Abstract
miRNAs are essential for self-renewal and differentiation of normal and malignant stem cells by regulating the expression of key stem cell regulatory genes. Here, we report evidence implicating the miR100 in self-renewal of cancer stem-like cells (CSC). We found that miR100 expression levels relate to the cellular differentiation state, with lowest expression in cells displaying stem cell markers. Utilizing a tetracycline-inducible lentivirus to elevate expression of miR100 in human cells, we found that increasing miR100 levels decreased the production of breast CSCs. This effect was correlated with an inhibition of cancer cell proliferation in vitro and in mouse tumor xenografts due to attenuated expression of the CSC regulatory genes SMARCA5, SMARCD1, and BMPR2. Furthermore, miR100 induction in breast CSCs immediately upon their orthotopic implantation or intracardiac injection completely blocked tumor growth and metastasis formation. Clinically, we observed a significant association between miR100 expression in breast cancer specimens and patient survival. Our results suggest that miR100 is required to direct CSC self-renewal and differentiation.
Introduction
There is increasing evidence that cancer stem cells (CSC) are resistant to chemotherapy and radiotherapy and, thus, may contribute to treatment resistance and relapse. The development and validation of breast cancer stem cell (BCSC) biomarkers, including CD24− CD44+, aldehyde dehydrogenase (ALDH), and assays including mammosphere formation and xenograft models by our laboratory and others (1–4) has facilitated studies demonstrating the relative resistance of BCSCs to radiation and chemotherapy.
Until recently, the function of noncoding regions of the genome was unknown. However, it is now clear that many of these regions code for miRNAs. Each miRNA is capable of regulating the expression of multiple proteins and, as a result, can have very potent effects on cellular functions. miRNAs mediate gene silencing through imperfect hybridization to 3′ untranslated regions (3′ UTR) in target mRNAs (5) and modulate a variety of cellular processes, including the regulation of mRNA stability and translation, cellular proliferation, and apoptosis (6). Many studies have demonstrated a link between dysregulated expression of miRNAs and carcinogenesis (7, 8). Emerging evidence demonstrates that miRNAs also play an essential role in stem cell self-renewal and differentiation by regulating the expression of certain key stem cell regulatory genes (9–12).
The Drosophila let-7-complex is a polycistronic locus encoding three ancient miRNAs: Let7, miR100, and fly lin-4 (mir25), which are cotranscribed as a single polycistronic transcript (13). These three miRNAs coordinately control gene expression to regulate developmental processes (13). It has been shown that miR100 is downregulated in ovarian cancer (14, 15), hepatocellular carcinomas (16), and head and neck squamous cell carcinoma (HNSCC; refs. 17, 18). Lower expression of let7 and mir-100 were significantly correlated with a poor prognosis in ovarian cancer (14). Gollin and colleagues reported that the let7-mir-100-mir25 cluster is located in intron 2 at chromosome 11q24 (19), and the downregulation of miR100 in HNSCC plays an important role in the development and/or progression of disease as well as contributing to resistance to radiotherapy (19). In mouse xenografts, expression of miR100 completely blocks tumor growth and metastasis. Our findings suggest that miR100 plays an important role in the regulation of BCSCs.
Materials and Methods
Cell culture
Breast cancer cell line SUM159 and SUM149 were from Asterland (20). MCF-7, T47D, and HCC1954 were purchased from ATCC. The culture medium for SUM159 and SUM149 is Ham F-12 (Invitrogen) supplemented with 5% FBS, 5 µg/mL insulin, and 1 µg/mL hydrocortisone (both from Sigma). MCF7, T47D, and HCC1954 cells were maintained in RPMI1640 medium (Invitrogen) supplemented with 10% FBS (ThermoFisher Scientific), 1% antibiotic-antimycotic (Invitrogen), and 5 µg/mL insulin (Sigma-Aldrich). All cell lines were recently obtained from ATCC or Asterland when the experiments were performed and their identity is routinely monitored by STR profiling.
Tumorigenicity in NOD/SCID mice
All NOD/SCID mice were bred and housed in Association for Assessment and Accreditation of Laboratory Animal Care-accredited specific pathogen-free rodent facilities at the University of Michigan (Ann Arbor, MI). Mice were housed in sterilized, ventilated microisolator cages and supplied with autoclaved commercial chow and sterile water. All mouse experiments were conducted in accordance with standard operating procedures approved by the University Committee on the Use and Care of Animals at the University of Michigan (Ann Arbor, MI). Tumorigenicity was determined by injecting breast cancer cells with Matrigel (BD Biosciences) into the #4 mammary fat pads of 6-week-old female NOD/SCID mice with 5 to 6 mice per cohort. The animals were euthanized when the tumors were 1.0–1.5 cm in diameter. A portion of each fat pad was fixed in formalin and embedded in paraffin for histologic analysis. Another portion was analyzed by the ALDH or CD24/CD44 cytometric staining. For the nanovector experiments, SUM 159 cells (1 × 106) were injected into the fat pads of NOD/SCID mice. Four weeks later, the tumors grew to approximately 40 to 60 mm3 at the injection site. Freshly prepared Lip-Tf-miR100 or LipA-Tf-NC containing 25µg of miRNA mimics in 300 µL of 5% glucose was intravenously injected per mouse via the tail vein three times a week for a total of nine injections. The tumor sizes were measured twice a week with a caliper and calculated as tumor volume = length × width2/2. All animal experiments were performed in accordance with the University of Kansas (Lawrence, Kansas) guidelines for the care and use of animals.
Chick chorioallantoic membrane invasion assays
Cell invasion by SUM159 (CTRL, miR100) in vivo was assessed using 11-day-old chick embryos in which an artificial air sac was created (21). SUM159 cells (CTRL, miR100) were labeled with DsRed (infected with DsRed-lentivirus). A total of 1 × 106 cells were inoculated atop the chick chorioallantoic membrane (CAM) for 3 days and the CAM was removed at the end of the incubation period. Tissues were fixed overnight in 4% paraformaldehyde and after an overnight incubation in 30% sucrose, CAM tissue was frozen in the optimum cutting temperature compound and cross sections were prepared for fluorescence microscopy. Invasion was quantified as a function of cell-associated fluorescence localized beneath the CAM surface (ImageQuant version 5.2; Molecular Dynamics, Inc.; ref. 21). To assess the distal metastasis of SUM159 (CTRL, miR100) cells, 1 × 105 cells were injected intravenously at upper CAM and cultured for 5 days. Lower CAM was isolated after culture period and metastatic growth was examined.
Statistical analysis
Results are presented as the mean ± SD for at least three repeated individual experiments for each group using Microsoft Excel. Statistical differences were determined by using ANOVA and Student t test for independent samples. For the clinical specimens, all statistical analyses were carried out using SPSS 13.0 (SPSS). Spearman order correlation was applied to analyze the association between pairs between the expression of ALDH1 and miR100. Survival curves were plotted by the Kaplan-Meier method and compared by the log-rank test. P < 0.05 in all cases was considered statistically significant.
Accession numbers
The GEO accession number for the gene expression of SUM159-miR100 ALDH+ and ALDH− cells from CTRL or doxycycline-treated groups reported in this article is GSE59361.
