Abstract
Notch signalling is implicated in stem and progenitor cell fate control in numerous organs. Using conditional in vivo genetic labelling we traced the fate of cells expressing the Notch2 receptor paralogue and uncovered the existence of two previously unrecognized mammary epithelial cell lineages that we term S (Small) and L (Large). S cells appear in a bead-on-a-string formation and are embedded between the luminal and basal/myoepithelial layers in a unique reiterative pattern, whereas single or paired L cells appear among ductal and alveolar cells. Long-term lineage tracing and functional studies indicate that S and L cells regulate ipsi- and contralateral spatial placement of tertiary branches and formation of alveolar clusters. Our findings revise present models of mammary epithelial cell hierarchy, reveal a hitherto undescribed mechanism regulating branching morphogenesis and may have important implications for identification of the cell-of-origin of distinct breast cancer subtypes.
Mammary epithelial cells (MECs) are classified into two lineages: basal/myoepithelial and luminal. The luminal lineage is further subdivided into ductal and alveolar cells. Classical MEC hierarchy models have been mainly inferred from transplantation and in vitro assays1–7. Recent in vivo lineage tracing study8 revealed, however, that under certain conditions transplanted MECs differentiate into non-physiological lineages, emphasizing the need to revisit and refine traditional hierarchy models using methodologies that preserve tissue architecture.
Key to lineage analysis is the use of appropriate markers that can trace the fate of progenitor cells. The Notch signalling pathway defines a fundamental cell fate controlling mechanism in metazoans, shown to be critical for the maintenance and differentiation of stem and progenitor cells in a variety of tissues, including mammary gland9–15. Among the four Notch receptor paralogues, Notch2 is the least studied in the normal mammary context and its role in tumorigenesis remains unclear16–22. Here, we used conditional in vivo genetic labelling in combination with functional assays to trace the fate of MECs expressing the Notch2 paralogue. Our analyses led to the discovery of two previously unrecognized lineages that we operationally name S (Small) and L (Large).
RESULTS
The Notch pathway is active in the luminal lineage in the pubertal mammary gland
To examine the involvement of Notch signalling in the pubertal mammary gland development, we used our Notch activity reporter strain Hes1emGFP23 (Supplementary Fig. S1a).
Analysis of tissue sections revealed that the Notch pathway is activated in the luminal lineage throughout the mammary ductal tree (Supplementary Fig. S1b,c). In all ducts examined, the emGFP signal intensity is strongest in actively growing terminal end buds (TEBs) and in budding lateral branches, gradually decreasing in the more mature, proximal regions of the ductal network (Supplementary Fig. S1b,c). TEBs that have reached the edge of the mammary fat pad show low or no detectable levels of emGFP (Supplementary Fig. S1d). Fluorescence-activated cell sorting (FACS) analysis indicates that Notch signalling is active in approximately half (52.1%) of all viable luminal cells (CD24+CD29low population; Supplementary Fig. S1e–i) and in a small fraction (4.1%) of the CD24+CD29high population, previously shown to contain myoepithelial and mammary stem cells1,2,4 (Supplementary Fig. S1j, values are the mean of two independent experiments). Our results corroborate previously published data obtained with the transgenic Notch activity reporter line TNR (ref. 15), and suggest that Notch activity may be critical in the subset of mammary cells that are actively involved in tissue remodelling.
The Notch2 receptor paralogue is expressed in distinct subsets of MECs at all stages of puberty
To analyse the distribution pattern of cells expressing the Notch2 receptor paralogue in pubertal mammary glands we crossed our N2-CreERT2SAT mice23 to the R26RLacZ reporter strain24 (Supplementary Fig. S1k). Bi-genic N2-CreERT2SAT/R26RLacZ and control N2-CreERT2SAT females ranging from 4 to 8 weeks of age (n = 3 mice per time point) were induced with a single dose of the tamoxifen metabolite 4-hydroxytamoxifen (4-OHT; 50 mg kg−1 mouse body weight) and euthanized after 24 h (24 h chase). Light microscopic analysis of x-gal-labelled mammary gland whole mounts revealed that during puberty, Notch2 is expressed in a distinct population of cells in virtually all end buds in a unique, discontinuous pattern (Fig. 1a). The same pattern was observed after 4-OHT inductions at different time points during puberty (4–8 weeks of age, data not shown). Induction with higher doses of 4-OHT did not result in an increased number of LacZ+ cells, confirming that this discontinuous pattern was not a result of insufficient Cre-mediated recombination.
Figure 1.
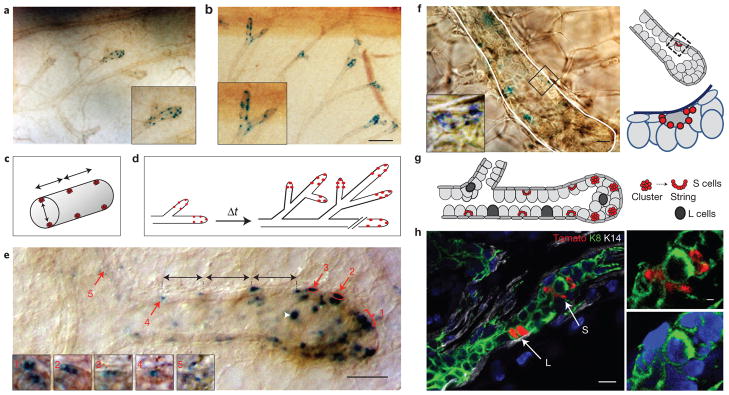
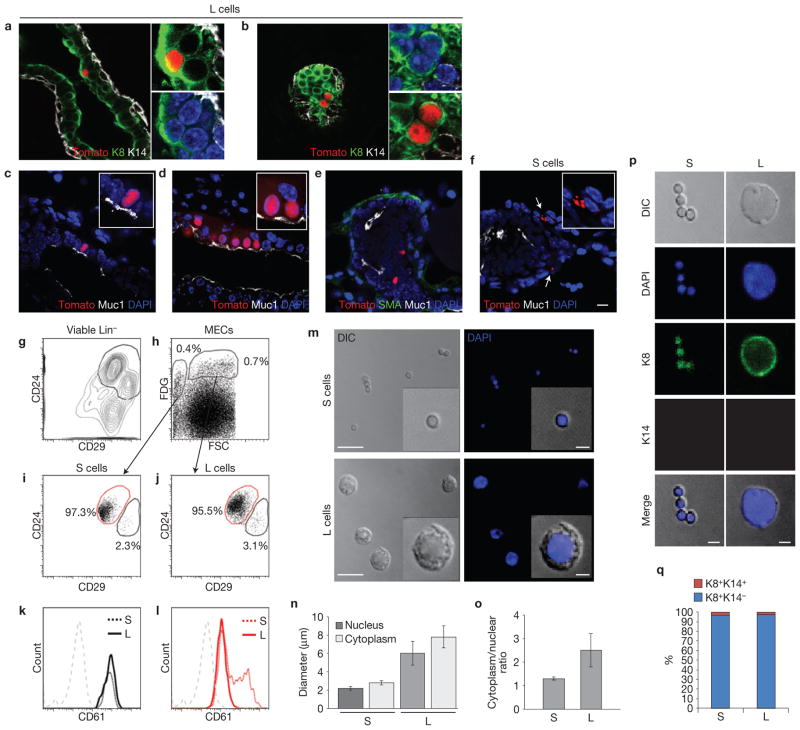
Notch2+ lineages define distinct subsets of MECs. (a,b) Representative x-gal-labelled mammary whole mounts from N2-CreERT2SAT/R26RLacZ littermates induced with 4-OHT at five weeks of age and euthanized after 24 h (a) or two weeks (b). n = 3 mice per time point. Insets in a and b are close-ups of the areas with LacZ+ cells. Scale bar, 200 μm. (c,d) Schematic presentation of the spatio-temporal distribution pattern of LacZ+ cells. (c) Topological arrangement of LacZ+ cells relative to the longitudinal axis and the diameter of the duct. (d) Distribution pattern of LacZ+ cells over time in actively growing pubertal end buds. (e) An x-gal-labelled whole mount of a typical, actively growing pubertal end bud. N2-CreERT2SAT/R26RLacZ female induced with 4-OHT at five weeks of age and euthanized after 48 h. Numbered inset panels (1–5) are close-ups of the groups of small LacZ+ cells from the regions labelled with red arrows, albeit at different focal planes, to better visualize individual cells. Groups of small LacZ+ cells are positioned at regular intervals along the longitudinal axis of the duct (black arrows). The white arrowhead marks a large LacZ+ cell. Scale bar, 50 μm. (f) High-power magnification image (left) and a schematic presentation (right) of an end bud that has reached the edge of the fat pad. x-gal-labelled whole-mount preparation from N2-CreERT2SAT/R26RLacZ female induced with 4-OHT at six weeks of age and euthanized after two weeks. Inset is a close-up of the indicated area in the trailing duct, showing a stereotypical arrangement of small LacZ+ cells in a beads on a string formation around a single large LacZ− luminal cell. Scale bars, 10 μm (main panel) and 2 μm (inset). (g) Schematic representation of the location of S and L cells during active ductal growth in puberty. (h) Representative immunofluorescence micrograph of a mature duct containing a pair of large oval Tomato+ cells (L) and a group of small Tomato+ cells (S) in a beads on a string formation around a single large Tomato− luminal cell. Mammary gland frozen section co-labelled with anti-K8 (green) and anti-K14 (white) antibody. Nuclei co-stained with DAPI. N2-CreERT2SAT/R26RTom female induced with 4-OHT at 7 weeks of age and euthanized after 5 weeks. Small panels are close-ups of S cells with different combinations of markers. Scale bars, 10 μm (main panel) and 2 μm (insets). Note that, owing to the process of dehydration and de-fatting, cells in mammary gland whole-mount preparations appear smaller than in frozen tissue sections.
The fate of Notch2+ MECs during pubertal growth
To follow the fate of Notch2+ cells during ductal tree expansion we performed a series of long-term chase experiments. Animals were induced with a single pulse of 4-OHT in mid-puberty (5–6 weeks of age) and euthanized at different times post-induction, ranging from 48 h to 2 weeks. Remarkably, irrespective of the length of the chase, ducts in every mammary gland exhibited the same distribution pattern of LacZ+ cells observed after 24 h chase (Fig. 1b). In virtually all end buds, LacZ+ cells are positioned at regular intervals along the longitudinal axis while maintaining a distinct spatial distribution relative to the diameter of the duct. Examination of the whole mounts at different focal depths revealed that they tend to occupy diametrically opposite positions around the circumference of the duct (Fig. 1c).
Considering that pubertal ducts elongate by proliferative expansion at the tips and bifurcate multiple times over the period of two weeks (Fig. 1d), the continuous presence of LacZ+ cells in virtually all end buds implies the existence of a Notch2+ progenitor within the most distal portion of each newly formed end bud. For such a progenitor cell, or cells, to give rise to the characteristic discontinuous distribution pattern, it would have to divide at regular intervals and at a much slower rate than the surrounding proliferating cell population. In addition, the progenitor would have to maintain its location within the most distal part of the proliferating tip either passively, that is, by being pushed forward within the mass of surrounding proliferating cells, or by active migration.
Closer inspection of the whole mounts revealed that LacZ+ areas distributed in regular intervals consist of groups of small LacZ+ cells flanked by longer stretches of LacZ− cells (Fig. 1e). In addition to the groups of small LacZ+ cells, rare single large LacZ+ cells can be found randomly scattered in the end buds and the mature ducts (Fig. 1e, white arrowhead). We will refer to LacZ+ small and large cells henceforth as S (Small) and L (Large) cells, respectively. In the most distal portion of each actively growing end bud (positions 1–3 in Fig. 1e), S cells form clusters, whereas in the trailing portion of the duct (positions 4 and 5 in Fig. 1e) they are less compact. High-power magnification imaging (×100) of x-gal-labelled whole mounts revealed that S cells in the trailing ducts are stereotypically arranged in a beads on a string formation around a single large LacZ− cell (Fig. 1f). S cells do not contact the lumen and are noticeably smaller than the surrounding luminal cells. All TEBs and lateral end buds that have stopped elongating, as they have either reached the end of the fat pad or abut another duct, no longer contain clusters of S cells in their most distal portion (Fig. 1f). In contrast, single L cells can be found in the most distal portion of both actively growing and growth-arrested end buds. We note that throughout the ductal network we also see single small cells dispersed in an apparently random fashion (data not shown). Figure 1g schematically summarizes the position of S and L cells in actively growing end buds.
It is noteworthy that large bulbous TEBs, which are abundant in early puberty (4–5 weeks of age), exhibit a random distribution of LacZ+ cells in contrast to small and medium-size TEBs, lateral end buds and subtending ducts (Supplementary Fig. S1l,m). After multiple rounds of bifurcation, the large bulbous TEBs become smaller and after the fifth week of age, virtually all end buds (TEBs and lateral end buds) exhibit the stereotypical LacZ distribution pattern described above.
Defining two classes of pubertal Notch2+ cell lineages
To obtain better visual resolution, corroborate our findings using the R26RLacZ reporter strain and perform marker analysis, we used a bi-genic N2-CreERT2SAT/R26RTomato strain that allowed us to fluorescently mark Notch2+ cell lineages. Mice were induced as described above and mammary gland frozen sections were labelled with MEC-lineage-specific markers.
All small (S) and large (L) Tomato+ cells are positive for the luminal marker cytokeratin 8 (K8) and negative for the basal marker cytokeratin 14 (K14; n = 61 and 90, respectively; Fig. 1h and Supplementary Fig. S2a–c). The L cells appear either as single cells or in pairs and are approximately the same size as neighbouring luminal cells, which are negative for the Notch2 lineage marker (Fig. 2a–c). In large ducts closest to the nipple, they occasionally appear in arrays (Fig. 2d). Only those L cells that are in contact with the lumen express the luminal marker mucin (Muc1; n = 96; Fig. 2c–e). S cells do not express Muc1 (n = 47; Fig. 2f). We note that inspection of x-gal-labelled whole-mount specimens in different focal depths is essential for deciphering the distinct three-dimensional spatial arrangement of the S cell strings. These topological features would be very difficult, if not impossible, to extrapolate from serial sections, where factors such as very small size of individual S cells, topological arrangement of the strings relative to the angle of the section and their scarcity confound the analysis.
