Abstract
Squamous cell carcinoma of the lung (SQCCL) remains a leading cause of cancer-related death. Unlike non-smoker adenocarcinoma of the lung, where highly efficient tyrosine kinase inhibitors are available for treating mutant EGFR or ALK-rearranged, no targetable biomarkers are available for SQCCL. The frequent and focal amplification of FGFR1 has generated great expectations in offering new therapeutical options in case of 16-22% of SQCCL patients.
Broad 3q chromosome amplification is widely recognized as the most common chromosomal aberration found in SQCCL, where PIK3CA, SOX2, ACK1, PRKCI, TP63, PLD1, ECT2, and others genes are located. Although SOX2 has been postulated as a key regulator of basal stem cells transformation and tumor progression, it seems to confer a good prognosis in SQCCL. It is known that each patient might carry a different length of 3q chromosome amplicon. Thus, we suggest that the number and the biological importance of the genes spanned along each patient’s 3q amplicon might help to explain inter-individual outcome variations of the disease and its potential predictive value, especially when relevant oncogenes such as those mentioned above are implicated.
Currently, there is no clinical predictive data available from clinical trials. In this review, we have focused on the potential role of FGFR1 in SQCCL prognosis. Additionally, we have explored recently available public data on the comprehensive genomic characterization of SQCCL, in relation to the protein-coding genes that have a strong gene copy number - mRNA correlation in 3q chromosome, that were previously described as potential driver oncogenes or its modifiers in SQCCL.
Key Words: FGFR1, Squamous cell carcinoma, 3q chromosome, amplification, lung cancer
Despite the decrease in the incidence of squamous cell carcinoma of the lung (SQCCL) in the last decades, it still represents 20-30% of non-small cell lung cancer (NSCLC) (1). Unlike non-smoker lung adenocarcinoma, where strong biomarkers of response to specific tyrosine kinase inhibitors (TKI) (such as activating mutations of EGFR or ALK rearrangements) are available, in SQCCL actionable alterations have only been partially characterized in recent years, without any breakthrough in treating such tumor entities (2).
Gene copy number (GCN), like other genetic structural variations, represents an event of strong evolutionary pressure within both normal cells and particularly in cancer cells, where genomic instability it is a hallmark. GCN gains, such as gene duplication or amplification, can cause an increase in protein levels. Nowadays, there are three molecular mechanisms that can potentially produce a gene amplification, including the double-stranded DNA repair pathways: non-homologous end-joining (NHEJ), non-allelic homologous recombination (NAHR) (3,4), and DNA re-replication. In DNA re-replication, license control of replication is lost and a single DNA molecule is replicated more than once, triggering GCN gains, amplification, genomic instability and tumorigenesis (5).
Fibroblast growth factor receptor 1 (FGFR1) is one of the four family members of the FGFR of transmembrane tyrosine kinase receptors (TKR) involved in regulation of embryonic development, differentiation and cell proliferation (6-8). The functional validation of FGFR1 gene amplification in SQCCL was initially described by Weiss et al. Their work placed this histological-tumor subtype on the edge of the wave, identifying new therapeutic options that could change the management of SQCCL patients (9).
Broad amplification at 3q chromosome is the most frequent chromosomal alteration in SQCCL tumors. It was initially reported using fluorescent in situ hybridization (FISH) (10). It is known that increasing frequency of 3q amplification can be found from dysplasia to metastatic squamous lesions (11). Moreover, the potential epidemiological relationship of 3q amplification and tobacco consumption has been suggested (2). A recent comprehensive genomic characterization of SQCCL reported that 3q amplicon covers 3q13 to 3q29 (12). They also showed a correlation of GCN and mRNA levels at single-gene resolution.
This review highlights the recent findings on the prognostic and/or predictive value of FGFR1, as well as other important genes targeted by the 3q chromosome amplification in SQCCL.
FGFR1 amplification
FGFR is a family of receptors tyrosine kinases (RTK) consisting of four family members (FGFR1-4). FGFR1, like other RTKs, has an extracellular domain, a transmembrane domain and an intracellular domain, where the catalytic tyrosine kinase domain is located. The FGFR1 gene resides at 8p12 cytoband and spans a genomic DNA fragment of 57.7 Kb in length. Upon receptor activation, it promotes cell proliferation, angiogenesis, survival and apoptotic resistance through the PLCγ/PKC, RAS/MAPK and PI3K-AKT pathways (13). The oncogenic potential of activated FGFR1 represents an attractive therapeutic target that is currently being clinically tested.