Results
miR100 expression is reduced in the ALDH+ population of breast cancer cells
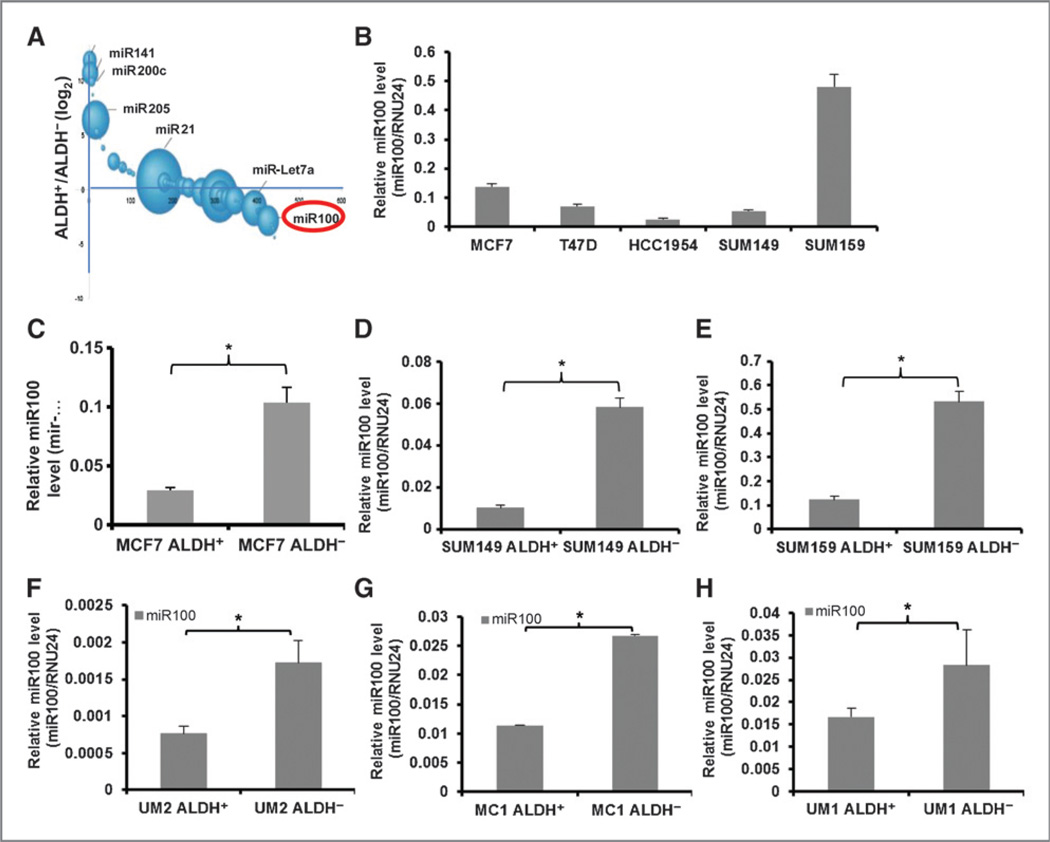
We have previously demonstrated that primary human breast cancers and established breast cancer cell lines contain subpopulations with stem cell properties that can be isolated by virtue of their expression of ALDH as assessed by the Aldefluor assay. These cells display increased tumor-initiating capacity and metastatic potential compared with corresponding Aldefluor-negative cells (3). ALDH+ and ALDH− populations were separated from a human breast carcinoma cell line SUM159 and miRNAs were quantitated by expression profiling. miR100 expression is significantly higher in the ALDH− population compared with the ALDH+ population as shown in Fig. 1A A “bubble plot” can be used to depict both the abundance of a particular miRNA (given as the sum of the reads in the two populations) and its relative expression (plotted as a log2 of the ratio of reads in each population). As assessed by qRT-PCR, miR100 expression was variable across different breast cancer cell lines and did not correlate with molecular subtypes (Fig. 1B) and the ALDH+ cells were also shown in Supplementary Fig. S1 utilizing the Aldefluor assay. However, within each cell line, miR100 expression was significantly increased in the ALDH− compared with ALDH+ cell population, including luminal (MCF7; Fig. 1C), basal (SUM149; Fig. 1D), and claudinlow (SUM159; Fig. 1E) cell lines. Similar findings were seen using cells isolated from primary human breast tumor xenografts (UM2, MC1, UM1), which were not passaged in vitro and directly established from patient tumors (Fig. 1F – H). MC1 and UM1 were derived from claudinlow and UM2 from a basal breast carcinoma (3). These studies demonstrate that in these breast cancer cell lines and primary xenografts, low miR100 expression is associated with the CSC phenotype characterized by increased ALDH expression.
Figure 1.
Comparison of miR100 expression in different cell populations. A, a bubble plot depicting the relative abundance and log2 ratio of miRNAs in SUM159 cells. B, miR100 expression level was measured indifferent cell lines by qRT-PCR. ALDH+ cells from MCF7 cells (C), SUM149 cells (D), SUM159 cells (E), or primary breast tumor xenografts UM2 (F), MC1 (G), and UM1 (H) show lower miR100 expression level in comparison with ALDH− cells from the same cell lines as accessed by qRT-PCR. * P < 0.05. Error bars, mean ± SD.
miR100 overexpression decreases the cancer stem/progenitor population and inhibits cancer cell proliferation in vitro
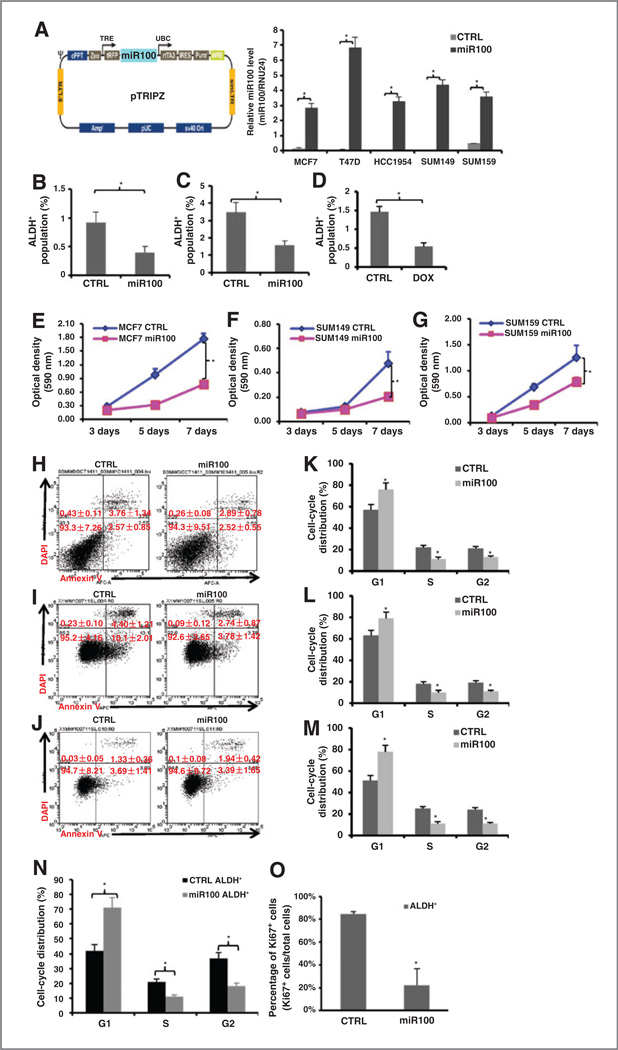
We utilized a tetracycline (doxycycline) inducible miR100 construct tagged with RFP (pTRIPZ-mir-100-RFP) to determine the functional role of miR100 in CSCs (Fig. 2A). miR100 levels were significantly increased by 10 hours after doxycycline induction and maintained at a high level in the presence of doxycycline in all transduced cell lines (Fig. 2A). Induction of miR100 resulted in a significant decrease in the proportion of CSCs as assessed by the Aldefluor assay, an effect seen in cell lines representing different breast cancer subtypes (Fig. 2B – D). In addition to its effect on the CSC population, induction of miR100 also inhibited cell proliferation in bulk tumor cells as assessed by MTT assay (Fig. 2E – G and Supplementary Fig. S2D–S2F), an effect not due to the induction of apoptosis as assessed by Annexin V staining (Fig. 2H – J). In addition, the differentiation assay was performed for each cell line and ALDH+ cells took about 7 to 10 days to differentiate back to the parental cell level (Supplementary Fig. S3A–S3C). To determine the relationship between miR100 expression and cell-cycle kinetics, we utilized the doxycycline-inducible miR100 construct to determine the effect of miR100 induction on cell-cycle distribution. Induction of miR100 increased the G1 cell population from 57% to 76% in MCF7, from 63% to 79% in SUM149 cells, and from 51% to 78% in SUM159 cells, with a concomitant decrease in the cycling population (S–G2–M). Furthermore, miR100 induces a G1 arrest in ALDH+ cell populations (Fig. 2K – N). This finding was confirmed by analysis of the cycling population as assayed by Ki67 staining (Fig. 2O). These experiments suggest that miR100 overexpression is able to reduce the CSC population as well inhibit growth of the bulk tumor population. Moreover, this is not due to the induction of apoptosis but due to the effect on the cell cycle. Furthermore, the decrease in the population of ALDH+ cells suggested a selective effect on the CSC population.
