Lehmann Syndrome in Females. Mol Syndromol. 2011;1(6):294–300. doi: 10.1159/000330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Van Vlierberghe P, Palomero T, Khiabanian H, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42(4):338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Van Vlierberghe P, Patel J, Abdel-Wahab O, et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25(1):130–134. doi: 10.1038/leu.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yoo NJ, Kim YR, Lee SH. Somatic mutation of PHF6 gene in T-cell acute lymphoblatic leukemia, acute myelogenous leukemia and hepatocellular carcinoma. Acta Oncol. 2012;51(1):107–111. doi: 10.3109/0284186X.2011.592148. [DOI] [PubMed] [Google Scholar]
- 143.Mavrakis KJ, Van Der Meulen J, Wolfe AL, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43(7):673–678. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Q, Qiu H, Jiang H, et al. Mutations of PHF6 are associated with mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214 in T-cell acute lymphoblastic leukemia. Haematologica. 2011;96(12):1808–1814. doi: 10.3324/haematol.2011.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Landais S, Quantin R, Rassart E. Radiation leukemia virus common integration at the Kis2 locus: simultaneous overexpression of a novel noncoding RNA and of the proximal Phf6 gene. J Virol. 2005;79(17):11443–11456. doi: 10.1128/JVI.79.17.11443-11456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lacorazza HD, Miyazaki Y, Di Cristofano A, et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17(4):437–449. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 147.Hedvat CV, Yao J, Sokolic RA, Nimer SD. Myeloid ELF1-like factor is a potent activator of interleukin-8 expression in hematopoietic cells. J Biol Chem. 2004;279(8):6395–6400. doi: 10.1074/jbc.M307524200. [DOI] [PubMed] [Google Scholar]
- 148.Seki Y, Suico MA, Uto A, et al. The ETS transcription factor MEF is a candidate tumor suppressor gene on the X chromosome. Cancer Res. 2002;62(22):6579–6586. [PubMed] [Google Scholar]
- 149.Lacorazza HD, Nimer SD. The emerging role of the myeloid Elf-1 like transcription factor in hematopoiesis. Blood Cells Mol Dis. 2003;31(3):342–350. doi: 10.1016/s1079-9796(03)00162-1. [DOI] [PubMed] [Google Scholar]
- 150.Totoki Y, Tatsuno K, Yamamoto S, et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet. 2011;43(5):464–469. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- 151.Lacorazza HD, Yamada T, Liu Y, et al. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell. 2006;9(3):175–187. doi: 10.1016/j.ccr.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 152.Yao JJ, Liu Y, Lacorazza HD, et al. Tumor promoting properties of the ETS protein MEF in ovarian cancer. Oncogene. 2007;26(27):4032–4037. doi: 10.1038/sj.onc.1210170. [DOI] [PubMed] [Google Scholar]
- 153.Sashida G, Liu Y, Elf S, et al. ELF4/MEF activates MDM2 expression and blocks oncogene-induced p16 activation to promote transformation. Mol Cell Biol. 2009;29(13):3687–3699. doi: 10.1128/MCB.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sashida G, Bae N, Di Giandomenico S, et al. The mef/elf4 transcription factor fine tunes the DNA damage response. Cancer Res. 2011;71(14):4857–4865. doi: 10.1158/0008-5472.CAN-11-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nagasaki K, Manabe T, Hanzawa H, Maass N, Tsukada T, Yamaguchi K. Identification of a novel gene, LDOC1, down-regulated in cancer cell lines. Cancer Lett. 1999;140(1–2):227–234. doi: 10.1016/s0304-3835(99)00087-7. [DOI] [PubMed] [Google Scholar]
- 156.Nagasaki K, Schem C, Von Kaisenberg C, et al. Leucine-zipper protein, LDOC1, inhibits NF-kappaB activation and sensitizes pancreatic cancer cells to apoptosis. Int J Cancer. 2003;105(4):454–458. doi: 10.1002/ijc.11122. [DOI] [PubMed] [Google Scholar]
- 157.Inoue M, Takahashi K, Niide O, Shibata M, Fukuzawa M, Ra C. LDOC1, a novel MZF-1-interacting protein, induces apoptosis. FEBS Lett. 2005;579(3):604–608. doi: 10.1016/j.febslet.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 158.Baffoe-Bonnie AB, Smith JR, Stephan DA, et al. A major locus for hereditary prostate cancer in Finland: localization by linkage disequilibrium of a haplotype in the HPCX region. Hum Genet. 2005;117(4):307–316. doi: 10.1007/s00439-005-1306-z. [DOI] [PubMed] [Google Scholar]
