Abstract
Aging leads to a number of disadvantageous changes in the cardiovascular system. Deterioration of vascular homoeostasis with increase in oxidative stress, chronic low-grade inflammation, and impaired nitric oxide bioavailability results in endothelial dysfunction, increased vascular stiffness, and compromised arterial-ventricular interactions. A chronic dietary supplementation with the micronutrient nitrate has been demonstrated to improve vascular function. Healthy dietary patterns may regulate gene expression profiles. However, the mechanisms are incompletely understood. The changes that occur at the gene expression level and transcriptional profile following a nutritional modification with nitrate have not been elucidated. To determine the changes of the vascular transcriptome, we conducted gene expression microarray experiments on aortas of old mice, which were treated with dietary nitrate. Our results highlight differentially expressed genes overrepresented in gene ontology categories. Molecular interaction and reaction pathways involved in the calcium-signaling pathway and the detoxification system were identified. Our results provide novel insight to an altered gene-expression profile in old mice following nitrate supplementation. This supports the general notion of nutritional approaches to modulate age-related changes of vascular functions and its detrimental consequences.
1. Introduction
Aging is considered to be the most important nonmodifiable cardiovascular risk factor, leading to increased cardiovascular morbidity and mortality [1]. It deteriorates vascular integrity and entails a number of disadvantageous structural and functional changes on the arterial system [2]. Structural alterations include increased luminal diameter, vascular wall remodeling with thickened intima and media, and altered extracellular matrix content [2]. Functional changes are characterized by increased aortic stiffness, endothelial dysfunction, and subsequent systolic hypertension. These alterations are regulated by signaling pathways activated by endogenous and exogenous factors [3–5]. On a cellular level, decreased protein synthesis, dysfunctional mitochondria, oxidative stress, impaired antioxidant defense mechanisms, disturbed calcium handling, and increased DNA and protein oxidation have been proposed [6–9]. On the genomic level, we recently identified differences in the transcriptional profile and the genes potentially responsible for the vascular aging process, with emphasis on the detoxification system [10].
Dietary interventions have emerged as novel options, counteracting age-related vascular dysfunction [11–14]. A definite composition of healthy diet is still elusive. Nevertheless, it is known, particularly since the DASH (dietary approaches to stop hypertension) trial, that certain dietary patterns, for example, the so-called Mediterranean diet, positively influence blood pressures and cardiovascular outcome [15–18].
Inorganic nitrate is abundant in our everyday diet and especially in leafy greens. The nutritional aspects of nitrate are intriguing since it represents a source for the endogenous generation of nitrite, nitric oxide (NO), and other reactive nitrogen oxides [19–21]. In experimental studies and first clinical observations, dietary inorganic nitrate interventions have emerged as novel nutrient options modulating cardiovascular functions. This relates to regenerative processes in ischemic tissues, enhancement of endothelial function, and limitation of mitochondrial reactive oxygen species in a cardiomyopathy model [22–24]. We have demonstrated that a chronic nitrate supplementation improves vascular stiffness and reduces systolic blood pressures and the level of proinflammatory cytokines in elderly volunteers [25, 26]. However, to comprehend the beneficial effects of a nitrate rich diet on the vasculature the underlying cellular and molecular changes have to be characterized.
Nitrate-derived nitrite and NO exert beneficial effects through second messengers, posttranslational modifications, and regulation of gene expression [27–29]. The genes potentially affected by a nitrate rich diet counteracting the vascular aging process have not been identified so far. We therefore set out to compare the transcriptional profile in aged aortas of old nitrate-treated mice with age-related endothelial dysfunction applying a high-throughput genomic screening approach. Using microarray analyses, identification of genes, and expression differences was simultaneously accomplished. Our study discloses alterations in gene expression following a dietary nitrate supplementation in the aged vascular system.
2. Material and Methods
2.1. Animals
Male C57BL/6 mice were obtained from Janvier (Saint Berthevin, France) and kept one week in the local animal house for acclimatization. Old (20 months) mice were housed in controlled temperature and light conditions with feeding (V1534-000, Sniff, Soest, Germany) and watering ad libitum. A group of old mice received 1 g/L sodium-nitrate (NaNO3, ~150 μmoles according to the measured daily drinking water consumption) in their drinking water for 8 weeks, as described previously [23, 26]. An equal concentration of sodium chloride served as control. For aortic vasoreactivity studies young (6 months) mice served as controls. For the gene-expression experiments different mice from the same supplier were used. The care and all experiments were performed according to the animal welfare regulations of the German local authorities conforming to NIH Guidelines.
2.2. Aortic Vascular Reactivity
Ex vivo endothelial functions were assessed using an aortic ring bioassay, as described [28, 30]. Briefly, three 2-3-mm wide aortic rings were prepared from each mouse, perivascular fat was removed and aortas were suspended in an organ bath containing 10 mL modified Krebs-Henseleit buffer and connected to an isometric force transducer (EMKA Technologies, Paris, France). Organ chambers were aerated continuously with 95% O2 and 5% CO2 to achieve pH of 7.4. Isometric tension was recorded continuously. The vascular rings with intact endothelium and similar dimensions equilibrated to a resting tension of 1.0 g. All rings were preconstricted with KCl (40 mM) and, after washout, phenylephrine (10 μM). Endothelium function was measured as the relaxation response to acetylcholine (from 10−9 to 10−4 M). Endothelium-independent relaxation was determined as the response to sodium nitroprusside (from 10−9 to 10−2 M).
2.3. Chemicals
All reagents were obtained from Sigma-Aldrich (Taufkirchen, Germany) unless indicated otherwise.
2.4. RNA Preparation and Microarray Assays
RNA preparation and microarray assays were conducted as described previously [10]. Briefly, the descending aorta of each mouse was used. The vessel was flushed thoroughly with ice-cold PBS, through the left ventricle of the heart, cleaned of periadventitial fat and connective tissues, snap-frozen in liquid nitrogen, and stored at −80°C. Extraction of total RNA from the tissue was performed (Qiagen RNeasy Mini Kit, Qiagen, Hilden, Germany). Removal of residual genomic DNA was achieved by treatment with DNase I. Total RNA preparations were checked for RNA integrity (Agilent 2100 Bioanalyzer, Agilent Technologies, Palo Alto, CA). All samples showed common high quality RNA Integrity Numbers (RIN 9.0–9.6) and RNA was quantified by photometric Nanodrop measurement.