Figure 2.
miR100 overexpression decreases ALDH+ population and inhibits cell proliferation. A, diagram of miR100-inducible lentiviral vector (left). Different cell lines were transduced with the pTRIPZ-miR100 lentivirus and selected with puromycin for 7 days. Cells were treated with (miR100) or without (CTRL) tetracycline (doxycycline; DOX). Total RNA was isolated and miR100 expression level were measured by qRT-PCR. B-D, transduced cells were treated with vehicle control or doxycycline (1µg/mL) for 7 days, and dissociated and utilized for Aldefluor assay by flow cytometry [MCF7 (B), SUM149 (C), SUM159 (D)]. E-G, 200–500 transduced cells were seeded in 96-well culture plates and cultured in the absence (CTRL) and presence (miR100) of doxycycline for 3,5, or 7 days. MTT assays were conducted following the manufacturer’s protocol and the optical density (OD) value was measured at 590 nm. H-O, transduced cells were treated with vehicle control or doxycycline (1µg/mL) for 7 days. Cells were dissociated and stained for Annexin V-APC and DAPI for apoptosis assay by flow cytometry (H-MCF7, I-SUM149, J-SUM159), and utilized for cell cycle by flow cytometry [MCF7 (K), SUM149 (L), SUM159 (M and N)]. Propidium iodide staining was used to analyze cell-cycle distribution. miR100 induction resulted in an increased proportion of cells in the G1 phase. O, ALDH+ cells were sorted from transduced SUM159 cells (CTRL and miR100) by Aldefluor assay, cytospun, and stained for Ki67 by immunohistochemical staining. Both Ki67+ tumor cells and Ki67− tumor cells were counted in at least five random fields. miR100 induction significantly decreased Ki67+ cells in ALDH+ cell populations. *, P< 0.05. Error bars, mean ± SD.
miR100 inhibits breast tumor growth in vivo
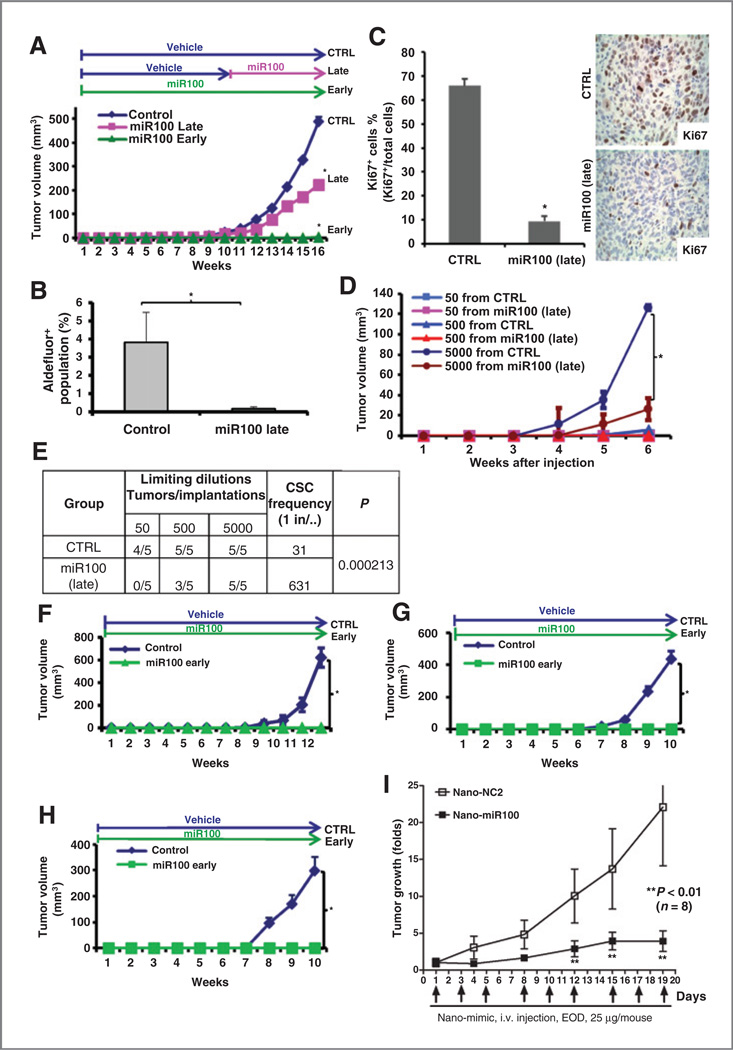
As the tetracycline-inducible miR100 system allows for the controlled regulation of CSC populations, it provides a valuable tool for assessing the role of CSCs in tumor growth in mouse xenograft models. Furthermore, the ability to regulate the CSC population during different phases of tumor growth allows for the assessment of the role of these cells in tumor initiation and maintenance. We first determined the effect of miR100 induction on the growth of established tumors. When tumors reached 0.2 to 0.3 cm in diameter, we induced miR100 with doxycycline treatment (hereafter “miR100 late”). Induction of miR100 significantly inhibited the growth of SUM159 (Fig. 3A), SUM149 (Supplementary Fig. S4A), MCF7 (Supplementary Fig. S4C), and T47D (Supplementary Fig. S4D) xenografts. After 6 to 10 weeks of treatment, animals were sacrificed and cell proliferation (Ki67 staining) and CSC populations were assessed. Induction of miR100 reduced the Aldefluor-positive population by more than 90% in SUM159 (Fig. 3B) and more than 30% in SUM149 (Supplementary Fig. S4B) compared with control. Cell proliferation as assessed by Ki67 expression was quantitated by immunohistochemistry in the tumor specimens from SUM159 xenografts. Consistent with retarded tumor growth, the proportion of Ki67-positive cells was significantly lower in the miR100 late group compared with the control group (Fig. 3C). miR100 expression was significantly higher in doxycycline-treated group compared with the control group at the end of treatment (Supplementary Fig. S5). To assess the effect of miR100 induction on the CSC population, we determined the ability of serial dilutions of cells obtained from primary tumors to form tumors in secondary NOD/SCID mice. Cells isolated from tumors with miR100 induction had markedly reduced tumor-initiating capacity in secondary mice with no tumors observed after the introduction of 50 cells from the miR100 late group (Fig. 3D). These functional assays allow us to calculate the frequency of tumor-initiating cells. miR100 expression decreases the CSC frequency (22) supporting the results from Aldefluor analysis (Fig. 3E). Interestingly, we found that the percentage of ALDH+ cells in both primary CTRL and secondary CTRL tumors were increased compared with the parental cells injected (Supplementary Fig. S6). These studies demonstrate that miR100 induction reduces the CSC population, reducing growth of established tumor xenografts.
