Abstract
Notch signalling links the fate of one cell to that of an immediate neighbour and consequently controls differentiation, proliferation and apoptotic events in multiple metazoan tissues. Perturbations in this pathway activity have been linked to several human genetic disorders and cancers. Recent genome-scale studies in Drosophila melanogaster have revealed an extraordinarily complex network of genes that can affect Notch activity. This highly interconnected network contrasts our traditional view of the Notch pathway as a simple linear sequence of events. Although we now have an unprecedented insight into the way in which such a fundamental signalling mechanism is controlled by the genome, we are faced with serious challenges in analysing the underlying molecular mechanisms of Notch signal control.
Notch signalling has been studied for nearly a century, but recent genome-wide and proteome-wide investigations in model organisms have provided a surge in Notch signalling information. Characterization of Notch signalling started with the discovery of the Notch gene in Drosophila melanogaster as a sex-linked mutation displaying a serrated wing margin phenotype1–3. Decades of detailed genetic research revealed a locus with notoriously complex allelic interactions4, which led to speculations regarding its biochemical nature5,6.
Cloning the Notch locus revealed that the Notch gene encoded a transmembrane surface receptor7, unveiling the existence of a cell interaction mechanism that relies on interactions between the Notch receptor expressed on the surface of one cell and membrane-bound ligands expressed on the surface of its neighbours. Detailed molecular genetic analyses have defined the central components and the canonical transduction of the signal. The canonical model requires the activation of the Notch receptor through a series of proteolytic events on binding to any of the Delta–Serrate–LAG2 (DSL) ligands. The crucial cleavage event for signalling depends on γ-secretase-mediated release of the Notch intracellular domain (NICD) from the cell membrane. This allows the translocation of NICD into the nucleus, where it directly participates in a core transcriptional complex with the DNA-binding protein Suppressor of Hairless (SU(H)) and the nuclear effector Mastermind (MAM), thereby activating transcription of target genes8,9 (Fig. 1; Supplementary information S1 (movie)). Investigations into the molecular signalling mechanisms have been accompanied by an explosion of studies dissecting the myriad roles of Notch signalling in all aspects of animal development.
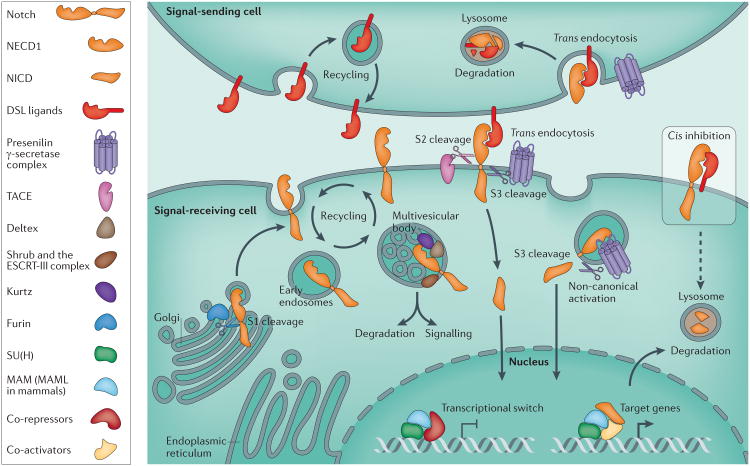
Figure 1. Summary of the main features of Notch signalling.
Three proteolytic cleavage steps are required for canonical Notch receptor signalling. The first proteolytic cleavage step (S1 cleavage) is mediated by Furin, occurs in the trans-Golgi and produces a heterodimer composed of a ligand-binding Notch extracellular domain (NECD) and a single-pass transmembrane signalling domain referred to as the Notch intracellular domain (NICD). The functional importance of this cleavage is still somewhat unclear130,131. The association of NECD and the transmembrane portion of the receptor heterodimer is dependent on non-covalent interactions. Pathway activation occurs when the NECD binds to Delta– Serrate–LAG2 (DSL) ligands that are expressed on the membrane of neighbouring cells. This trans interaction results in the second proteolytic event (S2 cleavage) of the Notch receptor, which clears most of the NECD from the outer portion of the membrane, a process mediated by the TACE (also known as ADAM17) metalloproteinase. The NECD is subsequently released and internalized through endocytosis by the ligand-expressing cell, where it undergoes lysosomal degradation. Subsequently, γ-secretase cleaves the tethered receptor near the inner leaflet of the membrane (S3 cleavage) in the Notch-expressing cell, producing the transcriptionally active NICD, which translocates to the nucleus through a poorly understood process. In the nucleus, the NICD interacts with Drosophila melanogaster Suppressor of Hairless (SU(H)) and the transcriptional co-activator, Mastermind (MAM) — the mammalian orthologues of which are CBF1–SU(H)–LAG1 (CSL) and Mastermind-like (MAML) proteins, respectively — thereby inducing transcription of target genes, by converting CSL into a transcription activator through the exchange of co-repressors for co-activators. Many Notch target genes encode transcriptional regulators, which influence cell-fate decisions through the regulation of basic helix–loop–helix hairy and enhancer of split (HES) proteins: Hairy and Enhancer of split (E(SPL)) in D. melanogaster and their mammalian orthologues HES1 and HES5. The HES proteins subsequently regulate the expression of genes involved in Notch-dependent cell-fate determination, such as apoptosis, proliferation or differentiation. By contrast, expression of ligands and the Notch receptor on the same cells results in cis inhibition of Notch signals and receptor degradation. Recycling the receptor through the endocytic pathway has been shown to be important for receptor and ligand maturation, non-canonical signalling and degradation (Box 2). Notch activity is regulated by ubiquitylation of nuclear NICD by the E3 ubiquitin ligases, SEL-10 in Caenorhabditis elegans and Suppressor of Deltex (SU(DX)) in D. melanogaster, leading to NICD degradation, thus allowing the cell to become ligand-competent once again. The ubiquitylation status of the receptor (by Kurtz, Deltex and Shrub) in the multi-vesicular bodies can also determine whether Notch continues to signal or undergoes proteasomal degradation. Additionally, signal attenuation is achieved through lysosomal degradation of the NICD.
Invertebrate and vertebrate studies indicate that the action of the pathway is highly pleiotropic, as it has a fundamental role in cell-fate determination in almost all developing tissues and organs10,11. The specific context of Notch activation dictates the particular process that is triggered: differentiation, proliferation or apoptotic events. Notch activity functions to drive multiple aspects of metazoan development and has recently been linked to stem cell fate and maintenance in adult tissues12,13. Such developmental pleiotropy is nevertheless based on a rather simple but pervasive developmental logic. Specifically, the acquisition of a fate by one cell affects the fate of its immediate cellular neighbour through signal transduction, in which, as we have argued previously, Notch signals are used to fine-tune differentiation in cellular fields during morphogenesis14. The classic loss of the Notch function ‘neurogenic’ phenotype in D. melanogaster embryos results from the failure of a field of cells properly to segregate two cell lineages that ultimately give rise to the cuticle (dermis) and the nervous system. This segregation depends on Notch receptor signalling in neuroblasts, thereby restricting the immediately neighbouring cells to adopt an epidermal fate.
Consistent with a role for Notch signalling in many aspects of differentiation and cell-fate determination, there is growing evidence that links perturbations in the pathway to various mammalian disease states, including several inherited syndromes and cancers11,15–17. Indeed, certain leukaemias have been causally associated with Notch, whereas Notch action has been implicated, directly or indirectly, with nearly all major solid tumours18. Thus, over the past few years, the emerging data justify the pursuit of the Notch pathway as a viable therapeutic target.
Here, we do not review the intricacies of the signalling mechanism, which have been discussed recently in excellent reviews10,11. Instead, we refer to some central mechanistic aspects of signal transduction, and we focus on regulatory mechanisms that are unique to Notch signalling, such as Notch dosage sensitivity and cis–trans interactions. In addition, we discuss several recent genome-wide studies, mainly from D. melanogaster, which indicate a vast Notch signalling network and extensive integration and crosstalk with other signalling pathways. Finally, we discuss the systems-level and proteomics approaches that can provide functional insights into this complex network. Given the high degree of conservation of the signalling pathways, we expect that insights from D. melanogaster will be directly applicable to all Metazoa and, importantly, to humans. Predicting all developmental outcomes of Notch function thus remains challenging because of the network's size and complexity. Nevertheless, understanding the biology and pathobiology of Notch signals rests on our ability comprehensively to uncover and to define the range of processes influencing Notch activity.
