Abstract
Background
Previous studies suggest that the X-ray repair cross-complementing group 3 gene (XRCC3) Thr241Met genetic variant could be potentially associated with the risk of prostate cancer. However, results from these published studies were conflicting rather than conclusive.
Objectives
his meta-analysis aimed to conduct a better understanding of the effects of XRCC3 Thr241Met genetic variant on prostate cancer risk.
Methods
We identified three eligible studies, 499 prostate cancer cases and 571 controls.
Results
Overall, significant associations were detected in the heterozygote comparison genetic model (CT versus (vs.) CC: OR = 0.71, 95% CI 0.53–0.94, Z =2.38, p= 0.017), and the dominant genetic model (TT/CT vs. CC: OR = 0.74, 95% CI 0.57–0.98, Z = 2.11, p =0.035). In the subgroup analysis by ethnicities, we found that this genetic variant was significantly associated with the decrease risk of prostate cancer in Caucasians for heterozygote comparison genetic model (CT vs. CC: OR = 0.66, 95% CI 0.44–0.98, Z = 2.04, p = 0.042).
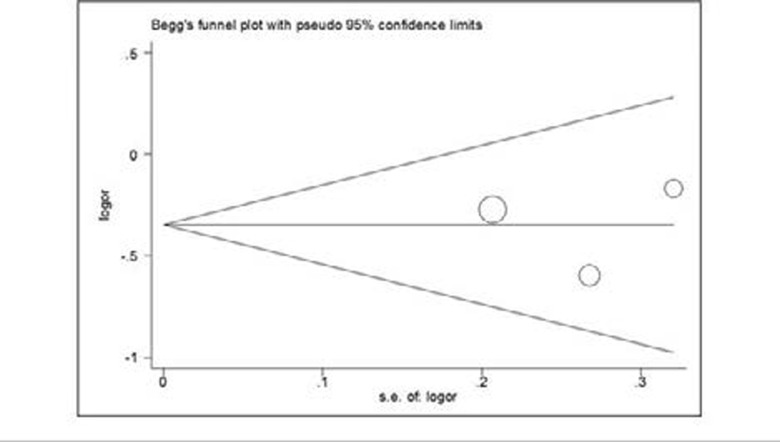
No publication bias was found in this study.
Conclusions
Results from this meta-analysis indicate that the XRCC3 Thr241Met genetic variant is associated with prostate cancer risk.
Keywords: Prostate cancer, XRCC3 gene, Genetic variant, Meta-analysis
Introduction
Prostate cancer is the most common malignancy of men in the world, accounting for 10% of men cancerrelated mortality1,2. The etiology of prostate cancer is largely unknown, although genetic and environmental factors might increase risk of prostate cancer2–6. The X-ray repair cross-complementing group 3 (XRCC3) is one of the DNA repair genes, and is an important candidate gene for mediating the genetic influence on prostate cancer7–13. The C18067T genetic variant in XRCC3 gene at exon 7(C>T, rs861539), one of the most studied functional genetic variants, results from a C to T mutation and causes the substitution of Threonine (Thr) to Methionine (Met) at codons 241 (p.Thr241Met), has been potentially associated with the risk of prostate cancer 7–10. However, results from published studies were conflicting rather than conclusive. Therefore, to clarify the effects of XRCC3 Thr241Met genetic variant on prostate cancer risk, we conducted a meta-analysis of all available published studies to date.
Materials and methods
Publication search
Pubmed, Excerpta Medica Database (EMBASE), and Chinese National Knowledge Infrastructure (CNKI) databases were searched using the search terms: “prostate cancer/neoplasm”, ȌXRCC3”,m“Thr241Met”, and Ȍrs8761539” (the last search was updated on June 2014). Publication searching was utilized without limitation on language and publication date. Two investigators searched the publication literature and extracted data independently.
Inclusion, exclusion criteria and Data extraction
For inclusion criteria in the present meta-analysis, the selected eligible articles had to provide information as follows:1 using a case-control design;2 evaluation of XRCC3 Thr241Met genetic variant with the risk of prostate cancer;3 offering enough data for estimating the odds ratios (ORs) and 95% confidence intervals (CIs);4 only full-text articles were included. The exclusion criteria of articles were as followed:1 duplication; 2 no usable data was provided;3 abstract, comment, letters, and review. For each eligible case-control articles, the following information was collected: the first author's name, publishing year, country, ethnicities, numbers of cases and controls, genotyping methods, numbers of allele and genotype.