Figure 3.
miR100 inhibits tumor growth of SUM159 cells in vivo. A, 50,000 pTRIPZ-SUM159-miR100 cells were injected into the fourth fat pads of NOD/SCID mice. The treatment started as indicated on the top of the growth curve. miR100 was induced by adding doxycycline (DOX, 1 mg/mL) in drinking water. miR100 late, doxycycline was added to the drinking water after the average tumor size reached to about 10 mm3; miR100 early, doxycycline was added to the drinking water right after SUM159 cells were inoculated to the fat pads. miR100 induction inhibits SUM159 tumor growth in advance setting and completely blocks tumor formation in adjuvant setting. B, tumors from CTRL and miR100 (late) were collected and cells were isolated from each tumor. ALDH was accessed by the Aldefluor assay on viable dissociated cells. C, Ki67 stainings were performed by immunohistochemistry on fixed sections; both Ki67+ cells (brown) and Ki67− tumor cells were counted in at least five random fields. D, serial dilutions of cells obtained from CTRL and miR100 (late) were implanted in the fourth fat pads of secondary mice, which received no further treatment. E, extreme limiting dilution analysis for the group CTRL or miR100 (late) was calculated on the website http://bioinf.wehi.edu.au/software/elda/. Briefly, utilizing the online calculation form, we input the number of cells injected and the number of mice used in each group and compared the CSC frequency between CTRL group and miR100 group. F-H, fresh isolated cells from human primary breast tumor xenografts (UM2, MC1, UM1) were infected with pTRIPZ-miR100 lentivirus in suspension and doxycycline was added to the medium for 1 to 2 days and RFP-positive cells were collected. A total of 100,000 cells from noninfected tumor cells or pTRIPZ-UM2 (F), MC1 (G), or UM1 (H) -miR100 cells was injected into the fourth fat pads of NOD/SCID mice. The treatment started right after injection as indicated on the top of the growth curve. miR100 was induced by adding doxycycline (1 mg/mL) in drinking water. *, P< 0.05; Error bars, mean ± SD. I, NOD/SCID mice bearing SUM159 xenografts were treated with either Tf-LipA-negative control (Nano-NC2) or Tf-LipA-miR100 (Nano-miR100; n = 8 mice per cohort) by tail vein injection at a dose of 25 µg miRNA mimics per mouse every other day (EOD). Treatment with miR100 anti-CD44 nanovector significantly inhibited tumor growth (P < 0.01) compared with the negative control group. Data, the tumor volume before initiation of treatment.
Preclinical models have suggested that CSCs play a role in tumor recurrence and metastasis following adjuvant therapy (23). This suggests that targeting of CSCs may have more dramatic effects with early treatments than with late treatments. To model this, we induced miR100 immediately after tumor cell implantation ("miR100 early"). Although tumors grew after 2 to 3 weeks after orthotopic induction in control animals of SUM159 cells, there was no observed tumor growth at 16 weeks after miR100 induction (Fig. 3A). Similar finding was seen in MCF7 and T47D xenografts (Supplementary Fig. S4C and S4D) and three additional primary breast tumor xenografts UM2, MC1, and UM1 (Fig. 3F – H). Utilizing qRT-PCR, we confirmed that miR100 level was significantly higher in the miR100 groups (late and early) compared with control (Supplementary Fig. S7). Furthermore, to simulate the adjuvant setting in clinic, we induced miR100 and/or administered docetaxel immediately after tumor cell implantation. Although tumors grew after 4 to 5 weeks in control animals, there was no observed tumor growth following miR100 induction and/or docetaxel treatments for 8 weeks (Supplementary Fig. S8). After 8 weeks, treatments were stopped and animals observed for an additional 4 weeks (except for the control group, which had to be sacrificed). In SUM159 xenografts, tumors developed in all mice who received 8 weeks of docetaxel alone. In contrast, there was minimal tumor growth in animals with previous miR100 induction, independent of whether they had received docetaxel (Supplementary Fig. S8). Together, these studies support the concept that inhibiting CSC immediately after implantation, simulating the adjuvant setting, has a profound effect on inhibiting subsequent tumor growth.
Recently, we developed tumor-specific, ligand-targeting, self-assembled nanoparticle-DNA lipoplex systems designed for systemic gene therapy of cancer (24–28). CD44 is preferentially expressed in breast CSCs, so we employed anti-CD44 monoclonal antibody H4C4 (29) and anti-CD44 scFv (30) in liposome–DNA complex to make nanovectors in our study, which has been tested both in vitro and in vivo. Our study showed that systemic miR100 delivery with anti-CD44 nano-vectors inhibits the tumor growth of SUM159 in NOD/SCID mice (Fig. 3I). This study further confirms the antitumor capability of miR100 in vivo.
To determine whether downregulation of endogenous miR100 in SUM159 cells promoted tumorigenesis, we utilized a mirZip antisense miRNA. qRT-PCR was utilized to confirm the efficient knockdown of miR100 (Supplementary Fig. S9A). miR100 knockdown significantly increased SUM159 cell proliferation as accessed by the MTT assay (Supplementary Fig. S9B). As shown in Supplementary Fig. S9C, knockdown of miR100 significantly promoted the growth of SUM159 cells in tumor xenografts as well as increased the CSC frequency (Supplementary Fig. S9D).
miR100 inhibits tumor metastasis in vivo
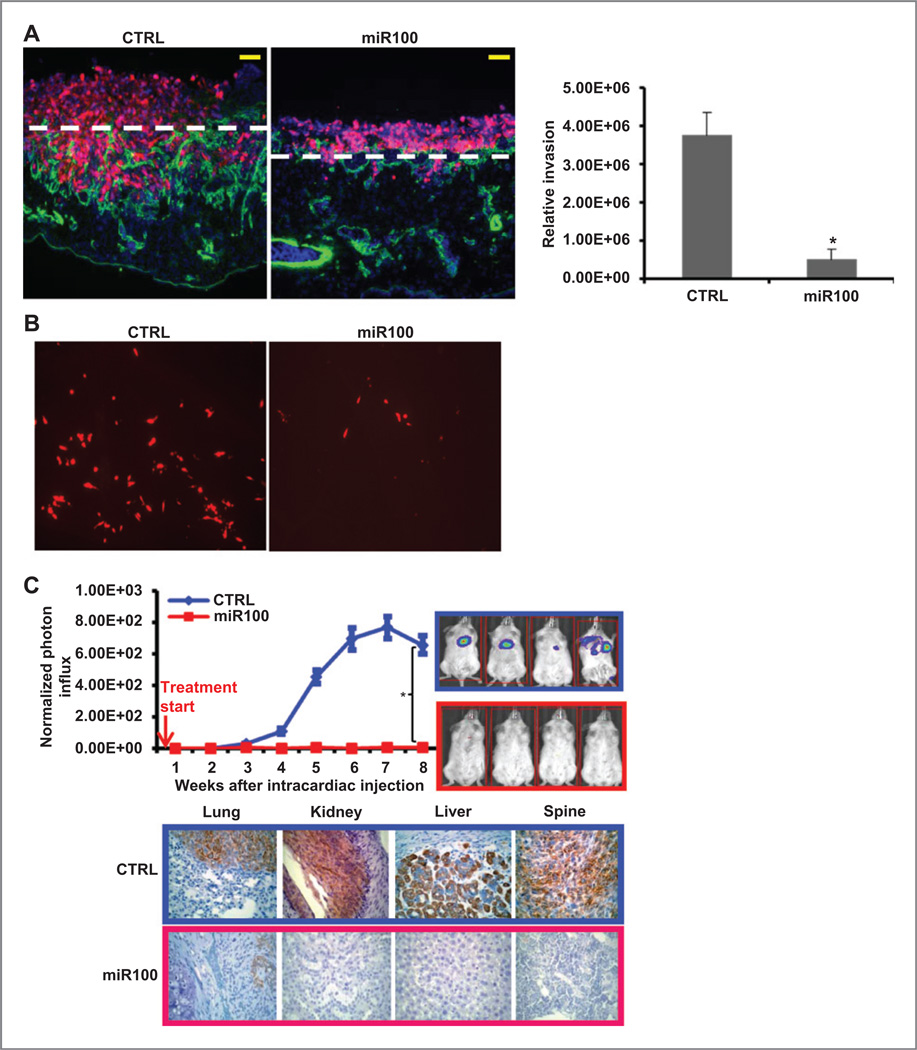
Previous studies have demonstrated that CSCs mediate invasion and metastasis. To determine the effect of miR100 expression on tumor invasion, we utilized the CAM invasion assay. SUM159 CTRL or SUM159 miR100 were labeled with DsRed and cultured atop the chick CAM, a tissue whose stromal compartment is rich in interstitial collagens (31). After a 3-day culture period, control SUM159 cells rapidly cross the CAM surface and infiltrate the underlying stromal tissues (Fig. 4A). SUM159 cells not only infiltrate the upper CAM surface but also access the chick vascular bed to travel to distant sites in the embryo (Fig. 4B). However, both invasion and intravasation of SUM159 cells were dramatically inhibited by miR100 overexpression (Fig. 4A, B).