The seminal work of Weiss et al. (9), demonstrated a growth dependency of a subset of SQCCL based on FGFR1 amplification that was abrogated both in lung cancer cell lines and in NCI-H1581 mice xenografts by PD173074, a specific TKI. No activating mutations were found. Twenty-two percent of squamous lung cancer tumors were carriers of FGFR1 focal amplification, as detected by FISH. Further studies confirmed that the percentage of amplification ranges from 16-22% (14-16) and independent in vitro studies confirmed that FGFR1-amplified cells are vulnerable when treated with a specific TKI (17). FGFR1 has also been reported to be amplified in other cancers, including 17.4% oral squamous cell carcinoma (18), 6% of esophageal squamous cell carcinoma (19), 10-17% of breast (20,21), 7.8% of ovarian (22,23), 3.4% of bladder (24) and 9% of prostate cancer (25).
Heist et al., in a retrospective cohort of 226 SQCLC, where almost 70% of the patients were staged as IA-IIB, detected 16% of FGFR1 amplification. They measured gene copy number by FISH, using for the threshold of gene amplification a FISH ratio equal to 2.2 or higher (14). In this study, amplification of FGFR1 was not correlated with age, sex, stage or smoking history. They found no correlation with overall survival. On the other hand, Weiss et al. reported a trend towards inferior survival among patients amplified for FGFR1 (9). In a recent work carried out by Kim et al. reported that patients, carriers of FGFR1 amplification, had significantly shorter disease-free survival and overall survival than diploid patients (wild type), regardless of sex, smoking status, adjuvant therapy and pathologic stage. These findings are in contradiction to those previously published by Heist and Weiss, and suggest FGFR1 amplification is an independent prognostic marker in this cohort of patients. Furthermore, in the same study, a positive association of FGFR1 amplification and smoking habit, in a dose-dependent manner, was reported. An interesting observation is that none of the 37 patients classified as never-smokers were carriers of amplified FGFR1 (26). Recently, a 100% concordance of FGFR1 amplification between primary SQCCL tumors and their lymph node metastatic tissue was described, suggesting an important role for FGFR1 in tumor prognosis and progression. So further studies are needed to validate whether the prognostic impact of FGFR1 amplification is a population-based phenomena or not (16).
Due to the important biological impact of FGFR activation in tumor cell growth, survival, tumor angiogenesis, progression and metastasis, the development and clinical testing of anti-FGFR compounds are currently major areas of research. There has been a great expectation as some reports have suggested FGFR1 amplification as a predictive biomarker of specific TKIs. There are two different types of FGFR inhibitors under development: small TKI molecules and ligand-competitor antibodies (see Table 1). Most of the small molecules exert their biological activity by binding into the ATP-binding pocket. This prevents either auto-phosphorylation of the receptor or proliferative signal transduction through transphosphorylation of receptor-dimers and their downstream adaptor proteins such as FSR2 (17,27). A clinical trial with BIBF1120, which inhibits FGFR1, will be developed in the Netherlands and in Spain in the second line treatment of SQCCL patients with FGFR1 amplification. Double methodological validation of FGFR1-amplified tumors will be carried out by FISH and multiplex ligation-dependent probe amplification (MLPA).
Table 1. Selected FGFR inhibitors currently used in clinical development and/or evaluation.
| DRUG | Company | TARGETS | Clinical development stage |
|---|---|---|---|
| Small-Molecule TKIs | |||
| Vargalef (BIBF1120) | Boehringer Ingelheim, Novartis | FGFRs, VEGFR and PDGFR | III |
| Ponatinib (AP24534) | Ariad | FGFR, VEGFR and IGF-1R | I |
| Dovotinib (TKI258) | Novartis | FGFRs, VEGFRs, KIT, FLT3, CSFR and PDGFRs | III |
| Brivanib (BMS582664) | Bristol Myers Squib | VEGFRs and FGFRs | II |
| AZD4547 | Astra Zeneca | FGFRs | I/II |
| Cediranib (AZ2171) | Astra Zeneca | VEGFRs, FGFRs and KIT | III |
| TSU68 (SU668) | Taiho Pahrmace | FGFRs, VEGFR and PDGFR | II |
| E7080 | Eisai | FGFRs, VEGFR and PDGFR | II |
| E3810 | Ethical Oncology Science | FGFRs, VEGFR | I |
| BGJ398 | Novartis | FGFRs | I |
| RG1507 | Roche, Genmab | FGFRs, VEGFR and PDGFR | II |
| LY2874455 | Lilly | FGFRs | n/a |
| FGFR antibodies | |||
| Figitumumab | Pfizer | FGFR, VEGFR and IGF-1R | III |
| Cixutumumab | ImClone Systems | FGFR, VEGFR and IGF-1R | II |
| AMG479 | Amgen | FGFR, VEGFR and IGF-1R | II/III |
| BIIB022 | Biogen Idec | FGFR, VEGFR and IGF-1R | I/II |
| FP1039 (Fusion protein) | Five Prime | FGFR1 | I/II |
| R3Mab | Genectech | FGFR3 | n/a |
Abbreviations: FGFRs, fibroblast growth factor receptors; VEGFRs, vascular endothelial growth factor receptor; PDFRs, platelet derived growth factor receptor; IGF-1R, Insulin Growth factor-1 receptor; KIT, mast/stem cell growth factor receptor; FLT3, fms-like tyrosine Kinase receptor 3; CSFR, colony stimulating factor receptor; n/a, not applicable
Taking advantage of what we have learned from gastrointestinal stromal tumors treated with imatinib/sunitinib (28,29), as well as from the history of erlotinib/gefitinib or crizotinib in lung cancers carriers of mutant EGFR (30,31) or ALK-rearrangements (32) respectively, we will need to identify the mechanisms of intrinsic, adaptive and acquired resistance to TKI treatment, as quickly as possible, and how to revert them clinically. The priority should be to analyze the presence of gain-of-functional mutations, amplification or overexpression of RTKs that activates redundant pro-survival pathways which bypass the drugged one (33,34). In addition, alterations in apoptotic pathways have also been demonstrated a key role in TKI resistance, and thus need to be analyzed (35-38).
