Abstract
Methamphetamine (METH) abuse is frequent in individuals infected with human immunodeficiency virus type-1 (HIV-1) and is suspected to aggravate HIV-associated neurocognitive disorders (HAND). METH is a psychostimulant that compromises several neurotransmitter systems and HIV proteins trigger neuronal injury but the combined effects of viral infection and METH abuse are incompletely understood. In this study we treated transgenic mice expressing the HIV envelope protein gp120 in the brain (HIV/gp120tg) at 3–4 months of age with an escalating-dose, multiple-binge METH regimen. The long-term effects were analyzed after 6–7 months of drug abstinence employing behavioral tests and analysis of neuropathology, electrophysiology and gene expression. Behavioral testing showed that both HIV/gp120tg and WT animals treated with METH displayed impaired learning and memory. Neuropathological analysis revealed that METH similar to HIV/gp120 caused a significant loss of neuronal dendrites and pre-synaptic terminals in hippocampus and cerebral cortex of WT animals. Electrophysiological studies in hippocampal slices showed that METH exposed HIV/gp120tg animals displayed reduced post-tetanic potentiation, whereas both gp120 expression and METH lead to reduced long-term potentiation. A quantitative reverse transcription-polymerase chain reaction array showed that gp120 expression, METH and their combination each caused a significant dysregulation of specific components of GABAergic and glutamatergic neurotransmission systems, providing a possible mechanism for synaptic dysfunction and behavioral impairment. In conclusion, both HIV-1/gp120 and METH caused lasting behavioral impairment in association with neuropathology and altered gene expression. However, combined METH exposure and HIV-1/gp120 expression resulted in the most pronounced, long lasting pre-and post-synaptic alterations coinciding with impaired learning and memory.
Keywords: Methamphetamine, NeuroAIDS, HIV-1 gp120, transgenic animal model, behavior, neuropathology, electrophysiology, gene expression
Introduction
Individuals infected with HIV-1 are at risk of developing HIV-associated neurocognitive disorders (HAND) despite the availability of combined antiretroviral therapy (Antinori et al., 2007; Heaton et al., 2010; Kraft-Terry et al., 2009). Clinical signs of HAND/dementia are closely associated with neuropathological evidence of glutamatergic excitotoxicity (Heyes et al., 1991), an increased number of microglia (Glass et al., 1995; Wesselingh et al., 1997), decreased synaptic and dendritic density (Masliah et al., 1997), and selective neuronal loss (Masliah et al., 1992). HIV infection of the brain also affects the dopaminergic system, and neurocognitive impairment is associated with reduced DA levels in cerebrospinal fluid (Berger et al., 1994; Nath et al., 2000).
Abuse of Methamphetamine (METH) increases the risk of HIV-1 infection (Kapadia et al., 2005; Mitchell et al., 2006; Urbina and Jones, 2004). The psychostimulant effect of METH apparently results from an elevated extracellular dopamine (DA) concentration in the striatum, but METH abuse also impairs attention, working memory and executive functions (Albertson et al., 1999; Scott et al., 2007; Sulzer et al., 1995; Theodore et al., 2007). The various affected brain structures and neural circuits suggest pathological consequences beyond the dopaminergic system (Cass, 1997; Thompson et al., 2004).
Similarly, animal studies confirmed that METH compromises several neurotransmitter systems, affecting dopaminergic, serotonergic, gamma-amino butyric acid (GABA)-ergic, and glutamatergic neural networks (Ferris et al., 2008; Krasnova and Cadet, 2009; Scott et al., 2007; Theodore et al., 2007). In the striatum, METH destroys specifically GABAergic neurons that express enkephalin (Jayanthi et al., 2005). Moreover, METH can trigger neuronal cell death in cortex, striatum, and hippocampus, and METH has been linked to glutamate-mediated excitotoxicity (Eisch and Marshall, 1998; Schmued and Bowyer, 1997; Yamamoto and Bankson, 2005).
In combination, METH and HIV-1 appear to cause more neurocognitive deficits and neuropathology than either agent alone (Cadet and Krasnova, 2007; Carey et al., 2006; Flora et al., 2003; Langford et al., 2003; Nath et al., 2000). In vivo and in vitro studies showed that acute exposure to viral envelope protein gp120 or transactivator of transcription (Tat) and METH causes oxidative stress, expression of inflammatory factors, such as TNFα and IL-1β and neurotoxicity (Flora et al., 2003; Maragos et al., 2002; Nath et al., 2000; Silverstein et al., 2011; Silverstein et al., 2012). In contrast, pre-treatment with low doses of METH seems to blunt acute toxic effects of high dosages while also resulting in changes of gene expression in the brain (Cadet et al., 2011; Cadet et al., 2009).
METH use is considered to be a co-morbidity of HIV-1 infection and since many patients develop HAND despite effective combined antiretroviral therapy the question arises in how far previous METH use may play a promoting role in neurocognitive deterioration (Antinori et al., 2007; Brew et al., 2009; Heaton et al., 2010; Kraft-Terry et al., 2009). However, the long-term effects of early and temporary METH abuse in combination with chronic viral infection are incompletely understood (Carey et al., 2006; Langford et al., 2003).
Transgenic mice expressing the envelope protein gp120 of HIV-1 in their brain under the control of the promotor for glial fibrillary acidic protein (HIV-1 gp120tg) manifest several neuropathological features observed in brains of AIDS patients, such as decreased synaptic and dendritic density, increased numbers of activated microglia and pronounced astrocytosis (Toggas et al., 1994). Recently, we showed that HIV-1 gp120tg mice are more sensitive than wild-type (WT) mice with regard to acute stereotypic effects of METH exposure, while being less differentially responsive to the locomotor stimulant effects of the drug (Roberts et al., 2010). In order to explore potential long-term effects of former METH use on HIV-associated neuronal injury we exposed in the present study 3–4 months old HIV-1 gp120tg mice and WT controls to an escalating-dose, multiple-binge METH regimen and analyzed 6–7 months later behavior, neuropathology, hippocampal long-term potentiation (LTP) and RNA expression of components of glutamatergic and GABAergic neurotransmission. Our findings indicate that METH and HIV-1 components in combination can aggravate their respective pathological effects on the brain in a long-lasting fashion.
Materials and Methods
Animals and drug treatment
Age- and sex matched 3–4 months old HIV-1 gp120tg and non-transgenic littermate control mice (WT) (Toggas et al., 1994) were s. c. injected with a sterile-filtered solution of METH ((+)-Methamphetamine hydrochloride, M-8750, Sigma-Aldrich, St. Louis, WA) or with saline (SAL, vehicle control) (Roberts et al., 2010). METH was given for 25 days in an established escalating-dose, multiple-binge regimen that was developed to recapitulate a human usage pattern and avoids hyperthermia (Henry et al., 2013; Kuczenski et al., 2007). Briefly, in the first 14 days, mice were injected 3 times a day, starting from 0.1 mg/kg and increasing step-wise by 0.1 mg/kg with each injection up to 4.0 mg/kg. This period was followed by an 11-day ‘binge’ with 4 injections per day of 6.0 mg/kg in 2-hour intervals and a 6–7 months long period without further METH exposure. All experimental procedures and protocols involving animals were in accordance with NIH guidelines and approved by the Institutional Animal Care and Use Committees of the Sanford-Burnham Medical Research Institute and The Scripps Research Institute.
Behavioral testing
At 10–11 months of age, a sex-matched cohort of each experimental group was subjected to behavioral testing (HIV-1 gp120tg SAL n = 11, HIV-1 gp120tg METH n = 10, WT SAL n = 8, WT METH n = 10). Tests were performed in the order below, separated by 3–7 days. i) Locomotor activity test: Locomotor activity was measured in polycarbonate cages placed into frames mounted with two levels of photocell beams at 2 and 7 cm above the bottom of the cage (San Diego Instruments, San Diego, CA). These two sets of beams allow for the recording of both horizontal (ambulation, center activity and total horizontal activity) and vertical (rearing) behavior. Mice were tested for 120 min. For the purpose of analysis and graphing, the 120 min period was divided into time blocks of 24 × 5 min epochs. Mean values were calculated for each epoch and experimental group. ii) Optomotor vision test: This test used a stationary elevated platform surrounded by a drum with black and white striped walls. The mouse was habituated on the platform to for one min and then the drum rotated at 2 rpm in one direction for one min, stopped for 30 sec, and then rotated in the other direction for one min. The total number of head tracks was recorded. iii) Barnes maze test: The Barnes maze is a spatial learning and memory test sensitive to impaired hippocampal function (Bach et al., 1995; Barnes, 1979; Holmes et al., 2002; Paylor et al., 2001). Distinct spatial cues were located all around the maze and were kept constant throughout the study. On the first day of testing, a training session was performed, which consisted of placing the mouse in the escape box for one min before the first session was started. At the beginning of each session, the mouse was placed in the middle of the maze in a 10 cm high start chamber. After 10 sec the start chamber was removed, a buzzer (80 dB) and a light (400 lux) were turned on, and the mouse was set free to explore the maze. The session ended when the mouse entered the escape tunnel or after 3 min elapsed. Mice were tested once a day for 9 days for the acquisition portion of the study. For the probe test (day 10), the escape tunnel was removed and the mouse was allowed to freely explore the maze for 3 min. The time spent in each quadrant was determined and the percent time spent in the target quadrant (the one originally containing the escape box) was compared with the average percent time in the other three quadrants. Each session was videotaped and scored by an experimenter blind to the genotype and group of the mouse. Measures recorded included the latency to escape the maze, the number of errors made per session, and the strategy employed by the mouse to locate the escape tunnel. Search strategies were classified according to three operationally defined categories: 1) Random search strategy - localized hole searches separated by crossings through the center of the maze, 2) Sequential search strategy - systematic hole searches (every hole or every other hole) in a clockwise or counterclockwise direction, or 3) Spatial search strategy - reaching the escape tunnel with both error and distance (number of holes between the first hole visited and the escape tunnel) scores of less than or equal to 3.
Immunostaining and deconvolution microscopy
Immunostainings of sagittal brain sections were performed as previously published (Kang et al., 2010; Maung et al., 2014; Maung et al., 2012) with minor modifications. Briefly, mice were terminally anesthetized and immediately transcardially perfused with 0.9% saline. Brains were quickly removed and fixed for 48 hours in 4% paraformaldehyde. 30 µm thick brain sections were stained with antibodies to MAP-2 (Sigma, M-4403; 1 : 250) and Synaptophysin (Syp; Dako, A0010; 1 : 250). Additional sections were stained with mouse IgG1 (MOPC21, Sigma, M-9269) serving as isotype control for the MAP-2 antibody, or secondary antibody alone. Secondary antibodies were goat-anti-mouse-Rhodamine Red X (Jackson ImmunoResearch; 1 : 125) and goat-anti-rabbit Alexa Fluor 488 (Invitrogen; 1 : 1,000). Nuclei were counterstained with Hoechst 33342 (Invitrogen). Stained brain slices were mounted on glass slides and covered with Vectashield mounting medium (Vector Labs, H-1000). Investigators blinded to treatment and genotype performed the microscopy analysis. Images were recorded with 0.5 µm steps along the Z-axis on a Zeiss Axiovert 200M microscope using a 40×/0.75 EC Plan-Neofluar objective and constant exposure times. The images were deconvolved and analyzed using Slidebook Software (Intelligent Imaging Innovations, Denver, CO). To assess neuronal injury, we estimated the percentage of MAP-2 and Syp-positive neuropil as a measure for neurites and presynaptic terminals, respectively, in layer III of the fronto-parietal cortex and the molecular layer of the CA1 region of the hippocampus. The volume of neuropil occupied by MAP-2-labeled processes or Syp-positive presynaptic terminals was estimated by threshold segmentation and calculated as the ratio of fluorescent to total volume of each image stack and expressed in percent.
Electrophysiology experiments
Hippocampal slices were prepared from male WT or HIV-1 gp120tg mice with a mean age of 10.5 months. Mice were killed by decapitation under deep terminal anesthesia with isoflurane. Brains were surgically exposed, gently scooped out of the skull and placed in a Petri dish filled with oxygenated ice-cold sucrose-substituted artificial cerebrospinal fluid (aCSF; in mM, NaCl, 125; KCl, 3; NaHCO3, 25; NaH2PO4, 1.25; Glucose, 10; MgCl2, 1; and CaCl2, 2, aerated with 95% O2 and 5% CO2). Slices (400°µm thick) containing the hippocampal formation and entorhinal cortex were kept in a holding chamber containing oxygenated aCSF at 32°C for a minimum of 2 hours before being transferred to the multielectrode array (MEA; MEA60 Multi Channel Systems, Reutlingen, Germany) for electrophysiological recording.