Synthesis of cDNA and subsequent fluorescent labeling of cRNA was done on six replicates of each group (old and old nitrate treated mice) according to the manufacturers protocol (One-Color Microarray-Based Gene Expression Analysis/Low Input Quick Amp Labeling; Agilent Technologies, Palo Alto, CA). Briefly, 100 ng of total RNA was converted to cDNA, followed by in vitro transcription and incorporation of Cy3-CTP into nascent cRNA. After fragmentation labeled cRNA was hybridized to Agilent SurePrint G3 Mouse GE 8x60K Microarrays for 17 h at 65°C. Quality control parameters of cRNA labeling and hybridization performance were found within the manufacturers specifications. Arrays were scanned as described by the manufacturer. Signal intensities on 20 bit tiff images were calculated by Feature Extraction software (FE, Vers. 10.7.1.1; Agilent Technologies, Palo Alto, CA).
GeneSpring GX software was used to analyze data (Vers. 12.5; Agilent Technologies, Palo Alto, CA). Probe signal intensities were quantile normalized across all samples to reduce interarray variability [31]. Input data preprocessing was concluded by baseline transformation to the median of all samples. Biological replicates were grouped according to their respective experimental condition. A given transcript had to be expressed above background (“detected” by GeneSpring GX) in at least five of six replicates in any one of two, or both conditions to be additional analyzed in pairwise comparisons of conditions. Hierarchical clustering of gene expression patterns identified no obvious outlier samples. Differential gene expression was determined statistically by a moderated t-test with genes showing P < 0.05 considered to be significantly differentially expressed [32].
The microarray data set as been deposited in the NCBI Gene Expression Omnibus (GEO) public database in compliance with MIAME (Minimum Information About a Microarray Experiment) guidelines (GEO series accession number GSE66572).
2.5. Functional Analysis by Gene Ontology and Gene Pathways
GeneSpring GX and Database for Annotation, Visualization, and Integrated Discovery (DAVID Bioinformatics database) was used to identify functional related categories of genes.
Differentially expressed genes (P < 0.05) with an additional fold change > 1.2 were considered relevant and were imported into DAVID Bioinformatics database [33, 34]. The DAVID software analysed significant enrichment of differentially expressed genes within gene ontology (GO) terms and additionally involves assessment of advanced pathway analysis [35, 36]. The background was chosen from the probes represented on the Agilent 8x60K mouse array, which were detected on either one of the three conditions to determine nitrate-associated genes in the aortic tissue. A modified Fisher's exact test assessed the significance of the association between the observed data and the data in the GO and canonical pathway. Each identified pathway was assigned a significance score, defined by the number of differentially regulated focus genes in the data set. This score was the negative logarithm of the P value, indicative of the likelihood that genes are found in a pathway randomly.
2.6. Statistical Analysis
The results of endothelial functions are presented as mean ± SEM. Data were analyzed by 1-way ANOVA and posthoc Bonferroni multiple comparison correction (for all pairwise tests) with GraphPad Prism 5 (GraphPad, California) software to compare differences between groups. A P value of <0.05 was considered significant. Agilent microarray probe sets were analyzed as described above.
3. Results and Discussion
3.1. Dietary Nitrate Reverses Age-Related Endothelial Dysfunction
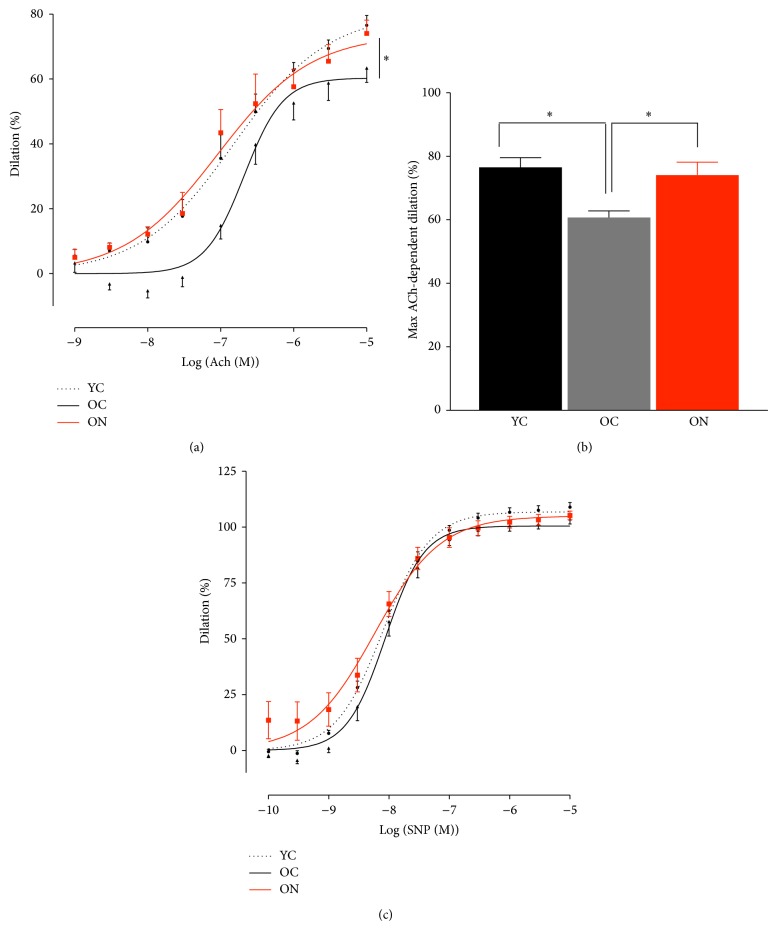
We studied old C57BL/6 mice and characterized age-related endothelial functions and the reversal through dietary nitrate. Using our model of aged mice, we were able to demonstrate an age-related vascular phenotype and associated alterations in vascular gene expression pattern [10]. Endothelial dysfunction is an important change that occurs in aged vessels, within increased cardiovascular risk and declining with advancing age [37, 38]. Decreases in endothelium-dependent dilator responses are believed to be a result of alterations in NO bioavailability, activity of the endothelial NO synthase, and increased formation of reactive oxygen species (ROS) and gene expression. We here show that aged mice have attenuated endothelial function as determined in isolated aortic bioassay studies compared to young mice (max vasodilator response to acetylcholine (ACh) young 76.6 ± 3.0% versus old 60.7 ± 2.1%, P = 0.003, Figures 1(a) and 1(b)). Importantly, following a chronic dietary nitrate supplementation a reversal of endothelial dysfunction was observed (max. vasodilator response to ACh old nitrate 60.7 ± 2.1% versus 74.1 ± 4.1%, P = 0.018, Figures 1(a) and 1(b)). No effect was observed for endothelium-independent function in young, old and old nitrate treated mice (Figure 1(c)). Our results are in line with previous findings showing decreased age-related endothelial functions and improvements through nitrate supplementation in elderly humans [25]. We have previously shown that a chronic nitrate supplementation regimen affects plasma nitrite and nitrate levels [23]. Although this dosage is relatively high as compared to what can be ingested from a diet, it yields the equivalent effect a diet rich in vegetables exerts in humans [25, 26]. In mice, a diet rich in nitrite led to a decrease in inflammatory cytokines, suggesting an enhancement in NO bioavailability through inhibition of inflammation and inactivation of ROS [11].