Currently, fluorescent in situ hybridization (FISH) is the standard method available for identification of gene amplification among cancer patients. The previous experience from ERBB2 in breast cancer has shown that a key point was the inter-laboratories standardization of FISH criteria (39,40). Recently it has been reported in a cohort of 307 squamous lung carcinomas a reference guide to classify the tumor entities with respect to their FGFR1 gene status by FISH (41). The authors defined low-level amplification by ≥5 FGFR1 signals in ≥50% of tumor cells, whereas high-level amplification is defined by an FGFR1/centromere 8 (CEN8) ratio ≥2.0, or by an average number of FGFR1 signals per tumor cell nucleus ≥6, or by the percentage of tumor cells containing ≥15 FGFR1 signals or large clusters ≥10%.
In order to establish an appropriate GCN threshold correlation between FGFR1 gene dosage and drug response in SQCCL patients, we propose to measure FGFR1 gene status by FISH along with, a secondary independent quantification of FGFR1 gene copy number by MLPA. In addition to clarify how FGFR1 amplification is translated at active-protein levels, we recommend measuring phospho FGFR1 and phospho FSR2 as indicators of FGFR1 signal transduction activity (17,27).
3q amplification
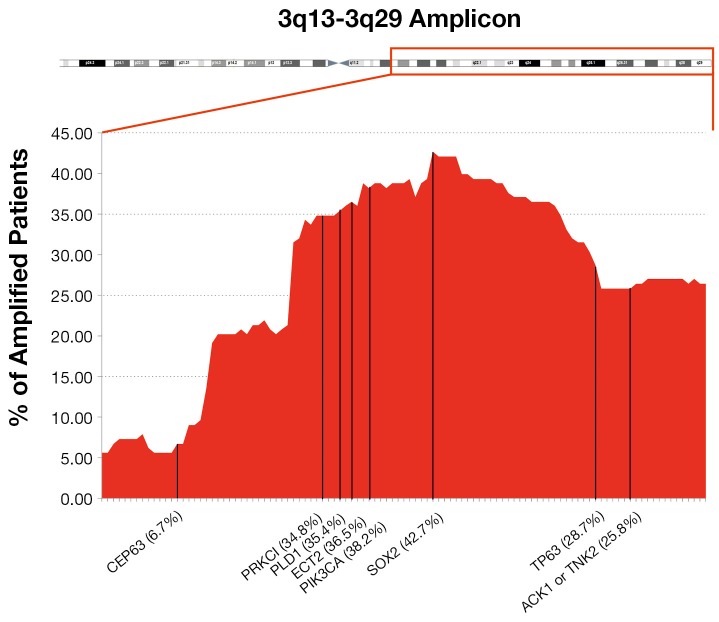
Over the recent decades, due to the great technical advancement in the field of molecular biology, there has been vast improvement towards the genetic characterization of tumors, in an effort to understand how their biology can be targeted to improve cancer patient care. One of the most frequent chromosomal aberrations found in NSCLC is the amplification at 3q chromosome, which can be present in up to 43% of cases. It can be found in squamous dysplasia, established carcinoma and also in metastatic tissue (42) and is suggested that 3q amplification frequency increases as disease progresses (43). It is known that each patient carries a different length of 3q chromosome amplicon (see Figure 1). We hypothesized that the number and the biological importance of the trapped genes in each patient’s 3q amplicon might be helpful to explain the inter-individual differences in disease outcome or its response towards specific targeted therapy.
Figure 1.
Percentage of patients, carriers of 3q chromosome amplification for each gene. A representative list of 3q chromosome region was sorted by chromosome position from 3q13 to 3q29.