While in the recording chamber, slices were continuously superfused with oxygenated aCSF at an estimated flow rate of 12 ml/min. Slices were left to recover for 15–30 min while the background spontaneous electrical activity was continuously monitored. We used a two-input protocol for orthodromic stimulation of Schäffer collateral axonal fibers (CA3→CA1) located in the stratum radiatum. The distance between the test and control inputs was between 600 to 800 µm. A single electrode located on the CA3 side was chosen as the test input while a second electrode located on the subicular side of the recording field was used as the control input. Electrical stimulation evoked field excitatory post synaptic potentials (fEPSP) in CA1 pyramidal neurons and long-term potentiation (LTP) was induced by a series of 4 high frequency stimuli (tetanic stimulation) following published protocols (Besl and Fromherz, 2002; Borkholder et al., 1997). The parameters used for data analysis were slope of the fEPSP amplitude of the presynaptic fiber volley and the intensity of the stimulus.
GABA and glutamate RT2 Profiler™ PCR Array
Mice were terminally anesthetized and immediately transcardially perfused with 0.9% saline. Brains were quickly removed, snap frozen in liquid nitrogen and stored at −80°C. RNA was purified from mouse hemibrains using a Qiagen RNeasy kit. Samples were analyzed for expression of 84 genes involved in the GABA and glutamate neurotransmitter systems by RT2 Profiler™ PCR Arrays (PAMM-152) following the supplier’s instructions (SABioscience/Qiagen). The arrays were run on a ViiA7™ Real-Time PCR System (Applied Biosystems/Life Technologies). The RT2 Profiler™ PCR Array Data Analysis software package (version 3.5) used 2−(ΔΔCT)–based fold change calculations (Livak and Schmittgen, 2001) and a modified Student’s t-test to compute two-tail, equal variance p-values. Data were normalized to the housekeeping genes hypoxanthine guanine phosphoribosyl transferase (Hprt), heat shock protein 90 alpha (cytosolic), class B member 1 (Hsp90ab1), and glyceraldehyde-3-phosphate dehydrogenase (Gapdh).
Bioinformatics analysis
A list of 20 genes out of 84 tested showed significant fold changes of RNA expression in at least one of the comparisons between the experimental groups and was further analyzed for identification of functional gene networks using Ingenuity Pathway Analysis (IPA; Ingenuity® Systems, www.ingenuity.com; build version: 261899; content version: 18030641; release date: 2013-12-06). The following settings were used: Reference set: Ingenuity Knowledge Base (Genes + Endogenous Chemicals); Relationship to include: Direct; Includes Endogenous Chemicals; Optional Analyses: My Pathways My List; Filter Summary: Consider only molecules and/or relationships where (confidence = Experimentally Observed) AND (tissues/cell lines = Cerebral Ventricles OR SF-539 OR Amygdala OR Parietal Lobe OR Brainstem OR Trigeminal Ganglion OR Spinal Cord OR U251 OR Sciatic Nerve OR Other Nervous System OR Olfactory Bulb OR Hypothalamus OR Brain OR Pituitary Gland OR Nucleus Accumbens OR Putamen OR Substantia Nigra OR SNB-75 OR U87MG OR Cerebellum OR Striatum OR Granule Cell Layer OR Other Neuroblastoma Cell Lines OR Thalamus OR Cerebral Cortex OR Nervous System not otherwise specified OR Other CNS Cell Lines OR SF-268 OR Dorsal Root Ganglion OR Caudate Nucleus OR Subventricular Zone OR Neuroblastoma Cell Lines not otherwise specified OR SNB-19 OR Medulla Oblongata OR Ventricular Zone OR Gray Matter OR Hippocampus OR SK-N-SH OR White Matter OR CNS Cell Lines not otherwise specified OR Choroid Plexus OR Corpus Callosum OR SF-295).
Statistical analysis
StatView software (version 5.0.1; SAS Institute Inc.) was used for statistical analysis. If not indicated otherwise, ANOVA with Fisher’s protected least significant difference (PLSD) post hoc tests were performed. Values indicate mean ± SEM. For the behavioral tests, ANOVAs included the factors genotype (WT vs. HIV-1 gp120tg), group (SAL vs. METH), sex (male vs. female) as well as a within subjects factor time (locomotor test) or trial (Barnes maze test). Where significant effects were observed, follow-up 2-way ANOVAs were performed. In addition, analyses determined a priori were performed in order to specifically examine the difference between WT and HIV-1 gp120tg mice treated with SAL, the effect of METH in each of the two genotypes, as well as a comparison of METH effects in the two genotypes.
Results
Behavioral changes due to HIV-1/gp120 expression and METH treatment
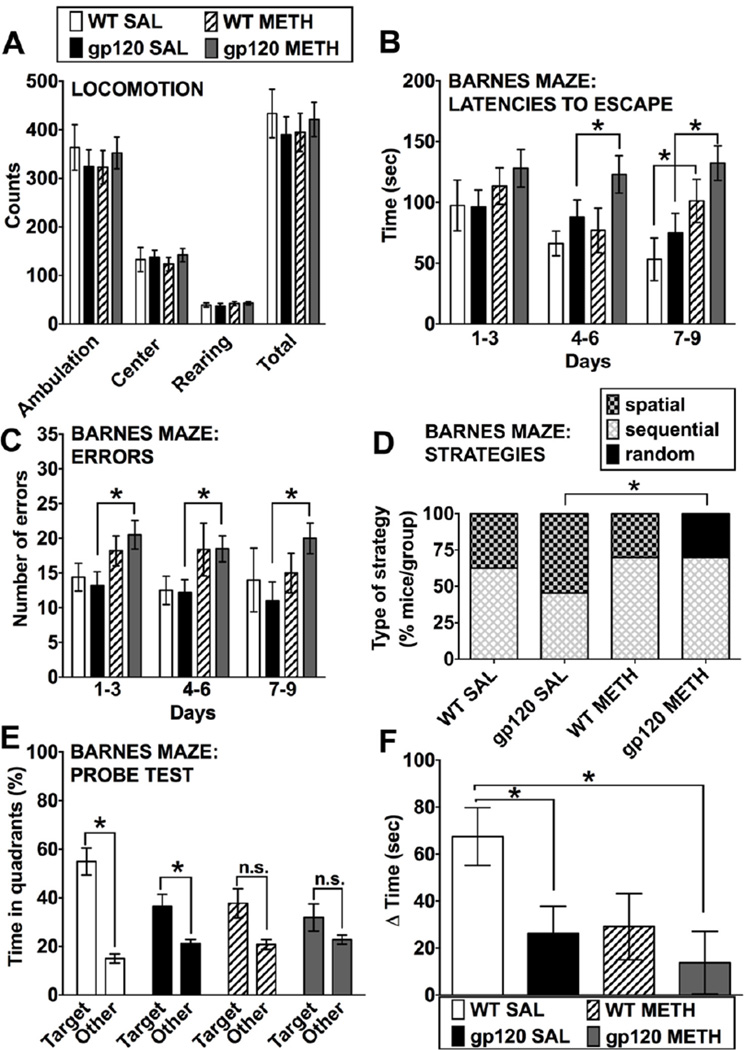
Seven months after exposure to the METH treatment, at 10–11 months of age, sex-matched groups of HIV-1 gp120tg mice and WT controls were tested for behavior. The capability of the animals to move (locomotion test, Figure 1A) and optomotor vision test (data not shown) was determined to be normal in all four experimental groups. The animals’ ability to move was determined by counting ambulation, rearing, center activity and the total horizontal activity of the mice. All mice were tested for 120 min and for the purpose of analysis and graphing, the 120 min period was divided into time blocks of 24 × 5 min epochs. During the 120 min time period the counts for all activities decreased in all animals of all experimental groups with no significant differences between the groups. Ambulation: While there were overall effects of time (F(23,736) = 61.1, p < 0.0001) and sex (F(1,32) = 4.24, p < 0.05), there were no effects of genotype (F(1,32) = 1.3, p > 0.05), group (F(1,32) = 0.98, p > 0.05) or their interaction (F(1,32) = 0.26, p > 0.05). Rearing: While there was an overall effect of time (F(23,736) = 36.5, p < 0.0001), there were no effects of genotype (F(1,32) = 0.10, p > 0.05), group (F(1,32) = 0.01, p > 0.05), sex (F(1,32) = 3.1, p > 0.05) or the interaction between genotype and group (F(1,32) = 0.04, p > 0.05). Center activity: While there were overall effects of time (F(23,736) = 41.1, p < 0.0001) and sex (F(1,32) = 6.52, p < 0.05), there were no effects of genotype (F(1,32) = 0.86, p > 0.05), group (F(1,32) = 0.32, p > 0.05) or their interaction (F(1,32) = 0.10, p > 0.05). Total horizontal activity: While there were overall effects of time (F(23,736) = 63.4, p < 0.0001) and sex (F(1,32) = 5.0, p < 0.05), there were no effects of genotype (F(1,32) = 1.2, p > 0.05), group (F(1,32) = 0.78, p > 0.05) or their interaction (F(1,32) = 0.18, p > 0.05). Figure 1A displays the mean values ± s.e.m. for the first 5 min epoch of the testing.
Figure 1.
Behavioral assessment of METH-treated WT and HIV-1 gp120tg mice after seven months of drug abstinence. At 10–11 months, an age- and sex-matched cohort of each group (WT SAL n = 8, WT METH n = 10, HIV-1 gp120tg SAL n = 11, HIV-1 gp120tg METH n = 10) was assessed for behavioral changes. All mice were subjected to all tests. (A) Locomotion tests included measures of ambulation, rearing, center activity and total horizontal activity of the mice. Values are shown for the first 5 min epoch of the testing. For a total of 9 days, shown in consecutive 3-day blocks, animals were assessed in the Barnes maze test: (B) The latencies to enter the escape hole over time (3 day blocks) were determined. (C) The number of errors made over time (3 day blocks) by each animal before finding the correct escape hole (Fig. 1 continued) was determined. (D) Strategies for escaping the maze shown as % of animals per group on the final acquisition trial revealed group differences. Specifically, the use of spatial strategy on the final acquisition trial was greater in HIV-1 gp120tg SAL vs. METH-treated mice. (E, F) The mean percentage of time spent in the target and non-target (‘other’) quadrants was determined in the probe test on day 10. (E): Examining the Target vs. Other difference separately in each group. (F) The differences between average times spent in the target and the other quadrants. All data represented are means ± SEM. Statistical analysis was performed as described in Materials and Methods. * p ≤ 0.05; ANOVAs and post hoc tests (A, B, E, F), Wald-Wolfowitz Test (D). For detailed outcomes of statistical analyses see the Results section. Note that for clarity significant changes between the trial/time blocks in (B) are only reported in the text.
Overall, sex differences (female > male) were observed only in the locomotor activity measures, but sex did not interact with genotype (gp120tg or non-tg) or group (METH or SAL). No sex differences were detected in head tracking behavior in the optomotor test or in any measure in the Barnes maze test. Therefore, we concluded that sex did not impact the behavioral effects gp120 expression, METH exposure, or the combination of these factors in our animal cohort and we collapsed across sex for post-hoc analyses in the Barnes maze test in which we found effects of genotype and/or group.
To assess cognitive brain functions and spatial reference memory, the animals were tested in a 20-hole Barnes maze (Figure 1B–F). Over an acquisition period of 9 days, the animals were monitored for latencies to escape, number of errors and strategy employed.
Latencies
There were significant effects of trial block (time; F(8,248) = 2.9, p < 0.01) and group (F(1,31) = 13.8, p < 0.001), but no effects of genotype (F(1,31) = 1.47, p > 0.05), sex (F(1,31) = 0.01, p > 0.05), or any interaction between factors. Overall, latencies decreased across trial blocks and METH increased latencies. More specifically, METH increased latencies in WT mice (F(1,16) = 5.9, p < 0.05) and in HIV-1 gp120tg mice (F(1,19) = 7.4, p < 0.05). Post-hoc Fisher’s PLSD tests for each block showed significant differences between SAL and METH HIV-1 gp120tg mice in the 4–6 day and 7–9 day blocks (p < 0.02); whereas METH only increased latencies in WT mice in the 7–9 day block ( p < 0.03; Figure 1B).
Errors
There was a significant effect of group (F(1,31) = 7.1, p < 0.05), but no effects of genotype (F(1,31) = 0.02, p > 0.05), sex (F(1,31) = 1.4, p > 0.05), or any interaction between factors. Further analysis of the group effect revealed a significant effect of METH in HIV-1 gp120tg mice (F(1,19) = 6.5, p = 0.02), but no effect in WT mice (F(1,16) = 1.5, p > 0.05). Posthoc testing supported this former effect across all three trial blocks (p < 0.05; Figure 1C).