Figure 2.
Expression of lineage-specific markers in L and S cells. (a–f) Immunofluorescence micrographs of typical pubertal mammary ducts containing single (a,c,e), a pair (b) and an array (d) of large Tomato+ (L) cells and two groups (arrows) of small Tomato+ (S) cells (f). Mammary gland frozen sections co-labelled with antibodies against luminal (K8, Muc1) and basal (SMA, K14) markers. Nuclei co-labelled with DAPI. N2-CreERT2SAT/R26RTom females induced with 4-OHT at 7 weeks of age and euthanized after 24 h. The small panels in a,b and the insets in c–f are enlarged details from the corresponding main panels. Scale bar, 10 μm. (g–l) Representative FACS analysis of Notch2 lineages in pubertal mammary glands. (g) Expression of CD24 and CD29 in a viable Lin− population. (h) FDG expression in a viable Lin−CD24+CD29+ population and gating strategy used to analyse FDG+ cells on the basis of size as defined by forward scatter (FSC). (i,j) Analysis of CD24 and CD29 expression in S cells (i; small Lin−CD24+CD29+FDG+ population) and L cells (j; large Lin−CD24+CD29+FDG+ population). (k,l) Expression of CD61 in S and L populations. The colours of the histograms correspond to the colours of the gates in i,j. Grey histograms represents the isotype control. The values are expressed as the mean from n = 3 independent experiments; individual values for all experiments are provided in Supplementary Table S1. (m–q) In vitro analysis of freshly sorted S and L cells gated in h. Sorted cells were plated for 2 h, fixed and co-labelled with DAPI (m), anti-K8 (green) and anti-K14 (red) antibody (p). (m) Representative confocal micrographs. Some S strings are partially disassociated and appear as quadruplets, triplets and doublets. (n,o) Cell size analysis. S cells: nuclear diameter 2.2 μm ± 0.2; cytoplasm (whole cell) diameter 2.8 μm ± 0.25; cytoplasm/nuclear ratio 1.3 ± 0.6. L cells: nuclear diameter 6 μm ± 1.3; cytoplasm diameter 7.8 μm ± 1.2; cytoplasm/nuclear ratio 2.5 ± 0.7 (mean ± s.d., n = 100 cells each, mammary glands pooled from 3 mice). The cytoplasm/nuclear ratio is calculated by dividing the whole cell area by the nuclear area. (p,q) Expression of lineage-specific markers in sorted S and L cells. Representative confocal micrographs (p) and a histogram (q) representing the percentage of K8+K14− and dual-positive K8+K14+ cells in the S and L populations (2.7% S and 3% L cells are dual-positive; n = 400 and 800 cells, respectively; also see Supplementary Fig. S2a). Scale bars, 10 μm and 2.5 μm in the insets (m); 2.5 μm (p).
To further characterize pubertal Notch2+ lineages and obtain more quantitative data we relied on FACS analysis using the β-gal substrate FDG and the 24 h pulse/chase protocol described above for the expression analysis. Analysis of viable FDG+ MECs (that is, DAPI−Lin−CD24+CD29+FDG+ subset) confirms the presence of two distinct populations based on cell size (Fig. 2g,h). Both populations are very rare, constituting approximately 1% of all MECs (small cells, 0.4% ± 0.15%; large cells 0.7% ± 0.26% of total viable Lin−CD24+CD29+ cells; mean ± s.d., n = 3). In agreement with lineage marker analysis of tissue sections, virtually all small (S) and large (L) cells (that is, viable Lin−CD24+CD29+FDG+ cells) are within the CD24+CD29low population, which contains luminal progenitors and mature luminal cells1 (Fig. 2i,j; red gates). A very small fraction of S (2.3% ± 0.18%) and L cells (3.1% ± 0.35%) resides within the CD24+CD29high population (Fig. 2i,j, black gates). Interestingly, the CD24+CD29high FDG+ subset is also highly enriched for CD61, a marker shown to be associated with luminal progenitor cells2,5 (Fig. 2k). S cells in the CD24+CD29low subset also contain a subpopulation enriched for CD61 (42% ± 2.94%; Fig. 2l). Results obtained with FDG reporter were corroborated using N2-CreERT2SAT/R26RTomato strain (Supplementary Fig. S2d,e).
To accurately measure the size of individual S and L cells, we sorted and analysed them in vitro using confocal microscopy (Fig. 2m). The cytoplasm (whole cell) diameter of S cells is 2.8 μm ± 0.25, and they have a relatively large nucleus and a narrow rim of cytoplasm (cytoplasm/nuclear ratio 1.3± 0.6), whereas the cytoplasm diameter of L cells is 7.8 μm± 1.2, with a cytoplasm/nuclear ratio of 2.5± 0.7 (mean± s.d., n = 100 cells each; Fig. 2n,o).
Analysis of freshly sorted S and L cells with lineage-specific markers, confirmed that 100% are positive for the luminal marker K8 (n = 400 and 800, respectively) and also revealed a very rare dual-positive K8+K14+ subpopulation (2.7% S and 3% L cells; Fig. 2p,q and Supplementary Fig. S3a). K8+K14+ cells are also positive for p63 (data not shown). Identification of the dual-positive population is consistent with the FACS analysis where ~3% of Notch2+ cells are found to reside in the CD24+CD29high population (Fig. 2i,j, black gates). Muc1 is expressed in 0% S and 67% L cells (n = 300 and 500, respectively; Supplementary Fig. S3b,c and Video).
To determine whether Notch signalling is active in S and L populations, we crossed N2-CreERT2SAT/R26RTomato bi-genic mice to the Hes1emGFP reporter line (Supplementary Fig. S3d). Tri-genic female pups were induced with 4-OHT at 6 weeks of age and euthanized two weeks later. As shown in Supplementary Fig. S3e,f, S and L cells express the Hes1emGFP reporter allele, indicating that Notch signalling is active in both. Our analysis however cannot distinguish whether signalling in the Notch2+ lineages is mediated through the Notch2 receptor, given that Hes1 could be activated by other receptor paralogues as indeed seems to be the case in the Tomato− subset of emGFP+ cells.
Pubertal Notch2+ cell lineages and alveologenesis
To follow the fate of pubertal Notch2+ lineages during adulthood, pregnancy, lactation and involution, we induced bi-genic N2-CreERT2SAT/R26RLacZ and control N2-CreERT2SAT females with a single dose of 4-OHT at seven weeks of age and euthanized them at appropriate stages (n = 3 mice per time point).