Only a few genes that are targeted by the 3q chromosome amplicon have been functionally validated as prognosis modifiers of cancer disease, and even fewer as biomarkers of cancer therapy. Among these genes are phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) (44-46), SRY-related HMG-box (SOX2) (47-50), tumor protein 63 (TP63) (42), atypical Protein kinase C iota (aPKCɩ) (51,52), eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) (53,54), member of RAS oncogene family RAP2B, and others.
PIK3CA encodes for the p110α catalytic subunit of phosphatidylinositol (PI) 3-kinase. A broad range of cancer-related functions have been associated with its activation, such as cell proliferation, survival, oncogenic RAS signaling and transformation, making this an attractive target for therapeutic intervention. PIK3CA was found to be amplified in up to 45% of SQCCL cancer patients (55-59) and, due the strong correlation between PIK3CA amplification and its increased activity through its downstream effectors such as AKT and mTOR, this gene also appeared as an oncogene candidate (44). Abnormalities including mutations and amplification of PIK3CA/AKT/mTOR/PTEN are more common in SQCCL than in adenocarcinoma of the lung (60-62). Similar results have been showed by Spoerke et al. in their study where they have evaluated the candidate predictive biomarkers of sensitivity to select PI3K/mTOR pathway inhibitors in lung cancer patients. They suggests that different predictive biomarker strategies might be needed for both squamous and non-squamous patient populations, due to their alteration patterns and frequency (46).
The transcription factor TP63 (TP73L) is a homologue of p53 that functions by transactivating p53-targeted genes. The TP63 gene is expressed as multiple isoforms with different functions, including a full length (TAp63) and a truncated amino-deleted isoform ΔNp63, also called p40 (63). TAp63 can induce cell cycle arrest and apoptosis in response to DNA damage (64), whereas ΔNp63 has opposite functions because of its competition with p53, with respect to cell cycle arrest, mobility, invasion (epithelial–mesenchymal transition) and senescence. The ratio of TAp63 and ΔNp63 regulates chemosensitivity. ΔNp63a is the most commonly expressed TP63 isoform in squamous cell carcinoma together with TP63 amplification (65). Massion et al. reported that 88% of SQCCLs have TP63 amplification by FISH (42). As an interesting finding, they observed that TP63 amplification was an early event in the development of squamous carcinoma along with overexpression by IHC which results in better survival. ΔNp63 has been demonstrated as a more specific maker of squamous cell carcinoma than full length TP63, in the differential diagnosis in comparison with other lung histologies (66,67).
The SOX2 gene is a key transcription factor that coordinates embryonic development, differentiation and self-renewal of normal non-alveolar epithelium of the airway. SOX2 amplification has been reported in 43-60% (11,48,50,68) of SQCCL and in 27% of SCLC (69). The biological and clinical impact of SOX2 in lung cancer is reviewed by Karachaliou et al. (doi: 10.3978/j.issn.2218-6751.2013.01.01).
CEP63 (centrosomal protein 63 kDa) plays a role in DNA damage response. Following DNA double strand breaks (DSBs) formation, it is delocalized from centrosomes and recruits CDK1, a regulator mitotic entry of the cell (70,71).
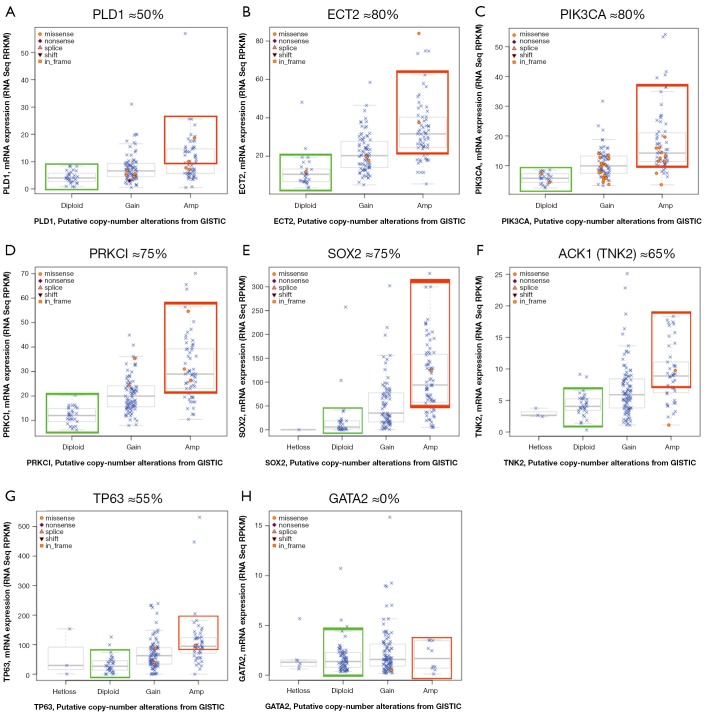
We took advantage of a recent report where authors performed a high resolution genomic characterization of SQCCL by RNA-seq, gene copy number and mRNA expression analysis (12) that is publicly available at the cBio cancer genomics portal (72). In this section, we will summarize the recent evidence of selected 3q-resident genes, where gene amplification might explain its contribution to malignant transformation, tumor progression or its role as a biomarker for targeted therapies. From protein-coding genes located at 3q, we only selected those were having strong correlation of GCN and mRNA. We defined strong GCN-mRNA correlation for a given gene, when at least 50% of the amplified tumors expressed higher levels of mRNA than diploid tumors (see Figure 2).