Multiple levels of signalling control
Gene dosage sensitivity
As we are considering the complexity of the network circuitry that modulates Notch signals, a few intricacies of the pathway are noteworthy. First is the exceptional sensitivity of Notch to gene dosage. Both Notch and Delta display haploinsufficiency, a property that is uncommon among genes in diploid organisms. More strikingly perhaps, Notch and the RNA helicase Triplolethal19 — a gene that is unrelated to the Notch pathway — are the only two genes in the D. melanogaster genome that are triplomutant. Females carrying a duplication of the locus — harbouring three, rather than two, normal copies of Notch — display a mutant wing vein phenotype4. Thus, the animal appears to ‘count’ Notch gene dosage, which is presumably associated with the intensity of the signal, such that either too much or too little signalling will result in altered function. This dosage sensitivity is not limited to D. melanogaster, as aberrant dosage of components of the Notch pathway in mammals (Box 1) also results in abnormal effects17,20, as demonstrated by several inherited human syndromes. For example, haploinsufficiency of either Jagged 1 (JAG1, a mammalian homologue of D. melanogaster Serrate) or NOTCH2 is associated with Alagille's syndrome21,22, whereas NOTCH1 haploinsufficiency is implicated in a subtype of inherited aortic disease23. Notch amplification and overexpression has been shown to be neoplastic, as gene amplification is one of the common mechanisms that activate oncogenes. For example, NOTCH3 copy number and therefore protein expression was positively correlated in 66% of ovarian serous carcinomas24 and NOTCH2 gain-of-function mutations and copy number increases are implicated in diffuse large-B-cell lymphomas25. Several studies have shown that cancer-related factors are differentially and crucially affected by low or high dosage of Notch signals20,26. Unlike other major signalling pathways, which rely on enzymatically amplifying the amount of signal, Notch signalling lacks such a step and instead relies on stoichiometric interactions between elements of the pathway. The lack of an enzymatic amplification step is a possible reason for the exquisite sensitivity of Notch signalling to gene dosage compared with other signalling pathways4. A small stoichiometric difference in receptor and ligand expression on the membrane becomes an important factor in restricting signalling to one of the two interacting cells. Studies in flies and worms had indicated that positive and negative transcriptional feedback mechanisms could amplify small differences in Notch and DSL ligand expression, causing directional changes in ligand or receptor expression in different cells27,28. However, this transcriptional feedback mechanism alone was insufficient to explain the robust signalling fidelity or observations in which Delta expression was found to be uniform among cells undergoing lateral inhibition during selection of the neural fate29,30. Therefore, mechanisms in addition to transcription feedback must exist to ensure fidelity in cell-fate decisions that are regulated by Notch signalling.
Box 1. Mammalian Notch signalling.
The core elements of the Notch signalling system in mammals include: the Notch receptors, the Delta-like (DLL) and Jagged (JAG) ligands (JAG ligands are the vertebrate orthologues of Drosophila melanogaster Serrate)) and CBF1-SU(H)-LAG1 (CSL) DNA-binding proteins (such as CBF1; also known as RBPJκ). Notch signalling is more complex in mammals owing to the presence of several paralogues and multiple regulatory components8. Four Notch paralogues, NOTCH1-4, and five Notch ligands (namely, JAG1 JAG2, DLL1, DLL3 and DLL4) have been identified in vertebrates9. Vertebrate Notch paralogues have different protein sequences, overlapping (yet individual) expression profiles and developmental functions and probably have interchangeable biochemical functions915. Different paralogues have been implicated in various diseases and individual paralogues may have opposite effects; for example, NOTCH1 and NOTCH2 have opposite effects on embryonic brain tumour growth114. Given that multiple paralogues exist in vertebrates for receptors, ligands and several of the pathway effectors, we expect Notch signalling to be profoundly more complex than what is observed in a relatively simple model organism such as D. melanogaster, which only encodes one receptor (Notch) and two ligands (Delta and Serrate).
Regulation in cis and in trans
The second noteworthy property of the pathway in the framework of our discussion is the recently recognized distinction between cis and trans interactions among the ligands and receptor31. Paradigmatic trans interactions occur between a ligand that is expressed on one cell and the receptor on a neighbouring cell, resulting in receptor activation. Although the presence of cis interactions (between the ligand and receptor in the same cell membrane) was inferred from genetic studies and experiments demonstrating colocalization of ligands and receptors on the same cell32, the importance and function of these interactions has remained enigmatic for some time. Recently, however, elegant studies33–35 have shown that this interaction is inhibitory, such that cis engagement of a surface-expressed receptor renders it refractory to trans-activating interactions (Supplementary information S2 (movie)). These ‘simple’ cis–trans relationships, the mechanisms of which remain to be fully elucidated, are crucial for the organism as the balance between them provides directionality to the signal. However, potential insights into the mechanism come from combining imaging approaches with mathematical models in mammalian cell culture systems33, which suggests that cis interaction generates a sensitive switch between mutually exclusive signalling states (a ‘sending’ state characterized by a high Delta/Notch ratio and a ‘receiving’ state characterized by a low Delta/Notch ratio). At the multicellular level, this switch can amplify small differences between neighbouring cells even without transcription-mediated feedback and thus can facilitate the formation of sharp boundaries and lateral inhibition patterns that are seen in development33. Therefore, the ratio of cis–trans interactions is an essential regulatory mechanism given that all of the cells within an equivalence group initially express both the receptor and the ligand. Hence, the decision of which of two neighbouring cells becomes the signal-sending versus the signal-receiving cell may be dictated by the dynamic competition between cis and trans interactions.
Additional regulatory mechanisms
By considering just the dosage sensitivity and cis–trans relationships of the Notch pathway, it can immediately be appreciated that any mechanism that influences the number and activity of Notch pathway components can potentially modulate the signal, hinting at the possible existence of a substantial and complex signalling circuitry. Indeed, factors affecting the biosynthesis, trafficking, degradation and many other molecular functions have been identified as modulators of the pathway36–39. In particular, post-translational modifications of the receptor or ligand, such as ubiquitylation, glycosylation and phosphorylation, have emerged as crucial steps in regulating the Notch signal40–43. Indeed, Notch ligands have been reported to be mono- and/or polyubiquitylated, but their functional consequences on Notch signalling are not well documented. Polyubiquitylation is usually associated with proteasome degradation, whereas both mono- and multi-mono-ubiquitylation can signal endocytosis of membrane proteins from the cell surface and can further influence intracellular trafficking44. It is also known that two structurally distinct RING-containing E3 ligases — namely, Neuralized (NEUR) and Mind bomb (MIB) — influence Notch signalling through interacting with and ubiquitylating DSL ligands to enhance their endocytosis45.
Some additional facts are worth taking into account in this context. Notch function is so prominent and ubiquitous throughout development that it is repeatedly required in multiple tissues. However, the specific cellular fate and/or developmental outcome, such as proliferation, differentiation or apoptosis, is invariably dependent on the context-specific integration of Notch signals with other signalling pathways (see below) and a growing list of cellular elements or tissue-specific cofactors. For example, Notch cooperates with the Twist transcription factor to activate target genes in myogenic progenitor cells, in which Twist appears to have the potential to confer specificity on Notch signalling at more than 100 gene enhancers46. Thus, a single cofactor can have a profound effect on the output of the Notch pathway.
Moreover, activation of the pathway can also be attained in a ‘non-canonical’, ligand-independent manner (Box 2; Supplementary information S3 (movie)), as the receptor traffics through endocytic compartments. Indeed, endocytosis has emerged as another important regulator of Notch signals, affecting its activity at multiple stages of the endocytic pathway (Box 2).
Box 2. Non-canonical Notch signalling.
DSL-independent Notch activation
Several lines of evidence indicate that the Notch pathway is regulated through several non-canonical mechanisms across species. Delta-Serrate-LAG2 (DSL)-independent activation of the pathway occurs by proteins — such as Delta-like 1 (DLK1), Delta- and Notch-like epidermal growth factor-related receptor (DNER) and Jedi — that resemble the Delta ligand but lack the DSL motif31,115. Compelling observations that are consistent with this notion indicate that two proteins (namely, contactin 1 (CNTN1; also known as F3) and CNTN6 (also known as NB3)) that are structurally unrelated to the DSL ligands interact with the epidermal growth factor (EGF) repeats of Notch and trigger activation through γ-secretase116,117, although these findings have not yet been confirmed by independent groups. Another ligand-independent and CBF1–SU(H)-LAG1 (CSL)-independent example includes the ability of the pathway to be activated in the Drosophila melanogaster wing imaginal disc by Deltex in late endosomes118. This mechanism does not require SU(H), which is consistent with observations of CSL-independent Notch activation during D. melanogaster muscle development119–121. Delta has been observed to colocalize with Notch in Notch-expressing cells and can cause the expression of Notch transcriptional targets; this indicates an independent mechanism by which Notch can be activated by Delta in the same cell118. This observation of Delta internalization contrasts the canonical signalling events in which the Notch extracellular domain (NECD) is internalized by Delta-expressing cells. Other groups have observed additional mechanisms of non-canonical activation that involves signal transduction without Notch cleavage; differential post-translational modifications; competition with or protection for a cofactor; and crosstalk with other signalling pathways (reviewed in REF. 115).