Figure 1.
Age-related endothelial dysfunction is reversed by dietary nitrate supplementation. (a) and (b) endothelium dependent function determined by dose-responses and maximum vasodilation to the endothelium-dependent dilator acetylcholine (ACh) in young, old, and nitrite-supplemented old male mice (YC, OC and ON). (c) Dose-responses to the endothelium-independent vasodilator sodium nitroprusside (SNP). (Values are mean ± SEM, n = 6, * P < 0.05.)
3.2. Microarrays, Gene Ontology, and Pathway Analysis
Dietary interventions are known to influence cardiovascular functions and impact gene expression patterns [39–42]. To explore the genes that underlie and are responsible for the beneficial effects of nitrate supplementation in aged mice we performed gene expression microarray experiments on whole thoracic aortas of old and old nitrate treated mice. Microarrays allow the simultaneous quantification of gene expression differences, obtaining insights in health and disease [43]. Aging as well as dietary interventions lead to subtle changes of the transcriptional profile and accordingly, we choose a statistical significance in differential gene expression with a P-value set to <0.05, not performing control for false discovery rate or family-wise error rate, as previously reported [10, 40, 44, 45].
Overall probe sets on the chip before filtering were 55682 and after exclusion of undetectable transcripts 26621 expressed transcripts were found. About 2900 transcripts were expressed differentially in aortic tissue of old and old nitrate treated mice, with totally 1565 upregulated and 1335 downregulated transcripts (Figure 2), which correspond to 2231 genes. Since the absolute numbers in gene expression differences do not provide explanations and comprehension of changes that occur through a dietary modification in aged mice, we performed GO and pathway analysis to gain insight into the transcriptional changes following nitrate supplementation.
Figure 2.

Differentially expressed transcripts in old male C57BL/6 aortas following nitrate treatment (n = 6; determined from total 26,774 transcripts; fold change > 1.2; P < 0.05). Old control versus old nitrate yielded 1565 upregulated and 1335 downregulated transcripts.
Pathway analysis was performed through DAVID Bioinformatics database to identify the biological processes associated to differential regulation in old nitrate treated mice. DAVIDs functional annotations tool identifies enriched GO annotations categorized in biological processes, molecular functions and cellular components, with highest GO terms displayed in Table 1.
Table 1.
Dietary nitrate induced enriched gene ontology terms in old mice. Enriched gene ontology (GO) terms according to biological processes, molecular functions and cellular components. GO terms are ordered by Fisher exact test (i.e., ease score) with highest enriched term at the top of the list. FDR, false discovery rate.
| Term | Count | % | P value | FDR |
|---|---|---|---|---|
| Biological process | ||||
| Brown fat cell differentiation | 15 | 0.7 | 1.3E − 5 | 2.4E − 2 |
| Rhythmic process | 19 | 0.9 | 1.4E − 3 | 2.4E0 |
| Extracellular structure organization | 30 | 1.5 | 2.1E − 3 | 3.7E0 |
| Negative regulation of lipid metabolic process | 6 | 0.3 | 2.3E − 3 | 4.1E0 |
| Signal transduction | 209 | 10.2 | 4.0E − 3 | 7.0E0 |
| Response to hormone stimulus | 29 | 1.4 | 4.2E − 3 | 7.3E0 |
| Fat cell differentiation | 17 | 0.8 | 6.1E − 3 | 1.1E1 |
| Regulation of cell proliferation | 77 | 3.7 | 6.9E − 3 | 1.2E1 |
| Negative regulation of multicellular organismal process | 19 | 0.9 | 9.7E − 3 | 1.6E1 |
| System development | 226 | 11.0 | 1.3E − 2 | 2.1E1 |
| Response to endogenous stimulus | 29 | 1.4 | 1.3E − 2 | 2.1E1 |
| Extracellular matrix organization | 20 | 1.0 | 1.4E − 2 | 2.3E1 |
| Response to hypoxia | 15 | 0.7 | 1.7E − 2 | 2.7E1 |
|
| ||||
| Cellular compartment | ||||
| Membrane | 703 | 34.2 | 6.1E − 6 | 8.8E − 3 |
| Integral to membrane | 492 | 24.0 | 9.8E − 6 | 1.4E − 2 |
| Extracellular region part | 98 | 4.8 | 1.0E − 5 | 1.5E − 2 |
| Extracellular region | 169 | 8.2 | 1.8E − 5 | 2.6E − 2 |
| Membrane part | 634 | 30.9 | 1.9E − 5 | 2.8E − 2 |
| Intrinsic to membrane | 507 | 24.7 | 3.6E − 5 | 5.2E − 2 |
| Extracellular matrix | 50 | 2.4 | 9.3E − 5 | 1.3E − 1 |
| Proteinaceous extracellular matrix | 48 | 2.3 | 1.0E − 4 | 1.5E − 1 |
| Extracellular matrix part | 22 | 1.1 | 3.9E − 4 | 5.6E − 1 |
| Basement membrane | 18 | 0.9 | 1.4E − 3 | 2.0E0 |
| Mitochondrial inner membrane | 56 | 2.7 | 2.0E − 3 | 2.8E0 |
| Plasma membrane | 291 | 14.2 | 4.8E − 3 | 6.6E0 |
| Mitochondrion | 190 | 9.3 | 6.1E − 3 | 8.4E0 |
|
| ||||
| Molecular function | ||||
| Antigen binding | 14 | 0.7 | 7.1E − 5 | 1.1E − 1 |
| Transmembrane receptor activity | 88 | 4.3 | 1.6E − 3 | 2.6E0 |
| Molecular transducer activity | 172 | 8.4 | 6.1E − 3 | 9.3E0 |
| Signal transducer activity | 172 | 8.4 | 6.1E − 3 | 9.3E0 |
| Protein tyrosine kinase activity | 28 | 1.4 | 9.9E − 3 | 1.5E1 |
| cAMP-dependent protein kinase regulator activity | 5 | 0.2 | 1.0E − 2 | 1.5E1 |
| Receptor activity | 142 | 6.9 | 1.1E − 2 | 1.6E1 |
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | 9 | 0.4 | 1.5E − 2 | 2.2E1 |
| Kinase regulator activity | 16 | 0.8 | 1.9E − 2 | 2.7E1 |
| Cation transmembrane transporter activity | 57 | 2.8 | 2.0E − 2 | 2.8E1 |
| Copper ion binding | 13 | 0.6 | 2.1E − 2 | 2.9E1 |
| Oxidoreductase activity | 10 | 0.5 | 2.6E − 2 | 3.4E1 |
| Potassium ion binding | 16 | 0.8 | 2.9E − 2 | 3.8E1 |
To determine the targets altered by a dietary nitrate supplementation in old mice we conducted DAVID's pathway analysis [33, 46]. The highest enriched databases of pathways that were in our study were KEGG, Panther, and Biocarta. Figure 3 depicts overrepresented pathways in aortas of old nitrate treated mice with the corresponding gene count. One of the highest enriched pathways was “calcium signaling” in the KEGG database and the “calcium/calmodulin dependent protein kinase activation” in Biocarta (Figure 3).