Statistical analysis
The strength of the association of XRCC3 Thr241Met genetic variant with the risk of prostate cancer was assessed by the pooled ORs with their 95% CIs. Subgroup analyses were evaluated by ethnicities.
The significance of pooled ORs was determined by the Z-test. The heterogeneity assumption was evaluated by the chi-square-based Q-test14,15 and the I2 index 16. I2 index < 50% and/or P-value > 0.10 for Q-test indicated a lack of heterogeneity among the studies17. The fixed effect model (the Mantel-Haenszel method) was utilized to calculate the pooled ORs when the heterogeneity was not significant among the studies18. Otherwise, the random-effects model (the DerSimonian and Laird method) was employed19. The Begg's funnel plot and Egger's linear regression methods were used to assess the publication bias20,21. All analyses were analyzed by the STATA software (version 11.0; STATA Corporation, College Station, TX, USA). P-values < 0.05 were defined as statistically significant level.
Results
Eligible studies
According to the inclusion and exclusion criteria listed above for the association of XRCC3 Thr241Met genetic variant with the risk of prostate cancer, three eligible studies with 499 prostate cancer cases and 571 controls were finally included in this meta-analysis7–9. There were two studies of subjects of Caucasians decent7–9, and one study of Asians decent8. The study characteristics were presented in Table 1. The polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), and Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF) methods were determined to investigate the genotypes of XRCC3 Thr241Met genetic variant in these included studies.
Table 1.
The characteristics of eligible studies included in this meta-analysis.
| First author | Year | Country | Ethnicity | Genotyping methods |
No. (cases/controls) |
Case (%) | Control (%) | ||||
| CC | CT | TT | CC | CT | TT | ||||||
| Ritchey | 2005 | USA | Caucasians | MALDI-TOF | 159/247 | 139 | 17 | 3 | 214 | 31 | 2 |
| Mandal | 2010 | India | Asians | PCR-RFLP | 224/192 | 137 | 78 | 9 | 103 | 77 | 12 |
| Dhillon | 2011 | Australia | Caucasians | PCR-RFLP | 116/132 | 60 | 44 | 12 | 54 | 72 | 6 |
MALDI-TOF, Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry; PCR-RFLP, Polymerase Chain Reaction-Restriction Fragment Length Polymorphism
Meta-analysis
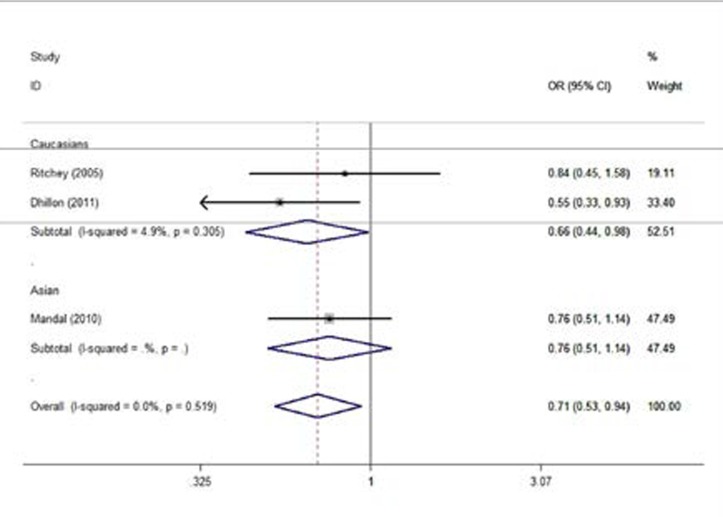
Table 2 summarized the association strength between XRCC3 Thr241Met genetic variant and the risk of prostate cancer. In the overall, significant associations were detected in heterozygote comparison genetic model (CT versus (vs.) CC: OR = 0.71, 95% CI 0.53–0.94, Z =2.38, P = 0.017, Table 2, Figure 1), and dominant genetic model (TT/CT vs. CC: OR = 0.74, 95% CI 0.57–0.98, Z =2.11, P = 0.035, Table 2).
Table 2.
