Abstract
Background
The colon plays a key role in regulating the homeostasis of bile acids.
Aim
The present study aims to evaluate the influence of colon cancer towards the homeostasis of bile acids.
Methods
The free and conjugated bile acids were determined using ultraperformance LC (UPLC) coupled with ABI 4000 QTRAP triple quadrupole instruments.
Results
The results showed that the free bile acids in serum of patients with colon cancers tend to increase, and the conjugated bile acids tended to decrease, especially for taurolithocholate (TLCA) (p<0.001).
Conclusion
The alteration of bile acids balance in colon cancers indicated the possibility of complicated diseases due to the disrupted balance of bile acids.
Keywords: Colon cancer, free bile acids, conjugated bile acids
Introduction
Bile acids, the main constituents of bile, are produced in the liver from cholesterol through a series of enzyme modification. Bile acids play a key role in solubilization and emulsification of fat to help digestion in the digestive tract1. Colon plays an important role in the modification of bile acids. For example, in the ileum, the enzymes released by intestinal flora can modify the deconjugated bile acids2. In the ileum and colon, bile acids will be re-absorbed into the liver for recycling. Therefore, the diseases influencing colon might disturb the homeostasis of bile acids.
Colon cancer, also known as colorectal cancer, has been defined as the cancer from uncontrolled cell growth in the colon3. Previous studies have demonstrated that 11 out of 26 serum amino acids significantly changed in colon cancer, including lysine, alanine, aspartic acid, glycine, histidine, leucine, methionine, sarcosine, threonine, tyrosine, and valine4, indicating the disruption of amino acids metabolism. Additionally, in the colon cancer, the lipid metabolic profile significantly changed5. The present study aims to compare the bile acids profile between normal individuals and patients with colon cancer.
Materials and methods
Reagents
Hyodeoxycholic acid (HDCA), lithocholic acid (LCA), sodium taurochenodeoxycholate (TCDCA), taurocholic acid sodium salt hydrate (TCA), sodium chenodeoxycholate (CDCA), sodium taurolithocholate (TLCA), ursodeoxycholic acid (UDCA), cholic acid (CA), dehydrocholic acid (DHCA), sodium deoxycholate (DCA), sodium tauroursodeoxycholate (TUDCA), sodium taurodeoxycholate hydrate (TDCA), and glycocholic acid hydrate (GCA) were purchased from Sigma-Aldrich (St Louis, MO). The purity of all these bile acids standards was above 95%, and they were dissolved in the dimethyl sulfoxide (DMSO) for utilization.
Determination of bile acids components in healthy volunteers and patients,5 healthy volunteers and 5 patients with colon cancers were enrolled in the Tangdu Hospital, Fourth Military Medical University. The blood was taken, and serum was prepared through centrifugation for 15 min at 8000×g in BD microtainer serum separator tubes. The serum was determined using UltraPerformance LC (UPLC) coupled with ABI 4000 QTRAP triple quadrupole instruments. 0.3 ml/min flow rate was used, and the elution phase contained water containing 0.2% formic acid (A) and methanol (B). The following conditions were used: 0–3 min, 65–75% B; 3–8 min, 75–80% B; 8–12 min, 80–95% B; 12–14 min, 65% B. MS source parameters were as follows: capillary voltage, 2.9 kV; cone voltage, 36 V; source temperature, 90 °C; and cone gas flow rate, 40 L/h at 4 psi.
Statistical analysis
The results were given as mean ± standard deviation (SD). Statistical differences were evaluated using the two-tailed Student's t-test and considered significant at the *p < 0.05, **p <0.01, ***p<0.001 level.
Results
Given the difficulty to separate numerous bile acids, multiple reaction monitoring (MRM) was performed. The ion pair (Q1→Q3, 391.1→391.1) was used to separate HDCA, CDCA, DCA and UDCA, and the retention time was 6 min, 8.55 min, 8.93 min, and 5.45 min, respectively. The ion pair (Q1→Q3, 375.1→375.1) was employed to separate LCA. The ion pair (Q1→Q3, 498.1→79.9) was used to separate TCDCA (Rt=8.95 min), TUDCA (Rt=5.51 min), and TDCA, respectively (Rt=9.68 min). The ion pair (Q1→Q3, 407.2→407.2) was employed to identify CA. GCA was separated using the ion pair 464.2→73.9 (Q1→Q3). The separation of TCA and TLCA used the ion pairs 514.2→79.8 (Q1→Q3) and 482.2→79.9 (Q1→Q3), respectively.
Using this monitoring method, the serum level of bile acids including free bile acids and conjugated bile acids was determined in five healthy volunteers and five patients with colon cancers. The relatively big difference was observed between the individuals (Fig. 1 & Fig. 2).
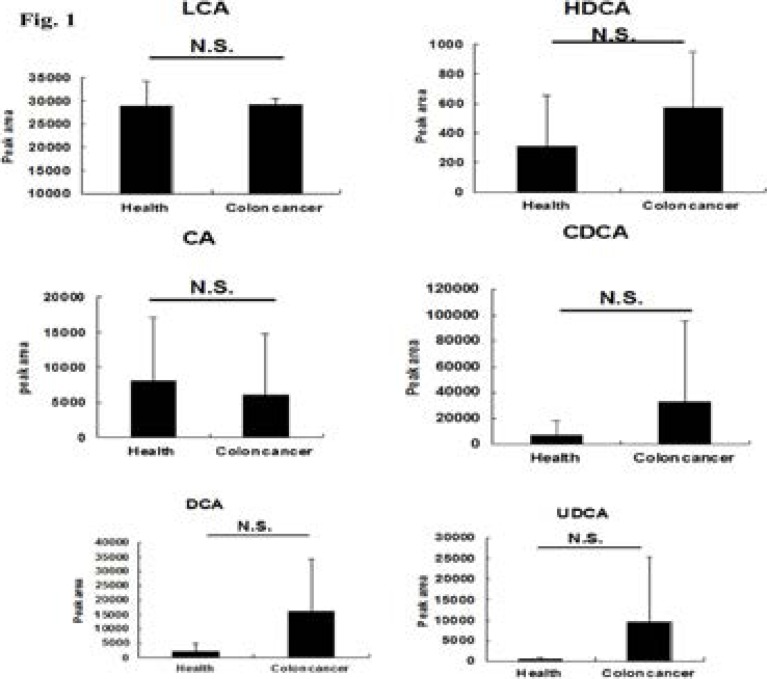
Fig. 1.
Comparison of free bile acids levels between healthy volunteers (n=5) and patients with colon cancers (n=5). The data were given as mean plus standard deviation (S.D.). N.S., not significant.
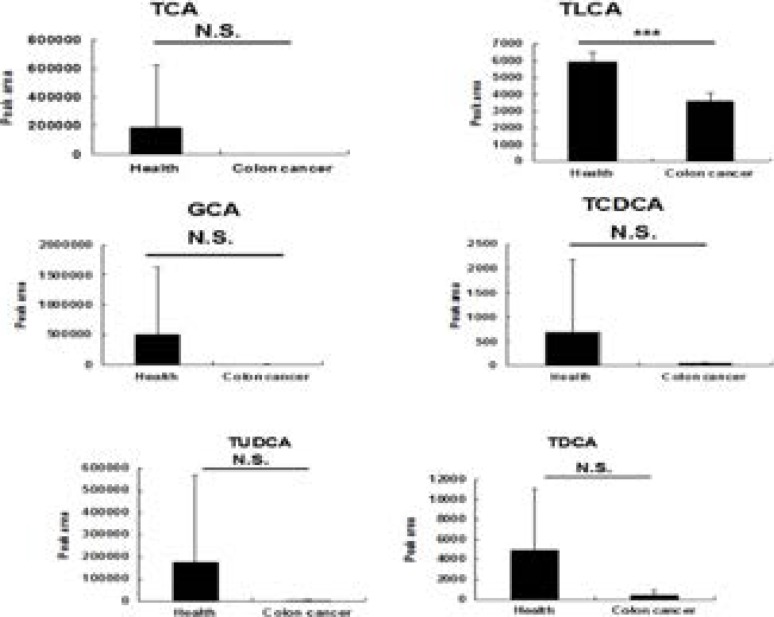
Fig. 2.
Comparison of conjugated bile acids levels between healthy volunteers (n=5) and patients with colon cancers (n=5). The data was given as mean plus standard deviation (S.D.). N.S., not significant, ***, p<0.001.
For free bile acids, compared with healthy volunteers, the increase trend was observed for HDCA, CDCA, DCA, and UDCA (although not significantly), respectively. The levels of the conjugated bile acids (TCA, TLCA, GCA, TCDCA, TUDCA, TDCA) decreased in patients with colon cancer, and the serum level of TLCA in patients with colon cancer significantly decreased in comparison with the health volunteers.