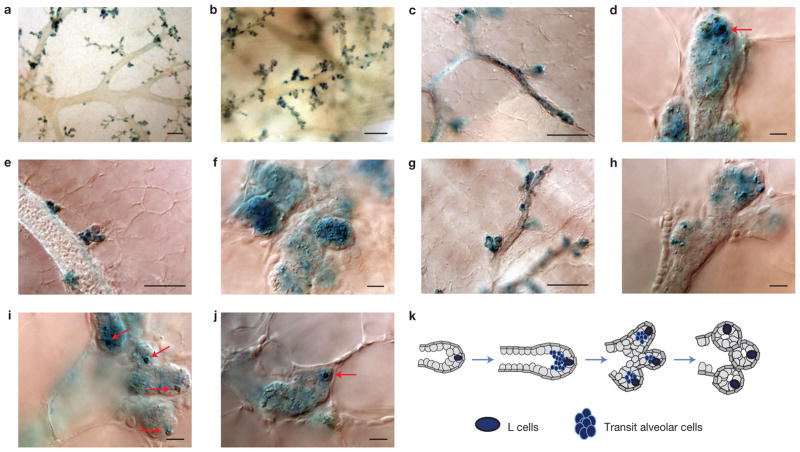
In adult virgin animals, S cell clusters can no longer be detected in end buds, whereas S cell strings and L cells retain the same topology as in the mature pubertal duct (see above). In early pregnancy, luminal space in tertiary branches and sprouting alveolar buds are transiently filled with LacZ+ cells (Fig. 3a–f). Gradually, the luminal space clears from transit LacZ+ alveolar cells, with the exception of one or two L cells that remain present in each sprouting bud until alveoli are fully formed (Fig. 3g–k).
Figure 3.
The fate of pubertal Notch2+ lineages in pregnancy. (a–j) Low(a,b) and high-power magnification images (c–j) of x-gal-labelled mammary whole-mount preparations from a N2-CreERT2SAT/R26RLacZ female induced with 4-OHT at 7 weeks of age and euthanized in early pregnancy (8 dpc; days post-coitus). (c–f) Early stage of alveologenesis. The lumen of tertiary branches (c) and alveolar buds (d–f) is filled with transit LacZ+ cells. The arrow in d points to a single large LacZ+ cell (L cell) positioned in the distal portion of a sprouting alveolar bud. (g–j) Advanced stage of alveolar bud formation. The luminal space is cleared, with the exception of one or two L cells (arrows) that remain present in each alveolar bud. (k) Schematic representation of LacZ+ cell distribution in early and late stages of alveologenesis. Scale bars, 200 μm (a,b); 100 μm (c,e,g); 20 μm (d,f,h–j).
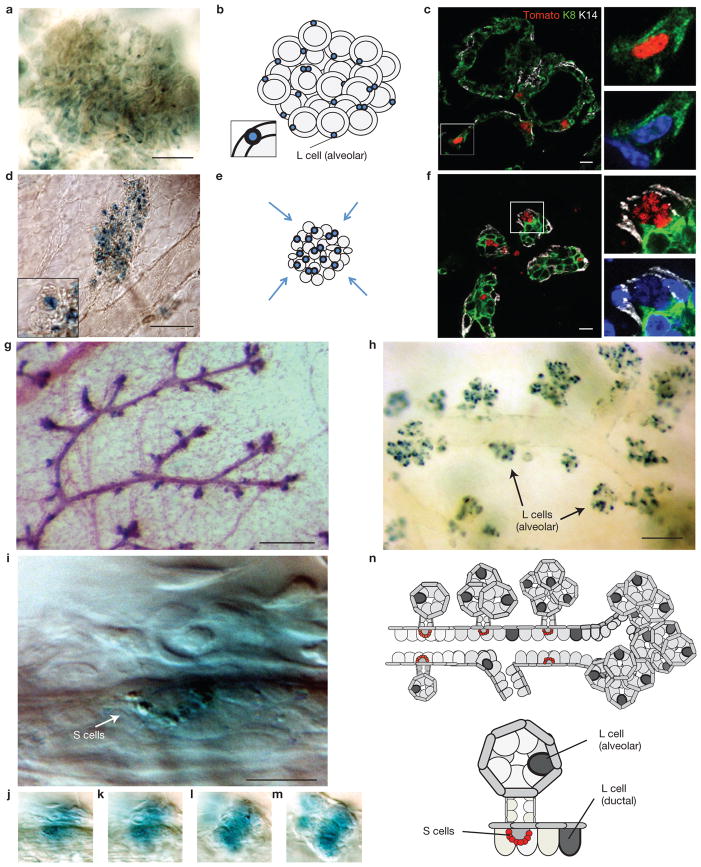
During lactation, each milk-secreting alveolus generally contains only one or two large LacZ+ (Tom+) cells embedded within the epithelial wall (n = 130 alveoli; Fig. 4a–c). Alveoli in close proximity to the nipple and large ducts occasionally contain multiple such cells (not shown). Marker analysis of N2-CreERT2SAT/R26RTomato glands reveals that all large Tom+ cells (n = 102) are positive for the luminal marker K8 and those contacting the lumen also express Muc1 (Fig. 4c and Supplementary Fig. S4a). Examination of lactating glands from tri-genic N2-CreERT2SAT/R26RTomato/Hes1emGFP reporter animals induced at 7 weeks of age revealed that the Notch signalling pathway is active in all large ductal and alveolar Tom+ cells (n = 100; Supplementary Fig. 4b,b′). Although the marker profile and the morphology of alveolar LacZ+ (Tom+) cells are indistinguishable from ductal L cells, we cannot equate them on the basis of these features alone; therefore, we operationally refer to them as L-alveolar cells.
Figure 4.
The fate of pubertal Notch2+ lineages during lactation and involution. (a–f) A typical alveolar cluster in lactating (a–c) and involuting (d–f) glands. N2-CreERT2SAT/R26RLacZ (a,d) and N2-CreERT2SAT/R26RTom (c,f) females induced with 4-OHT at 7 weeks of age and euthanized during lactation (a–c) or 4 days after forced weaning (d–f). x-gal-labelled whole-mount preparations (a,d), schematic presentations of alveolar cluster (b,e) and immunofluorescence micrographs of sections labelled with anti-K8 and anti-K14 antibodies showing several individual alveoli. Nuclei co-labelled with DAPI (c,f). The inset in b is a close-up of the alveolar L cell. Small panels in c and f are close-ups of the respective areas marked with the rectangle. The inset in d is a close-up of the collapsed alveolus. (g–m) x-gal-labelled mammary gland whole-mount preparations from N2-CreERT2SAT/R26RLacZ females induced at 7 weeks of age and euthanized in early pregnancy (8 dpc; g,i–m) or 4 days after forced weaning (h). (g,h) Budding tertiary branches (g) and alveolar clusters originating from primary and secondary ducts (h) are positioned at regular intervals alongside ductal longitudinal axis. (i) High-magnification image of an S cell string (arrow) in a large duct. (j–m) Different focal depths of the image shown in i. Beneath the S cells is an alveolar cluster consisting of several sprouting alveolar buds all of which are transiently filled with LacZ+ cells. (n) Schematic representation of the location of L (ductal and alveolar) and S cells in the lactating mammary gland. Gland in g is co-stained with carmine alum. Scale bars, 100 μm (a,d,h); 10 μm (c,f); 200 μm (g); 20 μm (i).
Four days after forced weaning, alveolar collapse associated with involution brings L-alveolar cells in close proximity to each other (Fig. 4d–f) giving an appearance that the entire alveolar cluster is LacZ+ (Supplementary Fig. S4c). However, higher magnification clearly reveals that each alveolus still retains individual L-alveolar cells (Fig. 4d, inset and Supplementary Fig. S4d). At this stage, most L-alveolar cells (90%, n = 100) are undergoing apoptosis as judged by cleaved caspase 3 staining (Supplementary Fig. S4e) and the appearance of apoptotic bodies (Fig. 4f). Comparison of end buds in the adult virgin and involuted glands 4 days and 8 days post-weaning indicates that some L-alveolar cells survive and remain incorporated within the involuted end buds (Supplementary Fig. S4f–h,i).