- 159.Mizutani K, Koike D, Suetsugu S, Takenawa T. WAVE3 functions as a negative regulator of LDOC1. J Biochem. 2005;138(5):639–646. doi: 10.1093/jb/mvi160. [DOI] [PubMed] [Google Scholar]
- 160.Duzkale H, Schweighofer CD, Coombes KR, et al. LDOC1 mRNA is differentially expressed in chronic lymphocytic leukemia and predicts overall survival in untreated patients. Blood. 2011;117(15):4076–4084. doi: 10.1182/blood-2010-09-304881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ruggero D, Grisendi S, Piazza F, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299(5604):259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 162.Yoon A, Peng G, Brandenburger Y, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312(5775):902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 163.Marrone A, Dokal I. Dyskeratosis congenita: molecular insights into telomerase function, ageing and cancer. Expert Rev Mol Med. 2004;6(26):1–23. doi: 10.1017/S1462399404008671. [DOI] [PubMed] [Google Scholar]
- 164.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29(11):1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bellodi C, Krasnykh O, Haynes N, et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70(14):6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Montanaro L, Calienni M, Bertoni S, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 2010;70(11):4767–4777. doi: 10.1158/0008-5472.CAN-09-4024. [DOI] [PubMed] [Google Scholar]
- 167.Dummler BA, Hauge C, Silber J, et al. Functional characterization of human RSK4, a new 90-kDa ribosomal S6 kinase, reveals constitutive activation in most cell types. The Journal of biological chemistry. 2005;280(14):13304–13314. doi: 10.1074/jbc.M408194200. [DOI] [PubMed] [Google Scholar]
- 168.Thakur A, Sun Y, Bollig A, et al. Anti-invasive and antimetastatic activities of ribosomal protein S6 kinase 4 in breast cancer cells. Clin Cancer Res. 2008;14(14):4427–4436. doi: 10.1158/1078-0432.CCR-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lopez-Vicente L, Pons B, Coch L, et al. RSK4 inhibition results in bypass of stress-induced and oncogene-induced senescence. Carcinogenesis. 2011;32(4):470–476. doi: 10.1093/carcin/bgr003. [DOI] [PubMed] [Google Scholar]
- 170.Lopez-Vicente L, Armengol G, Pons B, et al. Regulation of replicative and stress-induced senescence by RSK4, which is down-regulated in human tumors. Clin Cancer Res. 2009;15(14):4546–4553. doi: 10.1158/1078-0432.CCR-08-3159. [DOI] [PubMed] [Google Scholar]
- 171.Dewdney SB, Rimel BJ, Thaker PH, et al. Aberrant methylation of the X-linked ribosomal S6 kinase RPS6KA6 (RSK4) in endometrial cancers. Clin Cancer Res. 2011;17(8):2120–2129. doi: 10.1158/1078-0432.CCR-10-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Braghetti A, Piazzi G, Lanfranco L, Mondello C. Multiple DNA-protein interactions at the CpG island of the human pseudoautosomal gene MIC2. Somatic cell and molecular genetics. 1993;19(1):51–63. doi: 10.1007/BF01233954. [DOI] [PubMed] [Google Scholar]
- 173.Hahn JH, Kim MK, Choi EY, et al. CD99 (MIC2) regulates the LFA-1/ICAM-1-mediated adhesion of lymphocytes, and its gene encodes both positive and negative regulators of cellular adhesion. J Immunol. 1997;159(5):2250–2258. [PubMed] [Google Scholar]
- 174.Schenkel AR, Mamdouh Z, Chen X, Liebman RM, Muller WA. CD99 plays a major role in the migration of monocytes through endothelial junctions. Nature immunology. 2002;3(2):143–150. doi: 10.1038/ni749. [DOI] [PubMed] [Google Scholar]
- 175.Manara MC, Bernard G, Lollini PL, et al. CD99 acts as an oncosuppressor in osteosarcoma. Mol Biol Cell. 2006;17(4):1910–1921. doi: 10.1091/mbc.E05-10-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Scotlandi K, Zuntini M, Manara MC, et al. CD99 isoforms dictate opposite functions in tumour malignancy and metastases by activating or repressing c-Src kinase activity. Oncogene. 2007;26(46):6604–6618. doi: 10.1038/sj.onc.1210481. [DOI] [PubMed] [Google Scholar]