Figure 4.
miR100 inhibits metastasis of SUM159 cells in vivo. A, SUM159 cells (CTRL, miR100) were labeled with DsRed (infected with DsRed lentivirus) and cultured atop the live chick CAM for 3 days. CAM sections were stained with anti-chicken type IV collagen antibody for basement membrane and blood vessels (green) and counterstained with DAPI. The CAM surface is marked by dashed white lines, and CAM invasion was quantified as described in Materials and Methods. Results are expressed as the means ± SEM (n = 3). * P < 0.01. Bars, 100 µm. B, fluorescence micrograph of cells metastasizing to the distant organ sites. C, 200,000 pTRIPZ-SUM159-miR100-Luc cells in 100 µL of PBS were injected into the left ventricle of NOD/SCID mice. The treatment started immediately after injection as indicated by the red arrow. Metastasis formation was monitored using bioluminescence imaging. Quantification of the normalized photon flux, measured at weekly intervals following inoculation. Histologic confirmation (right) by pan-cytokeratin (AE1/AE3) staining (brown) of metastasis in bone and soft tissues resulting from the CTRL or miR100 group of mice. The metastasis was suppressed by miR100 overexpression. * P < 0.05. Error bars, mean ± SD.
To determine whether the expression of miR100 affects development of tumor metastasis in a mouse model, SUM159 cells cotransfected with the inducible miR100 vector and luciferase were introduced into NOD/SCID mice by intracardiac injection, and metastasis formation was monitored by bioluminescence imaging. Doxycycline treatment was initiated after intracardiac injection. As shown in Fig. 4C, miR100 induction completely suppressed metastasis formation, which was confirmed by histologic examination with pan-cytokeratin staining (Fig. 4C). To determine whether miR100 induction in ALDH+ cells alters their metastatic capacity, we sorted ALDH+ cells and ALDH− cells, and introduced them into NOD/SCID mice by intracardiac injection. Doxycycline treatment was initiated after intracardiac injection. We found that the miR100 induction completely blocked metastasis of ALDH+ cells in addition to the unsorted total population in these mice (Supplementary Fig. S10). Furthermore, we confirmed our previous findings that ALDH+ cells had much higher metastasis capability than the unsorted total population, but ALDH− cells barely metastasized (Supplementary Fig. S10). Together, these studies suggest miR100 expression inhibits CSC invasion as well as growth at metastatic sites.
miR100 downregulates stem cell regulatory and cell proliferation genes
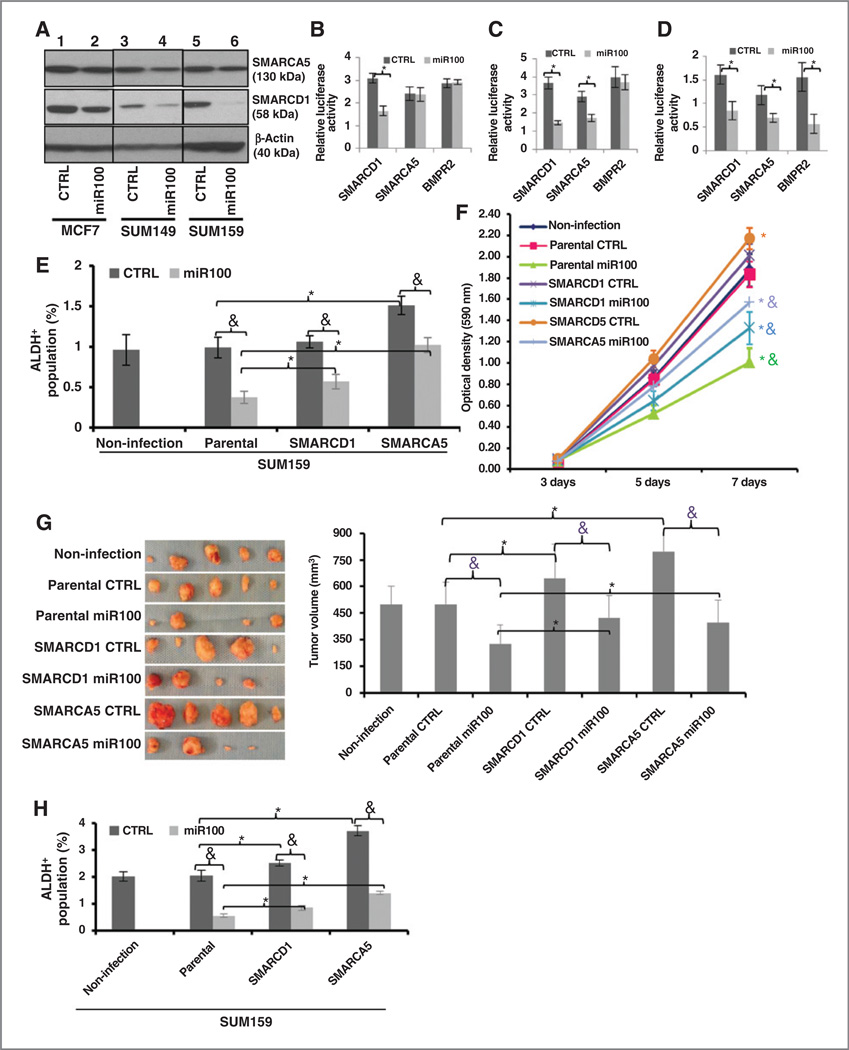
To determine the cellular targets of miR100 in BCSCs and non-BCSCs, ALDH+ and ALDH− populations of pTRIPZ-SUM159-miR100 cells were separated and cultured in suspension for 10 hours in the presence or absence of doxycycline. Gene expression profiles of the four populations were determined utilizing Affymetrix microarrays (Supplementary Fig. S11A). Of the 6,900 genes downregulated at least 2-fold upon doxycycline treatment in the ALDH+ population, 18 overlapped with the 40 predicted target sequences of miR100 from TargetScan including genes known to be involved in stem cell regulation and cell proliferation (Supplementary Fig. S11A), including SMARCA5, SMARCD1, BMPR2, FGFR, FZD5, and IFGR. The downregulation of SMARCA5, SMARCD1, and BMPR2 in the ALDH+ population of MCF7, SUM149, and SUM159 cells after miR100 induction was confirmed with qRT-PCR (Supplementary Fig. S11B–S11D). SMARCA5 was downregulated in SUM159 and SUM149 cells and BMPR2 was only downregulated in SUM159 cells after miR100 induction. Downregulation of SMARCD1 and SMARCA5 protein by miR100 induction was confirmed by Western blot analysis (Fig. 5A). In contrast, only 560 genes were significantly downregulated by doxycycline in the ALDH− population (Supplementary Fig. S11A) with three of these genes overlapping with predicted miR100 targets. These studies suggest that miR100 regulates the CSC population and inhibits cell proliferation by simultaneously targeting a number of stem cell regulatory genes and cell proliferation genes. To confirm these findings, we utilized a luciferase reporter assay to determine the effect of miR100 on the expression of the stem cell regulatory genes SMARCD1, SMARCA5, and BMPR2 selected from the expression profiling data. Expression of miR100 reduced the activities of SMARCD1 in MCF7 (Fig. 5B), SMARCD1 and SMARCA5 in SUM149 (Fig. 5C), SMARCD1, SMARCA5, and BMPR2 in SUM159 cells (Fig. 5D).