The overall spatial arrangement of the tertiary branches and the alveolar clusters emulates the stereotypical distribution pattern of the S cell strings. They tend to be distributed at regular intervals along the longitudinal axis while maintaining diametrically opposite positions relative to the diameter of the duct (Fig. 4g,h). High-magnification images of x-gal-stained whole mounts confirm that S cell strings are localized at the point of origin of tertiary branches (Fig. 4i–m). Figure 4n schematically summarizes the position of L (ductal and alveolar) and S cells in the lactating gland. We note that unique cis and trans spatial arrangement of tertiary structures relative to the ducts cannot be appreciated in whole mounts of lactating glands where numerous alveolar clusters block the view of the ductal tree. These features can be visualized only in early pregnancy when alveoli are not yet fully formed or during involution when they are collapsed (Fig. 4g,h).
We followed the distribution pattern of L (ductal and alveolar) and S cells through repeated pregnancies (up to 3) and determined that their distribution emulates the same pattern as described above for the first pregnancy (n = 3 mice). We cannot conclude on the basis of our analyses, however, whether L-alveolar cells in repeated pregnancies originate from long-lived LacZ+ L-alveolar progenitors or whether they are in fact residual L-alveolar cells remaining from the first pregnancy, which then re-populate new sprouting alveolar buds.
Functional characterization of pubertal Notch2+ lineages
The lineage tracing experiments demonstrate that Notch2+ cells are not progenitors of classical luminal and myoepithelial lineages. To corroborate this observation we performed standard mammary reconstitution assays and found that Notch2+ cells do not yield outgrowths in virgin recipients (0/6), confirming that S cells, L cells and their progenitors are neither multipotent nor myoepithelial stem cells. In contrast, all control glands containing a mixture of Notch2+ and Notch2− cells develop outgrowths (12/12) with S and L cells assuming the same topology as described for the adult glands (see above). Notch2− outgrowths contain no LacZ+ cells (Supplementary Fig. S5). We note that the very low abundance of Notch2+ MECs in virgin donors (~1%) required a prohibitive number of age-matched bi-genic donor animals to obtain larger numbers of FDG+ transplants; thus, a more rigorous analysis was not possible.
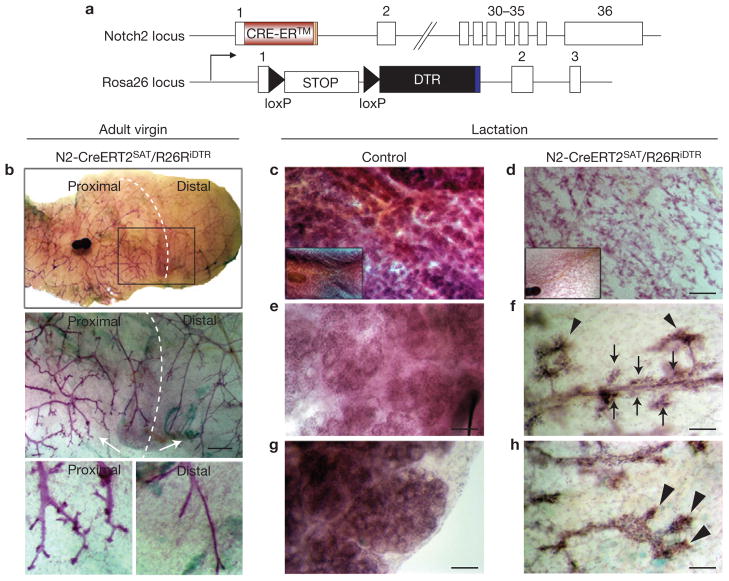
To complement the transplantation assays and gain more insight into the functional properties of Notch2+ lineages in their physiological environment, we performed conditional in vivo cell ablation using the Cre-inducible diphtheria toxin receptor (iDTR) transgenic mice25. To specifically and conditionally express iDTR in Notch2+ lineages, we crossed the R26RiDTR (iDTR) transgenic line to the N2-CreERT2SAT line (Fig. 5a). Bi-genic N2-CreERT2SAT/R26RiDTR and control R26RiDTR females were induced with a single pulse of 4-OHT in mid-to-late puberty (6–7 weeks of age) and treated with two doses of diphtheria toxin 48 and 96 h after 4-OHT induction. Mammary gland whole mounts were analysed in adult virgin and lactating females.
Figure 5.
Ablation of pubertal Notch2+ lineages impairs formation of tertiary branches and alveolar clusters. (a) Schematic representation summarizing the genetic strategy used to induce expression of diphtheria toxin receptor (DTR) specifically in Notch2-expressing cells and their progeny. Tamoxifen-inducible Cre-recombinase (Cre-ERT2) is expressed under the control of the Notch2 promoter. Treatment with Tamoxifen or 4-OHT results in the excision of the STOP cassette and the expression of DTR in bi-genic N2-CreERT2SAT/R26RiDTR mice. White rectangles represent exons. (b–h) Representative carmine-alum-stained mammary whole mounts from adult virgin N2-CreERT2SAT/R26RiDTR female 1 year after ablation of Notch2+ lineages (b) and lactating N2-CreERT2SAT/R26RiDTR and control R26RiDTR females 1 day post-partum (c–h). All females were induced with 4-OHT and diphtheria toxin at 6 weeks of age. (b) Tertiary branches are absent from the distal segment of the gland. The middle panel is a close-up of the area marked with a rectangle in the upper panel. Bottom panels are close-ups of branches marked with an arrow in the middle panel. (d,f,h) Alveolar clusters are absent from the entire gland. Hyperplastic outgrowths emulate the stereotypical topology of tertiary structures. (f) Proximal segment. Hyperplastic outgrowths are located on the distal tips of tertiary branches (arrowheads) and alongside ducts (arrows). (h) Distal segment. Tertiary branches are absent. End buds harbour hyperplastic papillary projections. Scale bars, 1 mm (b, middle panel); 500 μm (c,d); 50 μm (e–h). Insets in (c,d) are low-magnification (×1) images of the corresponding glands.
In adult virgins, depletion of pubertal Notch2+ lineages resulted in two contrasting phenotypes in proximal and distal segments of each mammary gland (n = 16 glands). Whereas proximal segments have an elaborate network of tertiary branches, distal segments exhibit virtually no tertiary branches (Fig. 5b). The margin between the two segments is well defined and its relative position correlates with the age of the animal at the time of diphtheria toxin administration. Females induced at 6 weeks of age have a smaller gland area with tertiary branches than those induced at 7 weeks of age. We therefore conclude that this boundary probably reflects the location of actively growing end buds at the time of diphtheria toxin administration (that is, Notch2+ lineage depletion) and that all branches distal to this margin represent the trailing ducts of such end buds (Supplemental Fig. S6a). Control mammary glands exhibit the same tertiary branching phenotype in the entire area of the gland (n = 16, data not shown).