- 177.Rocchi A, Manara MC, Sciandra M, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J Clin Invest. 2010;120(3):668–680. doi: 10.1172/JCI36667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Guo Y, Yuan F, Deng H, Wang HF, Jin XL, Xiao JC. Paranuclear dot-like immunostaining for CD99: a unique staining pattern for diagnosing solid-pseudopapillary neoplasm of the pancreas. Am J Surg Pathol. 2011;35(6):799–806. doi: 10.1097/PAS.0b013e318219c036. [DOI] [PubMed] [Google Scholar]
- 179.Li L, Li J, Hao C, et al. Immunohistochemical evaluation of solid pseudopapillary tumors of the pancreas: the expression pattern of CD99 is highly unique. Cancer Lett. 2011;310(1):9–14. doi: 10.1016/j.canlet.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 180.Xuan Y, Kim S, Lin Z. Protein expression and gene promoter hypermethylation of CD99 in transitional cell carcinoma of urinary bladder. J Cancer Res Clin Oncol. 2011;137(1):49–54. doi: 10.1007/s00432-010-0858-z. [DOI] [PubMed] [Google Scholar]
- 181.Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647(1–2):3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.De La Fuente R, Baumann C, Viveiros MM. Role of ATRX in chromatin structure and function: implications for chromosome instability and human disease. Reproduction. 2011;142(2):221–234. doi: 10.1530/REP-10-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Villard L, Toutain A, Lossi AM, et al. Splicing mutation in the ATR-X gene can lead to a dysmorphic mental retardation phenotype without alpha-thalassemia. Am J Hum Genet. 1996;58(3):499–505. [PMC free article] [PubMed] [Google Scholar]
- 184.Borgione E, Sturnio M, Spalletta A, et al. Mutational analysis of the ATRX gene by DGGE: a powerful diagnostic approach for the ATRX syndrome. Hum Mutat. 2003;21(5):529–534. doi: 10.1002/humu.10183. [DOI] [PubMed] [Google Scholar]
- 185.Nan X, Hou J, Maclean A, et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci U S A. 2007;104(8):2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Haas PS, Roy NB, Gibbons RJ, et al. The role of X-inactivation in the gender bias of patients with acquired alpha-thalassaemia and myelodysplastic syndrome (ATMDS) Br J Haematol. 2009;144(4):538–545. doi: 10.1111/j.1365-2141.2008.07505.x. [DOI] [PubMed] [Google Scholar]
- 187.Lugtenberg D, De Brouwer AP, Oudakker AR, et al. Xq13.2q21.1 duplication encompassing the ATRX gene in a man with mental retardation, minor facial and genital anomalies, short stature and broad thorax. Am J Med Genet A. 2009;149A(4):760–766. doi: 10.1002/ajmg.a.32742. [DOI] [PubMed] [Google Scholar]
- 188.Bo H, Ghazizadeh M, Shimizu H, et al. Effect of ionizing irradiation on human esophageal cancer cell lines by cDNA microarray gene expression analysis. J Nihon Med Sch. 2004;71(3):172–180. doi: 10.1272/jnms.71.172. [DOI] [PubMed] [Google Scholar]
- 189.Coutinho-Camillo CM, Miracca EC, Dos Santos ML, Salaorni S, Sarkis AS, Nagai MA. Identification of differentially expressed genes in prostatic epithelium in relation to androgen receptor CAG repeat length. Int J Biol Markers. 2006;21(2):96–105. doi: 10.1177/172460080602100205. [DOI] [PubMed] [Google Scholar]
- 190. Elsasser SJ, Allis CD, Lewis PW. Cancer. New epigenetic drivers of cancers. Science. 2011;331(6021):1145–1146. doi: 10.1126/science.1203280. **A recent article detailing epigenetic mechanisms of tumorigenesis.
- 191.Roy D, Guida P, Zhou G, Echiburu-Chau C, Calaf GM. Gene expression profiling of breast cells induced by X-rays and heavy ions. Int J Mol Med. 2008;21(5):627–636. doi: 10.3892/ijmm.21.5.627. [DOI] [PubMed] [Google Scholar]
- 192.Serrano E, Lasa A, Perea G, et al. Acute myeloid leukemia subgroups identified by pathway-restricted gene expression signatures. Acta Haematol. 2006;116(2):77–89. doi: 10.1159/000093636. [DOI] [PubMed] [Google Scholar]
- 193.Heaphy CM, De Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yachida S, Vakiani E, White CM, et al. Small Cell and Large Cell Neuroendocrine Carcinomas of the Pancreas are Genetically Similar and Distinct From Well-differentiated Pancreatic Neuroendocrine Tumors. Am J Surg Pathol. 2012;36(2):173–184. doi: 10.1097/PAS.0b013e3182417d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012 doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 196. Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. The Journal of cell biology. 2001;153(4):773–784. doi: 10.1083/jcb.153.4.773. **A comprehensive paper tying together the various mechanisms of X-inactivation maintenance.