Figure 2.
Boxplots of mRNA expression vs. gene copy number in SQCCL selected oncogenes. Only protein-coding genes were considered for screening as candidate genes. Gene data was retrieved from the cBio portal (http://www.cbioportal.org/public-portal/) based on their GCN- mRNA correlation and previous available bibliography. Those genes in which ≥50% of the amplified tumors expressed higher mRNA levels than diploid were selected for further bibliographic review. A-G, selected genes showing an strong correlation of GCN and mRNA expression levels. H, an example of a discarded gene due to its low correlation value.
Atypical protein kinase C iota (aPKCɩ)
aPKCɩ belongs to the atypical subgroup within the protein kinase C family of structurally related serine/threonine kinases. Different PKC isoenzymes are involved in different functions, such as: cellular differentiation, proliferation, polarity and apoptosis. Atypical PKCs, unlike most of the members of the family, can be activated independently of Ca2+, diacyglycerol or phosphatidylserine (73). High aPKCɩ expression has been found in several human tumors, including squamous carcinomas of head and neck (64), esophageal (74,75) and lung (52), but also in lung adenocarcinoma (76). Recent data suggests that aPCKɩ activity is required by the oncogenic RasG12D mice model to progress from bronchial hyperplasia to lung tumor (77). In the same study, bronchoalveolar stem cells that lacked Prkci, the mice gene that encodes for aPKCɩ, were unable to transform neither in vitro nor in vivo.
aPKCɩ interacts with PAR6α, forming a complex that triggers the activation of RAC1-PAK-MEK-ERK pro-survival pathway. Interestingly in NSCLC, the ECT2 oncogene, which also localizes at 3q amplicon, is mislocalized in the cytoplasm, where it is a target of phosphorylation at Thr-328 by aPKCɩ (78) for a proper oncogenic signaling through the RAC1 small GTPase pathway (79).
Taking into consideration, the importance of aPKCɩ in KRAS-mediated lung tumors, the prognostic and/or predictive role of PRKCI amplification and aPKCɩ overexpression needs to be evaluated in oncogene “addicted” lung tumors, such as lung adenocarcinoma induced by EGFR-activating mutations or oncogenic rearrangements of ALK, where targeted therapies have a strong impact on patient survival and quality of care. Of course, it might be also interested to address the same question in SQCCL carriers of FGFR1 amplification treated with FGFRs inhibitors.
Activated CDC42 kinase 1 (ACK1)
ACK1, also known as TNK2, is a non-receptor tyrosine and serine/threonine protein kinase which functions as transducer of multiple ligand-activated RTKs including EGFR (80,81), AXL (82), MERTK (53), HER2 (83) and PDGFR (84) by activating cytosolic or nuclear effectors such as AKT and AR respectively to promote cell growth and survival (85,86). EGF ligand stimulation activates the ACK1 activity, which at the same time prevents EGFR from ubiquitination (87). AKT activation by ACK1 happens in a PI3K-independent manner. When phosphorylated by ACK1 at Tyr-176, unlike the PI3K-activated AKT, it is confined to the membrane phosphatidic acid phospholipid. Once the phospho-activated AKT/ACK1 complex is located at the plasma membrane, it then translocates into the nucleus where it phosphorylates FoxO3a, preventing the expression of the BIM-1 pro-apoptotic gene, the GADD45 DNA repair gene and p21 and p27 inducers of cell cycle arrest. Moreover it can also activate the mitotic progression (88). In addition, the E3 ubiquitin ligase Nedd4-2 is a negative regulator of ACK1 when co-expressed (87,89), and can be rescued by treatment with MG132, a proteasomal inhibitor. Xenografts of prostate LNCaP cells are usually poorly tumorigenic in nude mice. But when LNCaP cells expressing a constitutively activated ACK1 were engrafted into nude mice, they rendered very large tumors within the first 24 days after injection. In prostate cancer, activated ACK1, phosphorylates androgen receptor (AR) either at Tyr-267 or Tyr-363 led to the nuclear translocation of AR/ACK1 complex, thus activating the transcription of AR target genes such as prostate-cancer proteins: prostate-specific antigen (PSA) and HK2, independently of androgen or testosterone, the Androgen receptor ligands (83). Interestingly, a hallmark of prostate cancer progression implies the acquisition of an androgen-resistant phenotype, which might be explained in some cases by the AR estrogen-independent activation by ACK1.