In lactating females (1 day post-partum, n = 12 glands), depletion of pubertal Notch2+ lineages resulted in the absence of alveolar clusters (Fig. 5c–h and Supplementary Fig. S6b–i). Instead, the ductal tree harbours hyperplastic outgrowths that emulate the stereotypical spatial arrangement of tertiary structures in wild-type glands (Fig. 5d and Supplementary Fig. S6d–i). Similarly to virgin glands, tertiary branches are located in the proximal segments of the glands and contain outgrowths on their distal tips (Fig. 5f and Supplementary Fig. S6d,f,h). In distal segments, the outgrowths are directly adjacent to primary and secondary ducts, whereas the end buds form papillary projections (Fig. 5h and Supplementary Fig. S6e,g,i). Glands contain very little or no milk in the ducts, consistent with the demonstration that proper formation of the alveolar lumen is a prerequisite for WAP gene expression followed by milk synthesis26.
Cell ablation results are therefore consistent with lineage tracing observations and imply that pubertal Notch2+ lineages play a critical role in the formation of tertiary branches and alveolar clusters.
DISCUSSION
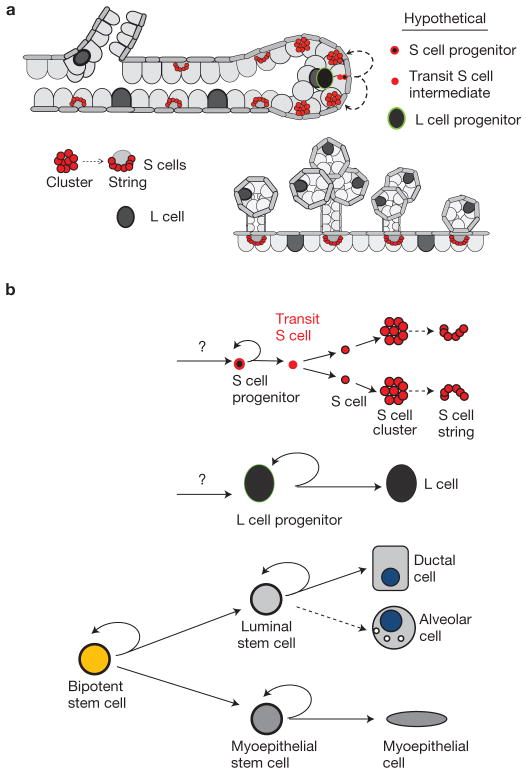
Using Notch2-specific genetic labelling we uncovered the existence of two previously unrecognized mammary epithelial cell populations, which we operationally name S (Small) and L (Large) cells. We provide evidence that S and L cells are morphologically, topologically, genetically, developmentally and functionally distinct from classical luminal and myoepithelial cells27,28. Furthermore, we demonstrate that they are neither direct descendants nor precursors of unipotent myoepithelial progenitors or luminal progenitors but rather distinct Notch2+ progenitors thus representing bona fide unique lineages.
On the basis of short- and long-term lineage tracing and functional data we postulate that during puberty Notch2 specifically labels two progenitor populations, S progenitors and L progenitors, that give rise to respective lineages (Fig. 6). Implicit in our model is the notion that both progenitors maintain their location in the most distal portion of the end bud during ductal tree expansion and asymmetrically divide at regular intervals but at a much slower rate than surrounding proliferating cell population. FACS data support the concept of two independent progenitors as both S and L cells contain a distinct subset residing within the CD24+CD29high population, which is also highly enriched for CD61. Although this working model accommodates our experimental observations, its validity can be rigorously examined only through functional experiments.
Figure 6.
Proposed revised model of the MEC lineages and differentiation hierarchy. (a) Schematic representation of the fate of S and L cells in the postnatal mammary gland. During puberty, the S cell progenitor gives rise to a self and a transit S cell intermediate, which in turn gives rise to two daughter S cells. These cells undergo several cell divisions resulting in the formation of two S cell clusters, which assume diametrically opposite positions relative to circumference of the duct. As the end bud continues forward growth, S cell clusters are left behind in the more mature, trailing duct, eventually assuming the bead on a string arrangement around a single large Notch2− cell. S cell strings remain embedded in a unique reiterative pattern between the luminal and basal/myoepithelial layer throughout adult life and mark the origins of tertiary branches. The L cell progenitor gives rise to a self and a committed L daughter cell, which is left behind embedded within ductal cells. Occasionally, L cells appear in pairs. In lactating glands, each milk-secreting alveolus contains one or two L cells. (b) Proposed revised model of the MEC lineages and differentiation hierarchy (adapted from ref. 8).
Cell ablation assays indicate that Notch2+ lineages are critical for the formation of tertiary branches. Long-term lineage tracing experiments demonstrate that S cell strings co-localize with the point of origin of tertiary branches and remain in their location throughout adulthood and repeated pregnancy cycles. Considering that L cells are randomly scattered throughout the ductal network in the adult gland and are therefore not likely to be directly involved in the genesis of tertiary branches, we propose that the functional role of S cell strings is to mark the site of origin of tertiary branches, thus ensuring that they do not encumber each other as they grow ipsi- and contra-laterally. Notably, not all sites with S cell strings eventually develop tertiary branches, implying that S strings may be a necessary but not a sufficient requirement for branch sprouting, as other factors have been shown to be critical29–33. Clearly, an important caveat in inferring the function of individual lineages from cell ablation experiments is that all Notch2+ cells and their descendants are depleted simultaneously, and it is therefore conceivable that some or all of the functional properties of S and L cells may be overlapping.
In early pregnancy we detected a distinct population of Notch2+ cells that we operationally refer to as transit alveolar cells (Fig. 3). These cells transiently fill in the lumen of actively growing tertiary branches and sprouting alveolar buds and are cleared as soon as these structures are shaped. It is not clear what Notch2+ cell type is the progenitor of this transient and possibly functionally distinct subset of alveolar lineage.
Interestingly, ablation of pubertal Notch2+ lineages does not seem to impair proliferation of alveolar cells in lactating glands, as evidenced by hyperplastic outgrowths, but rather formation of alveolar lumen and consequently alveolar clusters. Considering that transit alveolar cells and L-alveolar cells have distinct spatio-temporal distributions during alveologenesis we propose that the population of transit alveolar cells may be critical for the formation of lumen in individual alveoli whereas L cells may be required for the arrangement of alveoli into complex three-dimensional structures (that is, alveolar clusters). Importantly, the lineage tracing assays clearly show that none of the Notch2+ lineages are classical alveolar progenitors as evidenced by the lack of LacZ+ monoclonal mature alveoli.
Although most L-alveolar cells undergo apoptosis during involution, some survive and remain embedded in the end buds. It is unclear whether they define a different cell subgroup from those that perish during involution or whether survivors are stochastically chosen. A similar cell fate has been previously described for another MEC population, the so-called parity-induced MECs (ref. 34). These cells appear during the first full-term pregnancy and do not undergo apoptosis during involution. However, unlike L-alveolar cells, parity-induced MECs are bona fide alveolar progenitors. Consistent with previous reports35,36, our analysis clearly demonstrates the existence of at least two functionally distinct luminal alveolar lineages (that is, L-alveolar and classical secretory luminal alveolar). It remains to be determined whether the transit alveolar population we observed in early pregnancy also represents a functionally distinct luminal alveolar lineage.