Figure 3.
Enriched pathways in aortas of old nitrate treated male mice. Highest enriched KEGG, Panther, and Biocarta pathways are displayed of old aortic tissue following nitrate supplementation. Individual pathways and corresponding gene count are sorted by negative log [P value].
The calcium signaling pathways were followed by pathways related to neurological and brain function “neuroactive ligand-receptor interaction,” “Alzheimer disease,” and “GABA-B receptor II signaling.” This might be of particular interest given a potentially impaired NO bioavailability in neurological diseases like Alzheimer's disease [47, 48]. The possible link gained from our study is of importance since dietary nitrate was shown to improve regional brain perfusion in older adults and to improve reaction time in diabetic patients [49, 50]. NO signaling was additionally linked to neuroprotective signaling pathways and to promote survival and inhibit apoptotic cell death in neuronal cell types [51]. On the contrary, a recent study showed no effects on cognitive performance in older adults after short-term beetroot juice ingestion [52]. However, since we used a chronic supplementation regimen in mice a direct conclusion remains unfeasible.
3.3. Calcium Signaling Pathway
Aging leads to alterations of vascular gene expression which ultimately lead to increased vascular tone perpetuating arterial stiffness and hypertension. Increased age-related stiffness has been demonstrated to be an independent predictor of CVD events [53]. Comparing old and old nitrate treated mice we found KEGGs “calcium signaling” and Biocartas “calcium/calmodulin dependent protein kinase activation” to be the highest enriched pathways in old mice following nitrate supplementation. Calcium signaling is fundamental to vascular functions and the vascular tone is maintained by a highly regulated calcium homeostasis. In smooth muscle cells calcium is the main trigger for the action potential, calcium-dependent phosphorylation of the myosin light chain leads to an increase in muscle cell tone and the changes in intracellular calcium parallel the changes in contractile forces. We and others have recently shown that a dietary nitrate supplementation reduces arterial stiffness and blood pressure in older adults and healthy volunteers [25, 54, 55]. While acute effects are exerted by nitrate to nitrite bioconversion and subsequent reduction to NO, the long-term effects are incompletely understood. NO is known to modulate calcium signaling in the cardiovascular system, regulating important calcium handling proteins, for example, SERCA (sarcoplamatic reticulum calcium pump), ryanodine receptor, and L-Type calcium hannel, which are responsible for smooth muscle cell relaxation [56–58]. Recently, it was shown that ingestion of nitrate over 7 days increased sarcoplasmatic reticulum calcium release and tetanic contractile force production via alterations in cellular calcium handling with altered calcium handling protein expressions in nitrate treated mice [59]. We now identified n = 30 differentially expressed genes related to calcium pathway in old versus old nitrate treated mice (Table 2 and Figure 4). Major players of the cellular calcium handling process were found, namely the L-type calcium channel (Cacna1d and Ppapdc2), the sarcoplasmatic calcium release channel (Ryanodine Receptor 2, Ryr2), the calcium/calmodulin-dependent protein kinase type II (Calm2, Camk2g, Camk4), and the inositol triphosphate receptor (Itpr2, Itpr3, Itpka; for detailed regulation and fold change expression difference refer to Table 2).
Table 2.
Differentially expressed genes related to calcium pathway in old versus old nitrate treated mice.