The meta-analysis of XRCC3 Thr241Met genetic variant and prostate cancer risk.
| Comparisons | Population | Test of association | Test of Heterogeneity | ||||||
| N | OR (95% CI) | Z | P-value | Model | χ2 | P-value | I2(%) | ||
| TT vs. CC | Overall | 3 | 1.05(0.56–1.94) | 0.14 | 0.887 | F | 3.58 | 0.167 | 44.1 |
| Asians | 1 | 0.56(0.23–1.39) | 1.25 | 0.213 | F | - | - | - | |
| Caucasians | 2 | 1.91(0.77–4.73) | 1.40 | 0.161 | F | 0.05 | 0.815 | 0 | |
| CT vs. CC | Overall | 3 | 0.71(0.53–0.94) | 2.38 | 0.017 | F | 1.31 | 0.519 | 0 |
| Asians | 1 | 0.76(0.51–1.14) | 1.32 | 0.188 | F | - | - | - | |
| Caucasians | 2 | 0.66(0.44–0.98) | 2.04 | 0.042 | F | 1.05 | 0.305 | 4.9 | |
| TT/CT vs. CC | Overall | 3 | 0.74(0.57–0.98) | 2.11 | 0.035 | F | 0.86 | 0.650 | 0 |
| Asians | 1 | 0.74(0.50–1.09) | 1.54 | 0.122 | F | - | - | - | |
| Caucasians | 2 | 0.75(0.51–1.11) | 1.44 | 0.150 | F | 0.85 | 0.355 | 0 | |
| TT vs. CT/CC | Overall | 3 | 1.37(0.51–3.73) | 0.62 | 0.534 | R | 4.43 | 0.109 | 54.8 |
| Asians | 1 | 0.63(0.26–1.52) | 1.03 | 0.304 | F | - | - | - | |
| Caucasians | 2 | 2.41(1.00–5.82) | 1.95 | 0.051 | F | 0 | 0.979 | - | |
| T vs. C | Overall | 3 | 0.85(0.68–1.06) | 1.47 | 0.142 | F | 0.91 | 0.634 | 0 |
| Asians | 1 | 0.76(0.56–1.05) | 1.65 | 0.100 | F | - | - | - | |
| Caucasians | 2 | 0.93(0.68–1.27) | 0.45 | 0.653 | F | 0.17 | 0.680 | 0 | |
N, number of comparisons; OR, odds ratio; CI, confidence interval; vs., versus; TT vs. CC: Homozygote comparison; CT vs. CC: Heterozygote comparison; TT/CT vs. CC: Dominant model; TT vs. CT/CC: Recessive model; T vs. C: Allele comparison; R, random effect model; F, fixed effect model; Random effect model was chosen when P-value < 0.10 and/or I2 > 50% for heterogeneity test; otherwise fixed effect model was used.
Figure 1.
Forest plots of the association between XRCC3 Thr241Met genetic variant and prostate cancer risk (Heterozygote comparison by ethnicities (CT versus. CC)).
In the subgroup analysis by ethnicities, we found that the XRCC3 Thr241Met genetic variant was significantly associated with the decrease risk of prostate cancer in Caucasians for heterozygote comparison genetic model (CT vs. CC: OR = 0.66, 95% CI 0.44–0.98, Z = 2.04, P = 0.042, Table 2). Our data indicated that there were no significant associations between XRCC3 Thr241Met genetic variant and prostate cancer risk in other genetic models (All P-values >0.05, Table 2). No evidence of publication bias was found in all comparison genetic models (All P-values > 0.05).
Discussion
Emerging evidence suggest that the XRCC3 is one of the most important candidate genes for influencing the risk of prostate cancer, and several studies have carried out to investigate the potential association of XRCC3 Thr241Met genetic variant with the risk of prostate cancer. Ritchey and colleagues reported that XRCC3 Thr241Met genetic variant showed no significant associations with the risk of prostate cancer, while a significant interaction was found for XRCC3 Thr241Met genetic variant and consumption of total preserved foods7. Mandal suggested that no significant association of XRCC3 Thr241Met genetic variant genotypes with the risk of prostate cancer was observed8. Dhillon demonstrated that there was no association between the XRCC3 Thr241Met genetic variant and prostate cancer risk 9. The present meta-analysis, including 499 prostate cancer cases and 571 controls concerning the XRCC3 Thr241Met genetic variant, explored the more reliable association between Thr241Met genetic variant in XRCC3 gene and the risk of prostate cancer. Overall, we detected that this genetic variant were significantly associated with the risk of prostate cancer. Besides, in the subgroup analysis by ethnicities, we found that this genetic variant was significantly associated with the decrease risk of prostate cancer only in Caucasians population. Thus, results from this meta-analysis indicate that the XRCC3 Thr241Met genetic variant is associated with prostate cancer risk.