The authors of ref. 37 proposed the existence of five distinct MEC subtypes based on the ultrastructural criteria: classical myoepithelial and luminal cells, undifferentiated and differentiated large light cells (ULLCs, DLLCs) and small light cells (SLCs). Comparison of the three last morphotypes with our Notch2+ lineages reveals marked similarities. Whether S and L cells represent the SLC and LLC populations, respectively, remains to be demonstrated experimentally.
We believe further lineage tracing analyses involving Notch receptor paralogues will probably allow further refinement of MEC hierarchy models, possibly offering valuable insights into the cellular origin of different breast cancer subtypes.
METHODS
Methods and any associated references are available in the online version of the paper.
METHODS
Mice
Mouse strains carrying conditional alleles for LacZ (R26RLacZ), tdTOMATO (R26RTom) and DTR (R26RiDTR) were obtained from the Jackson Laboratory. The generation of N2-CreERT2SAT and Hes1emGFPSAT knock-in mice was previously described23. All strains were kept on the C57BL/6 background. All procedures involving animals were approved by the Harvard Institutional Animal Care and Use Committee (IACUC).
Conditional labelling of Notch2 lineages
To conditionally label cells expressing the Notch2 paralogue in pubertal mammary glands, N2-CreERT2SAT/R26RLacZ, N2-CreERT2SAT/R26RTom and control N2-CreERT2SAT females ranging from 4 to 8 weeks of age were induced with a single intraperitoneal (i.p.) injection of 4-hydroxytamoxifen (4-OHT, Sigma, H7904; 50 mg kg−1 mouse body weight). We determined empirically that this dose of 4-OHT is sufficiently high to induce recombination in most cells expressing CreERT2. Similarly, we established that 24 h is the optimal length of chase to allow for Cre-recombination and LacZ expression in most induced cells, yet short enough to exclude the possibility of detecting their progeny. For expression analysis (that is, detection of Notch2-paralogue-expressing cells), animals were euthanized 24 h post-induction. For lineage tracing experiments (that is, following the fate of Notch2-paralogue-expressing cells and their putative descendants), animals were euthanized at different time points ranging from 48 h to 1 year post-induction. For each time point, at least 3 mice were analysed (minimum 4 glands per mouse).
Mammary whole-mount preparation and x-gal staining
The thoracic and inguinal mammary glands were dissected from induced N2-CreERT2SAT/R26RLacZ and control N2-CreERT2SAT females, spread thin on a tissue paper, immersed immediately in cold fixative (2.5% paraformaldehyde (pH 7.65), 0.05% glutaraldehyde and 0.1% Triton X-100 in PBS) and fixed overnight at 4 °C. Tissues were then removed from the paper, washed for a minimum of 4 h in washing buffer (2 mM MgCl2, 0.1% Triton X-100 and 0.01% Na-deoxycholate in PBS), transferred to x-gal solution (Active Motif β-gal staining kit, #100443) and incubated at room temperature for 14–16 h. Glands were then visually inspected under a stereoscope to assess the intensity of the LacZ staining in individual MECs. x-gal incubation was stopped when these cells turned dark blue or if the colour of the mammary fat pad started turning light green. To minimize nonspecific staining, glands were never incubated for more than 20 h or at temperatures higher than 25 °C. Tissues were then immersed in 70% ethanol, to inhibit β-galactosidase activity. Mammary glands were then dehydrated through ethanol gradients (70%, 95% and 100%, 1 h each), cleared in xylene, flattened on microscopy glass slides and mounted with permount. Alternatively, x-gal-labelled glands that had been incubated in 70% ethanol for 1 h, were stained in carmine alum overnight, dehydrated, cleared and mounted as described above. All incubations were done on a rocking platform. The volume of fixative, washing buffer and ethanol gradients was at least 10 volumes of the mammary gland (for example, 50 ml per 4 non-lactating mammary glands). For lactating glands, the washing step was extended to 6 h and the washing buffer was changed twice.
Histology and immunofluorescence labelling
Dissected mammary glands from induced N2-CreERT2SAT/R26RTom and control N2-CreERT2SAT females were embedded in OCT (VWR) without fixation, snap-frozen and stored at −80 °C. Thin (12 μm) or thick (20 μm) frozen sections were cut using a HM560 Microm cryostat (Mikron Instruments). Slides were air-dried for 5 min at room temperature and tissue sections were circled with a hydrophobic pen. Sections were then fixed for 10 min, at room temperature, in different fixatives depending on the primary antibody. After rinsing slides three times in TPBS (0.1% Triton X-100 in PBS) for 5 min, sections were incubated in blocking buffer (5% goat serum, 0.1% Triton X-100 in PBS) for 1 h at room temperature. The different primary antibody combinations were incubated for 1 h at room temperature or overnight at 4 °C. Sections were then rinsed three times for 5 min in TPBS and incubated with secondary antibodies diluted at 1:400 in blocking buffer for 1 h at room temperature. The following primary antibodies were used: anti-K8 (TROMA-1, Developmental Studies Hybridoma Bank; 1:150), anti-K14 (Covance, PRB-155P; 1:1,000), anti-SMA (Sigma, A5228; 1:300), anti-Muc1 (Abcam, ab15481; 1:200) and cleaved caspase3 (Cell Signaling, no. 9661; 1:400). The following secondary antibodies were used: anti-rabbit, anti-rat and anti-mouse conjugated to AlexaFluor 488 or AlexaFluor 647 (Molecular Probes; dilution 1:400). Nuclei were stained with DAPI and the slides were mounted in Vectashield mounting medium (Vector Laboratories).
Microscope image acquisition
X-gal-labelled whole-mount mammary specimens were imaged on a Zeiss stereo microscope equipped with an Achromat 5 lens and an AxioCam digital colour camera using the AxioVison software; or a Nikon 80i upright microscope with a Nikon Digital Sight DS-Fi1 colour camera using the NIS-Elements acquisition software. Immunofluorescence micrographs were acquired on a Nikon Ti w/A1R confocal inverted microscope with the Perfect Focus System using Nikon Elements acquisition software. Raw images for each channel were merged and adjusted for brightness and contrast using the ImageJ software.
Mammary cell preparation, flow cytometry and transplantation assays
Bi-genic N2-CreERT2SAT/R26LacZ females were induced with 4-OHT at 7 weeks of age and euthanized after 24 h. Mammary glands were dissected and single-cell suspensions were prepared as described previously1. Cells were then labelled with the β-gal substrate FDG (Sigma, F2756) according to the manufacturer’s protocol and incubated with antibodies as previously described1. The following antibodies were used: AlexaFluor 647 anti-mouse CD29 (Biolegend, 102214), APC-Cy7-conjugated streptavidin (eBioscience, 17-4317-82), PE-conjugated anti-mouse CD24 (eBioscience, 12-0241-82), biotin-conjugated anti-mouse CD61 (eBioscience, 13-0611-82), PE-Cy7-conjugated anti-mouse TER119 (eBioscience, 25-5921-81), PE-Cy7-conjugated anti-mouse CD31 (eBioscience, 25-0311-81) and PE-Cy7-conjugated anti-mouse CD45 (eBioscience, 25-0451-81). DAPI was added to exclude non-viable cells. FACS analysis was carried out on a FACS Aria (Becton Dickinson). Three independent experiments were performed, each included glands pooled from three mice. Data analysis was performed using the FlowJo software.