Strategies
Specifically, the use of spatial strategy on the final acquisition trial was greater in HIV-1 gp120tg SAL vs. METH-treated mice (Wald-Wolfowitz Test (z (2.68), p = 0.007). On the final acquisition trial, previous METH exposure was associated with a complete lack of spatial strategy use in HIV-1 gp120tg animals whereas use of spatial strategy was not affected in WT mice (Figure 1D).
For further determination of spatial memory, mice were subjected to the probe test 24 hours after the last acquisition test, on day 10. Probe test: There was an overall effect of genotype when Target and Other times were collapsed (F(1,31) = 4.8, p < 0.05); however, there were no overall effects of group (F(1,31) = 3.3, p > 0.05), sex (F(1,31) = 0.35, p > 0.05), or any interaction between these factors. There was an overall within-subjects effect of quadrant (Target > Other; F(1,31) = 29.1, p < 0.0001) and a quadrant × genotype interaction (F(1,31) = 4.8, p < 0.05). WT mice had greater Target vs. Other differences than HIV-1 gp120tg mice. Examining the Target vs. Other difference separately in each group, only the WT SAL (ANOVA: F(1,7) = 29.0, p = 0.001) and HIV-1 gp120tg SAL (F(1,10) = 5.42, p = 0.042) groups spent significantly more time in the target quadrant than the average of the other quadrants, an indicator of spatial learning and memory (Figure 1E), whereas neither METH-treated group did. The differences between average times spent in the target and the other quadrants were significantly lower for both HIV-1 gp120tg SAL and METH than WT SAL animals (Fisher’s PLSD: p = 0.035 for WT SAL vs. HIV-1 gp120tg SAL; p = 0.008 for WT SAL vs. HIV-1 gp120tg). Thus, METH-treated HIV-1 gp120tg mice displayed the smallest difference between the average time spent in the target and other quadrants (Figure 1 F) indicating the most compromised memory performance.
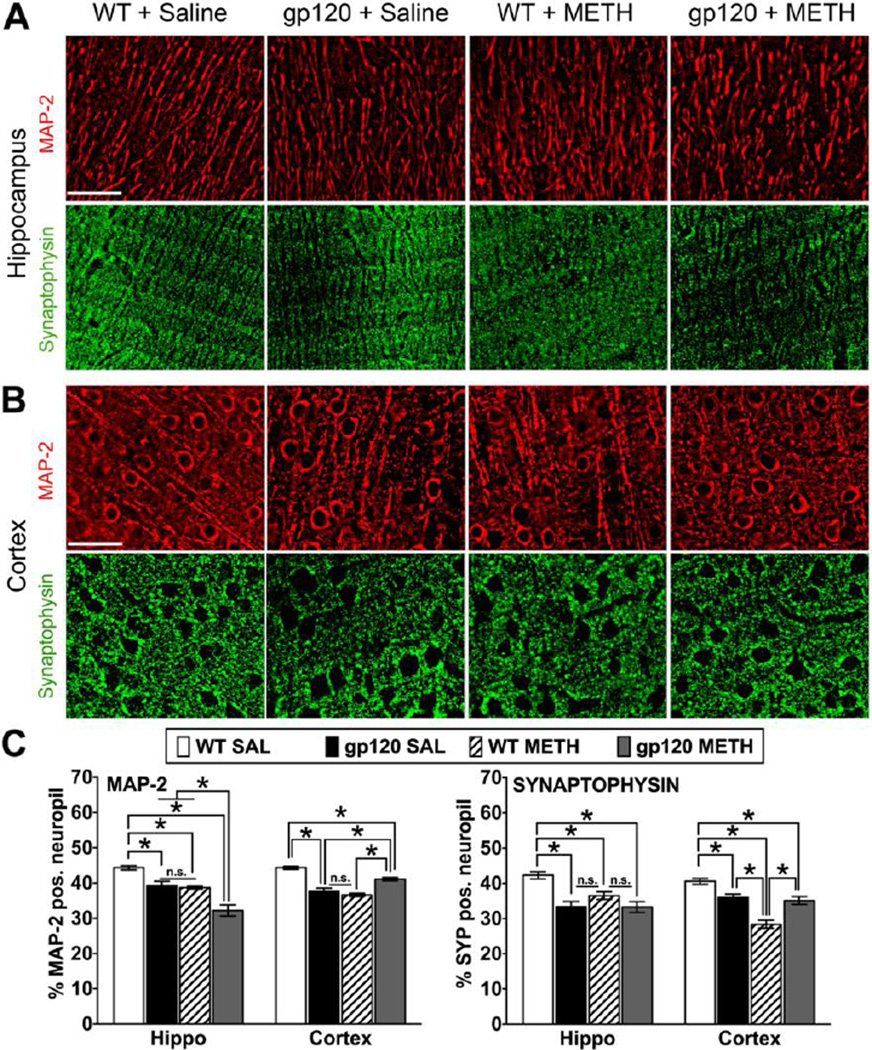
Neuronal injury associated with METH exposure and HIV-1 gp120 expression
A separate cohort of HIV-1 gp120tg and WT animals were sacrificed seven months after METH exposure at an age of 10–11 months, and brain tissues were harvested. To assess neuronal injury, we estimated by quantitative microscopy the percentage of MAP-2 and Synaptophysin-positive (Syp+) neuropil as a measure for neurites and presynaptic terminals, respectively, in hippocampus and fronto-parietal cortex (Figure 2). We observed a significant reduction of MAP-2+ neuronal dendrites in the HIV-1 gp120tg SAL, WT METH, and HIV-1 gp120tg METH-treated animals, with no differences between the HIV-1 gp120tg SAL and WT METH groups in both hippocampus and cortex. However, the loss of MAP-2+ neuropil was increased in the hippocampus but unexpectedly diminished in the cortex in the HIV-1 gp120tg METH group compared to HIV-1 gp120tg SAL and WT METH samples (Figure 2C). In contrast, WT animals treated with METH showed a significant larger reduction of Syp+ neuropil in cortex but less loss in hippocampus compared to the HIV-1 gp120tg SAL and METH groups. Finally, all three groups displayed a significant reduction of Syp+ neuropil compared to SAL-injected WT mice (Figure 2C).
Figure 2.
Pathological changes in neuronal dendrites and presynaptic terminals of METH-treated WT and HIV-1 gp120tg mice after seven months of drug abstinence. At 10–11 months of age and seven months of abstinence from METH, gp120tg and WT animals were sacrificed and brain tissues harvested. Immunostaining of sagittal brain sections for neuronal markers and quantitative microscopic analysis was performed as described in the methods section. Deconvolved images of MAP-2 and Synaptophysin (Syp) staining on brain sections from hippocampus CA1 region, molecular layer (A), and fronto-parietal cortex, layer III (B). Scale bar: 40 µm. (C) Percentage of neuropil positive for neuronal MAP-2 and Syp estimated by deconvolution microscopy in hippocampus and fronto-parietal cortex of METH- or SAL-treated gp120tg and WT mice. Graphs show mean ± SEM; * p ≤ 0.0007 (ANOVA and Fisher’s PLSD post hoc test, n = 4–6 animals per group; n.s., not significant).
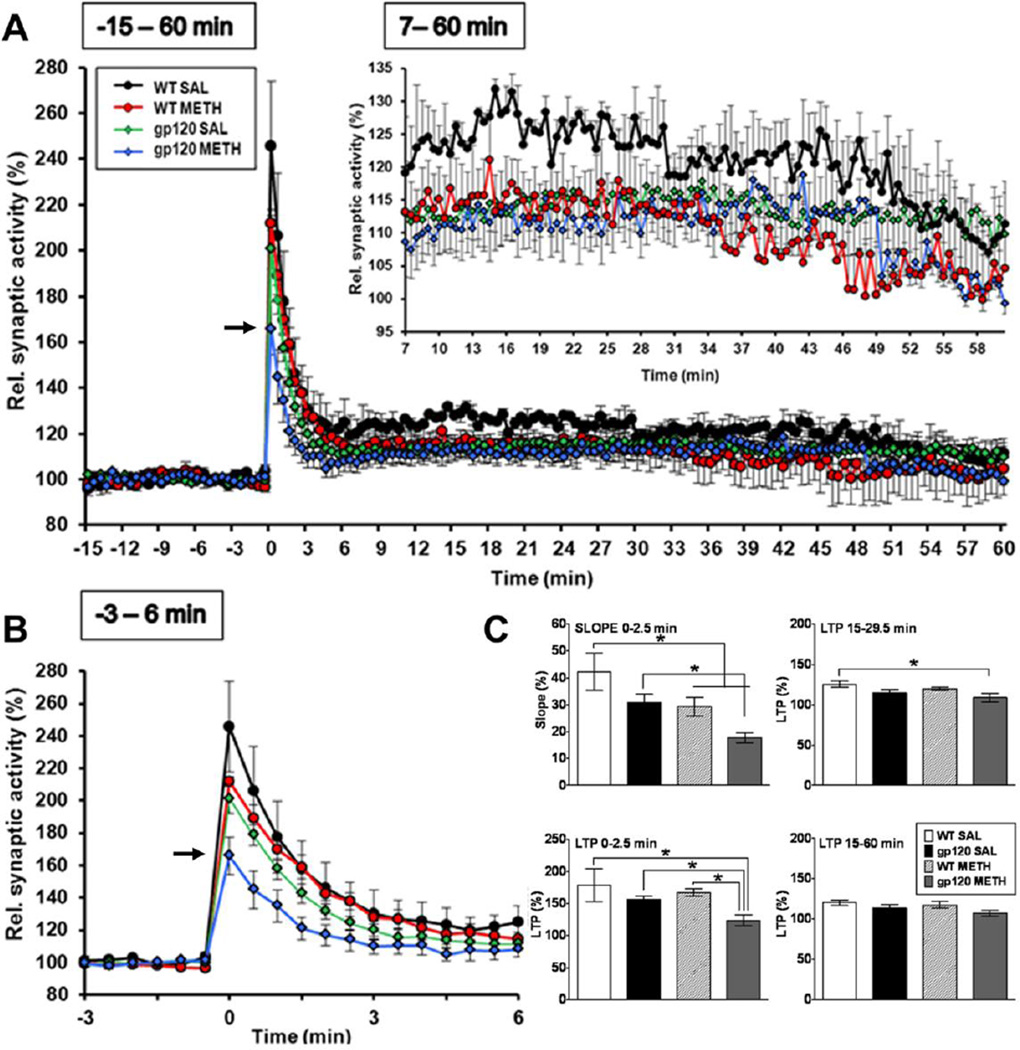
METH and HIV-1 gp120 in combination reduce post-tetanic potentiation (PTP) and LTP
Six months after exposure to METH hippocampal slices were prepared from HIV-1 gp120tg mice and WT controls. The hippocampal slices were laid down on a multielectrode array for electrophysiological experiments assessing LTP as described in the methods section. Prior METH exposure and gp120 expression were associated with reduced LTP (Figure 3). However, only METH-treated HIV-1 gp120tg mice showed a significantly reduced PTP (p = 0.017 for slope 0–2.5 min of HIV-1 gp120tg METH vs. SAL control, and p = 0.001 for HIV-1 gp120tg METH vs. WT SAL control; Figure 3, upper left panel). This significant change was not observed in saline-treated transgenic animals or METH- or SAL-injected WT controls, although the slope describing the time-dependent reduction of synaptic activity following tetanic potentiation was also significantly reduced in METH-treated WT mice (p = 0.05 for slope 0–2.5 min of WT METH vs. SAL control (Figure 3C, upper left panel). The combination of METH exposure and gp120 expression only had a significant effect on LTP early after tetanic stimulation (0–2.5 min; p = 0.011 for early LTP of WT METH vs. HIV-1 gp120tg METH, p = 0.026 for HIV-1 gp120tg METH vs. SAL control, and p = 0.004 for HIV-1 gp120tg METH vs. WT SAL control; Figure 3C, lower left panel) and between 15–29.5 min (p = 0.04 for LTP of HIV-1 gp120tg METH vs. WT SAL control; Figure 3C, upper right panel).
Figure 3.
Electrophysiological studies on hippocampal slices. Hippocampal slices were prepared and electrophysiological experiments performed as described in Material and Methods. (A, B) Only METH-treated HIV-1 gp120tg animals displayed reduced post tetanic potentiation (PTP, 0–2.5 min) in hippocampal CA1 neurotransmission (arrow), but not saline-injected HIV-1 gp120tg or METH and saline exposed WT control mice. Both METH and gp120 reduced long-term potentiation (LTP) following tetanic stimulation (inset in (A), 7–60 min). (C) Analysis of slope and different stages of LTP. * p ≤ 0.05 (ANOVA with Fisher’s PLSD post hoc test; WT METH (n = 5), WT SAL (n = 3), HIV-1 gp120tg METH (n = 5), HIV-1 gp120tg SAL (n = 5).