Figure 5.
SMARCA5 and SMARCD1 are direct and functional targets of miR100. A, targets (SMARCA5 and SMARCD1) are verified by Western blot analysis in all three cell lines. B–D, activity of the luciferase gene linked to the 3′ UTR of SMARCD1, SMARCA5, or BMPR2. The pMIR-REPORT or LightSwitch 3′ UTR luciferase reporter plasmids were transiently transfected into pTRIPZ-miR100 cell lines [MCF7 (B); SUM149 (C); SUM159 (D)]and an internal control ACTB luciferase reporter was cotransfected for normalization. The relative luciferase activity was calculated as the ratio of the results from the cells transfected by individual reporter/the results from the cells transfected by the internal control in the same cell group. The data are mean and SD of separate transfections (n = 4). E and F, overexpression of human SMARCA5 and SMARCD1 lacking 3′ UTR partially overcome the decrease of ALDH+ cells and the inhibition of cell proliferation by miR100 induction in SUM159 cells. G, overexpression of human SMARCA5 and SMARCD1 lacking 3′ UTR partially overcome the inhibition of tumor growth by miR100 induction in SUM159 cells. Fifty thousand cells from different groups were injected into the fourth fat pads of NOD/SCID mice. The treatment was started when the tumors reached to the average size of 10 mm3 in the control group and the treatment lasted for 8 weeks. miR100 was induced by adding doxycycline (DOX, 1 mg/mL) in drinking water. At the end of the treatment, the tumors were taken out and the tumor images (left) and tumor size (right) are shown. H, overexpression of human SMARCA5 and SMARCD1 lacking 3′ UTR partially overcome the decrease of ALDH+ cells by miR100 induction in SUM159 tumors. Cells were isolated from each tumor collected in G and ALDH was accessed by the Aldefluor assay on viable dissociated cells. * P < 0.05 (compared with the corresponding parental group); &, P < 0.05 (compared with the corresponding CTRL group). Error bars, mean ± SD.
To determine whether SMARCA5, SMARCD1, and BMPR2 represent functionally important targets of miR100, we knocked down these genes individually or in combination in SUM159 cells and confirmed knockdown by qRT-PCR for BMPR2 and Western blot analysis for SMARCD1 and SMARCA5 (Supplementary Fig. S12A). Knocking down any of these genes individually inhibited cell proliferation, whereas knockdown of all three genes together had most dramatic effect on the inhibition of cell proliferation (Supplementary Fig. S12B). Furthermore, knockdown of SMARCD1 and SMARCA5 significantly decreases the proportion of ALDH+ SUM159 cells, suggesting these genes play a role in the regulation of CSC self-renewal (Supplementary Fig. S12C). To determine whether knockdown of these genes inhibited tumorigenesis, we injected serial dilutions of SUM159 control, SUM159 SMARCD1-SMARCA5-shRNAs, and SUM159 SMARCD1-SMARCA5-BMPR2-shRNAs into the mammary fat pads of NOD/SCID mice. Knockdown of SMARCA5 and SMARCD1 significantly retarded tumor growth and decreased the proportion of BCSCs in these tumors (Supplementary Fig. S12D). "Rescue" experiments where either SMARCA5 or SMARCD1 was overexpressed using a cDNA that lacked the 3′-UTR containing the miR100-binding sites, which was verified by Western blot analysis as shown in Supplementary Fig. S13, partially abrogated miR100-mediated decrease of ALDH+ cell population (Fig. 5E), and partially abrogated miR100-mediated inhibition of cell proliferation (Fig. 5F) as well as inhibition of tumor growth of SUM159 cells (Fig. 5G) by partially abrogating miR100-mediated decrease of ALDH+ cell population in the tumors. Similar results were also seen in another breast cancer cell line SUM149 in vitro (Supplementary Fig. S14). Together, these results strongly support the role of SMARCA5 and SMARCD1 as direct and functional targets of miR100. Furthermore, to understand the clinical significance of both SMARCA5 and SMARCD1 in breast cancer, we examined the relationship between expression of SMARCA5 or SMARCD1 and overall survival of patients in publically available datasets (32, 33). Utilizing the Kaplan–Meier method, we determined that breast tumors with high SMARCA5 or SMACD1 expression had a significantly shorter survival time, compared with those with low expression (Supplementary Fig. S15). This supports the clinical relevance of our in vitro and mouse studies.