For transplantation assays, bi-genic N2-CreERT2SAT/R26LacZ females were induced with 4-OHT at 7 weeks of age and euthanized after 24 h. Mammary single-cell suspensions were labelled with the β-gal substrate FDG and the following antibodies against endothelial and haematopoietic antigens (Lin+ population): PE-Cy7-conjugated anti-mouse TER119 (eBioscience, 25-5921-81), PE-Cy7-conjugated anti-mouse CD31 (eBioscience, 25-0311-81) and PE-Cy7-conjugated anti-mouse CD45 (eBioscience, 25-0451-81). Dead cells were excluded with DAPI. To obtain a sufficient number of Lin− FDG+ cells, glands were pooled from 6 and 16 mice per experiment in two independent experiments. Viable Lin− FDG+, Lin− FDG− and control Lin− populations were sorted using a FACS Aria (Becton Dickinson) and transplanted into cleared mammary fat pads of 20-day-old syngeneic virgin females as previously described1. Recipient virgin females were euthanized 9 weeks post-transplantation. One Lin− FDG− recipient was mated and euthanized one day post-partum. All recipient glands containing Lin− FDG+ and Lin− FDG− populations as well as the three control recipient glands containing Lin− cells were labelled with x-gal. An outgrowth was defined as an epithelial structure comprising ducts and terminal end buds.
For in vitro analysis of sorted cells, sorted FDG+ MECs were plated in chamber slides in growth media for 2 h to allow attachment, then rinsed with PBS and immediately fixed in 4% PFA. Immunofluorescence labelling with lineage-specific markers was done as described above.
For cell size analysis, z-stack sections (0.1 μm, ×100) of individual cells were taken using a confocal microscope. DIC and DAPI channels were used to visualize whole-cell and nuclear area, respectively. For each cell, the z-section with the largest cell diameter was used to calculate size. The following parameters were measured and analysed using Metamorph: nuclear diameter, cytoplasm (whole cell) diameter and cytoplasm/nuclear ratio (whole-cell area divided by nuclear area).
Conditional in vivo ablation of Notch2+ lineages
The diphtheria toxin dosing schedule was optimized using x-gal-labelled mammary gland whole mounts from tri-genic N2-CreERT2SAT/R26RLacZ/R26RiDTR females. Experimental bi-genic N2-CreERT2SAT/R26RiDTR and control R26RiDTR females were induced with a single i.p. injection of 4-hydroxytamoxifen (4-OHT, Sigma; 50 mg kg−1 mouse body weight) at 6 or 7 weeks of age followed by two injections of diphtheria toxin (Sigma, PRB-155P; 50 μg kg−1 mouse weight) 48 and 96 h after 4-OHT induction. Carmine-alum-stained mammary glands were analysed 1 year post diphtheria toxin administration in virgin females (n = 4) or one day post-partum in lactating females (n = 3).
Supplementary Material
Acknowledgments
The authors would like to thank J. S. Brugge for her long-standing and very generous technical and theoretical advice throughout this project as well as for critical reading of the manuscript. We thank A. Shiang-Ru Kaanta for technical advice and critical reading of the manuscript and also A. Louvi for critical reading of the manuscript. In addition, we would also like to thank the Nikon Imaging Center at Harvard Medical School for help with light microscopy and the Animal Facility at Harvard Center for Comparative Medicine for helping in the maintenance and care of the transgenic animals. This work was supported by grants from the NIH to S.A-T.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
S.S. conceptualized, designed and performed all experiments and data analysis; D.L. contributed to transplantation experiments; S.A-T. conceived the study. S.S. and S.A-T. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at www.nature.com/reprints
References
- 1.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 2.Asselin-Labat ML, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 3.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 4.Stingl J, Raouf A, Eirew P, Eaves CJ. Deciphering the mammary epithelial cell hierarchy. Cell Cycle. 2006;5:1519–1522. doi: 10.4161/cc.5.14.2983. [DOI] [PubMed] [Google Scholar]
- 5.Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villadsen R, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visvader JE, Lindeman GJ. The unmasking of novel unipotent stem cells in the mammary gland. EMBO J. 2011;30:4858–4859. doi: 10.1038/emboj.2011.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 9.Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- 10.Politi K, et al. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004;5:341–7. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- 12.Bouras T, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Buono KD, et al. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 14.Raouf A, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Yalcin-Ozuysal O, et al. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 2010;17:1600–1612. doi: 10.1038/cdd.2010.37. [DOI] [PubMed] [Google Scholar]
- 16.Raafat A, et al. Expression of Notch receptors, ligands, and target genes during development of the mouse mammary gland. J Cell Physiol. 2011;226:1940–1952. doi: 10.1002/jcp.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill CF, et al. Notch2 signaling induces apoptosis and inhibits human MDA-MB-231 xenograft growth. Am J Pathol. 2007;171:1023–1036. doi: 10.2353/ajpath.2007.061029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mole Med. 2004;14:779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 19.Florena AM, et al. Associations between Notch-2, Akt-1 and HER2/neu expression in invasive human breast cancer: a tissue microarray immunophenotypic analysis on 98 patients. Pathobiol: J Immunopathol Mol Cell Biol. 2007;74:317–322. doi: 10.1159/000110024. [DOI] [PubMed] [Google Scholar]
- 20.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Gene. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu YP, et al. NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. Mol Cancer. 2010;9:113. doi: 10.1186/1476-4598-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson DR, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fre S, et al. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PloS One. 2011;6:e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Gene. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 25.Saito M, et al. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol. 2001;19:746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- 26.Chen LH, Bissell MJ. A novel regulatory mechanism for whey acidic protein gene expression. Cell Regul. 1989;1:45–54. doi: 10.1091/mbc.1.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller PJ, Arendt LM, Kuperwasser C. Stem cell maintenance of the mammary gland: it takes two. Cell Stem Cell. 2011;9:496–497. doi: 10.1016/j.stem.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Gene Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simian M, et al. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiseman BS, et al. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J Cell Biol. 2003;162:1123–1133. doi: 10.1083/jcb.200302090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 32.Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sternlicht MD, Kouros-Mehr H, Lu P, Werb Z. Hormonal and local control of mammary branching morphogenesis. Diff; Res Biol Diversity. 2006;74:365–381. doi: 10.1111/j.1432-0436.2006.00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner KU, et al. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129:1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 35.Oakes SR, et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Gene Dev. 2008;22:581–586. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliver CH, Khaled WT, Frend H, Nichols J, Watson CJ. The Stat6-regulated KRAB domain zinc finger protein Zfp157 regulates the balance of lineages in mammary glands and compensates for loss of Gata-3. Gene Dev. 2012;26:1086–1097. doi: 10.1101/gad.184051.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith GH, Chepko G. Mammary epithelial stem cells. Microsc Res Techn. 2001;52:190–203. doi: 10.1002/1097-0029(20010115)52:2<190::AID-JEMT1005>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.