Discussion
After synthesis, most of bile acids immediately undergo the conjugation process with amino acids (glycine and taurine) to form the corresponding conjugates catalyzed by bile acid coenzyme A synthase (BACS) and bile acid amino acid transferase (BAAT)6.
Like drug conjugations, bile acids conjugates prevent Ca2+ precipitation, minimize passive absorption, and greatly prepare bile acids for efficient transport and detoxification6. In the intestine, the conjugated bile acids can be deconjugated. Colorectal cancer (CRC), the third most common cancer in both men and women, has been regarded as the second leading cause of cancer death in the United States7. In the patients with colon cancers, many normal functions might be damaged. For example, the iron homeostasis will be disrupted through the influence towards the expression of iron uptake and export proteins, such as divalent metal transporter-1 (DMT-1), ferroportin (FPN), and hephaestin (HEPH)8.
In the present study, the conjugation reaction of bile acids was demonstrated to be strongly affected in colon cancer, resulting the increased levels of free bile acids and decreased levels of bile acids conjugates in serum. This alteration of bile acids homeostasis can result in the change of some physiological function due to their important roles as cell signaling molecules9. For example, the activity of farnesoid X receptor (FXR) can be activated by chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), and lithocholic acid (LCA), and the activation of FXR enhances hepatocyte chemoprotection and liver tumor chemoresistance against genotoxic compounds10,11. Therefore, the increased levels of free bile acids in colon cancers might significantly induce the chemoresistance towards the anti-tumor drugs for colon cancers. It should be noted that the enzyme-catalyzed synthesis of bile acids might be affected, besides the conjugation reaction of bile acids which might be mainly affected.
Conclusion
The present study determined the serum level of bile acids in healthy volunteers and patients with colon cancers. Compared with the healthy volunteers, the free bile acids were detected to increase, and the conjugated bile acids were observed to decrease in patients with colon cancers. These data provide a new mechanism explanation and the potential biomarkers for colon cancers.
References
- 1.Poupon R, Chignard N, Rosmorduc O, Barbu V, Housset C. Biliary function and its regulation. Med Sci. 2004;20:1096–1099. doi: 10.1051/medsci/200420121096. [DOI] [PubMed] [Google Scholar]
- 2.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Sabel MS, Terjimanian M, Conlon AS, Griffith KA, Morris AM, Mulholland MW, Englesbe MJ, Holcombe S, Wang SC. Analytic morphometric assessment of patients undergoing colectomy for colon cancer. J Surg Oncol. 2013;108(3):169–175. doi: 10.1002/jso.23366. [DOI] [PubMed] [Google Scholar]
- 4.Leichtle AB, Nuoffer JM, Ceglarek U, Kase J, Conrad T, Witzigmann H, Thiery J, Fiedler GM. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics. 2012;8(4):643–653. doi: 10.1007/s11306-011-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fhaner CJ, Liu S, Ji H, Simpson RJ, Reid GE. Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal Chem. 2012;84(21):8917–8926. doi: 10.1021/ac302154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, Chiang JY. Nuclear receptors in bile acid metabolism. Drug Metab Rev. 2013;45(1):145–155. doi: 10.3109/03602532.2012.740048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 8.Xue X, Shah YM. Intestinal iron homeostasis and colon tumorigenesis. Nutrients. 2013;5:2333–2351. doi: 10.3390/nu5072333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaquero J, Monte MJ, Dominguez M, Muntané J, Marin JJ. Differential activation of the human farnesoid X receptor depends on the pattern of expressed isoforms and the bile acid pool composition. Biochem Pharmacol. 2013;pii doi: 10.1016/j.bcp.2013.07.022. S0006-2952(13)00501-7. [DOI] [PubMed] [Google Scholar]
- 10.Rizzo G, Renga B, Mencarelli A, Pellicciari R, Fiorucci S. Role of FXR in regulating bile acid homeostasis and relevance for human diseases. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5(3):289–303. doi: 10.2174/1568008054863781. [DOI] [PubMed] [Google Scholar]
- 11.Vaquero J, Briz O, Herraez E, Muntané J, Marin JJ. Activation of the nuclear receptor FXR enhances hepatocyte chemoprotection and liver tumor chemoresistance against genotoxic compounds. Biochim Biophys Acta. 2013;1833(10):2212–2219. doi: 10.1016/j.bbamcr.2013.05.006. (2013) [DOI] [PubMed] [Google Scholar]