| Gene symbol | Description | Regulation | Fold change | P value |
|---|---|---|---|---|
| Atp2a1 | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 | Up | 1.2 | 0.010 |
| Atp2b2 | ATPase, Ca++ transporting, plasma membrane 2 | Up | 1.3 | 0.018 |
| Ptk2b | PTK2 protein tyrosine kinase 2 beta | Down | 1.2 | 0.008 |
| Adcy7 | Adenylate cyclase 7 | Up | 1.4 | 0.001 |
| Adra1a | Adrenergic receptor, alpha 1a | Down | 1.5 | 0.002 |
| Adrb3 | Adrenergic receptor, beta 3 | Down | 1.9 | 0.004 |
| Cacna1d | Calcium channel, voltage-dependent, L type, alpha 1D subunit | Up | 1.4 | 0.013 |
| Ppapdc2 | Calcium channel, voltage-dependent, L type, alpha 1S subunit | Down | 1.3 | 0.010 |
| Camk4 | Calcium/calmodulin-dependent protein kinase IV | Up | 1.4 | 0.009 |
| F2r | Coagulation factor II (thrombin) receptor | Up | 1.3 | 0.001 |
| Ednrb | Endothelin receptor type B | Down | 1.2 | 0.045 |
| Gna14 | Guanine nucleotide binding protein, alpha 14 | Up | 1.6 | 0.031 |
| Itpr2 | Inositol 1,4,5-triphosphate receptor 2 | Down | 1.3 | 0.001 |
| Itpr3 | Inositol 1,4,5-triphosphate receptor 3 | Up | 1.2 | 0.024 |
| Itpka | Inositol 1,4,5-trisphosphate 3-kinase A | Down | 1.4 | 0.017 |
| Nos2 | Nitric oxide synthase 2, inducible | Down | 1.2 | 0.019 |
| Oxtr | Oxytocin receptor | Down | 1.4 | 0.042 |
| Plcb4 | Phospholipase C, beta 4 | Up | 1.4 | 0.005 |
| Plcd1 | Phospholipase C, delta 1 | Up | 1.2 | 0.001 |
| Phka2 | Phosphorylase kinase alpha 2 | Down | 1.2 | 0.037 |
| Pdgfrb | Platelet derived growth factor receptor, beta polypeptide | Up | 1.2 | 0.007 |
| Calm2 | Calmodulin 2 | Up | 1.2 | 0.020 |
| Ptger1 | Prostaglandin E receptor 1 (subtype EP1) | Up | 1.2 | 0.023 |
| Ptger3 | Prostaglandin E receptor 3 (subtype EP3) | Down | 1.9 | 0.020 |
| Prkx | Protein kinase, X-linked | Down | 1.2 | 0.031 |
| Ppp3cc | Protein phosphatase 3, catalytic subunit, gamma isoform | Down | 1.3 | 0.025 |
| P2rx3 | Purinergic receptor P2X, ligand-gated ion channel, 3 | Up | 1.3 | 0.035 |
| Ryr2 | Ryanodine receptor 2, cardiac | Up | 1.4 | 0.007 |
| Camk2g | Similar to Calcium/calmodulin-dependent protein kinase type II gamma chain | Up | 1.3 | 0.010 |
| Sphk2 | Sphingosine kinase 2 | Down | 1.2 | 0.021 |
Figure 4.
Calcium signaling pathway in old nitrate treated aorta. KEGG-pathway analysis determined by DAVID Bioinformatics resources showing differentially expressed genes highlighted with red stars (for up-/downregulation refer to Table 2).
3.4. Detoxification, Antioxidation, and Free Radical Removal
The vascular aging process is characterized by increased oxidative stress and impaired antioxidant defense mechanisms [6, 60]. We have recently shown a distinctive transcriptomic profile with altered expression patterns in the old aorta with emphasis on the detoxification system [10]. Oxidative stress progresses due to excessive generation of ROS, to reduced antioxidant capacity or due to uncoupled nitric oxide synthase, amongst others. We have demonstrated that nitrite-derived NO impacts ROS generation and additionally reduces protein damage in myocardial ischemia/reperfusion injury [30, 61]. It was evidenced that NO regulates cardiovascular cell signaling and modulates cellular energetics [62, 63]. While in the acute setting nitrite is reduced by nitrite reductases and exerts vascular effects, the consequences of a long-term administration are unclear [64–66]. Recently, decreased inflammatory cytokines, inhibition of inflammation and inactivation of ROS following a nitrite rich diet in mice and healthy volunteers with subsequent improved age-related vascular functions were observed [11, 26, 67]. We now determined the enriched genes in aortas of old nitrate treated mice associated with the biological process of detoxification, antioxidation, and free radical removal as defined by Panther GO terms (Table 3). Most of the genes altered by nitrate in the old aorta were related to glutathione metabolism, the major antioxidant system. Remarkably, a downregulation of inducible nitric oxide synthase was noted, which could suggest a reduced inflammation in nitrate treated animals. Of note, the diversity of transcriptional regulation is observed through the bilateral regulation of superoxide dismutases (Table 3).
Table 3.
Enriched genes associated with the biological process of detoxification, antioxidation, and free radical removal, according to Panther Gene Ontology in old versus old nitrate treated mice.
| Gene symbol | Description | Regulation | Fold change | P value |
|---|---|---|---|---|
| pAbcc1 | ATP-binding cassette, subfamily C (CFTR/MRP), member 1 | Up | 1.2 | 0.007 |
| Asna1 | arsA arsenite transporter, ATP-binding | Down | 1.2 | 0.025 |
| Ces1d | Carboxyl esterase 3 | Down | 1.4 | 0.002 |
| Ces1f | Expressed sequence AU018778 | Down | 1.6 | 0.010 |
| Ephx1 | Epoxide hydrolase 1, microsomal | Up | 1.3 | 0.015 |
| Gpx3 | Glutathione peroxidase 3 | Down | 1.5 | 0.001 |
| Gpx4 | Heterogeneous nuclear ribonucleoprotein L-like; glutathione peroxidase 4 | Down | 1.4 | 0.003 |
| Gsta3 | Glutathione S-transferase, alpha 3 | Down | 2.1 | 0.039 |
| Gsta4 | Glutathione S-transferase, alpha 4 | Down | 1.5 | 0.010 |
| Gstm4 | Glutathione S-transferase, mu 4 | Down | 1.3 | 0.009 |
| Gstt3 | Glutathione S-transferase, theta 3 | Up | 1.2 | 0.020 |
| Gstz1 | Glutathione transferase zeta 1 | Down | 1.3 | 0.034 |
| Haghl | Hydroxyacyl glutathione hydrolase-like | Up | 1.2 | 0.006 |
| Mgst1 | Microsomal glutathione S-transferase 1 | Down | 1.3 | 0.044 |
| Mgst2 | Microsomal glutathione S-transferase 2 | Down | 1.7 | 0.015 |
| Mgst3 | Microsomal glutathione S-transferase 3 | Down | 1.2 | 0.044 |
| Mpv17l | Mpv17 transgene, kidney disease mutant-like | Up | 1.3 | 0.001 |
| Nos2 | Nitric oxide synthase 2, inducible | Down | 1.2 | 0.019 |
| Pnkd | Paroxysmal nonkinesiogenic dyskinesia | Down | 1.4 | 0.030 |
| Pon3 | Paraoxonase 3 | Down | 1.2 | 0.002 |
| Prdx4 | Peroxiredoxin 4 | Up | 1.2 | 0.001 |
| Prdx5 | Peroxiredoxin 5 | Down | 1.2 | 0.011 |
| Pxmp2 | Peroxisomal membrane protein 2 | Down | 1.2 | 0.011 |
| Sod1 | Superoxide dismutase 1, soluble | Down | 1.2 | 0.015 |
| Sod3 | Superoxide dismutase 3, extracellular | Up | 1.2 | 0.015 |
| Tst | Thiosulfate sulfotransferase, mitochondrial | Down | 1.3 | 0.037 |
| Txndc16 | Thioredoxin domain containing 16 | Up | 1.2 | 0.009 |
| Txnl4a | Thioredoxin-like 4A | Down | 1.2 | 0.002 |
3.5. Limitations
We used the whole thoracic aorta instead of a single cell type. Although a specific cell might provide fundamental information, cell-cell interactions are imperative for coordinated organ functions and we thus intended to examine the complete transcriptional vascular changes that occur following nitrate supplementation. As we used only male mice our results might not apply to female mice and this should be taken into account for further studies. Moreover, alternative splicing variants are inadequately reflected by our analysis. Verification of microarray data is needed in forthcoming studies to confirm the beneficial effects of dietary nitrate on vascular tissue. Important targets to confirm would be the family of superoxide dismutases (SOD 1 and 3) the nitric oxide synthase 2, the platelet-derived growth factor beta (PDGFR-beta), and the adrenergic receptor. Also, measurement of markers of oxidative stress or inflammation would be helpful to verify the presented findings. Finally, it has to be noted that gene expression data does not always correspond to protein expression and activity.