Low miR100 expression in primary breast cancer tissues correlates with high ALDH1 expression and poor patient survival
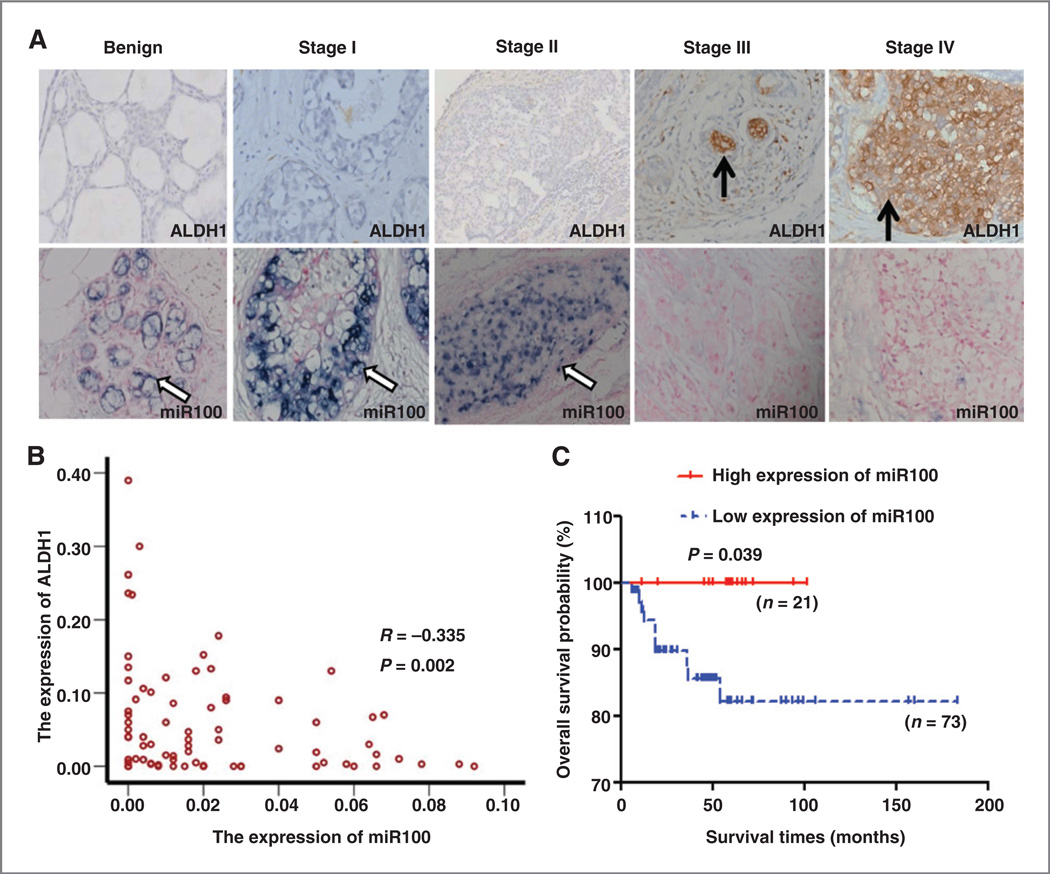
To investigate the clinical significance of miR100 expression, we determined the relationship between expression of miR100, ALDH1, and patient survival in a cohort of 94 patients with breast cancer. We utilized immunohistochemical staining to assess ALDH1 expression and ISH with a digoxigenin-labeled miR100 probe to assess miR100 expression in primary breast cancer tissues of patients with different molecular subtypes of breast cancer (Supplementary Table S1) as well as in normal breast tissue (benign). As shown in Fig. 6A, ALDH1 expression was very low in normal breast and in stages I and II and dramatically increased in invasive breast cancer, whereas previously reported ALDH expression correlated with tumor grade (Fig. 6A and Supplementary Table S2).In contrast, miR100 level was very high in normal breast and ductal carcinoma in situ tumors, and was decreased in invasive tumors where its expression was inversely related to tumor stage (Fig. 6A). Expression of miR100 and ALDH1 were inversely correlated (R = −0.334; P = 0.001) with low expression of miR100 positively related to high expression of ALDH1 (Fig. 6B). Kaplan–Meier survival curves demonstrated that the overall survival of the patients with high miR100 expression was significantly longer than those with low miR100 expression (Fig. 6C; P < 0.05).
Figure 6.
Expression of ALDH1 and miR100 in breast carcinoma. A, representative example of ALDH1 (DAB-IHC, 400×; top, brown) and miR100 (NBT/BCIP-ISH, 400×; bottom, blue) in normal breast and breast tumor (stage I, II, III, IV). Black arrow, ALDH1; white arrow, miR100. B, the correlation analysis of miR100 and ALDH1 expressions: the expressions of miR100 and ALDH1 are negatively correlated (R = −0.335; P = 0.002). The low expression of miR100 was positively related to the high expression of ALDH1. C, Kaplan-Meier curves with log rank tests show statistical difference in overall survival: the red full line and the blue dotted line represent the high and low expression of miR100, respectively. It shows that the 5-year survival rates were shorter in patients with low expression of miR100 (P = 0.039).
Discussion
Accumulating evidence suggests that more effective therapies of cancer will require the successful targeting of CSC populations. However, it is still not fully understood how these CSCs are regulated. Previous studies show that both miRNA Let7 and mir200c regulates self-renewal of breast stem cells (34, 35). These miRNAs offer great promise for cancer therapy as they might have potential to target the CSCs. Thus, miRNA therapy could be a powerful tool to address CSC dysregulation and its resulting self-renewal and cancer progression in patients.
In this study, we demonstrate that miR100 is capable of modulating BCSC self-renewal and inhibiting breast cancer cell proliferation through targeting SMARCA5, SMARCD1, and BMPR2 both in vitro and in vivo in addition to the Wnt/β-catenin pathway. Enforced miR100 expression in breast cancer cell lines and primary xenografts significantly reduced the tumor growth by reducing CSC populations in vivo. These studies are consistent with one recent finding (36); however, they mainly used one immortalized transformed HMLE in this study. CSC models predict that the efficacy of CSC-targeting agents should be most pronounced in the early setting where tumor growth from micrometastasis is dependent on stem cell self-renewal (37). Consistent with this model, induction of miR100 immediately after fat pad implantation or after development of micrometastasis by intracardiac injection completely blocked tumor initiation and metastasis. These studies suggest that miR100 inhibits tumor growth and metastasis by inhibiting BCSC self-renewal and cell proliferation. Our studies differ from a recent report suggesting that miR100 regulates cancer cell proliferation by targeting IGF2 (38). Our studies did not show IGF2 was significantly affected in either stem cell population or non–stem cell population by overexpression of miR100. Instead, we found that a majority of genes downregulated by miR100 overexpression were seen in ALDH+ population, including genes known to be involved in stem cell self-renewal and cell proliferation, including SMARCA5, SMARCD1, BMPR2, and FZD5. It has been shown that miR100 was required for proper differentiation of mouse ESCs and that it functions in part by targeting SMARCA5 (39). SMARCA5 (hSNF2H) is a member of SWI/SNF family, containing helicase and ATPase activities. It is overexpressed in ovarian cancer where it promotes tumor growth (40). SMARCD1 (BAF60a) is also a member of SWI/SNF family of proteins and regulates cell proliferation. In this study, we confirmed that both SMARCA5 and SMARCD1 are direct targets of miR100 and demonstrated that the miR100 represses the expression of SMARCD1 and SMARCA5 at both mRNA and protein level, and that both are involved in mediating the effects of miR100 on CSC self-renewal and cancer cell proliferation. Furthermore, previous studies showed that SWI/SNF family members interact with the components of the Wnt signaling pathway, resulting in decreased Wnt signaling (41). Previous work indicates that telomerase catalytic subunit [human telomerase reverse transcriptase (hTERT)] regulates stem cell homeostasis independent of its function at telomeres (42) and that TERT, together with the SWI/SNF complex protein brahma-related gene 1 (BRG1, also called SMARCA4), modulates stem cell homeostasis by modulating the WNT/β-catenin signaling pathway (43). In that system, telomerase directly modulates Wnt/β-catenin signaling by serving as a cofactor in a β-catenin transcriptional complex where the telomerase protein component TERT interacts with BRG1 activating Wnt signaling. Furthermore, chromatin immunoprecipitation of the endogenous TERT protein from mouse gastrointestinal tract demonstrated that TERT physically occupies gene promoters of Wnt-dependent genes (43). Our previous studies showed that the Wnt/β-catenin pathway is an important regulator of BCSC self-renewal (44). The observation that miR100 expression leads to a decrease in both proportion and absolute number of BCSCs indicates an inhibitory effect on CSC self-renewal, resulting in a depletion of this cell population. In addition to SWI/SNF proteins, our profiling data also demonstrated that miR100 targets the Wnt receptor FZD5, the significance of which requires further investigation. Together, these studies suggest that miR100 inhibits BCSC self-renewal primarily by targeting SWI/SNF family members. A recent report suggested that the tumor-inhibiting properties of miR100 were not due to depletion of the stem-like population (34). However, these studies assessed CSCs solely by expression of the CSC marker ALDH1. In contrast, our studies utilized functional CSC assays as well as the Aldefluor assay, which measures total ALDH activity and is not limited to the ALDH1 isoform.