Alterations at RNA level in GABAergic and glutamatergic circuitries associated with METH exposure and HIV-1 gp120 expression
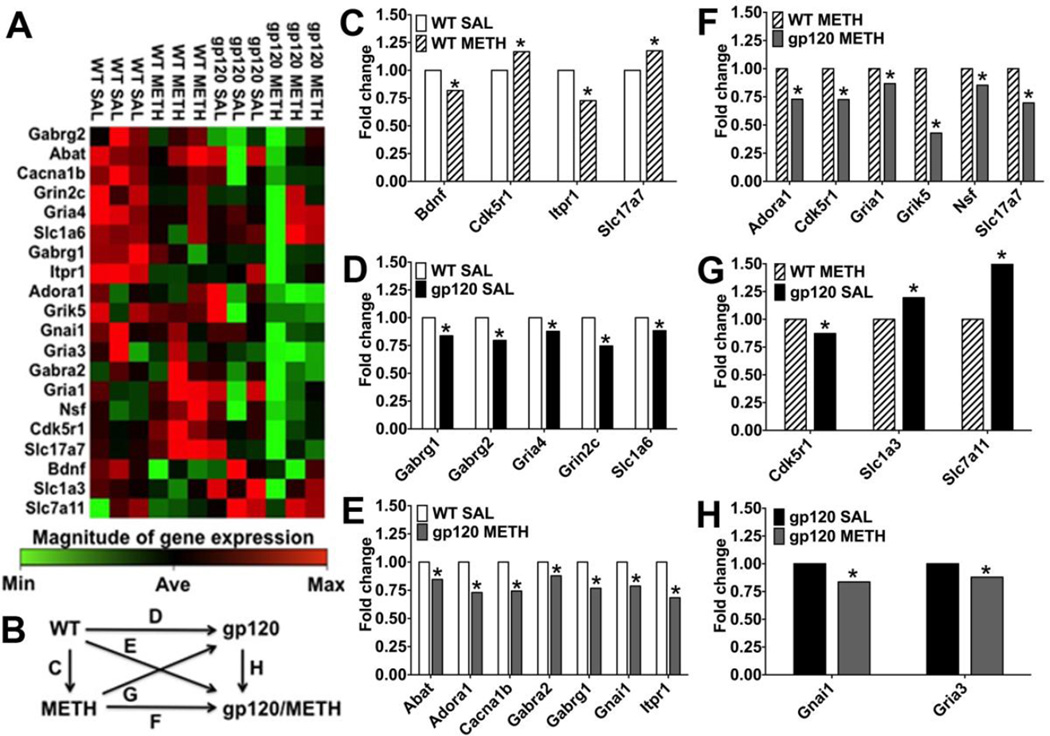
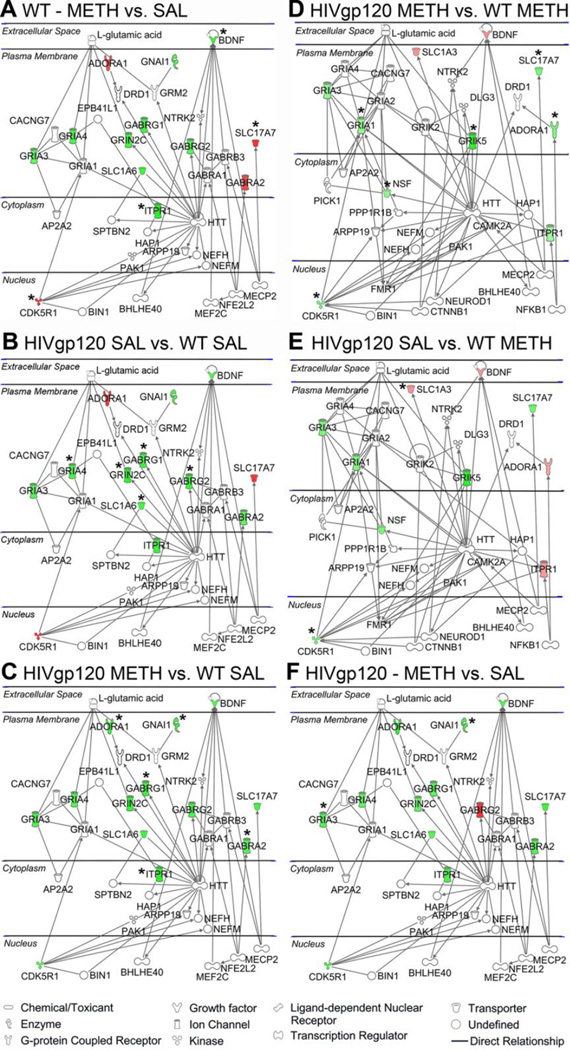
LTP prominently involves glutamatergic excitatory neurotransmission (Kauer et al., 1988) and PTP presumably engages both pre- and post–synaptic mechanisms (Bao et al., 1997; Hughes, 1958). To explore potential molecular substrates of our behavioral, neuropathological and electrophysiological observations, we next analyzed components of the GABA and glutamate neurotransmitter systems at RNA level. The results of the GABA and glutamate RT2 Profiler PCR Arrays showed that about seven months after exposure to METH, HIV-1 gp120tg and WT mice displayed significant changes in RNA expression of 20 components of the glutamatergic and GABAergic neurotransmission systems (Figure 4, Table 1). The affected genes were primarily down-regulated in the METH and gp120tg groups compared to WT SAL except for METH-treated WT brains, where Cdk5r1 and Slc17a7 were significantly up-regulated. The pattern of genes significantly affected by gp120 and METH exposure strongly suggested a disturbance in the composition of excitatory and inhibitory synapses (Abel and Kohli, 1999; Barde et al., 1982; Bellocchio et al., 2000; Block et al., 1988; Buckle et al., 1989; Coppola et al., 1994; Ebralidze et al., 1996; Johansson et al., 2001; Maeda et al., 1988; Puckett et al., 1991; Sato et al., 1999; Sato et al., 1993; Sommer et al., 1991; Sullivan et al., 1986; Szpirer et al., 1994; Takai et al., 1995; Takamori et al., 2000; Tsai et al., 1994; Wilcox et al., 1992; Yamada et al., 1997). Therefore, the genes which showed significant fold changes of RNA expression in at least one of the comparisons shown in Figure 4B were further analyzed for enrichment in biologically related genes using the bioinformatics software IPA. The bioinformatic analysis predicted two highly significant functional networks of directly interacting components of neurotransmission systems that were affected by METH, gp120 or the combination of both. The first network (score 26; Figure 5 A, B, C, F) emerged from comparisons of differential gene expression in which SAL-treated WT or gp120tg brains served as control. The second network (score 19; Figure 5 D, E) was identified in comparisons using WT METH as control. Each network consisted of 48 directly interacting factors, 47 genes and glutamic acid. The two networks shared 22 components (21 genes + glutamate) and each network comprised 13 specific genes (Table 2). Therefore, while METH and gp120 shared differential regulation of components of both the glutamatergic and GABergic neurotransmissions systems, the viral protein and the psychostimulant each also regulated a number of distinct genes.
Figure 4.
METH and HIV-1/gp120 induce gene expression changes in the GABA and glutamate neurotransmitter systems. Differentially regulated expression of genes of the GABAergic and glutamatergic neurotransmission systems in brains of WT or HIV-1 gp120tg mice after METH exposure. Whole brain RNA was analyzed using the GABA and glutamate RT2 Profiler PCR Array and the associated Data Analysis software package (Qiagen), n = 3 animals per group. (A) Gene expression changes representing 20 significant differentially regulated genes out of the 84 genes analyzed with the GABA and glutamate RT2 Profiler PCR Array shown as clustergram heat map, showing the three biological replicates. WT SAL was set as control, and genes that were relatively highly expressed are colored in red; genes that were expressed at low level are shown in green; and genes that showed average expression (Ave) are indicated in black. (B) Overview diagram of the following comparisons shown in graphs C-H; depicted is fold change for the various comparisons. (C) Differentially regulated genes for METH-treated WT animals with SAL-treated WT animals set as control (Bdnf, * p = 0.034; Cdk5r1, p = 0.018; Itpr1, p = 0.0002; Slc17a7, p = 0.037). (D) Differentially regulated genes for SAL-treated HIV-1 gp120tg animals with SAL- treated WT animals set as control (Gabrg1, * p = 0.002; Gabrg2, p = 0.028; Gria4, p = 0.014; Grinc2c, p = 0.015; Slc1a6, p = 0.028). (E) Differentially regulated genes for METH-treated HIV-1 gp120tg animals with SAL-treated WT animals set as control (Abat, * p = 0.036; Adora1, p = 0.037; Cacna1b, p = 0.005; Gabra2, p = 0.037; Gabrg1, p = 0.01; Gnai1, p = 0.03; Itpr1, p = 0.016). (F) Differentially regulated genes for METH-treated HIV-1 gp120tg animals with METH-treated WT animals set as control (Adora1, * p = 0.027; Cdk5r1, p = 0.02; Gria1, p = 0.025; Grik5, p = 0.0001; Nsf, p = 0.026; Slc17a7, p = 0.014). (G) Differentially regulated genes for SAL-treated HIV-1 gp120tg animals with METH-treated WT animals set as control (Cdk5r1, * p = 0.03; Slc1a3, p = 0.049; Slc7a11, p = 0.035). (H) Differentially regulated genes for METH-treated HIV-1 gp120tg animals with SAL-treated HIV-1 gp120tg set as control; Gnai1, * p = 0.035; Gria3, p = 0.017. RT2 Profiler™ PCR Array Data Analysis software package (version 3.5) was used for 2−(ΔΔCT)–based fold change calculations and a modified Student’s t-test to compute two-tail, equal variance p-values.
Table 1.
Differentially regulated genes in brains of METH-treated gp120tg and WT mice and vehicle-treated gp120tg mice.