4. Conclusions
Aging is the most important nonmodifiable cardiovascular risk factor, which leads to disadvantageous changes of the vasculature. A dietary intervention with inorganic nitrate has been evidenced to improve vascular functions. We determined changes that occur at a gene expression level and transcriptional profile following a chronic nitrate supplementation in aged mice. Our results highlight differentially expressed genes overrepresented in GO categories, pathways related to the calcium-signaling, the detoxification and antioxidation system. Our results contribute to existing knowledge of nitrate's beneficial effects. This supports the concept of nutritional approaches to modulate age-related changes of vascular functions and its detrimental consequences.
Acknowledgments
The authors would like to thank Sebastian Wohlfromm, Ph.D., BMFZ Genomics and Transcriptomics Laboratory (GTL), University of Düsseldorf, for conducting the microarray experiments. This study was supported by grants from the Forschungskommission of the Heinrich-Hein-University (Matthias Totzeck) and the German Research Foundation (DFG) to Tienush Rassaf (RA969/4-2) and to Malte Kelm (Ke405/5-1). Tienush Rassaf is a Heisenberg professor funded by DFG (Ra969/7-2).
Conflict of Interests
The authors declare no conflict of interests.
References
- 1.Lakatta E. G., Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: part I: aging arteries: a ‘set up’ for vascular disease. Circulation. 2003;107(1):139–146. doi: 10.1161/01.cir.0000048892.83521.58. [DOI] [PubMed] [Google Scholar]
- 2.Lakatta E. G. Arterial and cardiac aging: Major shareholders in cardiovascular disease enterprises. Part III: cellular and molecular clues to heart and arterial aging. Circulation. 2003;107(3):490–497. doi: 10.1161/01.cir.0000048894.99865.02. [DOI] [PubMed] [Google Scholar]
- 3.Barzilai N., Guarente L., Kirkwood T. B. L., Partridge L., Rando T. A., Eline Slagboom P. E. The place of genetics in ageing research. Nature Reviews Genetics. 2012;13(8):589–594. doi: 10.1038/nrg3290. [DOI] [PubMed] [Google Scholar]
- 4.Kenyon C. J. The genetics of ageing. Nature. 2010;464(7288):504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 5.Greer E. L., Brunet A. Signaling networks in aging. Journal of Cell Science. 2008;121(4):407–412. doi: 10.1242/jcs.021519. [DOI] [PubMed] [Google Scholar]
- 6.Ungvari Z., Kaley G., de Cabo R., Sonntag W. E., Csiszar A. Mechanisms of vascular aging: new perspectives. Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2010;65(10):1028–41. doi: 10.1093/gerona/glq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawabe M. Vascular aging: from molecular mechanism to clinical significance. Geriatrics and Gerontology International. 2010;10(supplement 1):S213–S220. doi: 10.1111/j.1447-0594.2010.00603.x. [DOI] [PubMed] [Google Scholar]
- 8.Kovacic J. C., Moreno P., Nabel E. G., Hachinski V., Fuster V. Cellular senescence, vascular disease, and aging: part 2 of a 2-part review: clinical vascular disease in the elderly. Circulation. 2011;123(17):1900–1910. doi: 10.1161/circulationaha.110.009118. [DOI] [PubMed] [Google Scholar]
- 9.Gems D., Doonan R. Antioxidant defense and aging in C. elegans: is the oxidative damage theory of aging wrong? Cell Cycle. 2009;8(11):1681–1687. doi: 10.4161/cc.8.11.8595. [DOI] [PubMed] [Google Scholar]
- 10.Rammos C., Hendgen-Cotta U. B., Deenen R., et al. Age-related vascular gene expression profiling in mice. Mechanisms of Ageing and Development. 2014;135(1):15–23. doi: 10.1016/j.mad.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Sindler A. L., Fleenor B. S., Calvert J. W., et al. Nitrite supplementation reverses vascular endothelial dysfunction and large elastic artery stiffness with aging. Aging Cell. 2011;10(3):429–437. doi: 10.1111/j.1474-9726.2011.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jablonski K. L., Racine M. L., Geolfos C. J., et al. Dietary sodium restriction reverses vascular endothelial dysfunction in middle-aged/older adults with moderately elevated systolic blood pressure. Journal of the American College of Cardiology. 2013;61(3):335–343. doi: 10.1016/j.jacc.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heffernan K. S., Fahs C. A., Ranadive S. M., Patvardhan E. A. Review article: L-arginine as a nutritional prophylaxis against vascular endothelial dysfunction with aging. Journal of Cardiovascular Pharmacology and Therapeutics. 2010;15(1):17–23. doi: 10.1177/1074248409354599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss E. P., Fontana L. Caloric restriction: powerful protection for the aging heart and vasculature. American Journal of Physiology: Heart and Circulatory Physiology. 2011;301(4):H1205–H1219. doi: 10.1152/ajpheart.00685.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appel L. J., Moore T. J., Obarzanek E., et al. A clinical trial of the effects of dietary patterns on blood pressure. The New England Journal of Medicine. 1997;336(16):1117–1124. doi: 10.1056/nejm199704173361601. [DOI] [PubMed] [Google Scholar]
- 16.Appel L. J., Sacks F. M., Carey V. J., et al. Effects of protein, monounsaturated fat, and carbohydrate intake on blood pressure and serum lipids: results of the OmniHeart randomized trial. Journal of the American Medical Association. 2005;294(19):2455–2464. doi: 10.1001/jama.294.19.2455. [DOI] [PubMed] [Google Scholar]
- 17.Sofi F., Cesari F., Abbate R., Gensini G. F., Casini A. Adherence to Mediterranean diet and health status: meta-analysis. British Medical Journal. 2008;337 doi: 10.1136/bmj.a1344.a1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estruch R., Ros E., Martínez-González M. A. Mediterranean diet for primary prevention of cardiovascular disease. The New England Journal of Medicine. 2013;369(7):676–677. doi: 10.1056/nejmc1306659. [DOI] [PubMed] [Google Scholar]