Conclusions
Taking into consideration, the potential biological and medical impact of FGFR1, its activation turned to be a major area of research interest. Although prognostic data on FGFR1 has only recently been reported, the results are contradictory. Larger studies are needed to clarify its prognostic role. Furthermore, FGFR1 inhibitors have entered clinical trials, and over the next few years its predictive role with targeted TKIs will be definitely clarified.
On the other hand, finding new predictive biomarkers in highly genetic heterogeneous tumors such as SQCCL might be challenging because of the coexistence of multiple driver oncogenes, both in the same cellular clone or in different ones. An example might be the 3q chromosome amplification in SQCCL.
Acknowledgements
Work in Dr. Rosell’s laboratory is partly supported by a grant from La Caixa Foundation, which had no role in writing the manuscript or in the decision of publication.
Disclosure: This manuscript has not been published so far or submitted for publication elsewhere. The authors declare no conflicts of interest.
References
- 1.Travis WD. Pathology of lung cancer. Clin Chest Med 2011;32:669-92. [DOI] [PubMed] [Google Scholar]
- 2.Drilon A, Rekhtman N, Ladanyi M, et al. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol 2012;13:e418-26. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F, Gu W, Hurles ME, et al. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 2009;10:451-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shlien A, Malkin D.Copy number variations and cancer. Genome Med 2009;1:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truong LN, Wu X. Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol 2011;3:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem 2011;149:121-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner N, Grose R.Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116-29. [DOI] [PubMed] [Google Scholar]
- 8.Heist RS, Sequist LV, Engelman JA. Genetic changes in squamous cell lung cancer: a review. J Thorac Oncol 2012;7:924-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kettunen E, el-Rifai W, Björkqvist AM, et al. A broad amplification pattern at 3q in squamous cell lung cancer--a fluorescence in situ hybridization study. Cancer Genet Cytogenet 2000;117:66-70. [DOI] [PubMed] [Google Scholar]
- 11.McCaughan F, Pole JC, Bankier AT, et al. Progressive 3q amplification consistently targets SOX2 in preinvasive squamous lung cancer. Am J Respir Crit Care Med 2010;182:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mason I.Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat Rev Neurosci 2007;8:583-96. [DOI] [PubMed] [Google Scholar]
- 14.Heist RS, Mino-Kenudson M, Sequist LV, et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol 2012;7:1775-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 2011;6:e20351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Göke F, Franzen A, Menon R, et al. Rationale for treatment of metastatic squamous cell carcinoma of the lung using fibroblast growth factor receptor inhibitors. Chest 2012;142:1020-6. [DOI] [PubMed] [Google Scholar]
- 17.Guagnano V, Kauffmann A, Wöhrle S, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov 2012;2:1118-33. [DOI] [PubMed] [Google Scholar]
- 18.Freier K, Schwaenen C, Sticht C, et al. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC). Oral Oncol 2007;43:60-6. [DOI] [PubMed] [Google Scholar]
- 19.Ishizuka T, Tanabe C, Sakamoto H, et al. Gene amplification profiling of esophageal squamous cell carcinomas by DNA array CGH. Biochem Biophys Res Commun 2002;296:152-5. [DOI] [PubMed] [Google Scholar]
- 20.Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 2010;70:2085-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courjal F, Cuny M, Simony-Lafontaine J, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res 1997;57:4360-7. [PubMed] [Google Scholar]
- 22.Gorringe KL, Jacobs S, Thompson ER, et al. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res 2007;13:4731-9. [DOI] [PubMed] [Google Scholar]
- 23.Theillet C, Adelaide J, Louason G, et al. FGFRI and PLAT genes and DNA amplification at 8p12 in breast and ovarian cancers. Genes Chromosomes Cancer 1993;7:219-26. [DOI] [PubMed] [Google Scholar]
- 24.Simon R, Richter J, Wagner U, et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res 2001;61:4514-9. [PubMed] [Google Scholar]
- 25.Edwards J, Krishna NS, Witton CJ, et al. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin Cancer Res 2003;9:5271-81. [PubMed] [Google Scholar]
- 26.Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol 2013;31:731-7. [DOI] [PubMed] [Google Scholar]
- 27.Marek L, Ware KE, Fritzsche A, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol 2009;75:196-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agulnik M, Giel JL. Understanding Rechallenge and Resistance in the Tyrosine Kinase Inhibitor Era: Imatinib in Gastrointestinal Stromal Tumor. Am J Clin Oncol 2012. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 29.Joensuu H, DeMatteo RP. The management of gastrointestinal stromal tumors: a model for targeted and multidisciplinary therapy of malignancy. Annu Rev Med 2012;63:247-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayo C, Bertran-Alamillo J, Molina-Vila MÁ, et al. Pharmacogenetics of EGFR in lung cancer: perspectives and clinical applications. Pharmacogenomics 2012;13:789-802. [DOI] [PubMed] [Google Scholar]