| Group | Gene | Description | Reference sequence |
Reference | Neuro- trans- mitter system |
Pre- or postsyna ptic location |
Fold change |
p value |
|---|---|---|---|---|---|---|---|---|
|
WT METH vs. WT SAL (ctl) |
Bdnf | Brain derived neurotrophic factor |
NM_007540 | Barde et al., 1982 | GABA, glutamate |
NA | 0.818 | 0.034 |
| Cdk5r1 | Cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
NM_009871 | Tsai et al., 1994 | glutamate | NA | 1.168 | 0.018 | |
| Itpr1 | Inositol 1,4,5-trisphosphate receptor 1 |
NM_010585 | Maeda et al., 1988 | glutamate | NA | 0.727 | 0.0002 | |
|
Slc17a7/ Vglut1 |
Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7; vesicular glutamate transporter 1 |
NM_182993 |
Takamori et al., 2000 Bellocchio et al., 2000 |
glutamate | pre | 1.176 | 0.037 | |
|
gp120 SAL vs. WT SAL (ctl) |
Gabrg1 | Gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
NM_010252 | Wilcox et al., 1992 | GABA | post | 0.837 | 0.002 |
| Gabrg2 | Gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
NM_008073 | Wilcox et al., 1992 | GABA | post | 0.796 | 0.028 | |
| Gria4 | Glutamate receptor, ionotropic, AMPA4 (alpha 4) |
NM_019691 | Sommer et al., 1991 | glutamate | post | 0.877 | 0.014 | |
| Grin2c | Glutamate receptor, ionotropic, NMDA2C (epsilon 3) |
NM_010350 | Ebralidze et al., 1996 | glutamate | post | 0.746 | 0.015 | |
|
Slc1a6/ Eaat4 |
Solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6; excitatory amino-acid transporter 4 |
NM_009200 | Yamada et al., 1997 | glutamate | post | 0.883 | 0.028 | |
|
gp120 METH vs. WT SAL (ctl) |
Abat | 4-aminobutyrate aminotransferase, GABA- transaminase |
NM_172961 | Abel et al., 1999 | GABA | pre | 0.846 | 0.036 |
| Adora1 | Adenosine A1 receptor | NM_001008533 | Johansson et al., 2001 | GABA, glutamate |
pre and post |
0.730 | 0.037 | |
| Cacna1b | Calcium channel, voltage- dependent, N type, alpha 1B subunit |
NM_007579 | Coppola et al., 1994 | GABA | pre | 0.743 | 0.005 | |
| Gabra2 | Gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
NM_008066 | Buckle et al., 1989 | GABA | post | 0.878 | 0.037 | |
| Gabrg1 | Gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
NM_010252 | Wilcox et al., 1992 | GABA | post | 0.768 | 0.010 | |
| Gnai1 | Guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
NM_010305 | Sullivan et al., 1986 | GABA, glutamate |
NA | 0.788 | 0.033 | |
| Itpr1 | Inositol 1,4,5-trisphosphate receptor 1 |
NM_010585 | Maeda et al., 1988 | GABA | NA | 0.684 | 0.016 | |
|
gp120 METH vs. WT METH (ctl) |
Adora1 | Adenosine A1 receptor | NM_001008533 | Johansson et al., 2001 | GABA, glutamate |
pre and post |
0.730 | 0.027 |
| Cdk5r1 | Cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
NM_009871 | Tsai et al., 1994 | glutamate | NA | 0.726 | 0.020 | |
| Gria1 | Glutamate receptor, ionotropic, AMPA1 (alpha 1) |
NM_008165 | Puckett et al., 1991 | glutamate | post | 0.868 | 0.025 | |
| Grik5 | Glutamate receptor, ionotropic, kainate 5 (gamma 2) |
NM_010350 | Szpirer et al., 1994 | glutamate | post | 0.429 | 0.00007 | |
| Nsf | N-ethylmaleimide sensitive fusion protein |
NM_008740 | Block et al., 1988 | GABA, glutamate |
pre | 0.854 | 0.026 | |
|
Slc17a7/ Vglut1 |
Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7; vesicular glutamate transporter 1 |
NM_182993 |
Takamori et al., 2000 Bellocchio et al., 2000 |
glutamate | pre | 0.697 | 0.014 | |
|
gp120 SAL vs. WT METH (ctl) |
Cdk5r1 | Cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
NM_009871 | Tsai et al., 1994 | glutamate | NA | 1.146 | 0.031 |
|
Slc1a3/ Eaat1/ Glast |
Solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6, EAAT1, GLAST |
NM_009200 | Takai et al., 1995 | glutamate | NA | 0.837 | 0.049 | |
| Slc7a11 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
NM_011990 | Sato et al., 1999 | glutamate | NA | 0.438 | 0.035 | |
|
gp120 METH vs. gp120 SAL (ctl) |
Gnai1 | Guanine nucleotide binding protein (G protein), alpha inhibiting 1 |
NM_010305 | Sullivan et al., 1986 | GABA and glutamate |
NA | 0.836 | 0.034 |
| Gria3 | Glutamate receptor, ionotropic, AMPA3 (alpha 3) |
NM_016886 | Sato et al., 1993 | glutamate | post | 0.880 | 0.017 |
Differentially regulated genes in brains of METH-treated gp120tg and WT mice and vehicle-treated gp120tg mice. List of 20 genes that were found to be significantly differentially regulated using a GABA and glutamate RT2 Profiler PCR Array. The following information is provided: experimental group (ctl = control condition in the given comparison) abbreviated and full gene name, sequence number in GenBank, references, indication to which neurotransmitter system the gene belongs, synaptic localization (if applicable), fold change (ctl condition = 1) and p-values for the respective comparisons.
Figure 5.
Prediction of functional neural gene networks of the GABAergic and glutamatergic neurotransmitter systems affected by METH and HIV-1 gp120. RNA expression data obtained with the GABA and glutamate RT2 Profiler PCR Array were analyzed using Ingenuity Pathway Analysis (IPA) software. Red and green color indicate up- and down-regulated genes, respectively, while components without color represent genes implicated by IPA for which expression levels were not experimentally determined. IPA predicted two highly specific networks based on direct interactions, one from differentially regulated genes in comparison to SAL-treated WT or gp20tg as control (score 26, shown in A, B, C and F) and one from comparisons with METH-treated WT as control (score 19, D and E). Network scores represent the –log10 of the calculated probability that the network occurred at random. Solid lines show direct interactions, arrows indicate direction of action (activation or induction). Note that glutamic acid/glutamate is shown in the extracellular space where it acts as neurotransmitter, but it exists also in intracellular compartments and is highly enriched in presynaptic vesicles of glutamatergic synapses. * Indicates genes for which differential regulation reached significance in the RT2 Profiler PCR Array.
Table 2.
Components of two functional biological networks identified by IPA based on comparisons of differentially regulated genes of the glutamatergic and GABAergic neurotransmission systems in METH-treated gp120tg and WT mice and vehicle-treated gp120tg mice.
| Symbol | Entrez Gene Name | Location | Family | Entrez Gene ID for Mouse |
Network |
|---|---|---|---|---|---|
| ADORA1 | adenosine A1 receptor | Plasma Membrane |
G-protein coupled receptor |
11539 | 1,2 |
| AP2A2 | adaptor-related protein complex 2, alpha 2 subunit |
Cytoplasm | transporter | 11772 | 1,2 |
| ARPP19 | cAMP-regulated phosphoprotein, 19kDa | Cytoplasm | transporter | 59046 | 1,2 |
| BDNF | brain-derived neurotrophic factor | Extracellular Space | growth factor | 12064 | 1,2 |
| BHLHE40 | basic helix-loop-helix family, member e40 | Nucleus | transcription regulator |
20893 | 1,2 |
| BIN1 | bridging integrator 1 | Nucleus | other | 30948 | 1,2 |
| CACNG7 | calcium channel, voltage-dependent, gamma subunit 7 |
Plasma Membrane |
ion channel | 81904 | 1,2 |
| CAMK2A | calcium/calmodulin-dependent protein kinase II alpha |
Cytoplasm | kinase | 12322 | 2 |
| CDK5R1 | cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
Nucleus | kinase | 12569 | 1,2 |
| CTNNB1 | catenin (cadherin-associated protein), beta 1, 88kDa |
Nucleus | transcription regulator |
12387 | 2 |
| DLG3 | discs, large homolog 3 (Drosophila) | Plasma Membrane |
kinase | 53310 | 2 |
| DRD1 | dopamine receptor D1 | Plasma Membrane |
G-protein coupled receptor |
13488 | 1,2 |
| EPB41L1 | erythrocyte membrane protein band 4.1-like 1 |
Plasma Membrane |
other | 13821 | 1 |
| FMR1 | fragile X mental retardation 1 | Nucleus | other | 14265 | 2 |
| GABRA1 | gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
Plasma Membrane |
ion channel | 14394 | 1 |
| GABRA2 | gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
Plasma Membrane |
ion channel | 14395 | 1 |
| GABRB3 | gamma-aminobutyric acid (GABA) A receptor, beta 3 |
Plasma Membrane |
ion channel | 14402 | 1 |
| GABRG1 | gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
Plasma Membrane |
ion channel | 14405 | 1 |
| GABRG2 | gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
Plasma Membrane |
ion channel | 14406 | 1 |
| GNAI1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
Plasma Membrane |
enzyme | 14677 | 1 |
| GRIA1 | glutamate receptor, ionotropic, AMPA 1 |
Plasma Membrane |
ion channel | 14799 | 1,2 |
| GRIA2 | glutamate receptor, ionotropic, AMPA 2 |
Plasma Membrane |
ion channel | 14800 | 2 |
| GRIA3 | glutamate receptor, ionotropic, AMPA 3 |
Plasma Membrane |
ion channel | 53623 | 1,2 |
| GRIA4 | glutamate receptor, ionotropic, AMPA 4 |
Plasma Membrane |
ion channel | 14802 | 1,2 |
| GRIK2 | glutamate receptor, ionotropic, kainate 2 |
Plasma Membrane |
ion channel | 14806 | 2 |
| GRIK5 | glutamate receptor, ionotropic, kainate 5 |
Plasma Membrane |
ion channel | 14809 | 2 |
| GRIN2C | glutamate receptor, ionotropic, N- methyl D-aspartate 2C |
Plasma Membrane |
ion channel | 14813 | 1 |
| GRM2 | glutamate receptor, metabotropic 2 | Plasma Membrane |
G-protein coupled receptor |
108068 | 1 |
| HAP1 | huntingtin-associated protein 1 | Cytoplasm | other | 15114 | 1,2 |
| HTT | huntingtin | Cytoplasm | transcription regulator |
15194 | 1,2 |
| ITPR1 | inositol 1,4,5-trisphosphate receptor, type 1 |
Cytoplasm | ion channel | 16438 | 1,2 |
| L-glutamic acid |
Other | chemical - endogenous mammalian |
1,2 | ||
| MECP2 | methyl CpG binding protein 2 (Rett syndrome) |
Nucleus | transcription regulator |
17257 | 1,2 |
| MEF2C | myocyte enhancer factor 2C | Nucleus | transcription regulator |
17260 | 1 |
| NEFH | neurofilament, heavy polypeptide | Cytoplasm | other | 380684 | 1,2 |
| NEFM | neurofilament, medium polypeptide | Cytoplasm | other | 18040 | 1,2 |
| NEUROD | neuronal differentiation 1 | Nucleus | transcription regulator |
18012 | 2 |
| NFE2L2 | nuclear factor, erythroid 2-like 2 | Nucleus | transcription regulator |
18024 | 1 |
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
Nucleus | transcription regulator |
18033 | 2 |
| NSF | N-ethylmaleimide-sensitive factor | Cytoplasm | transporter | 18195 | 2 |
| NTRK2 | neurotrophic tyrosine kinase, receptor, type 2 |
Plasma Membrane |
kinase | 18212 | 1,2 |
| PAK1 | p21 protein (Cdc42/Rac)-activated kinase 1 |
Cytoplasm | kinase | 18479 | 1,2 |
| PICK1 | protein interacting with PRKCA 1 | Cytoplasm | enzyme | 18693 | 2 |
| PPP1R1B | protein phosphatase 1, regulatory (inhibitor) subunit 1B |
Cytoplasm | phosphatase | 19049 | 2 |
| SLC17A7 | solute carrier family 17 (vesicular glutamate transporter), member 7 |
Plasma Membrane |
transporter | 72961 | 1,2 |
| SLC1A3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
Plasma Membrane |
transporter | 20512 | 2 |
| SLC1A6 | solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
Plasma Membrane |
transporter | 20513 | 1 |
| SPTBN2 | spectrin, beta, non-erythrocytic 2 | Cytoplasm | other | 20743 | 1 |
Components of two functional biological gene networks identified by IPA. List of 48 factors (47 genes and glutamic acid) that constitute two predicted biologically functional networks identified by IPA. Network 1 (score 26; Figure 5 A, B, C, F) emerged from comparisons of differential gene expression in which SAL-treated WT or gp120tg brains served as control. Network 2 (score 19; Figure 5 D, E) was identified in comparisons using WT METH as control. The score is the –log10 of the probability that the network components assemble at random. Note that the two networks only include directly interacting factors and share 22 components (21 genes + glutamate) whereas each network comprises 13 specific genes.
Discussion
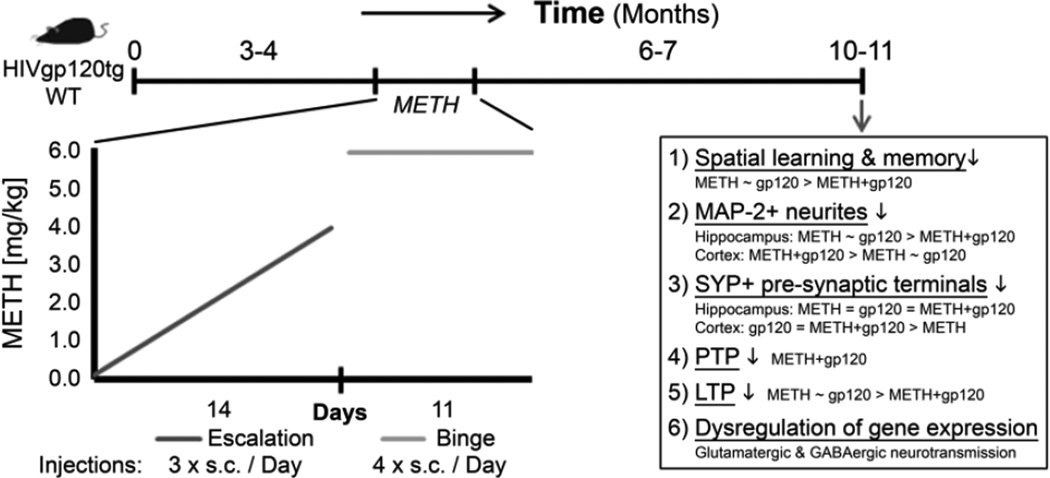
METH use seems to be most prevalent in young adults and increases the risk of HIV infection and the associated development of neurocognitive impairment (Kapadia et al., 2005; Mitchell et al., 2006; Rippeth et al., 2004). Here, we investigated the consequences of a single 25-day long escalating-dose, multiple-binge regimen of METH applied to young adult HIV-1 gp120tg mice and what the long-lasting effects may be, if any, after several months of drug abstinence. Acute effects of METH have been assessed in mice at 12 hours to 21 days after application of 2.5 – 40 mg/kg (reviewed in (Krasnova and Cadet, 2009)). Neuronal damage or death, such as an increase in apoptotic cells, was reported for METH dosages ranging from 5 to- 40 mg/kg. However, 5 mg/kg were injected 4 times at 2-hour intervals with no prior exposure to lower concentrations (Ladenheim et al., 2000). Thus, the METH dosages causing acute neuronal injury were higher than in our present study. However, a recent study found that the pharmacokinetics of METH in HIV-1 gp120tg and non-tg control mice were indistinguishable (Kesby et al., 2012). The established escalating-dose, multiple-binge METH regimen used in the present study avoided hyperthermia and was originally developed to mimic a human usage pattern (Henry et al., 2013; Kuczenski et al., 2007). The regimen was first applied in rats where neuropathological changes were only detected at 30 days but not 3 days after METH exposure (Kuczenski et al., 2007). Acute and short term effects of the escalating-dose, multiple-binge METH regimen in mice are still being investigated. So far two studies analyzed behavior 7 days after the last METH injection and found that novel object interactions and pre-pulse inhibition were increased (Henry et al., 2013; Henry et al., 2014). Using behavioral testing, deconvolution microscopy-based assessment of neuronal injury, targeted gene expression analysis and electrophysiology we detected 6–7 months after a single application of the METH regimen significant alterations in the brains of HIV-1 gp120tg and WT mice (Figure 6).