- 19.Lundberg J. O., Weitzberg E., Gladwin M. T. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nature Reviews Drug Discovery. 2008;7(2):156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 20.Lundberg J. O., Carlström M., Larsen F. J., Weitzberg E. Roles of dietary inorganic nitrate in cardiovascular health and disease. Cardiovascular Research. 2011;89(3):525–532. doi: 10.1093/cvr/cvq325. [DOI] [PubMed] [Google Scholar]
- 21.Lundberg J. O., Gladwin M. T., Ahluwalia A., et al. Nitrate and nitrite in biology, nutrition and therapeutics. Nature Chemical Biology. 2009;5(12):865–869. doi: 10.1038/nchembio.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu S.-G., Kukreja R. C., Das A., Chen Q., Lesnefsky E. J., Xi L. Dietary nitrate supplementation protects against doxorubicin-induced cardiomyopathy by improving mitochondrial function. Journal of the American College of Cardiology. 2011;57(21):2181–2189. doi: 10.1016/j.jacc.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendgen-Cotta U. B., Luedike P., Totzeck M., et al. Dietary nitrate supplementation improves revascularization in chronic ischemia. Circulation. 2012;126(16):1983–1992. doi: 10.1161/CIRCULATIONAHA.112.112912. [DOI] [PubMed] [Google Scholar]
- 24.Heiss C., Meyer C., Totzeck M., et al. Dietary inorganic nitrate mobilizes circulating angiogenic cells. Free Radical Biology and Medicine. 2012;52(9):1767–1772. doi: 10.1016/j.freeradbiomed.2012.02.051. [DOI] [PubMed] [Google Scholar]
- 25.Rammos C., Hendgen-Cotta U. B., Sobierajski J., Bernard A., Kelm M., Rassaf T. Dietary nitrate reverses vascular dysfunction in older adults with moderately increased cardiovascular risk. Journal of the American College of Cardiology. 2014;63(15):1584–1585. doi: 10.1016/j.jacc.2013.08.691. [DOI] [PubMed] [Google Scholar]
- 26.Rammos C., Hendgen-Cotta U. B., Pohl J., et al. Modulation of circulating macrophage migration inhibitory factor in the elderly. BioMed Research International. 2014;2014:8. doi: 10.1155/2014/582586.582586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendgen-Cotta U. B., Merx M. W., Shiva S., et al. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(29):10256–10261. doi: 10.1073/pnas.0801336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Totzeck M., Hendgen-Cotta U. B., Luedike P., et al. Nitrite regulates hypoxic vasodilation via myoglobin-dependent nitric oxide generation. Circulation. 2012;126(3):325–334. doi: 10.1161/circulationaha.111.087155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryan N. S., Fernandez B. O., Bauer S. M., et al. Nitrite is a signaling molecule and regulator of gene expression in mammalian tissues. Nature Chemical Biology. 2005;1(5):290–297. doi: 10.1038/nchembio734. [DOI] [PubMed] [Google Scholar]
- 30.Totzeck M., Hendgen-Cotta U. B., Kelm M., Rassaf T., Jourd'heuil D. Crosstalk between nitrite, myoglobin and reactive oxygen species to regulate vasodilation under hypoxia. PLoS ONE. 2014;9(8) doi: 10.1371/journal.pone.0105951.e105951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolstad B. M., Irizarry R. A., Åstrand M., Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 32.Smyth G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1):1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 33.Huang D. W., Sherman B. T., Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Huang D. W., Sherman B. T., Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40(1):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandes R. P., Fleming I., Busse R. Endothelial aging. Cardiovascular Research. 2005;66(2):286–294. doi: 10.1016/j.cardiores.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 38.Rammos C., Hendgen-Cotta U. B., Sobierajski J., et al. Macrophage migration inhibitory factor is associated with vascular dysfunction in patients with end-stage renal disease. International Journal of Cardiology. 2013;168(6):5249–5256. doi: 10.1016/j.ijcard.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Grayson M. Nutrigenomics. Nature. 2010;468(7327, article S1) doi: 10.1038/468s1a. [DOI] [PubMed] [Google Scholar]
- 40.Park S.-K., Kim K., Page G. P., Allison D. B., Weindruch R., Prolla T. A. Gene expression profiling of aging in multiple mouse strains: identification of aging biomarkers and impact of dietary antioxidants. Aging Cell. 2009;8(4):484–495. doi: 10.1111/j.1474-9726.2009.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee C. K., Weindruch R., Prolla T. A. Gene-expression profile of the ageing brain in mice. Nature Genetics. 2000;25(3):294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 42.Raat N. J. H., Noguchi A. C., Liu V. B., et al. Dietary nitrate and nitrite modulate blood and organ nitrite and the cellular ischemic stress response. Free Radical Biology and Medicine. 2009;47(5):510–517. doi: 10.1016/j.freeradbiomed.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhodes D. R., Yu J., Shanker K., et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bodyak N., Kang P. M., Hiromura M., et al. Gene expression profiling of the aging mouse cardiac myocytes. Nucleic Acids Research. 2002;30(17):3788–3794. doi: 10.1093/nar/gkf497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu H., Tian B., Resuello R. G., et al. Sex-specific regulation of gene expression in the aging monkey aorta. Physiological Genomics. 2007;29(2):169–180. doi: 10.1152/physiolgenomics.00229.2006. [DOI] [PubMed] [Google Scholar]