As determined by ISH, we found that miR100 was expressed at the highest level in normal breast tissue with progressively less expression in advanced invasive breast carcinoma. In these tumors, there was a strong inverse correlation between miR100 expression and the expression of CSC marker ALDH1 (P = 0.002). We also found a significant association (P = 0.039) between low miR100 expression and decreased patient overall survival. Our results are consistent with a previous report showing that miR100 expression was decreased in advanced and metastatic prostate cancer relative to normal prostate epithelium (45, 46) and in invasive human breast tumors compared with benign patient samples (38).
In conclusion, these data demonstrate that miR100 modulates BCSCs and cancer cell proliferation via targeting SMARCA5 and SMARCD1, in addition to BMPR2 signaling pathways. The tetracycline-inducible miR100 system allows for controlled regulation of the CSC population, providing a valuable model to simulate the effects of CSC-directed therapies on breast cancer growth and metastasis. Furthermore, pathways regulated by miR100 may provide novel targets for CSC-directed therapies.
Our miR100 anti-CD44 nanovector studies provide novel technique that can deliver miR100 to breast CSCs and inhibit their self-renewal and tumor initiation, which will provide an important impetus to develop the anti-CD44-nanovector-miR100 as a novel and more effective therapy for human breast cancer by modulating BCSCs. Furthermore, pathways regulated by miR100 may provide novel targets for CSC-directed therapies.
Supplementary Material
Acknowledgments
The authors thank Dr. Stephen Ethier for generously providing the breast cancer cell lines SUM159.
Grant Support
This work was supported by CAS stem cell grant XDA01040410, NSFC grants 81472741 and 81322033, Susan G Komen Career Catalyst award KG110316 and the Fundamental Research Funds for the Central Universities WK2070000034 (S. Liu). M.S. Wicha and G.J. Hannon were supported by a Stand Up to Cancer Dream Team Translational Research Grant, Grant Number SU2C–AACR-DT0409. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
M.S. Wicha has ownership interest (including patents) in Oncomed and is a consultant/advisory board member for Verastem. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: L.Deng, J.Chen, I.Ibarra, G.J.Hannon, M.S. Wicha, S.Liu
Development of methodology: L. Deng, L. Shang, X. He, T.K. Luther, L. Xu, E. Song, S.J. Weiss, M.S. Wicha, S. Liu
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): L. Deng, L. Shang, S. Bai, J. Chen, S. Chen, X. Li, X. Meng, B. Yu, X. Wang, S.P. McDermott, A.E. Ariazi, C. Ginestier, J. Ke, T.K. Luther, S.G. Clouthier, L. Xu, H. Yao, S.J. Weiss, S. Liu
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): L. Deng, L. Shang, S. Bai, S. Chen, X. Li, X. Wang, S.P. McDermott, H. Yao, S.J. Weiss, M.S. Wicha, S. Liu
Writing, review, and/or revision of the manuscript: S.G. Clouthier, M.S. Wicha, S. Liu
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): X. He, R.M. Trevino, S. Chen, X. Meng, Y. Liu, A.E. Ariazi, C. Ginestier, T.K. Luther, S.G. Clouthier, H. Yao, M.S. Wicha, S. Liu
Study supervision: S.G. Clouthier, T.K. Luther, S.G. Clouthier, L. Xu, G. Shan, M.S. Wicha, S. Liu
References
- 1.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;1:59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are micro-RNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Kato M, Slack FJ. microRNAs: small molecules with big roles - C. elegans to human cancer. Biol Cell. 2008;100:71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- 7.Lowery AJ, Miller N, McNeill RE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin Cancer Res. 2008;14:360–365. doi: 10.1158/1078-0432.CCR-07-0992. [DOI] [PubMed] [Google Scholar]
- 8.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331:357–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 11.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci U S A. 2012;109:14470–14475. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 16.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 17.Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O'Brien C, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 18.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 19.Henson BJ, Bhattacharjee S, O'Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48:569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowe RG, Li XY, Hu Y, Saunders TL, Virtanen I, Garcia de Herreros A, et al. Mesenchymal cells reactivate Snail1 expression to drive three-dimensional invasion programs. J Cell Biol. 2009;184:399–408. doi: 10.1083/jcb.200810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28:4006–4012. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirollo KF, Rait A, Zhou Q, Hwang SH, Dagata JA, Zon G, et al. Materializing the potential of small interfering RNA via a tumor-targeting nanodelivery system. Cancer Res. 2007;67:2938–2943. doi: 10.1158/0008-5472.CAN-06-4535. [DOI] [PubMed] [Google Scholar]
- 25.Xu L, Pirollo KF, Chang EH. Tumor-targeted p53-gene therapy enhances the efficacy of conventional chemo/radiotherapy. J Control Release. 2001;74:115–128. doi: 10.1016/s0168-3659(01)00324-8. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Huang CC, Huang W, Tang WH, Rait A, Yin YZ, et al. Systemic tumor-targeted gene delivery by anti-transferrin receptor scFv-immunoliposomes. Mol Cancer Ther. 2002;1:337–346. [PubMed] [Google Scholar]
- 27.Xu L, Frederik P, Pirollo KF, Tang WH, Rait A, Xiang LM, et al. Self-assembly of a virus-mimicking nanostructure system for efficient tumor-targeted gene delivery. Hum Gene Ther. 2002;13:469–481. doi: 10.1089/10430340252792594. [DOI] [PubMed] [Google Scholar]
- 28.Xu L, Tang WH, Huang CC, Alexander W, Xiang LM, Pirollo KF, et al. Systemic p53 gene therapy of cancer with immunolipoplexes targeted by anti-transferrin receptor scFv. Mol Med. 2001;7:723–734. [PMC free article] [PubMed] [Google Scholar]
- 29.Belitsos PC, Hildreth JE, August JT. Homotypic cell aggregation induced by anti-CD44(Pgp-1) monoclonal antibodies and related to CD44(Pgp-1) expression. J Immunol. 1990;144:1661–1670. [PubMed] [Google Scholar]
- 30.Hauptschein RS, Sloan KE, Torella C, Moezzifard R, Giel-Moloney M, Zehetmeier C, et al. Functional proteomic screen identifies a modulating role for CD44 in death receptor-mediated apoptosis. Cancer Res. 2005;65:1887–1896. doi: 10.1158/0008-5472.CAN-04-3571. [DOI] [PubMed] [Google Scholar]
- 31.Lu C, Li XY, Hu Y, Rowe RG, Weiss SJ. MT1-MMP controls human mesenchymal stem cell trafficking and differentiation. Blood. 2010;115:221–229. doi: 10.1182/blood-2009-06-228494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self-renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 35.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen D, Sun Y, Yuan Y, Han Z, Zhang P, You MJ, et al. miR-100 induces epithelial-mesenchymal transition but suppresses tumorigenesis, migration and invasion. PLoS Genet. 2014;10:e1004177. doi: 10.1371/journal.pgen.1004177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea - a paradigm shift. Can Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 38.Gebeshuber CA, Martinez J. miR-100 suppresses IGF2 and inhibits breast tumorigenesis by interfering with proliferation and survival signaling. Oncogene. 2013;32:3306–3310. doi: 10.1038/onc.2012.372. [DOI] [PubMed] [Google Scholar]
- 39.Tarantino C, Paolella G, Cozzuto L, Minopoli G, Pastore L, Parisi S, et al. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB J. 2010;24:3255–3263. doi: 10.1096/fj.09-152207. [DOI] [PubMed] [Google Scholar]
- 40.Sheu JJ, Choi JH, Yildiz I, Tsai FJ, Shaul Y, Wang TL, et al. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68:4050–4057. doi: 10.1158/0008-5472.CAN-07-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc Natl Acad Sci U S A. 2011;108:2282–2287. doi: 10.1073/pnas.1013751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009;7:e1000121. doi: 10.1371/journal.pbio.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, Evans C, et al. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71:1313–1324. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.