- 31.Santarpia M, Altavilla G, Salazar MF, et al. Tyrosine kinase inhibitors for non-small-cell lung cancer: finding patients who will be responsive. Expert Rev Respir Med 2011;5:413-24. [DOI] [PubMed] [Google Scholar]
- 32.Camidge DR, Doebele RC. Treating ALK-positive lung cancer--early successes and future challenges. Nat Rev Clin Oncol 2012;9:268-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yano S, Takeuchi S, Nakagawa T, et al. Ligand-triggered resistance to molecular targeted drugs in lung cancer: roles of hepatocyte growth factor and epidermal growth factor receptor ligands. Cancer Sci 2012;103:1189-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson TR, Fridlyand J, Yan Y, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012;487:505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med 2007;4:1669-79; discussion 1680. [DOI] [PMC free article] [PubMed]
- 36.Garofalo M, Romano G, Di Leva G, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med 2011;18:74-82. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ng KP, Hillmer AM, Chuah CT, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med 2012;18:521-8. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto K, Okamoto I, Okamoto W, et al. Role of survivin in EGFR inhibitor-induced apoptosis in non-small cell lung cancers positive for EGFR mutations. Cancer Res 2010;70:10402-10. [DOI] [PubMed] [Google Scholar]
- 39.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med 2007;131:18-43. [DOI] [PubMed] [Google Scholar]
- 40.Gruver AM, Peerwani Z, Tubbs RR. Out of the darkness and into the light: bright field in situ hybridisation for delineation of ERBB2 (HER2) status in breast carcinoma. J Clin Pathol 2010;63:210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol 2012;25:1473-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massion PP, Taflan PM, Jamshedur Rahman SM, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res 2003;63:7113-21. [PubMed] [Google Scholar]
- 43.Pelosi G, Del Curto B, Trubia M, et al. 3q26 Amplification and polysomy of chromosome 3 in squamous cell lesions of the lung: a fluorescence in situ hybridization study. Clin Cancer Res 2007;13:1995-2004. [DOI] [PubMed] [Google Scholar]
- 44.Massion PP, Kuo WL, Stokoe D, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res 2002;62:3636-40. [PubMed] [Google Scholar]
- 45.Massion PP, Taflan PM, Shyr Y, et al. Early involvement of the phosphatidylinositol 3-kinase/Akt pathway in lung cancer progression. Am J Respir Crit Care Med 2004;170:1088-94. [DOI] [PubMed] [Google Scholar]
- 46.Spoerke JM, O’Brien C, Huw L, et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin Cancer Res 2012;18:6771-83. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, et al. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol 2007;20:474-81. [DOI] [PubMed] [Google Scholar]
- 48.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gen Y, Yasui K, Zen Y, et al. SOX2 identified as a target gene for the amplification at 3q26 that is frequently detected in esophageal squamous cell carcinoma. Cancer Genet Cytogenet 2010;202:82-93. [DOI] [PubMed] [Google Scholar]
- 50.Wilbertz T, Wagner P, Petersen K, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol 2011;24:944-53. [DOI] [PubMed] [Google Scholar]
- 51.Regala RP, Weems C, Jamieson L, et al. Atypical protein kinase C iota plays a critical role in human lung cancer cell growth and tumorigenicity. J Biol Chem 2005;280:31109-15. [DOI] [PubMed] [Google Scholar]
- 52.Regala RP, Weems C, Jamieson L, et al. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res 2005;65:8905-11. [DOI] [PubMed] [Google Scholar]
- 53.Mahajan NP, Whang YE, Mohler JL, et al. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res 2005;65:10514-23. [DOI] [PubMed] [Google Scholar]
- 54.van der Horst EH, Degenhardt YY, Strelow A, et al. Metastatic properties and genomic amplification of the tyrosine kinase gene ACK1. Proc Natl Acad Sci U S A 2005;102:15901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet 1999;21:99-102. [DOI] [PubMed] [Google Scholar]
- 56.Ma YY, Wei SJ, Lin YC, et al. PIK3CA as an oncogene in cervical cancer. Oncogene 2000;19:2739-44. [DOI] [PubMed] [Google Scholar]
- 57.Redon R, Muller D, Caulee K, et al. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res 2001;61:4122-9. [PubMed] [Google Scholar]
- 58.Massion PP, Taflan PM, Rahman SM, et al. Role of p63 amplification and overexpression in lung cancer development. Chest 2004;125:102S. [PubMed] [Google Scholar]