Some advantages of this meta-analysis should be addressed. First, a strict searching strategy to enroll all the possible eligible articles as much as possible was conducted. Second, all included articles had acceptable quality. Third, the whole pooled findings are unbiased. However, some limitations of this meta-analysis should be addressed. Firstly, only three eligible articles were eventually enrolled in this meta-analysis. Secondly, the enrolled articles only concerned about Asians and Caucasians, not mentioned about other ethnicities. Thirdly, only published articles were enrolled, unpublished articles were not enrolled in this study.
Conclusion
This meta-analysis provided evidence of the association of XRCC3 Thr241Met genetic variant with risk of prostate cancer. More well-designed studies in large populations should be carried out to confirm these findings.
Figure 2.
Begg's funnel plot for publication bias test (Heterozygote comparison (CT versus. CC)).
Acknowledgements
This work was supported by National Natural Science Foundation of China (31101148).
Conflict of Interest
None.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Geng J, Zhang Q, Zhu C, Wang J, Chen L. XRCC1 genetic polymorphism Arg399Gln and prostate cancer risk: a meta-analysis. Urology. 2009;74:648–653. doi: 10.1016/j.urology.2009.02.046. [DOI] [PubMed] [Google Scholar]
- 3.Pienta KJ, Esper PS. Risk factors for prostate cancer. Ann Intern Med. 1993;118:793–803. doi: 10.7326/0003-4819-118-10-199305150-00007. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13 Spec No 1:R103–R121. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- 6.Wei B, Zhou Y, Xu Z, Ruan J, Zhu M, Jin K, Zhou D, Hu Q, Wang Q, Wang Z, Yan Z. XRCC1 Arg399Gln and Arg194Trp polymorphisms in prostate cancer risk: a meta-analysis. Prostate Cancer Prostatic Dis. 2011;14:225–231. doi: 10.1038/pcan.2011.26. [DOI] [PubMed] [Google Scholar]
- 7.Ritchey JD, Huang WY, Chokkalingam AP, Gao YT, Deng J, Levine P, Stanczyk FZ, Hsing AW. Genetic variants of DNA repair genes and prostate cancer: a population-based study. Cancer Epidemiol Biomarkers Prev. 2005;14:1703–1709. doi: 10.1158/1055-9965.EPI-04-0809. [DOI] [PubMed] [Google Scholar]
- 8.Mandal RK, Kapoor R, Mittal RD. Polymorphic variants of DNA repair gene XRCC3 and XRCC7 and risk of prostate cancer: a study from North Indian population. DNA Cell Biol. 2010;29:669–674. doi: 10.1089/dna.2010.1047. [DOI] [PubMed] [Google Scholar]
- 9.Dhillon VS, Yeoh E, Fenech M. DNA repair gene polymorphisms and prostate cancer risk in South Australia—results of a pilot study. Urol Oncol. 2011;29:641–646. doi: 10.1016/j.urolonc.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Ben Salah G, Fendri-Kriaa N, Kamoun H, Kallabi F, Mkaouar-Rebai E, Fourati A, Ayadi H, Fakhfakh F. An interethnic variability and a functional prediction of DNA repair gene polymorphisms: the example of XRCC3 (p.Thr241>Met) and XPD (p.Lys751>Gln) in a healthy Tunisian population. Mol Biol Rep. 2012;39:9639–9647. doi: 10.1007/s11033-012-1829-z. [DOI] [PubMed] [Google Scholar]
- 11.Burri RJ, Stock RG, Cesaretti JA, Atencio DP, Peters S, Peters CA, Fan G, Stone NN, Ostrer H, Rosenstein BS. Association of single nucleotide polymorphisms in SOD2, XRCC1 and XRCC3 with susceptibility for the development of adverse effects resulting from radiotherapy for prostate cancer. Radiat Res. 2008;170:49–59. doi: 10.1667/RR1219.1. [DOI] [PubMed] [Google Scholar]
- 12.Fachal L, Gomez-Caamano A, Peleteiro P, Carballo A, Calvo-Crespo P, Sanchez-Garcia M, Lobato-Busto R, Carracedo A, Vega A. Association of a XRCC3 polymorphism and rectum mean dose with the risk of acute radio-induced gastrointestinal toxicity in prostate cancer patients. Radiother Oncol. 2012;105:321–328. doi: 10.1016/j.radonc.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 14.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 15.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 16.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 18.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 20.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]