Vesicle-mediated transport
Vesicle-mediated transport has an important role in Notch activation mechanism in which the intracellular location of Notch is an important determinant of its activity. Mutations in elements of the endosomal-sorting machinery have been shown to be capable of triggering non-canonical signalling in the early endosome122–125 or late endosome118,126. Endocytosis of Notch can downregulate or activate signalling, and this depends on sorting Notch into different destinations45,127. A recent report indicates that Numb might regulate the endocytosis of Notch and prevent Notch from interacting with its ligand during sensory organ precursor cell development128. But there is little consensus regarding the location of the compartment that is involved in activation of Notch, or whether it is the early or late endosome or in the lysosome. It is also not clear whether there is just one universal mechanism of Notch activation or whether there are different mechanisms that can bring about activation at different cellular locations. It needs to be determined whether endocytosis has a role only in ligand-independent activation or whether it is also involved in the canonical ligand-induced Notch signalling and how this might relate to the cellular location in which Notch is activated.
Thus, considering just these properties of the Notch pathway, it may be predicted that the genetic circuitry involved in regulating Notch activity will be complex: a conclusion that is supported by the evidence elaborated below.
The genetics of Notch signal modulation
Classical genetic approaches have been exceptionally useful in defining elements of the Notch pathway by identifying modulators of Notch-related phenotypes. All core pathway components were uncovered using genetic screens in D. melanogaster and Caenorhabditis elegans, but such classical genetic approaches are limited in mammalian systems. However, molecular approaches such as genome-wide transcriptional profiling studies from mammalian cells and tissues, which we do not describe in detail here, also contribute to the roster of Notch interactors47,48. Several studies have shown a correlation between Notch signalling and many genetic disorders and cancers with a subset of syndromes resulting from specific mutations in elements the Notch pathway (Box 1).
Notch genetic interactors in D. melanogaster
The current availability of genome-wide molecularly characterized mutant collections in D. melanogaster49,50 has vastly improved the value of genetic analyses by providing a systems-level perspective that was not previously possible. These collections allow us to survey the genome systematically for loci that alter phenotypes resulting from mutations that affect any feature of the pathway, thereby identifying genes that are functionally relevant to Notch.
The first genome-wide systematic screen for Notch modifiers36 was feasible owing to the then recent availability of the Exelixis collection of D. melanogaster strains49, which harbours molecularly characterized transposon insertion mutations in ∼50% of the genes. A noteworthy characteristic of this collection is that it contains both gain- as well as loss-of-function mutations in approximately equal amounts. This screen was designed to uncover modifiers of a Notch loss-of-function wing phenotype that are associated with the inhibition of MAM activity. As MAM is a downstream element of the pathway, the screen captured modulators that affect targets in many parts of the pathway36, resulting in the recovery of 408 genes that affected Notch signals. In addition to recovering several known members of the pathway (thus validating the screen), more than half of the identified genes were not previously associated with Notch activity. A similar screen in the developing D. melanogaster eye identified modifiers of cell-fate alterations caused by overexpression of the Notch ligand Delta37. Of the 274 genes recovered, many defined novel Notch genetic interactors. These two screens interrogated different developmental contexts and identified 50 overlapping genes (Fig. 2a), which represents a tenfold increase in overlap compared to what would be expected at random. Thus, these two independent screens, which used the same mutant collection but distinct developmental phenotypic parameters, identified a diverse range of genes that modify Notch activity. These findings demonstrate that the modulation of the developmental outcome of Notch signals in diverse tissues and developmental events must be attained through distinct pathways or branches of the network.
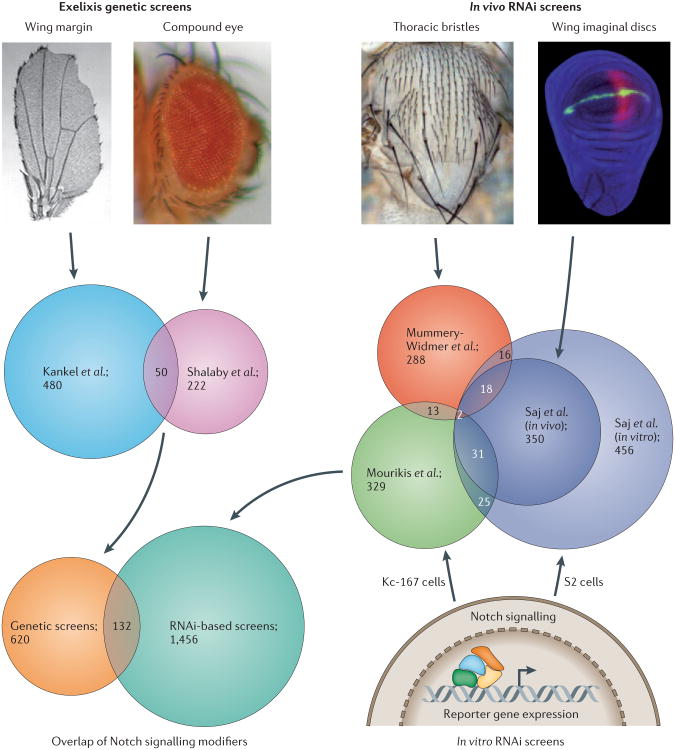
Figure 2. Overlap of Notch genetic modifiers from different genome-wide screens in Drosophila melanogaster.
The first screen to take advantage of the Exelixis collection of insertional mutations was carried out by Kankel et al.36, and the authors screened for modifiers of a Drosophila melanogaster wing margin phenotype that resulted from ectopic expression of dominant negative version of Mastermind (MAM). The second screen to use the Exelixis collection was carried out by Shalaby et al.37 and isolated modifiers of the cell-fate alterations in the D. melanogaster eye that are caused by overexpression of the Notch ligand Delta. Among the genes identified from these two independent genetic screens, 50 genes were observed in both studies. Genome-wide RNAi screens were carried out by several different groups using in vivo developmental phenotypes associated with the Notch pathway or in vitro cell-based Notch-signalling-dependent reporter assays. Mummery-Widmer et al.39 used external sensory organ development during thoracic bristle formation as a phenotypic parameter to isolate genes that affect Notch signalling, whereas Saj et al.38 carried out a genome-wide screen in S2 cells for regulators of Notch activity. Nearly half of the genes identified in the primary S2-cell-based screen were subsequently in vivo validated in developing wing imaginal discs. Mourikis et al.51 measured the transcriptional response of a luciferase-reporter (m3-luc) in Kc-167 cells to identify modifiers of Notch activity. However, the overlap of genes identified in all of the in vivo and in vitro RNAi screens is small (consisting of only two genes). From these screens, a total of 2,208 genes (14.25% of 15,494 genes based on FlyBase release 5.43) were found to affect Notch signalling (Supplementary information S4 (table)), 132 of which were identified in both Exelixis-based and RNAi-based screens. The ‘Wing margin’ image is reproduced, with permission, from REF. 36 © (2007) Genetics Society of America. The ‘Thoracic bristles’ image is reproduced, with permission, from REF. 132. The ‘Wing imaginal discs’ image is reproduced from REF. 38 © (2010) Cell Press.
Complementing the above studies, independent RNAi screens38,39 involving inducible short hairpin RNA (shRNA) constructs covering nearly 88% of the D. melanogaster genome, the Vienna Drosophila RNAi Center (VDRC) and Fly Stocks of National Institute of Genetics (NIG-FLY)50 uncovered yet more genes that are involved in Notch signalling. The first screen for modifiers affecting external sensory organ development39 identified almost 340 modifiers, including two partially overlapping sets, one with 226 genes involved in asymmetric cell division and another with 233 genes involved in lateral inhibition. Another independent screen38 identified nearly 900 candidate Notch regulators in a cell-based genome-wide RNAi screen, approximately 400 of which were genetically validated in the developing wing and eye. A third, cell-based RNAi screen examined Notch pathway transcriptional response using a luciferase reporter (m3-luc) assay in Kc-167 cells51 and found 399 putative genes that were capable of promoting (189) or antagonizing (210) Notch signalling.
These independent genome-scale RNAi screens in different developmental contexts (both in vivo and in vitro) also provided largely non-overlapping sets of genes (Fig. 2b): a finding that is consistent with results obtained in the genetic screens. By comparing results from all of these novel exploratory studies, the size and complexity of the genetic circuitry that affects Notch activity is clearly much larger than would initially be anticipated (Fig. 2c) even if we presume some fraction of these genes to be false positives. These results also provide further evidence supporting the notion of the diversity of context-specific developmental output of Notch signals. Taken together, studies in D. melanogaster identified several hundred genes, with limited overlap, encompassing almost 14% of the genome (Supplementary information S4 (table)), which appear to participate in seemingly diverse molecular functions, revealing an extraordinary degree of complexity that includes the surprisingly large number of mechanisms and circuitries that this organism has evolved to regulate Notch activity.