Figure 6.
Transgenic mice expressing the HIV envelope protein gp120 in the brain (HIVgp120tg) and WT controls were treated at 3–4 months of age with an escalating-dose, multiple-binge METH regimen. The long-term effects were analyzed after 6–7 months of drug abstinence employing behavioral tests and analysis of neuropathology, electrophysiology and gene expression. The METH regimen induced long-lasting pre- and postsynaptic neuronal injury in association with impaired learning and memory in WT and HIV-1 gp120tg mice. The combination of METH and HIV-1 gp120 expression resulted in the most pronounced pathological alterations.
The Barnes maze test revealed that both METH and gp120 affected spatial learning and memory, but the combination resulted in the most errors, longest latencies lowest average time spent in the target quadrant relative to the other quadrants, and a complete lack of spatial strategy use, thus the most compromised performance.
In contrast, we did not detect any long-term effects of METH or gp120 on locomotion, although we found overall sex differences in the locomotor activity measures. However, sex did not interact with genotype or group (METH or SAL) indicating that sex did not impact the behavioral effects of gp120 expression, METH exposure, or the combination of these factors in our animal cohort. Indeed, we detected no sex differences in head tracking behavior in the optomotor test or in any measure in the Barnes maze test. In contrast, other studies using different behavioral paradigms (inhibition, reward) or acutely higher METH concentrations or mice of a different genetic background or age reported sex differences in some of the behavioral measures or neuronal injury (Bourque et al., 2011; Henry et al., 2013; Kesby et al., 2012). Thus, whether or not sex-dependent effects of METH can be detected may depend on the dosage regimen, behavioral test, age and genetic background of the animals.
Analysis of neuronal injury using immunofluorescence and deconvolution microscopy confirmed for HIV-1 gp120tg mice a significant loss of MAP-2+ neurites and Syp+ presynaptic terminals in hippocampus and cerebral cortex in comparison to WT controls (Kang et al., 2010; Toggas et al., 1994). However, METH caused comparable injury in WT mice and the drug surprisingly ameliorated the loss of MAP-2+ neurites, but not Syp+ presynaptic terminals, in the cortex of gp120tg brains for reasons that remain to be explored. One possible explanation is that only combined exposure to HIV-1 protein and METH results in aberrant sprouting of neurites. However, that aberrant expansion of MAP-2+ neurites could be triggered independently of the order in which the brain is exposed to METH and HIV gp120 as long as they overlap at some time to deliver the initiating signal. Overall, the effects of gp120, METH, and their combination differed for pre-synaptic markers and neuronal dendrites, and for hippocampus and cortex. However, the additive reducing effect of combined METH exposure and gp120 expression on the density of MAP-2+ neuronal dendrites in hippocampus was in line with recent reports by others (Cisneros and Ghorpade, 2012; Ferris et al., 2008; Silverstein et al., 2011). While the reason for the ameliorated loss of MAP-2+ neurites in METH-treated gp120tg mice remains to be revealed, the increased damage in the hippocampus may, at least in part, result from an impaired neural repair in the presence of gp120. In fact, we previously found that neurogenesis is reduced in the hippocampus of gp120tg mice at 4–5 months of age (Okamoto et al., 2007), thus at a time that overlapped with the exposure to the escalating dose, multiple binge METH regimen. Hence, the METH treatment could have had an acutely aggravating effect on the disturbed hippocampal neurogenesis.
The detrimental effect of METH on spatial learning and memory in HIV-1 gp120tg mice and the loss of hippocampal neurites and pre-synaptic terminals coincided with reduced hippocampal LTP. LTP of synaptic transmission can last for long periods of time and is widely accepted as a key process of learning and memory (Lisman et al., 2012; Martin et al., 2000). In our 10.5 months old animals, LTP was less pronounced than others have reported for 6–9 weeks old HIV-1 gp120tg and WT controls or 2–4 months old C57BL/6 mice (Krucker et al., 1998; Swant et al., 2010). However, METH exposure and HIV-1 gp120 expression were associated with reduced LTP thus confirming for 10.5 months old mice what has been observed by others in younger animals (Krucker et al., 1998; Swant et al., 2010). Most prominent was the significant reduction of PTP in METH-exposed HIV-1 gp120tg brains. PTP can involve both pre- and post-synaptic mechanisms (Bao et al., 1997; Hughes, 1958). Diminished PTP has been observed in other models of neurodegenerative diseases, such as mice carrying a Huntington’s Disease mutation (Usdin et al., 1999). Furthermore, impaired LTP and PTP have also been observed in brains of animals injected with HIV-1 infected human macrophages, a model of HIV-induced encephalitis (HIVE) (Zink et al., 2002). Altogether, the most prominent and specific reduction of PTP and LTP in METH-exposed HIV-1 gp120tg brains supported again the notion of a lasting combined effect of the two injurious factors.
Many previous animal studies have focused on effects of METH on the dopamine and serotonin pathways, although METH also injures GABAergic neurons, causes glutamate release in the brain and thus produces oxidative stress and glutamate-mediated excitotoxicity (Flora et al., 2003; Krasnova and Cadet, 2009; Langford et al., 2003; Lee et al., 2002; Mark et al., 2004; Nath et al., 2000; Theodore et al., 2007). Similarly, HIV infection has been linked to aberrant glutamate release and excitotoxic neuronal damage but also perturbs the dopaminergic system (Berger et al., 1994; Kaul et al., 2001; Kaul et al., 2005; Nath et al., 2000). Since previous studies of METH-induced brain injury by others have established the alteration of the DA system, we investigated in the present study the glutamate and GABA neurotransmitter systems in our model (Cass, 1997; Ferris et al., 2008; Jayanthi et al., 2005; Krasnova and Cadet, 2009; McCann et al., 1998; Schmued and Bowyer, 1997; Sekine et al., 2006; Theodore et al., 2007; Thompson et al., 2004; Volkow et al., 2001; Wilson et al., 1996; Yamamoto and Bankson, 2005). The function of neuronal networks relies on excitatory and inhibitory synapses, and the pattern of genes significantly affected by gp120 and METH exposure strongly suggested a disturbance in the composition of both systems (Enna and Gallagher, 1983; Mehta et al., 2013). Therefore, the findings provided a possible mechanism for the observations of altered PTP and LTP. BDNF, which signals through its high affinity receptor TrkB and low-affinity co-receptor p75NTR, was significantly down-regulated in METH-exposed WT brains. BDNF regulates both excitatory and inhibitory synapses, although by different mechanisms (Bramham, 2008; Minichiello, 2009; Seil and Drake-Baumann, 2000). BDNF-TrkB signaling has been suggested as a general regulatory mechanism for synapse formation, stabilization and function (Wuchter et al., 2012). In the adult hippocampus, BDNF is essential for LTP and thus is involved in spatial learning and memory formation (Korte et al., 1995).
The bioinformatics tool IPA uses a knowledge database that comprises information curated from biomedical scientific literature and various available databases, such as those for chemical compounds and protein interactions. The IPA knowledge database enabled us to analyze our gene expression data set for connections to diseases, physiological, cellular and molecular functions, and biological pathways and networks. We used the RT2 Profiler™ PCR Array to analyze alterations in the glutamatergic and GABAergic neurotransmission systems. Both, glutamate and GABA play significant roles in the normal performance of cortical and hippocampal neuronal circuitries, such as required for executive functions and learning and memory. IPA enabled us to link the components of the neurotransmission systems that were significantly changed in RNA expression to other known neuronal components, such as those shown in Figure 5 and Table 2. Those proteins and molecules included pre- and post-synaptic components as well as factors affecting neuronal maintenance and repair, such as BDNF. The histopathological analysis indicated loss of neurites and presynaptic terminals triggered by gp120 and METH. The electrophysiological experiments showed for the hippocampus that gp120 and METH alter LTP and, in combination, PTP. The behavioral tests showed impaired spatial learning and memory due to gp120 and METH. Therefore, the gene expression data and IPA complemented the picture by linking significantly altered components of two neurotransmission systems to pre- and postsynaptic compartments, and pointing to other factors that may be affected by gp120 and METH even if their expression remained unchanged. Using IPA we identified two gene networks of high significance in which components of the glutamatergic and GABAergic systems are directly interacting. The highest scoring network emerged from analysis of genes that were significantly up- or down-regulated in experimental conditions compared to WT SAL or HIV-1 gp120 SAL as controls. Thus the changes primarily reflected effects driven by METH or similarly by METH and gp120. The second network was predicted based on comparisons of gene expression that used WT METH as control. Thus, this network mostly indicated components affected by viral gp120. Interestingly, both functional networks implicated Huntingtin (Htt), BDNF, CDKR5R1 and glutamate as central factors as well as DA receptor D1 (DRD1). Although our present study did not specifically investigate the DA and serotonergic systems, our findings must be interpreted in the context of everything that is known about the effects of HIV and METH on those other neurotransmission systems. Therefore, the central position of Htt in the predicted networks is in line with a report that PTP is diminished in mice carrying the HD mutation and provides in addition to DRD1 a link to the DAergic system (Usdin et al., 1999). Moreover, mutations in Htt have been shown to affect dopamine release and the response to METH (Cuesta et al., 2012; Johnson et al., 2006).
In conclusion, a single escalating-dose, multiple-binge METH regimen induced long-lasting pre-and postsynaptic neuronal injury in association with impaired learning and memory in WT and HIV-1 gp120tg mice. The combination of METH and HIV-1 gp120 expression resulted in the most pronounced pathological alterations, at least in the hippocampus, thus supporting the hypothesis that early and temporary METH use can aggravate neurocognitive sequelae and brain injury in HIV patients in a long-lasting fashion.
Highlights.
Methamphetamine binge impairs learning and memory of mice in a long-lasting fashion.
Methamphetamine worsens impaired learning and memory in HIV-1 gp120 transgenic mice.
HIV-1 gp120 and methamphetamine injure neuronal dendrites and synapses.
Methamphetamine-exposed HIV-1 gp120tg mice show reduced post-tetanic potentiation.
Methamphetamine and HIV-1 gp120 in combination dysregulate specific synaptic genes.