- 197.Robertson KD. DNA methylation and human disease. Nature reviews. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 198. Sirchia SM, Tabano S, Monti L, et al. Misbehaviour of XIST RNA in breast cancer cells. PLoS One. 2009;4(5):e5559. doi: 10.1371/journal.pone.0005559. *An overview of aberrant XIST expression in breast cancer.
- 199.Lin H, Huber R, Schlessinger D, Morin PJ. Frequent silencing of the GPC3 gene in ovarian cancer cell lines. Cancer Res. 1999;59(4):807–810. [PubMed] [Google Scholar]
- 200.Xiang YY, Ladeda V, Filmus J. Glypican-3 expression is silenced in human breast cancer. Oncogene. 2001;20(50):7408–7412. doi: 10.1038/sj.onc.1204925. [DOI] [PubMed] [Google Scholar]
- 201.Chen CM, Chen HL, Hsiau TH, et al. Methylation target array for rapid analysis of CpG island hypermethylation in multiple tissue genomes. Am J Pathol. 2003;163(1):37–45. doi: 10.1016/S0002-9440(10)63628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Busque L, Mio R, Mattioli J, et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996;88(1):59–65. [PubMed] [Google Scholar]
- 203.Amos-Landgraf JM, Cottle A, Plenge RM, et al. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am J Hum Genet. 2006;79(3):493–499. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Medema RH, Burgering BM. The X factor: skewing X inactivation towards cancer. Cell. 2007;129(7):1253–1254. doi: 10.1016/j.cell.2007.06.008. *A review of skewed X-inactivation in cancer.
- 205.Kristiansen M, Langerod A, Knudsen GP, Weber BL, Borresen-Dale AL, Orstavik KH. High frequency of skewed X inactivation in young breast cancer patients. J Med Genet. 2002;39(1):30–33. doi: 10.1136/jmg.39.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lose F, Duffy DL, Kay GF, Kedda MA, Spurdle AB. Skewed X chromosome inactivation and breast and ovarian cancer status: evidence for X-linked modifiers of BRCA1. J Natl Cancer Inst. 2008;100(21):1519–1529. doi: 10.1093/jnci/djn345. [DOI] [PubMed] [Google Scholar]
- 207.Jazaeri AA, Chandramouli GV, Aprelikova O, et al. BRCA1-mediated repression of select X chromosome genes. J Transl Med. 2004;2(1):32. doi: 10.1186/1479-5876-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Silver DP, Dimitrov SD, Feunteun J, et al. Further evidence for BRCA1 communication with the inactive X chromosome. Cell. 2007;128(5):991–1002. doi: 10.1016/j.cell.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 209.Ross MT, Grafham DV, Coffey AJ, et al. The DNA sequence of the human X chromosome. Nature. 2005;434(7031):325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Reinius B, Shi C, Hengshuo L, et al. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics. 2010;11:614. doi: 10.1186/1471-2164-11-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11(6):213. doi: 10.1186/gb-2010-11-6-213. *This paper discusses regions of the X chromosome that escape inactivation.
- 212.Cohen DE, Lee JT. X-chromosome inactivation and the search for chromosome-wide silencers. Curr Opin Genet Dev. 2002;12(2):219–224. doi: 10.1016/s0959-437x(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 213. Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20(14):1848–1867. doi: 10.1101/gad.1422906. *A review of dosage compensation in mammals.
- 214.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 215. Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. **An excellent in-depth profile of gene expression on the X chromosome.
- 216. Liu Y, Wang Y, Li W, Zheng P. Activating transcription factor 2 and c-Jun-mediated induction of FoxP3 for experimental therapy of mammary tumor in the mouse. Cancer Res. 2009;69(14):5954–5960. doi: 10.1158/0008-5472.CAN-09-0778. *This paper details a possible therapeutic mechanism for inducing expression of an X-linked tumor suppressor gene.
- 217. Heinze E, Baldwin S, Chan G, et al. Antibody-mediated FOXP3 protein therapy induces apoptosis in cancer cells in vitro and inhibits metastasis in vivo. Int J Oncol. 2009;35(1):167–173. doi: 10.3892/ijo_00000325. *This paper details a possible therapeutic mechanism for inducing expression of an X-linked tumor suppressor gene.