Studies in D. melanogaster have also revealed a growing number of microRNAs (miRNAs) that are associated with Notch signalling. Their roles are yet to be fully elucidated, but these findings add to the complexity of Notch pathway regulation. The Notch pathway is a major target of miRNA-mediated regulation. The Enhancer of split complex (E(spl)-C) and the Bearded complex (Brd-C) family of genes, which are Notch target genes, are directly regulated by three different families of D. melanogaster miRNAs52. Moreover, the wing and bristle phenotypes that result from ectopic expression of these miRNAs resemble characteristic Notch loss-of-function. These include wing margin defects, thickened wing veins, increased bristle density and tufted bristles52. Several miRNAs modulate Notch signalling activity by targeting the Notch pathway transcripts and other miRNAs affect genes that alter the cellular response to Notch signals, whereas a few behave as nodes in crosstalk with other signalling pathways53–55. It is only a matter of time before large-scale screens will be designed to identify and systematically to characterize the miRNAs that can modulate Notch signalling.
Notch genetic interactors in C. elegans
Elegant genetic screens in C. elegans, in which signals are transmitted through the two Notch receptor paralogues LIN-12 and GLP-1, have contributed substantially to our understanding of the Notch genetic circuitry56. As was the case in D. melanogaster, these screens also identified several pathway components, including the canonical ligands, receptor cleavage proteins, members of the nuclear transcriptional activation complex or mediators of protein trafficking and degradation (a series of genes called sel, spr, sog, ego and teg)56. Importantly, the first evidence of a link between a Notch paralogue (LIN-12) and β-amyloid precursor protein (APP) transmembrane cleavage events came from the recovery of sel-12 as a Notch suppressor57. The gene sel-12 was shown to encode a C. elegans form of mammalian presenilin — the catalytic subunit of the γ-secretase protease complex58 — which was identified as a prominent locus in human early-onset familial Alzheimer's disease59. Genetic analysis in C. elegans established that SEL-12 activity is crucial for LIN-12 signalling60,61. Interestingly, functional Notch and APP proteins undergo a similar maturation process in which cleavage by γ-secretase occurs near the inner leaflet of the membrane62. Not surprisingly, the extent of functional categories that are associated with the vast number of genes identified as interactors63 also indicates that regulation of the Notch pathway in worms is highly complex.
Making sense of genetic screens
Together, these functional genomics approaches reveal an exceptionally complex genetic circuitry that is capable of affecting Notch activity such that we prefer to consider a Notch signalling ‘system’ rather than a ‘pathway’. Although genetic links clearly indicate biologically important functional relationships, the mechanisms underlying such links tend to be indirect rather than direct, and as a result they are quite challenging to dissect at a molecular level. Clearly, modifier screens that identify tens or hundreds of genes pose formidable challenges, given that it is impractical to examine every interaction pair in depth. Thus, secondary assays and other considerations must be applied to ‘filter’ and prioritize genes for detailed functional characterization.
As far as Notch is concerned, there are no general rules for such prioritization, but two important, although certainly not absolute, parameters have been used successfully. First, a gene that is to be classified as high priority for further investigations must interact genetically with various loss- and gain-of-function mutations in bona fide pathway components. These Notch pathway components include: the ligands (Delta or Serrate); the receptor (Notch); intracellular effectors such as deltex (a ubiquitin ligase) or kurtz (the non-visual β-arrestin in D. melanogaster), which together regulate Notch activity through receptor protein degradation64; or the nuclear factors SU(H) and MAM36. An unexpected outcome of such prioritization studies was the identification of two distinct groups of dominant negative MAM modifiers, with one group genetically interacting with multiple Notch pathway alleles, and another group only interacting with MAM36. Second, genes that are recovered as modifiers in more than one independent screen carried out in different cellular and/or developmental contexts are often of considerable interest. Such an approach is useful for further studies, as 50% of the already-known Notch pathway genes (as listed in the Kyoto Encyclopedia of Genes and Genomes (KEGG)) were identified in more than one independent screen (see Supplementary information S4 (table)). Of course, identification of a gene in only one screen does not rule out its importance to the pathway, as individual screens have different sensitivity levels.
Owing to the nature of the experimental designs, there are no accurate ways to estimate the rates of false-positive hits, which may contribute to the limited overlap between gene lists from different screens. Nevertheless, a large proportion of the D. melanogaster genome appears to be capable of affecting or modulating Notch activity — and by extension other signalling pathways — thus greatly extending the limited repertoire of genes associated with these pathways from traditional, more focused studies. As mentioned above, future studies to elucidate the molecular mechanisms between Notch and these genes will further delineate pathway dynamics.
Signalling crosstalk and nodes of integration
Notch signal integration with other pathways
The cell signalling framework in Metazoa is defined by a small number of conserved pathways that govern nearly all aspects of morphogenesis. These include the Notch, receptor tyrosine kinase (RTK)–RAS, WNT, Hedgehog, transforming growth factor-β (TGFβ) and Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signalling pathways, as well as a few others65. Their repeated use throughout development points to a profound biological problem: how can the same genetic framework drive such a diverse range of morphogenetic programs? Undoubtedly, part of the answer must be based on how these conserved signals integrate their action. Thus, exploring signal integration or crosstalk is of fundamental importance, as identifying points of integration may unveil rules that govern morphogenesis and, ultimately, pathogenesis. The means to study integration in a systematic way have only begun to emerge through the various genomic approaches that we now have at our disposal48,66–69. Transcriptome analyses, which have now been enhanced by chromatin immunoprecipitation followed by microarrays (ChIP–chip) or ChIP followed by high-throughput sequencing (ChIP–seq)48,70 methods, have been instrumental in improving our ability to identify both transcriptional targets of Notch signalling, as well as genes that direct Notch function. Relative to genetic approaches, ChIP-based methods have emerged as more efficient and cost-effective approaches for identifying nodes of signal integration, as they can easily identify genes for which transcriptional regulation is strictly dependent on the activity of two distinct signals and thus a target of signal integration. It seems likely that the activity of such genes may ultimately control a particular developmental outcome. These methods provide a useful list of candidate genes to validate by molecular genetics approaches in model organisms.
Notch signalling, consistent with its pleiotropic nature, can exhibit diametrically opposed action in distinct developmental contexts. For example, activation of mammalian Notch signalling is oncogenic in the haematopoietic system but tumour suppressive in the skin71. The underlying mechanisms of such context specificity are obscure yet are of obvious importance. The manner in which the Notch signal is integrated with the signals from other pathways is one potential way to affect Notch action and may provide context specificity. Previous work has shown that in different contexts, the targets of Notch signals include the Hedgehog, JAK– STAT, TGFβ and WNT pathway ligands, and reciprocally Notch ligands are targets of Hedgehog, RTK, TGFβ and WNT signalling72. Such reciprocity of crosstalk suggests that feedback loops represent important interlinking mechanisms that function to combine individual signals into an interconnected network. Moreover, in the past decade there has been an explosion of studies focusing on signal integration, which can occur at any point along the pathway72. Although the degree of crosstalk affecting multiple processes is beginning to be appreciated, the general themes or the precise mechanisms of all such interactions are yet to be determined.
Notch–RTK crosstalk
Although Notch signals integrate with all major signalling pathways (FIG. 3), the crosstalk between Notch and RTK signalling has been the subject of several studies. Often, Notch activation restricts cells to an uncommitted fate15,73–75, whereas RTK signalling promotes developmental commitment and acts to induce cells to follow specific differentiation programs76,77. The Notch–RTK crosstalk also serves as a paradigm to demonstrate the pervasiveness and importance of signal crosstalk and also underscores our limited knowledge of these integration mechanisms. The developmental consequences of Notch–RTK crosstalk are well shown by the cooperative, sequential and antagonistic relationships displayed between these pathways, which mediate the spatially and temporally regulated processes that generate the D. melanogaster eye78. For example, Notch and RTK pathway antagonism is observed in photoreceptor development in the D. melanogaster eye76,79 and specification in sensory organ precursor develop-ment80,81, whereas Notch and RTK signalling act cooperatively during accessory cell specification82,83. Similarly, in C. elegans, vulval development is also dependent on the interplay between Notch and RAS84.