Acknowledgements
We thank Jasmin Buenrostro, Lili Lacarra, Elizabeth Chappell, Sara Streiner, Jorge Morales, Kenny Venegas, Buddy Charbono for excellent technical assistance with animal maintenance. We are grateful for the support of this work by NIH grants R25 MH081482 (IRFN Fellowship to MMH and ABS), P30 NS057096 to JPC, DZ and AJR, R01 MH087332, R03 DA02948 and P50 DA026306 (TMARC Project 5) to MK. The Translational Methamphetamine AIDS Research Center (TMARC) is supported by Center award P50 DA026306 from the National Institute on Drug Abuse (NIDA) and is affiliated with the University of California, San Diego (UCSD) and the Sanford-Burnham Medical Research Institute (SBMRI). The TMARC is comprised of: Director – Igor Grant, M.D.; Co-Directors – Ronald J. Ellis, M.D., Ph.D., Scott L. Letendre, M.D., and Cristian L. Achim, M.D., Ph.D.; Center Manager – Steven Paul Woods, Psy.D.; Assistant Center Manager – Aaron M. Carr, B.A.; Clinical Assessment and Laboratory (CAL) Core: Scott L. Letendre, M.D. (Core Director), Ronald J. Ellis, M.D., Ph.D., Rachel Schrier, Ph.D.; Neuropsychiatric (NP) Core: Robert K. Heaton, Ph.D. (Core Director), J. Hampton Atkinson, M.D., Mariana Cherner, Ph.D., Thomas D. Marcotte, Ph.D., Erin E. Morgan, Ph.D.; Neuroimaging (NI) Core: Gregory Brown, Ph.D. (Core Director), Terry Jernigan, Ph.D., Anders Dale, Ph.D., Thomas Liu, Ph.D., Miriam Scadeng, Ph.D., Christine Fennema-Notestine, Ph.D., Sarah L. Archibald, M.A.; Neurosciences and Animal Models (NAM) Core: Cristian L. Achim, M.D., Ph.D. (Core Director), Eliezer Masliah, M.D., Stuart Lipton, M.D., Ph.D., Virawudh Soontornniyomkij, M.D.; Administrative Coordinating Core (ACC) – Data Management and Information Systems (DMIS) Unit: Anthony C. Gamst, Ph.D. (Unit Chief), Clint Cushman, B.A. (Unit Manager); ACC – Statistics Unit: Ian Abramson, Ph.D. (Unit Chief), Florin Vaida, Ph.D., Reena Deutsch, Ph.D., Anya Umlauf, M.S.; ACC – Participant Unit: J. Hampton Atkinson, M.D. (Unit Chief), Jennifer Marquie-Beck, M.P.H. (Unit Manager); Project 1: Arpi Minassian, Ph.D. (Project Director), William Perry, Ph.D., Mark Geyer, Ph.D., Brook Henry, Ph.D., Jared Young, Ph.D.; Project 2: Amanda B. Grethe, Ph.D. (Project Director), Martin Paulus, M.D., Ronald J. Ellis, M.D., Ph.D.; Project 3: Sheldon Morris, M.D., M.P.H. (Project Director), David M. Smith, M.D., M.A.S., Igor Grant, M.D.; Project 4: Svetlana Semenova, Ph.D. (Project Director), Athina Markou, Ph.D., James Kesby, Ph.D.; Project 5: Marcus Kaul, Ph.D. (Project Director).
The views expressed in this article are those of the authors and do not reflect the official policy or position of the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare that they have no competing conflict of interest.
Contributor Information
Melanie M. Hoefer, Email: mhoefer@sanfordburnham.org.
Ana B. Sanchez, Email: asanchez@sanfordburnham.org.
Ricky Maung, Email: rmaung@sanfordburnham.org.
Cyrus M. de Rozieres, Email: cderozieres@sanfordburnham.org.
Irene C. Catalan, Email: cata6498@pacificu.edu.
Cari C. Dowling, Email: cari.j.cox78@gmail.com.
Victoria E. Thaney, Email: vthaney@sanfordburnham.org.
Juan Piña-Crespo, Email: pina-crespo@sanfordburnham.org.
Dongxian Zhang, Email: dzhang@sanfordburnham.org.
Amanda J. Roberts, Email: aroberts@scripps.edu.
Marcus Kaul, Email: mkaul@sanfordburnham.org.
References
- Abel MS, Kohli N. GABA-transaminase antisense oligodeoxynucleotide modulates cocaine- and pentylenetetrazol-induced seizures in mice. Metab Brain Dis. 1999;14:253–263. doi: 10.1023/a:1020737125843. [DOI] [PubMed] [Google Scholar]
- Albertson TE, et al. Methamphetamine and the expanding complications of amphetamines. West J Med. 1999;170:214–219. [PMC free article] [PubMed] [Google Scholar]
- Antinori A, et al. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69:1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach ME, et al. Impairment of spatial but not contextual memory in CaMKII mutant mice with a selective loss of hippocampal LTP in the range of the theta frequency. Cell. 1995;81:905–915. doi: 10.1016/0092-8674(95)90010-1. [DOI] [PubMed] [Google Scholar]
- Bao JX, et al. Involvement of pre- and postsynaptic mechanisms in posttetanic potentiation at Aplysia synapses. Science. 1997;275:969–973. doi: 10.1126/science.275.5302.969. [DOI] [PubMed] [Google Scholar]
- Barde YA, et al. Purification of a new neurotrophic factor from mammalian brain. EMBO J. 1982;1:549–553. doi: 10.1002/j.1460-2075.1982.tb01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CA. Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat. J Comp Physiol Psychol. 1979;93:74–104. doi: 10.1037/h0077579. [DOI] [PubMed] [Google Scholar]
- Bellocchio EE, et al. Uptake of glutamate into synaptic vesicles by an inorganic phosphate transporter. Science. 2000;289:957–960. doi: 10.1126/science.289.5481.957. [DOI] [PubMed] [Google Scholar]
- Berger JR, et al. Cerebrospinal fluid dopamine in HIV-1 infection. AIDS. 1994;8:67–71. doi: 10.1097/00002030-199401000-00010. [DOI] [PubMed] [Google Scholar]
- Besl B, Fromherz P. Transistor array with an organotypic brain slice: field potential records and synaptic currents. Eur J Neurosci. 2002;15:999–1005. doi: 10.1046/j.1460-9568.2002.01943.x. [DOI] [PubMed] [Google Scholar]
- Block MR, et al. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc Natl Acad Sci U S A. 1988;85:7852–7856. doi: 10.1073/pnas.85.21.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkholder DA, et al. Microelectrode arrays for stimulation of neural slice preparations. J Neurosci Methods. 1997;77:61–66. doi: 10.1016/s0165-0270(97)00112-x. [DOI] [PubMed] [Google Scholar]
- Bourque M, et al. Sex differences in methamphetamine toxicity in mice: Effect on brain dopamine signaling pathways. Psychoneuroendocrinology. 2011;36:955–969. doi: 10.1016/j.psyneuen.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Bramham CR. Local protein synthesis, actin dynamics, and LTP consolidation. Curr Opin. Neurobiol. 2008;18:524–531. doi: 10.1016/j.conb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Brew BJ, et al. Neurodegeneration and ageing in the HAART era. J Neuroimmune Pharmacol. 2009;4:163–174. doi: 10.1007/s11481-008-9143-1. [DOI] [PubMed] [Google Scholar]
- Buckle VJ, et al. Chromosomal localization of GABAA receptor subunit genes: relationship to human genetic disease. Neuron. 1989;3:647–654. doi: 10.1016/0896-6273(89)90275-4. [DOI] [PubMed] [Google Scholar]
- Cadet JL, et al. Methamphetamine preconditioning causes differential changes in striatal transcriptional responses to large doses of the drug. Dose.Response. 2011;9:165–181. doi: 10.2203/dose-response.10-011.Cadet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet JL, Krasnova IN. Interactions of HIV and methamphetamine: cellular and molecular mechanisms of toxicity potentiation. Neurotox.Res. 2007;12:181–204. doi: 10.1007/BF03033915. [DOI] [PubMed] [Google Scholar]
- Cadet JL, et al. Methamphetamine preconditioning alters midbrain transcriptional responses to methamphetamine-induced injury in the rat striatum. PLoS One. 2009;4:e7812. doi: 10.1371/journal.pone.0007812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey CL, et al. Additive deleterious effects of methamphetamine dependence and immunosuppression on neuropsychological functioning in HIV infection. AIDS Behav. 2006;10:185–190. doi: 10.1007/s10461-005-9056-4. [DOI] [PubMed] [Google Scholar]
- Cass WA. Decreases in evoked overflow of dopamine in rat striatum after neurotoxic doses of methamphetamine. J Pharmacol.Exp.Ther. 1997;280:105–113. [PubMed] [Google Scholar]
- Cisneros IE, Ghorpade A. HIV-1, methamphetamine and astrocyte glutamate regulation: combined excitotoxic implications for neuro-AIDS. Curr HIV Res. 2012;2012:392–406. doi: 10.2174/157016212802138832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola T, et al. Molecular cloning of a murine N-type calcium channel alpha 1 subunit. Evidence for isoforms, brain distribution, and chromosomal localization. FEBS Lett. 1994;338:1–5. doi: 10.1016/0014-5793(94)80105-3. [DOI] [PubMed] [Google Scholar]
- Cuesta M, et al. The methamphetamine-sensitive circadian oscillator is dysfunctional in a transgenic mouse model of Huntington's disease. Neurobiol Dis. 2012;45:145–155. doi: 10.1016/j.nbd.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Ebralidze AK, et al. Modification of NMDA receptor channels and synaptic transmission by targeted disruption of the NR2C gene. J Neurosci. 1996;16:5014–5025. doi: 10.1523/JNEUROSCI.16-16-05014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisch AJ, Marshall JF. Methamphetamine neurotoxicity: dissociation of striatal dopamine terminal damage from parietal cortical cell body injury. Synapse. 1998;30:433–445. doi: 10.1002/(SICI)1098-2396(199812)30:4<433::AID-SYN10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Enna SJ, Gallagher JP. Biochemical and electrophysiological characteristics of mammalian GABA receptors. Int Rev Neurobiol. 1983;24:181–212. doi: 10.1016/s0074-7742(08)60222-6. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, et al. Neurotoxic profiles of HIV, psychostimulant drugs of abuse, and their concerted effect on the brain: current status of dopamine system vulnerability in NeuroAIDS. Neurosci Biobehav.Rev. 2008;32:883–909. doi: 10.1016/j.neubiorev.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora G, et al. Methamphetamine potentiates HIV-1 Tat protein-mediated activation of redox-sensitive pathways in discrete regions of the brain. Exp Neurol. 2003;179:60–70. doi: 10.1006/exnr.2002.8048. [DOI] [PubMed] [Google Scholar]
- Glass JD, et al. Immunocytochemical quantitation of human immunodeficiency virus in the brain: correlations with dementia. Ann.Neurol. 1995;38:755–762. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- Heaton RK, et al. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–2096. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry BL, et al. Behavioral effects of chronic methamphetamine treatment in HIV-1 gp120 transgenic mice. Behav Brain Res. 2013;236:210–220. doi: 10.1016/j.bbr.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry BL, et al. Prepulse inhibition in HIV-1 gp120 transgenic mice after withdrawal from chronic methamphetamine. Behav Pharmacol. 2014;25:12–22. doi: 10.1097/FBP.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes MP, et al. Quinolinic acid in cerebrospinal fluid and serum in HIV-1 infection: relationship to clinical and neurological status. Ann.Neurol. 1991;29:202–209. doi: 10.1002/ana.410290215. [DOI] [PubMed] [Google Scholar]
- Holmes A, et al. Behavioral profiles of inbred strains on novel olfactory, spatial and emotional tests for reference memory in mice. Genes Brain Behav. 2002;1:55–69. doi: 10.1046/j.1601-1848.2001.00005.x. [DOI] [PubMed] [Google Scholar]
- Hughes JR. Post-tetanic potentiation. Physiol Rev. 1958;38:91–113. doi: 10.1152/physrev.1958.38.1.91. [DOI] [PubMed] [Google Scholar]
- Jayanthi S, et al. Calcineurin/NFAT-induced up-regulation of the Fas ligand/Fas death pathway is involved in methamphetamine-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 2005;102:868–873. doi: 10.1073/pnas.0404990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson B, et al. Hyperalgesia, anxiety, and decreased hypoxic neuroprotection in mice lacking the adenosine A1 receptor. Proc Natl Acad Sci U S A. 2001;98:9407–9412. doi: 10.1073/pnas.161292398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, et al. Dopamine release is severely compromised in the R6/2 mouse model of Huntington's disease. J Neurochem. 2006;97:737–746. doi: 10.1111/j.1471-4159.2006.03762.x. [DOI] [PubMed] [Google Scholar]
- Kang YJ, et al. Erythropoietin plus insulin-like growth factor-I protects against neuronal damage in a murine model of human immunodeficiency virus-associated neurocognitive disorders. Ann.Neurol. 2010;68:342–352. doi: 10.1002/ana.22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia F, et al. The role of substance abuse in HIV disease progression: reconciling differences from laboratory and epidemiologic investigations. Clin.Infect.Dis. 2005;41:1027–1034. doi: 10.1086/433175. [DOI] [PubMed] [Google Scholar]
- Kauer JA, et al. NMDA application potentiates synaptic transmission in the hippocampus. Nature. 1988;334:250–252. doi: 10.1038/334250a0. [DOI] [PubMed] [Google Scholar]