- 46.Ashburner M., Ball C. A., Blake J. A., et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuiper M. A., Visser J. J., Bergmans P. L. M., Scheltens P., Wolters E. C. Decreased cerebrospinal fluid nitrate levels in Parkinson's disease, Alzheimer's disease and multiple system atrophy patients. Journal of the Neurological Sciences. 1994;121(1):46–49. doi: 10.1016/0022-510X(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 48.Austin S. A., Santhanam A. V., Hinton D. J., Choi D.-S., Katusic Z. S. Endothelial nitric oxide deficiency promotes Alzheimer's disease pathology. Journal of Neurochemistry. 2013;127(5):691–700. doi: 10.1111/jnc.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Presley T. D., Morgan A. R., Bechtold E., et al. Acute effect of a high nitrate diet on brain perfusion in older adults. Nitric Oxide. 2011;24(1):34–42. doi: 10.1016/j.niox.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilchrist M., Winyard P. G., Fulford J., Anning C., Shore A. C., Benjamin N. Dietary nitrate supplementation improves reaction time in type 2 diabetes: development and application of a novel nitrate-depleted beetroot juice placebo. Nitric Oxide. 2014;40:67–74. doi: 10.1016/j.niox.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Estévez A. G., Spear N., Anthony Thompson J., et al. Nitric oxide-dependent production of cGMP supports the survival of rat embryonic motor neurons cultured with brain-derived neurotrophic factor. The Journal of Neuroscience. 1998;18(10):3708–3714. doi: 10.1523/JNEUROSCI.18-10-03708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelly J., Fulford J., Vanhatalo A., et al. Effects of short-term dietary nitrate supplementation on blood pressure, O2 uptake kinetics, and muscle and cognitive function in older adults. The American Journal of Physiology—Regulatory Integrative and Comparative Physiology. 2013;304(2):R73–R83. doi: 10.1152/ajpregu.00406.2012. [DOI] [PubMed] [Google Scholar]
- 53.Mattace-Raso F. U. S., van der Cammen T. J. M., Hofman A., et al. Arterial stiffness and risk of coronary heart disease and stroke: the Rotterdam Study. Circulation. 2006;113(5):657–663. doi: 10.1161/circulationaha.105.555235. [DOI] [PubMed] [Google Scholar]
- 54.Larsen F. J., Ekblom B., Sahlin K., Lundberg J. O., Weitzberg E. Effects of dietary nitrate on blood pressure in healthy volunteers. The New England Journal of Medicine. 2006;355(26):2792–2793. doi: 10.1056/nejmc062800. [DOI] [PubMed] [Google Scholar]
- 55.Kapil V., Milsom A. B., Okorie M., et al. Inorganic nitrate supplementation lowers blood pressure in humans: role for nitrite-derived no. Hypertension. 2010;56(2):274–281. doi: 10.1161/hypertensionaha.110.153536. [DOI] [PubMed] [Google Scholar]
- 56.Adachi T., Weisbrod R. M., Pimentel D. R., et al. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nature Medicine. 2004;10(11):1200–1207. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 57.Lima B., Forrester M. T., Hess D. T., Stamler J. S. S-nitrosylation in cardiovascular signaling. Circulation Research. 2010;106(4):633–646. doi: 10.1161/circresaha.109.207381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barouch L. A., Harrison R. W., Skaf M. W., et al. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature. 2002;416(6878):337–339. doi: 10.1038/416005a. [DOI] [PubMed] [Google Scholar]
- 59.Hernández A., Schiffer T. A., Ivarsson N., et al. Dietary nitrate increases tetanic [Ca2+ ]i and contractile force in mouse fast-twitch muscle. Journal of Physiology. 2012;590(15):3575–3583. doi: 10.1113/jphysiol.2012.232777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bachschmid M. M., Schildknecht S., Matsui R., et al. Vascular aging: chronic oxidative stress and impairment of redox signaling—consequences for vascular homeostasis and disease. Annals of Medicine. 2013;45(1):17–36. doi: 10.3109/07853890.2011.645498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rassaf T., Totzeck M., Hendgen-Cotta U. B., Shiva S., Heusch G., Kelm M. Circulating nitrite contributes to cardioprotection by remote ischemic preconditioning. Circulation Research. 2014;114(10):1601–1610. doi: 10.1161/CIRCRESAHA.114.303822. [DOI] [PubMed] [Google Scholar]
- 62.Rassaf T., Lauer T., Heiss C., et al. Nitric oxide synthase-derived plasma nitrite predicts exercise capacity. British Journal of Sports Medicine. 2007;41(10):669–673. doi: 10.1136/bjsm.2007.035758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rassaf T., Flögel U., Drexhage C., Hendgen-Cotta U., Kelm M., Schrader J. Nitrite reductase function of deoxymyoglobin: oxygen sensor and regulator of cardiac energetics and function. Circulation Research. 2007;100(12):1749–1754. doi: 10.1161/circresaha.107.152488. [DOI] [PubMed] [Google Scholar]
- 64.Hendgen-Cotta U. B., Kelm M., Rassaf T. Myoglobin's novel role in nitrite-induced hypoxic vasodilation. Trends in Cardiovascular Medicine. 2014;24(2):69–74. doi: 10.1016/j.tcm.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 65.Rassaf T., Heiss C., Hendgen-Cotta U., et al. Plasma nitrite reserve and endothelial function in the human forearm circulation. Free Radical Biology and Medicine. 2006;41(2):295–301. doi: 10.1016/j.freeradbiomed.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 66.Hendgen-Cotta U. B., Kelm M., Rassaf T. A highlight of myoglobin diversity: the nitrite reductase activity during myocardial ischemia-reperfusion. Nitric Oxide. 2010;22(2):75–82. doi: 10.1016/j.niox.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 67.Sindler A. L., DeVan A. E., Fleenor B. S., Seals D. R. Inorganic nitrite supplementation for healthy arterial aging. Journal of Applied Physiology. 2014;116(5):463–477. doi: 10.1152/japplphysiol.01100.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]