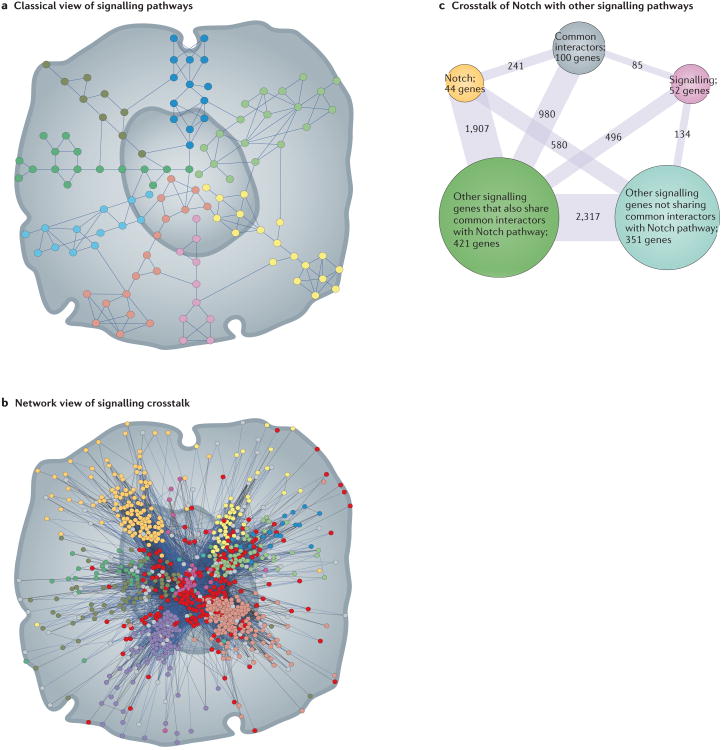
Figure 3. Summary of crosstalk between Notch and other signalling pathways.
a,b | A schematic representation depicting classical and current views of signalling pathways and how these pathways are thought to integrate their signals (crosstalk) within a cell. Panel a portrays signal transduction pathways as quite distinct sets of systems or cascades that transmit information by a step-by-step or linear mechanism with minimal crosstalk. Proteins are shown as circles and their physical interactions as blue lines. Pathways are distinguished by different coloured circles. Panel b displays a more representative and current perspective of signalling crosstalk with different signalling pathways as complex, highly interconnected networks with several shared members (red circles). The network was generated using human signal transduction pathway members from KEGG database133 and queried in GeneMania134 for genetic and physical interactions with the Notch pathway, as reported in the literature. Network visualization was created using Cytoscape135. c| A summary version of the network in panel b, highlighting the fact that Notch pathway genes (yellow circle) interact genetically or physically with most genes that are associated with other signalling pathways. The number of genes in each category is listed in its corresponding coloured circle, and the number shown on the blue line indicates the number of genetic and physical interactions between categories. Only a small subset (52 genes, pink circle) appears to connect to the Notch pathway indirectly. The common interactors (grey circle) are 100 genes that are not classified as signalling genes by KEGG or GeneMania and may represent potential points of signal integration or crosstalk between Notch and other canonical signalling pathways.
Microarray-based transcriptome analyses66,68,72,85 also indicate a profound interrelationship between the two pathways. This is exemplified by the surprising finding that 65% of transcriptional targets of RTK signalling in D. melanogaster are also targets of Notch signalling, whereas the effects on common RTK and Notch transcriptional targets are equally distributed between synergistic (acting in the same direction) and antagonistic (having opposing effects). Furthermore, distinct groups of genes have been identified that respond to both pathways either simultaneously or sequentially, acting as nodes to integrate their effects on development66. Transcriptional profiling approaches also provide a tool that allows us to examine the consequences of different exposures to Notch activation, such as short-term versus long-term activation, in a particular cell or experimental system and also provide evaluation of the quantitative effects of the signal. Such approaches have been instrumental in vertebrate systems48, in which disruptions to Notch signalling can result in clinically relevant pathologies, but in which genetic approaches are far more limited.
Conservation of nodes of crosstalk
An understanding of how Notch signals integrate their activity with other signals to affect specific genes, which in turn influence developmental outcomes and associated pathologies, is of great importance. Identifying nodes of integration will allow the characterization of whether they integrate the same signals in the same way in different tissue types and whether these relationships are observed in more than one species. Phylogenetic conservation of such nodes may imply the existence of a more universal signal integration logic and network architecture, whereas absence of conservation may indicate recently evolved tissue-specific and organism-specific modes of integration. What has emerged is the evidence for Notch crosstalk with several conserved pathways, such as p53 in apoptosis and cancer86,87, Hedgehog, WNT and growth factors in breast cancer88, TGFβ in tumour cell invasion89, Hedgehog in pancreatic cancer90, vascular endothelial growth factor (VEGF) in tumour angiogenesis91 and vascular morphogenesis92, androgen-dependent signals in prostate cancer93 and, finally, WNT in intestinal epithelia94. Investigating these relationships requires multifaceted approaches and sophisticated molecular genetic tools, which are more amenable in genetically tractable model organisms than in mammalian systems.
Systems-level and proteomics approaches
The data derived from genome-wide studies clearly indicate the confounding complexity of the genetic circuitry that affects Notch signalling. A vast number of genes that affect Notch signalling have been identified by these analyses, but the molecular mechanisms that underpin these relationships are difficult to assess and are largely uncharacterized. However, large-scale proteomics studies, such as yeast two-hybrid screens95–101 and protein complex analyses102–104 are increasingly accessible owing to the development of mass spectrometry techniques and advanced bioinformatics tools105,106 (Box 3) and therefore provide the possibility of general insights. Thus, it is likely that examining how genes that interact genetically with Notch are interconnected within the physical protein–protein interaction network will allow us to formulate specific molecular hypotheses that may explain underlying molecular principles of the relationships that have been revealed by genetics. Such analyses have been carried out for individual screens38,39 by incorporating different functional information available from the literature to establish connections and also to predict functional categories and modules among the genetic modifiers. The network analyses and integration of such maps have been pioneered in yeast, in which the proteomic and synthetic genetic interaction landscapes are well defined107–111. By comparison, genetic interactions at a genome-wide level are yet to be established in D. melanogaster, with the first large-scale synthetic genetic interaction analysis by RNAi being used recently to map the RAS–MAPK signalling networks in D. melanogaster112.
Box 3. Databases and bioinformatic tools for systems-level studies.
Although the fundamental importance of complex signalling networks is well recognized, until recently most studies focused on just individual components, as the tools were not available to address the Notch pathway as a system. The recent genomics and proteomics approaches have provided a wealth of new information. Advances in bioinformatics approaches will allow for more sophisticated analyses of such enormous data sets and provide yet more insights into the complexity of signalling networks. Most of the current databases (see Further information) are good repositories of large-scale data sets; however, the overall quality and analytical methods originally used to define those original data sets are highly variable. Currently, such databases are also not well adapted to track and update all of the genetic, physical and biochemical interactions that are defined in the thousands of individual, detailed, small-scale studies. Integrating such diverse data sets (and removing redundancies) is necessary to obtain a comprehensive understanding of how signalling pathways integrate their signals with other pathways. Commercial data-mining and integrated knowledge-base software suites such as Ingenuity Pathway Analysis and MetaCore adequately address this need, but they are not widely accessible. The existence of open-source, open-access, manually curated, peer-reviewed databases of pathways and processes, such as Reactome129, provide a reasonable alternative.
The availability of high-quality paradigmatic protein complex maps, such as the recently published Drosophila Protein Interaction Map (DPiM)113, allows us to connect modifiers that have been identified from different screens into one unified network that represents a static baseline of the proteome. A key attribute of the DPiM, which is the first comprehensive metazoan proteome map, is that it defined 556 protein complexes, most of which show a high degree of interconnectivity113. Several lines of evidence, predominantly accumulated by genetic analyses, support the notion that most of the protein complexes are functionally interconnected. Although mechanistic characterization of such connections is challenging, the existing map offers the opportunity to examine whether the disruption of a complex can affect its network neighbourhood and hence its functionality. The importance of complex interconnectedness is in our view profound, as it will allow us to explore protein interaction dynamics at a system-wide level in various contexts, such as different genetic backgrounds, physiological conditions or metabolic states.
Placing genetic and other functional interactions affecting Notch activity onto a physical interaction map presents an unprecedented and unified view of the network of Notch-associated genes and how they relate in the context of all protein interactions within a cell (Fig. 4). Additionally, such a map allows us to observe potential physical interactions between the products of genes that are known to interact with Notch. Moreover, a genetic modifier within the map that is associated with other proteins that were previously unlinked to Notch raises the possibility of functional links between these previously unlinked proteins and Notch signalling. These analyses provide potential functional context to hundreds of modifiers identified in different genetic studies while generating numerous testable hypotheses. Although discussion of individual relationships as determined by this map is beyond the scope of this Review, it is important to note that this approach raises new questions and allows us to design experiments that can address some — in our opinion — insightful questions regarding the Notch signalling system.
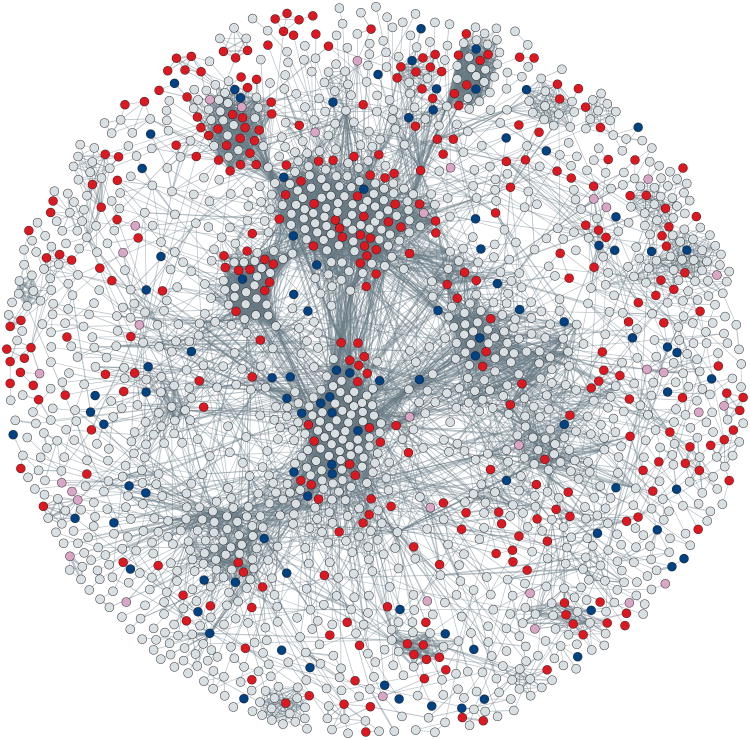
Figure 4. Mapping genetic modifiers on proteome map.