- 59.Tonon G, Wong KK, Maulik G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A 2005;102:9625-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji M, Guan H, Gao C, et al. Highly frequent promoter methylation and PIK3CA amplification in non-small cell lung cancer (NSCLC). BMC Cancer 2011;11:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okudela K, Suzuki M, Kageyama S, et al. PIK3CA mutation and amplification in human lung cancer. Pathol Int 2007;57:664-71. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res 2008;68:6913-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Melino G.p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ 2011;18:1487-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frederick MJ, VanMeter AJ, Gadhikar MA, et al. Phosphoproteomic analysis of signaling pathways in head and neck squamous cell carcinoma patient samples. Am J Pathol 2011;178:548-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hibi K, Trink B, Patturajan M, et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci U S A 2000;97:5462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bishop JA, Teruya-Feldstein J, Westra WH, et al. p40 (ΔNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol 2012;25:405-15. [DOI] [PubMed] [Google Scholar]
- 67.Nonaka D.A study of ΔNp63 expression in lung non-small cell carcinomas. Am J Surg Pathol 2012;36:895-9. [DOI] [PubMed] [Google Scholar]
- 68.Hussenet T, Dali S, Exinger J, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One 2010;5:e8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Löffler H, Fechter A, Matuszewska M, et al. Cep63 recruits Cdk1 to the centrosome: implications for regulation of mitotic entry, centrosome amplification, and genome maintenance. Cancer Res 2011;71:2129-39. [DOI] [PubMed] [Google Scholar]
- 71.Smith E, Dejsuphong D, Balestrini A, et al. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat Cell Biol 2009;11:278-85. [DOI] [PubMed] [Google Scholar]
- 72.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murray NR, Kalari KR, Fields AP. Protein kinase Cι expression and oncogenic signaling mechanisms in cancer. J Cell Physiol 2011;226:879-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu SG, Wang BS, Jiang YY, et al. Atypical protein kinase Cι (PKCι) promotes metastasis of esophageal squamous cell carcinoma by enhancing resistance to Anoikis via PKCι-SKP2-AKT pathway. Mol Cancer Res 2011;9:390-402. [DOI] [PubMed] [Google Scholar]
- 75.Yang YL, Chu JY, Luo ML, et al. Amplification of PRKCI, located in 3q26, is associated with lymph node metastasis in esophageal squamous cell carcinoma. Genes Chromosomes Cancer 2008;47:127-36. [DOI] [PubMed] [Google Scholar]
- 76.Erdogan E, Klee EW, Thompson EA, et al. Meta-analysis of oncogenic protein kinase Ciota signaling in lung adenocarcinoma. Clin Cancer Res 2009;15:1527-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Regala RP, Davis RK, Kunz A, et al. Atypical protein kinase C{iota} is required for bronchioalveolar stem cell expansion and lung tumorigenesis. Cancer Res 2009;69:7603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Justilien V, Jameison L, Der CJ, et al. Oncogenic activity of Ect2 is regulated through protein kinase C iota-mediated phosphorylation. J Biol Chem 2011;286:8149-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene 2009;28:3597-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kelley LC, Weed SA. Cortactin is a substrate of activated Cdc42-associated kinase 1 (ACK1) during ligand-induced epidermal growth factor receptor downregulation. PLoS One 2012;7:e44363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin Q, Wang J, Childress C, et al. The activation mechanism of ACK1 (activated Cdc42-associated tyrosine kinase 1). Biochem J 2012;445:255-64. [DOI] [PubMed] [Google Scholar]
- 82.Pao-Chun L, Chan PM, Chan W, et al. Cytoplasmic ACK1 interaction with multiple receptor tyrosine kinases is mediated by Grb2: an analysis of ACK1 effects on Axl signaling. J Biol Chem 2009;284:34954-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mahajan NP, Liu Y, Majumder S, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc Natl Acad Sci U S A 2007;104:8438-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galisteo ML, Yang Y, Ureña J, et al. Activation of the nonreceptor protein tyrosine kinase Ack by multiple extracellular stimuli. Proc Natl Acad Sci U S A 2006;103:9796-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mahajan K, Mahajan NP. Shepherding AKT and androgen receptor by Ack1 tyrosine kinase. J Cell Physiol 2010;224:327-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mahajan K, Mahajan NP. PI3K-independent AKT activation in cancers: a treasure trove for novel therapeutics. J Cell Physiol 2012;227:3178-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan W, Tian R, Lee YF, et al. Down-regulation of active ACK1 is mediated by association with the E3 ubiquitin ligase Nedd4-2. J Biol Chem 2009;284:8185-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mahajan K, Coppola D, Challa S, et al. Ack1 mediated AKT/PKB tyrosine 176 phosphorylation regulates its activation. PLoS One 2010;5:e9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin Q, Wang J, Childress C, et al. HECT E3 ubiquitin ligase Nedd4-1 ubiquitinates ACK and regulates epidermal growth factor (EGF)-induced degradation of EGF receptor and ACK. Mol Cell Biol 2010;30:1541-54. [DOI] [PMC free article] [PubMed] [Google Scholar]