- Kaul M, et al. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature. 2001;410:988–994. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- Kaul M, et al. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ. 2005;12(Suppl 1):878–892. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- Kesby JP, et al. Expression of HIV gp120 protein increases sensitivity to the rewarding properties of methamphetamine in mice. Addict.Biol. 2012 doi: 10.1111/adb.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte M, et al. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft-Terry SD, et al. A coat of many colors: neuroimmune crosstalk in human immunodeficiency virus infection. Neuron. 2009;64:133–145. doi: 10.1016/j.neuron.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnova IN, Cadet JL. Methamphetamine toxicity and messengers of death. Brain Res Rev. 2009;60:379–407. doi: 10.1016/j.brainresrev.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krucker T, et al. Transgenic mice with cerebral expression of human immunodeficiency virus type-1 coat protein gp120 show divergent changes in short- and long-term potentiation in CA1 hippocampus. NeuroScience. 1998;83:691–700. doi: 10.1016/s0306-4522(97)00413-2. [DOI] [PubMed] [Google Scholar]
- Kuczenski R, et al. Escalating dose-multiple binge methamphetamine exposure results in degeneration of the neocortex and limbic system in the rat. Exp Neurol. 2007;207:42–51. doi: 10.1016/j.expneurol.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladenheim B, et al. Methamphetamine-induced neurotoxicity is attenuated in transgenic mice with a null mutation for interleukin-6. Mol.Pharmacol. 2000;58:1247–1256. doi: 10.1124/mol.58.6.1247. [DOI] [PubMed] [Google Scholar]
- Langford D, et al. Patterns of selective neuronal damage in methamphetamine-user AIDS patients. J Acquir.Immune.Defic.Syndr. 2003;34:467–474. doi: 10.1097/00126334-200312150-00004. [DOI] [PubMed] [Google Scholar]
- Lee YW, et al. Methamphetamine activates DNA binding of specific redox-responsive transcription factors in mouse brain. J Neurosci Res. 2002;70:82–89. doi: 10.1002/jnr.10370. [DOI] [PubMed] [Google Scholar]
- Lisman J, et al. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maeda N, et al. Purification and characterization of P400 protein, a glycoprotein characteristic of Purkinje cell, from mouse cerebellum. J Neurochem. 1988;51:1724–1730. doi: 10.1111/j.1471-4159.1988.tb01151.x. [DOI] [PubMed] [Google Scholar]
- Maragos WF, et al. Human immunodeficiency virus-1 Tat protein and methamphetamine interact synergistically to impair striatal dopaminergic function. J Neurochem. 2002;83:955–963. doi: 10.1046/j.1471-4159.2002.01212.x. [DOI] [PubMed] [Google Scholar]
- Mark KA, et al. High-dose methamphetamine acutely activates the striatonigral pathway to increase striatal glutamate and mediate long-term dopamine toxicity. J Neurosci. 2004;24:11449–11456. doi: 10.1523/JNEUROSCI.3597-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, et al. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Masliah E, et al. Selective neuronal vulnerability in HIV encephalitis. J Neuropathol Exp Neurol. 1992;51:585–593. doi: 10.1097/00005072-199211000-00003. [DOI] [PubMed] [Google Scholar]
- Masliah E, et al. Dendritic injury is a pathological substrate for human immunodeficiency virus-related cognitive disorders. HNRC group. The HIV Neurobehavioral Research Center. Ann Neurol. 1997;42:963–972. doi: 10.1002/ana.410420618. [DOI] [PubMed] [Google Scholar]
- Maung R, et al. CCR5 Knockout Prevents Neuronal Injury and Behavioral Impairment Induced in a Transgenic Mouse Model by a CXCR4-Using HIV-1 Glycoprotein 120. J Immunol. 2014;193:1895–1910. doi: 10.4049/jimmunol.1302915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maung R, et al. Genetic knockouts suggest a critical role for HIV co-receptors in models of HIV gp120-induced brain injury. J Neuroimmune Pharmacol. 2012;7:306–318. doi: 10.1007/s11481-011-9328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann UD, et al. Reduced striatal dopamine transporter density in abstinent methamphetamine and methcathinone users: evidence from positron emission tomography studies with [11C]WIN-35,428. J Neurosci. 1998;18:8417–8422. doi: 10.1523/JNEUROSCI.18-20-08417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, et al. Excitotoxicity: bridge to various triggers in neurodegenerative disorders. Eur J Pharmacol. 2013;698:6–18. doi: 10.1016/j.ejphar.2012.10.032. [DOI] [PubMed] [Google Scholar]
- Minichiello L. TrkB signalling pathways in LTP and learning. Nat Rev.Neurosci. 2009;10:850–860. doi: 10.1038/nrn2738. [DOI] [PubMed] [Google Scholar]
- Mitchell SJ, et al. Methamphetamine use and sexual activity among HIV-infected patients in care--San Francisco, 2004. AIDS Patient.Care STDS. 2006;20:502–510. doi: 10.1089/apc.2006.20.502. [DOI] [PubMed] [Google Scholar]
- Nath A, et al. Neurotoxicity and dysfunction of dopaminergic systems associated with AIDS dementia. J Psychopharmacol. 2000;14:222–227. doi: 10.1177/026988110001400305. [DOI] [PubMed] [Google Scholar]
- Okamoto S, et al. HIV/gp120 decreases adult neural progenitor cell proliferation via checkpoint kinase-mediated cell-cycle withdrawal and G1 arrest. Cell Stem Cell. 2007;1:230–236. doi: 10.1016/j.stem.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Paylor R, et al. Learning impairments and motor dysfunctions in adult Lhx5-deficient mice displaying hippocampal disorganization. Physiol Behav. 2001;73:781–792. doi: 10.1016/s0031-9384(01)00515-7. [DOI] [PubMed] [Google Scholar]
- Puckett C, et al. Molecular cloning and chromosomal localization of one of the human glutamate receptor genes. Proc Natl Acad Sci U S A. 1991;88:7557–7561. doi: 10.1073/pnas.88.17.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippeth JD, et al. Methamphetamine dependence increases risk of neuropsychological impairment in HIV infected persons. J Int.Neuropsychol.Soc. 2004;10:1–14. doi: 10.1017/S1355617704101021. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, et al. Alteration of Methamphetamine-induced stereotypic behaviour in transgenic mice expressing HIV-1 envelope protein gp120. J Neurosci Methods. 2010;186:222–225. doi: 10.1016/j.jneumeth.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, et al. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J Biol Chem. 1999;274:11455–11458. doi: 10.1074/jbc.274.17.11455. [DOI] [PubMed] [Google Scholar]
- Sato K, et al. The differential expression patterns of messenger RNAs encoding non-N-methyl-D-aspartate glutamate receptor subunits (GluR1-4) in the rat brain. NeuroScience. 1993;52:515–539. doi: 10.1016/0306-4522(93)90403-3. [DOI] [PubMed] [Google Scholar]
- Schmued LC, Bowyer JF. Methamphetamine exposure can produce neuronal degeneration in mouse hippocampal remnants. Brain Res. 1997;759:135–140. doi: 10.1016/s0006-8993(97)00173-x. [DOI] [PubMed] [Google Scholar]
- Scott JC, et al. Neurocognitive effects of methamphetamine: a critical review and meta-analysis. Neuropsychol.Rev. 2007;17:275–297. doi: 10.1007/s11065-007-9031-0. [DOI] [PubMed] [Google Scholar]
- Seil FJ, Drake-Baumann R. TrkB receptor ligands promote activity-dependent inhibitory synaptogenesis. J Neurosci. 2000;20:5367–5373. doi: 10.1523/JNEUROSCI.20-14-05367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, et al. Brain serotonin transporter density and aggression in abstinent methamphetamine abusers. Arch.Gen.Psychiatry. 2006;63:90–100. doi: 10.1001/archpsyc.63.1.90. [DOI] [PubMed] [Google Scholar]
- Silverstein PS, et al. Methamphetamine toxicity and its implications during HIV-1 infection. J Neurovirol. 2011 doi: 10.1007/s13365-011-0043-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein PS, et al. HIV-1 gp120 and Drugs of Abuse: Interactions in the Central Nervous System. Curr HIV.Res. 2012;10:369–383. doi: 10.2174/157016212802138724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, et al. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Sullivan KA, et al. Inhibitory and stimulatory G proteins of adenylate cyclase: cDNA and amino acid sequences of the alpha chains. Proc Natl Acad Sci U S A. 1986;83:6687–6691. doi: 10.1073/pnas.83.18.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D, et al. Amphetamine redistributes dopamine from synaptic vesicles to the cytosol and promotes reverse transport. J Neurosci. 1995;15:4102–4108. doi: 10.1523/JNEUROSCI.15-05-04102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swant J, et al. Methamphetamine reduces LTP and increases baseline synaptic transmission in the CA1 region of mouse hippocampus. PLoS.ONE. 2010;5:e11382. doi: 10.1371/journal.pone.0011382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpirer C, et al. The genes encoding the glutamate receptor subunits KA1 and KA2 (GRIK4 and GRIK5) are located on separate chromosomes in human, mouse, and rat. Proc Natl Acad Sci U S A. 1994;91:11849–11853. doi: 10.1073/pnas.91.25.11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai S, et al. Localization of the gene (SLC1A3) encoding human glutamate transporter (GluT-1) to 5p13 by fluorescence in situ hybridization. Cytogenet Cell Genet. 1995;69:209–210. doi: 10.1159/000133965. [DOI] [PubMed] [Google Scholar]
- Takamori S, et al. Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature. 2000;407:189–194. doi: 10.1038/35025070. [DOI] [PubMed] [Google Scholar]
- Theodore S, et al. Progress in understanding basal ganglia dysfunction as a common target for methamphetamine abuse and HIV-1 neurodegeneration. Curr.HIV.Res. 2007;5:301–313. doi: 10.2174/157016207780636515. [DOI] [PubMed] [Google Scholar]
- Thompson PM, et al. Structural abnormalities in the brains of human subjects who use methamphetamine. J Neurosci. 2004;24:6028–6036. doi: 10.1523/JNEUROSCI.0713-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toggas SM, et al. Central nervous system damage produced by expression of the HIV-1 coat protein gp120 in transgenic mice. Nature. 1994;367:188–193. doi: 10.1038/367188a0. [DOI] [PubMed] [Google Scholar]
- Tsai LH, et al. p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature. 1994;371:419–423. doi: 10.1038/371419a0. [DOI] [PubMed] [Google Scholar]
- Urbina A, Jones K. Crystal methamphetamine, its analogues, and HIV infection: medical and psychiatric aspects of a new epidemic. Clin.Infect.Dis. 2004;38:890–894. doi: 10.1086/381975. [DOI] [PubMed] [Google Scholar]
- Usdin MT, et al. Impaired synaptic plasticity in mice carrying the Huntington's disease mutation. Hum.Mol Genet. 1999;8:839–846. doi: 10.1093/hmg/8.5.839. [DOI] [PubMed] [Google Scholar]
- Volkow ND, et al. Low level of brain dopamine D2 receptors in methamphetamine abusers: association with metabolism in the orbitofrontal cortex. Am.J Psychiatry. 2001;158:2015–2021. doi: 10.1176/appi.ajp.158.12.2015. [DOI] [PubMed] [Google Scholar]
- Wesselingh SL, et al. Cellular localization of tumor necrosis factor mRNA in neurological tissue from HIV-infected patients by combined reverse transcriptase/polymerase chain reaction in situ hybridization and immunohistochemistry. J Neuroimmunol. 1997;74:1–8. doi: 10.1016/s0165-5728(96)00160-9. [DOI] [PubMed] [Google Scholar]
- Wilcox AS, et al. Human chromosomal localization of genes encoding the gamma 1 and gamma 2 subunits of the gamma-aminobutyric acid receptor indicates that members of this gene family are often clustered in the genome. Proc Natl Acad Sci U S A. 1992;89:5857–5861. doi: 10.1073/pnas.89.13.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, et al. Striatal dopamine nerve terminal markers in human, chronic methamphetamine users. Nat Med. 1996;2:699–703. doi: 10.1038/nm0696-699. [DOI] [PubMed] [Google Scholar]
- Wuchter J, et al. A comprehensive small interfering RNA screen identifies signaling pathways required for gephyrin clustering. J Neurosci. 2012;32:14821–14834. doi: 10.1523/JNEUROSCI.1261-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, et al. Changes in expression and distribution of the glutamate transporter EAAT4 in developing mouse Purkinje cells. Neurosci Res. 1997;27:191–198. doi: 10.1016/s0168-0102(96)01148-0. [DOI] [PubMed] [Google Scholar]
- Yamamoto BK, Bankson MG. Amphetamine neurotoxicity: cause and consequence of oxidative stress. Crit Rev Neurobiol. 2005;17:87–117. doi: 10.1615/critrevneurobiol.v17.i2.30. [DOI] [PubMed] [Google Scholar]
- Zink WE, et al. Impaired spatial cognition and synaptic potentiation in a murine model of human immunodeficiency virus type 1 encephalitis. J Neurosci. 2002;22:2096–2105. doi: 10.1523/JNEUROSCI.22-06-02096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]