The highly interconnected major component of the Drosophila Protein Interaction Map (DPiM)113, showing Notch signalling modifiers that were identified from Exelixis screens (blue), RNAi screens (red) or both (pink). This overlay of genetic data on a proteomics map helps to combine independent and seemingly disparate data sets into one integrated network while providing potential mechanistic insights into the basic biochemistry of these interactions. Grey lines indicate protein–protein interactions, the thickness being proportional to the interaction score. In many cases, it is clear that several members of a protein complex were identified in independent screens, suggesting that the complex, as a functional unit, is important for Notch signalling. Such integrative analyses provide numerous experimentally testable hypotheses into the links between Notch signalling and these modifiers, as well as their associated functions, as defined in DPiM. Within DPiM, connections between uncharacterized proteins to protein complexes with defined Gene Ontological terms provide a potential function to the uncharacterized proteins. A high-resolution version of this figure is available in Supplementary information S5 (figure).
Initially, we can ask whether first- and second-degree physical interaction neighbours of any genetic modifier are indicative of functionally relevant relationships between these neighbours, beyond merely protein–protein binding. If several members of a particular complex are identified in genetic screens, do other complex members that are not identified by the screens behave in a functionally similar manner? Do clusters that are enriched for Notch pathway modifiers represent protein complexes that are functionally relevant for Notch signalling? If a particular protein complex is determined to be important, how does complex disruption — for example, as achieved through RNAi for select members — affect Notch signalling?
We can also determine whether the observed relationships in D. melanogaster are valid across species by performing a series of analogous proteomics experiments using the corresponding human orthologues in human cells, as was carried out for selected cases in the DPiM113, which indicated that most of the interactions were conserved across species. Importantly, we are now able to examine the molecular dynamics of protein interactions involving Notch-related genes and to probe relationships under conditions of aberrant Notch signals or by manipulating other pathways that integrate their activity with Notch. Such different cellular contexts can determine the dynamics of the interaction under different Notch signalling conditions. These studies can also be enhanced by using quantitative proteomics, which will provide a unique perspective, as different cellular signalling states may not reflect qualitative changes to the Notch signalling network but may instead reflect differences in the relative amounts of the particular complex component (or components). Such quantitative information is crucial to delineate the stochastic interactions that eventually determine the final signal output of cells. Such an integrative analysis will certainly provide insights into what appears to be a highly complex network of genes that can regulate Notch activity, and these insights are valuable owing to the important developmental and pathological roles of Notch signalling.
Conclusions and future perspectives
Since the discovery of the Notch locus in D. melanogaster around a century ago, Notch biology and genetics have at no point ceased to provide important insights while raising numerous difficult questions. The notoriously perplexing complementation analysis of the locus is now surpassed and replaced with the task of interpreting the complexity of the circuitry that functions to modulate Notch activity. However, as the genetics of the Notch locus was elucidated through molecular analyses, we are certain that the intricate circuitry that regulates the pleiotropic action of the signals through the Notch receptor will be deciphered through the combined use of large-scale genome-wide and proteome-wide approaches. Such multipronged approaches are necessary, as no single study has reached saturation in terms of identifying all of the possible modifiers of Notch signalling. Not only are the overlapping genes from different screens limited, the genetic screens that use the Exelixis collection interrogated ∼50% of the D. melanogaster genome. Nevertheless, although capturing its own ‘snapshot’ of the pathway in a specific cellular or developmental context, each approach also contains certain experimental limitations. Only integration of all of these studies, owing to their complementary nature, will provide a more complete understanding of Notch signalling.
Given the central role that Notch biology has in the development and homeostasis of organisms, understanding its function and how it relates to the normal cellular and organismal physiology is valuable, both in terms of fundamental biology as well as from a translational perspective. Because Notch signalling has an extensive role in normal development, including in adult stem cell function, and its disruption is associated with cancer, the pharmaceutical industry considers Notch to be a highly desirable therapeutic target. This is despite numerous challenges owing to the intricate and complex controls governing its activity that can no longer be addressed solely by examining pairwise interactions. Even small modulations of Notch signalling through pharmacological interventions would be expected to have profound effects on multiple cellular targets, and this is likely to be the case whether the intervention is targeted to the canonical Notch signalling machinery or to more peripheral molecules that indirectly interact with the Notch signalling system. The numerous genome-wide investigations that are focused on the Notch signalling pathway have revealed a vast number of genes and proteins that affect Notch developmental outputs. Moreover, these studies indicate that Notch should no longer be considered in isolation as a strictly linear pathway; rather, its effects are highly interconnected with other conserved signalling pathways. A comprehensive understanding of Notch during development and various disease states requires a more thorough understanding of the genes that can impinge on the Notch signalling system in specific contexts.
Supplementary Material
Acknowledgments
We would like to thank A. Louvi, D. Dimlich, K. Hori, B. Obar and A. Sen for critically reading the manuscript. Special thanks to J. Iwasa for the Notch pathway animations. We also apologize to our colleagues whose original work was not discussed or cited here in the interest of space. Work in the Artavanis-Tsakonas laboratory is supported by grants from the US National Institutes of Health.
Glossary
- Haploinsufficiency
A genetic condition in a diploid organism in which a single functional copy of a gene fails to generate sufficient gene product, leading to an abnormal or diseased state.
- Triplomutant
A genetic variant that carries three copies of a single gene, as opposed to the normal two copies, and displays a specific mutant phenotype that may include lethality or morphological defects. This mutant is distinct from aneuploidy, which alters copy numbers for large numbers of genes owing to chromosomal aberrations.
- Alagille's syndrome
An inherited autosomal dominant genetic disorder that can affect multiple vital organs, such as the liver, heart and other body parts. The disorder may also affect the blood vessels within the brain, spinal cord and the kidneys. The estimated prevalence of Alagille's syndrome is 1 in 70,000 newborns.
- Transcriptional feedback
A regulatory loop in which the gene products positively or negatively regulate the expression or activity of other members of the same pathway and therefore regulate themselves.
- Quantitative proteomics
Quantitative proteomics is identical to general (qualitative) proteomics but includes quantification as an additional dimension. Information about differences between two or more protein samples is obtained with the use of isotopes or mass tags that are distinguishable in mass spectrometry
- Equivalence group
A group of unspecified cells that have the same developmental potential to adopt various fates. Typically, these are cells from an equivalence group that receive a signal take on fates that are distinct from those cells that do not receive a signal and therefore adopt a default fate
Footnotes
Competing interests statement: The authors declare no competing financial interests.
Further Information: Spyros Artavanis-Tsakonas's homepage: https://artavanis-tsakonas.med.harvard.edu/index.html
BioGRID: http://thebiogrid.org
Drosophila Protein Interaction Map: https://interfly.med.harvard.edu
DroID: http://www.droidb.org
Exelixis: https://drosophila.med.harvard.edu
Fly Stocks of National Institute of Genetics (NIG-FLY): http://www.shigen.nig.ac.jp/fty/nigfty/index.jsp
Proteomics Identifications database (PRIDE): http://www.ebi.ac.uk/pride
MINT: http://mint.bio.uniroma2.it/mint/Welcome.do
modEncode: http://www.modencode.org
Pathguide: http://www.pathguide.org
Reactome: http://www.reactome.org/ReactomeGWT/entrypoint.html
Vienna Drosophila RNAi Center (VDRC): http://stockcenter.vdrc.at/control/main
Supplementary Information: See online article: S1 (movie) | S2 (movie) | S3, (movie) | S4 (table) | S5 (figure)
References
- 1.Dexter JS. The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am Nat. 1914;48:712–758. [Google Scholar]
- 2.Morgan TH, Bridges CB. Sex-Linked Inheritance in Drosophila. Carnegie Institute of Washington; 1916. [Google Scholar]
- 3.Mohr OL. Character changes caused by mutation of an entire region of a chromosome in Drosophila. Genetics. 1919;4:275–282. doi: 10.1093/genetics/4.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- 5.Thorig GE, Heinstra PW, Scharloo W. The action of the notch locus in Drosophila melanogaster. II Biochemical effects of recessive lethals on mitochondrial enzymes. Genetics. 1981;99:65–74. doi: 10.1093/genetics/99.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorig GE, Heinstra PW, Scharloo W. The action of the notch locus in Drosophila melanogaster. I Effects of the Notch8 deficiency on mitochondrial enzymes. Mol Gen Genet. 1981;182:31–38. doi: 10.1007/BF00422763. [DOI] [PubMed] [Google Scholar]
- 7.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. This was the original description of the molecular cloning and structure of the Notch receptor in D. melanogaster that revealed an epidermal growth factor (EGF)-repeat-containing transmembrane protein, suggesting a role for Notch in intracellular communication. [DOI] [PubMed] [Google Scholar]
- 8.Bray SJ. Notch signalling: a simple pathway becomes complex. Nature Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 9.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopan R, editor. Notch Signaling. 1st. Academic Press; 2010. [Google Scholar]
- 11.Fortini ME. Introduction—Notch in development and disease. Semin Cell Dev Biol. 2012;23:419–420. doi: 10.1016/j.semcdb.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Bigas A, Espinosa L. Hematopoietic stem cells: to be or Notch to be. Blood. 2012;119:3226–3235. doi: 10.1182/blood-2011-10-355826. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 14.Artavanis-Tsakonas S. The molecular biology of the Notch locus and the fine tuning of differentiation in Drosophila. Trends Genet. 1988;4:95–100. doi: 10.1016/0168-9525(88)90096-0. [DOI] [PubMed] [Google Scholar]
- 15.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 16.Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12(Suppl. 1):R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 17.Louvi A, Artavanis-Tsakonas S. Notch and disease: a growing field. Semin Cell Dev Biol. 2012;23:473–480. doi: 10.1016/j.semcdb.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nature Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 19.Dorer DR, Christensen AC. A recombinational hotspot at the triplo-lethal locus of Drosophila melanogaster. Genetics. 1989;122:397–401. doi: 10.1093/genetics/122.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazzone M, et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci USA. 2010;107:5012–5017. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oda T, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nature Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 22.McDaniell R, et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 24.Park JT, et al. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 25.Lee SY, et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 2009;100:920–926. doi: 10.1111/j.1349-7006.2009.01130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Greenwald I, Rubin GM. Making a difference: the role of cell–cell interactions in establishing separate identities for equivalent cells. Cell. 1992;68:271–281. doi: 10.1016/0092-8674(92)90470-w. [DOI] [PubMed] [Google Scholar]
- 28.Seugnet L, Simpson P, Haenlin M. Transcriptional regulation of Notch and Delta: requirement for neuroblast segregation in Drosophila. Development. 1997;124:2015–2025. doi: 10.1242/dev.124.10.2015. This study evaluated the role of transcriptional regulation during lateral inhibition within the proneural group and how one cell overcomes Notch-mediated repression. [DOI] [PubMed] [Google Scholar]
- 29.Kooh PJ, Fehon RG, Muskavitch MA. Implications of dynamic patterns of Delta and Notch expression for cellular interactions during Drosophila development. Development. 1993;117:493–507. doi: 10.1242/dev.117.2.493. [DOI] [PubMed] [Google Scholar]
- 30.Kopczynski CC, Muskavitch MA. Complex spatio-temporal accumulation of alternative transcripts from the neurogenic gene Delta during Drosophila embryogenesis. Development. 1989;107:623–636. doi: 10.1242/dev.107.3.623. [DOI] [PubMed] [Google Scholar]
- 31.D'Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fehon RG, et al. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. This paper was the first to show that the Notch and Delta proteins physically interact at the cell surface through their extracellular domains in a calcium-dependent manner. [DOI] [PubMed] [Google Scholar]
- 33.Sprinzak D, et al. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. This study showed that cis interaction of Notch– Delta generates an ultrasensitive switch between mutually exclusive (sending versus receiving) signalling states and results in the amplification of small differences in expression levels between neighbouring cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glittenberg M, Pitsouli C, Garvey C, Delidakis C, Bray S. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 2006;25:4697–4706. doi: 10.1038/sj.emboj.7601337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordle J, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nature Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kankel MW, et al. Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics. 2007;177:2493–2505. doi: 10.1534/genetics.107.080994. This was the first large-scale genetic screen for Notch pathway modifiers in D. melanogaster using the Exelixis mutant collection, more than doubling the number of genes known to interact with Notch, revealing that a highly complex network of genes and functionalities are involved in mediating Notch activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shalaby NA, et al. A screen for modifiers of notch signaling uncovers Amun, a protein with a critical role in sensory organ development. Genetics. 2009;182:1061–1076. doi: 10.1534/genetics.108.099986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saj A, et al. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive Notch interaction network. Dev Cell. 2010;18:862–876. doi: 10.1016/j.devcel.2010.03.013. A comprehensive, genome-wide cell-based RNAi screen dissecting Notch regulation and its connections to cellular pathways identified candidate Notch regulators. Many candidates were validated in vivo by using transgenic D. melanogaster RNAi strains. [DOI] [PubMed] [Google Scholar]
- 39.Mummery-Widmer JL, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. This was the first genome-wide RNAi screen for Notch pathway modifiers during external sensory organ development in D. melanogaster, uncovering hundreds of genes involved in lateral inhibition and asymmetric cell division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- 41.Stanley P, Okajima T. Roles of glycosylation in Notch signaling. Curr Top Dev Biol. 2010;92:131–164. doi: 10.1016/S0070-2153(10)92004-8. [DOI] [PubMed] [Google Scholar]
- 42.Le Bras S, Loyer N, Le Borgne R. The multiple facets of ubiquitination in the regulation of notch signaling pathway. Traffic. 2011;12:149–161. doi: 10.1111/j.1600-0854.2010.01126.x. [DOI] [PubMed] [Google Scholar]
- 43.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto S, Charng WL, Bellen HJ. Endocytosis and intracellular trafficking of Notch and its ligands. Curr Top Dev Biol. 2010;92:165–200. doi: 10.1016/S0070-2153(10)92005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernard F, Krejci A, Housden B, Adryan B, Bray SJ. Specificity of Notch pathway activation: twist controls the transcriptional output in adult muscle progenitors. Development. 2010;137:2633–2642. doi: 10.1242/dev.053181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Hibbs MA, Gard AL, Shylo NA, Yun K. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem Cells. 2012;30:741–752. doi: 10.1002/stem.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Artavanis-Tsakonas S. Accessing the Exelixis collection. Nature Genet. 2004;36:207. doi: 10.1038/ng1316. [DOI] [PubMed] [Google Scholar]
- 50.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 51.Mourikis P, Lake RJ, Firnhaber CB, DeDecker BS. Modifiers of notch transcriptional activity identified by genome-wide RNAi. BMC Dev Biol. 2010;10:107. doi: 10.1186/1471-213X-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, et al. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. 2010;292:141–148. doi: 10.1016/j.canlet.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nature Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 55.Ichimura A, Ruike Y, Terasawa K, Tsujimoto G. miRNAs and regulation of cell signaling. FEBS J. 2011;278:1610–1618. doi: 10.1111/j.1742-4658.2011.08087.x. [DOI] [PubMed] [Google Scholar]
- 56.Greenwald I. LIN-12/Notch signaling in C. elegans. WormBook. 2005 Aug 8; doi: 10.1895/wormbook.1.10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levitan D, Greenwald I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature. 1995;377:351–354. doi: 10.1038/377351a0. By screening for suppressors of a Notch gain-of-function mutation in C. elegans, presenilin was identified as a regulator of Notch activity and was thus the first study to link the presenilin complex — which is implicated in Alzheimer's disease — with Notch signalling. [DOI] [PubMed] [Google Scholar]
- 58.Kopan R, Goate A. A common enzyme connects Notch signaling and Alzheimer's disease. Genes Dev. 2000;14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 59.Sherrington R, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 60.Li X, Greenwald I. HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc Natl Acad Sci USA. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westlund B, Parry D, Clover R, Basson M, Johnson CD. Reverse genetic analysis of Caenorhabditis elegans presenilins reveals redundant but unequal roles for sel-12 and hop-1 in Notch-pathway signaling. Proc Natl Acad Sci USA. 1999;96:2497–2502. doi: 10.1073/pnas.96.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kopan R, Ilagan MX. γ-secretase: proteasome of the membrane? Nature Rev Mol Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 63.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nature Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 64.Mukherjee A, et al. Regulation of Notch signalling by non-visual β-arrestin. Nature Cell Biol. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- 65.Krauss G. Biochemistry of Signal Transduction and Regulation. 3rd. Wiley-VCH; 2003. [Google Scholar]
- 66.Hurlbut GD, Kankel MW, Artavanis-Tsakonas S. Nodal points and complexity of Notch–Ras signal integration. Proc Natl Acad Sci USA. 2009;106:2218–2223. doi: 10.1073/pnas.0812024106. A microarray-based approach indicating the integration of Notch signals with other major signalling pathways is described in this paper. Importantly, this study also showed that most genes that are responsive to RTK signalling are also responsive to Notch signalling, showing that the Notch and RTK pathways are highly interconnected. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flaherty MS, Zavadil J, Ekas LA, Bach EA. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of Notch signaling by the JAK/STAT pathway. Dev Dynam. 2009;238:2235–2253. doi: 10.1002/dvdy.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krejci A, Bernard F, Housden BE, Collins S, Bray SJ. Direct response to Notch activation: signalling crosstalk and incoherent logic. Sci Signal. 2009;2:ra1. doi: 10.1126/scisignal.2000140. This study catalogued the immediate cellular consequences of Notch activation in cultured cells using mRNA expression and CBF1–SU(H)–LAG1 (CSL) occupancy at enhancers. [DOI] [PubMed] [Google Scholar]
- 69.Hegde A, et al. Genomewide expression analysis in zebrafish Mind bomb alleles with pancreas defects of different severity identifies putative Notch responsive genes. PLoS ONE. 2008;3:e1479. doi: 10.1371/journal.pone.0001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamidi H, Gustafason D, Pellegrini M, Gasson J. Identification of novel targets of CSL-dependent Notch signaling in hematopoiesis. PLoS ONE. 2011;6:e20022. doi: 10.1371/journal.pone.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Semin Cell Dev Biol. 2012;23:458–464. doi: 10.1016/j.semcdb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 73.Fortini ME, Rebay I, Caron LA, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 74.Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- 75.Yeo SY. Zebrafish CiA interneurons are late-born primary neurons. Neurosci Lett. 2009;466:131–134. doi: 10.1016/j.neulet.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 76.Sundaram MV. The love–hate relationship between Ras and Notch. Genes Dev. 2005;19:1825–1839. doi: 10.1101/gad.1330605. [DOI] [PubMed] [Google Scholar]
- 77.Rebay I. Keeping the receptor tyrosine kinase signaling pathway in check: lessons from Drosophila. Dev Biol. 2002;251:1–17. doi: 10.1006/dbio.2002.0806. [DOI] [PubMed] [Google Scholar]
- 78.Doroquez DB, Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- 79.Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dynam. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- 80.Culi J, Martin-Blanco E, Modolell J. The EGF receptor and N signalling pathways act antagonistically in Drosophila mesothorax bristle patterning. Development. 2001;128:299–308. doi: 10.1242/dev.128.2.299. [DOI] [PubMed] [Google Scholar]
- 81.zur Lage P, Jarman AP. Antagonism of EGFR and notch signalling in the reiterative recruitment of Drosophila adult chordotonal sense organ precursors. Development. 1999;126:3149–3157. doi: 10.1242/dev.126.14.3149. [DOI] [PubMed] [Google Scholar]
- 82.Carmena A, et al. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- 83.Price JV, Savenye ED, Lum D, Breitkreutz A. Dominant enhancers of EGFR in Drosophila melanogaster: genetic links between the Notch and EGFR signaling pathways. Genetics. 1997;147:1139–1153. doi: 10.1093/genetics/147.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sundaram MV. Vulval development: the battle between Ras and Notch. Curr Biol. 2004;14:R311–R313. doi: 10.1016/j.cub.2004.03.052. [DOI] [PubMed] [Google Scholar]
- 85.Charlton-Perkins M, et al. Prospero and Pax2 combinatorially control neural cell fate decisions by modulating Ras- and Notch-dependent signaling. Neural Dev. 2011;6:20. doi: 10.1186/1749-8104-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roemer K. Notch and the p53 clan of transcription factors. Adv Exp Med Biol. 2012;727:223–240. doi: 10.1007/978-1-4614-0899-4_17. [DOI] [PubMed] [Google Scholar]
- 87.Dang TP. Notch, apoptosis and cancer. Adv Exp Med Biol. 2012;727:199–209. doi: 10.1007/978-1-4614-0899-4_15. [DOI] [PubMed] [Google Scholar]
- 88.Guo S, Liu M, Gonzalez-Perez RR. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim Biophys Acta. 2011;1815:197–213. doi: 10.1016/j.bbcan.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fuxe J, Vincent T, Garcia de Herreros A. Transcriptional crosstalk between TGF-β and stem cell pathways in tumor cell invasion: role of EMT promoting Smad complexes. Cell Cycle. 2010;9:2363–2374. doi: 10.4161/cc.9.12.12050. [DOI] [PubMed] [Google Scholar]
- 90.Ristorcelli E, Lombardo D. Targeting Notch signaling in pancreatic cancer. Expert Opin Ther Targets. 2010;14:541–552. doi: 10.1517/14728221003769895. [DOI] [PubMed] [Google Scholar]
- 91.Li JL, Harris AL. Crosstalk of VEGF and Notch pathways in tumour angiogenesis: therapeutic implications. Frontiers Biosci. 2009;14:3094–3110. doi: 10.2741/3438. [DOI] [PubMed] [Google Scholar]
- 92.Holderfield MT, Hughes CC. Crosstalk between vascular endothelial growth factor, notch, and transforming growth factor-β in vascular morphogenesis. Circul Res. 2008;102:637–652. doi: 10.1161/CIRCRESAHA.107.167171. [DOI] [PubMed] [Google Scholar]
- 93.Villaronga MA, Bevan CL, Belandia B. Notch signaling: a potential therapeutic target in prostate cancer. Curr Cancer Drug Targets. 2008;8:566–580. doi: 10.2174/156800908786241096. [DOI] [PubMed] [Google Scholar]
- 94.Nakamura T, Tsuchiya K, Watanabe M. Crosstalk between Wnt and Notch signaling in intestinal epithelial cell fate decision. J Gastroenterol. 2007;42:705–710. doi: 10.1007/s00535-007-2087-z. [DOI] [PubMed] [Google Scholar]
- 95.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 96.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. This study presented the first and the largest binary interaction (yeast-two hybrid) map for D. melanogaster. This work marks one of the earliest efforts at modelling multicellular organisms using a systems-biology approach. [DOI] [PubMed] [Google Scholar]
- 98.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 100.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 101.Stanyon CA, et al. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 103.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 104.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 105.Cramer R. Editorial for “advances in biological mass spectrometry and proteomics”. Methods. 2011;54:349–350. doi: 10.1016/j.ymeth.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 106.Gavin AC, Maeda K, Kuhner S. Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr Opin Biotechnol. 2011;22:42–49. doi: 10.1016/j.copbio.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 107.Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput Biol. 2008;4:e1000065. doi: 10.1371/journal.pcbi.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Wageningen S, et al. Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell. 2010;143:991–1004. doi: 10.1016/j.cell.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li B, Cao W, Zhou J, Luo F. Understanding and predicting synthetic lethal genetic interactions in Saccharomyces cerevisiae using domain genetic interactions. BMC Systems Biol. 2011;5:73. doi: 10.1186/1752-0509-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Linden RO, Eronen VP, Aittokallio T. Quantitative maps of genetic interactions in yeast — comparative evaluation and integrative analysis. BMC Systems Biol. 2011;5:45. doi: 10.1186/1752-0509-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Horn T, et al. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nature Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- 113.Guruharsha KG, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. This study presents the first metazoan protein complex map and defined more than 500 protein complexes involving several thousand proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fan X, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 115.Heitzler P. Biodiversity and noncanonical Notch signaling. Curr Top Dev Biol. 2010;92:457–481. doi: 10.1016/S0070-2153(10)92014-0. [DOI] [PubMed] [Google Scholar]
- 116.Hu QD, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- 117.Cui XY, et al. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J Biol Chem. 2004;279:25858–25865. doi: 10.1074/jbc.M313505200. [DOI] [PubMed] [Google Scholar]
- 118.Hori K, et al. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- 119.Shawber C, et al. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 120.Rusconi JC, Corbin V. Evidence for a novel Notch pathway required for muscle precursor selection in Drosophila. Mech Dev. 1998;79:39–50. doi: 10.1016/s0925-4773(98)00170-1. [DOI] [PubMed] [Google Scholar]
- 121.Martinez Arias A, Zecchini V, Brennan K. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr Opin Genet Dev. 2002;12:524–533. doi: 10.1016/s0959-437x(02)00336-2. [DOI] [PubMed] [Google Scholar]
- 122.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 123.Thompson BJ, et al. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 124.Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol. 2008;180:755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vaccari T, et al. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J Cell Sci. 2009;122:2413–2423. doi: 10.1242/jcs.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wilkin MB, et al. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol. 2004;14:2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 127.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J Cell Biol. 2011;195:1005–1015. doi: 10.1083/jcb.201104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Couturier L, Vodovar N, Schweisguth F. Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nature Cell Biol. 2012;14:131–139. doi: 10.1038/ncb2419. [DOI] [PubMed] [Google Scholar]
- 129.Croft D, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gordon WR, et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS ONE. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lake RJ, Grimm LM, Veraksa A, Banos A, Artavanis-Tsakonas S. In vivo analysis of the Notch receptor S1 cleavage. PLoS ONE. 2009;4:e6728. doi: 10.1371/journal.pone.0006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Qurashi A, et al. HSPC300 and its role in neuronal connectivity. Neural Dev. 2007;2:18. doi: 10.1186/1749-8104-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.