Homozygous and compound heterozygous mutations in ABCA4 are associated with multiple phenotypes, including bull’s-eye maculopathy (BEM).1 The BEM fundus patterning can also be conferred by other genes such as the retinitis pigmentosa guanosine triphosphatase regulator gene (RPGR), a gene that encodes a protein involved in trafficking to the outer segment.2 The carrier frequency of disease-causing mutations in ABCA4 is 1 in 20. Even after complete sequencing of the ABCA4 exons and adjacent intronic sequences in patients with a clinical diagnosis of Stargardt disease 1 (STGD1), 15% to 20% of cases still have only 1 identified disease-causing mutation and no mutations are found in 10% to 15% of individuals.3 Thus, the phenotypic and allelic variability of ABCA4-related disease remains a challenge.
Due to reduced protein function, ABCA4 mutations trigger elevated accumulation of bisretinoid lipofuscin in retinal pigment epithelial cells. This increase is revealed in most but not all cases as augmented fundus autofluorescence (AF), the inherent AF of the retina that is emitted with 488-nm excitation.4 Using a standardized method for quantification of fluorescence intensities in fundus AF images (quantitative fundus AF[ qAF]), increased qAF levels have been observed in patients with STGD14 even at young ages and in fundus areas that qualitatively appear to be unaffected. Herein, we show that qAF is a clinical tool that may help in the management of inherited retinal diseases.
Report of a Case
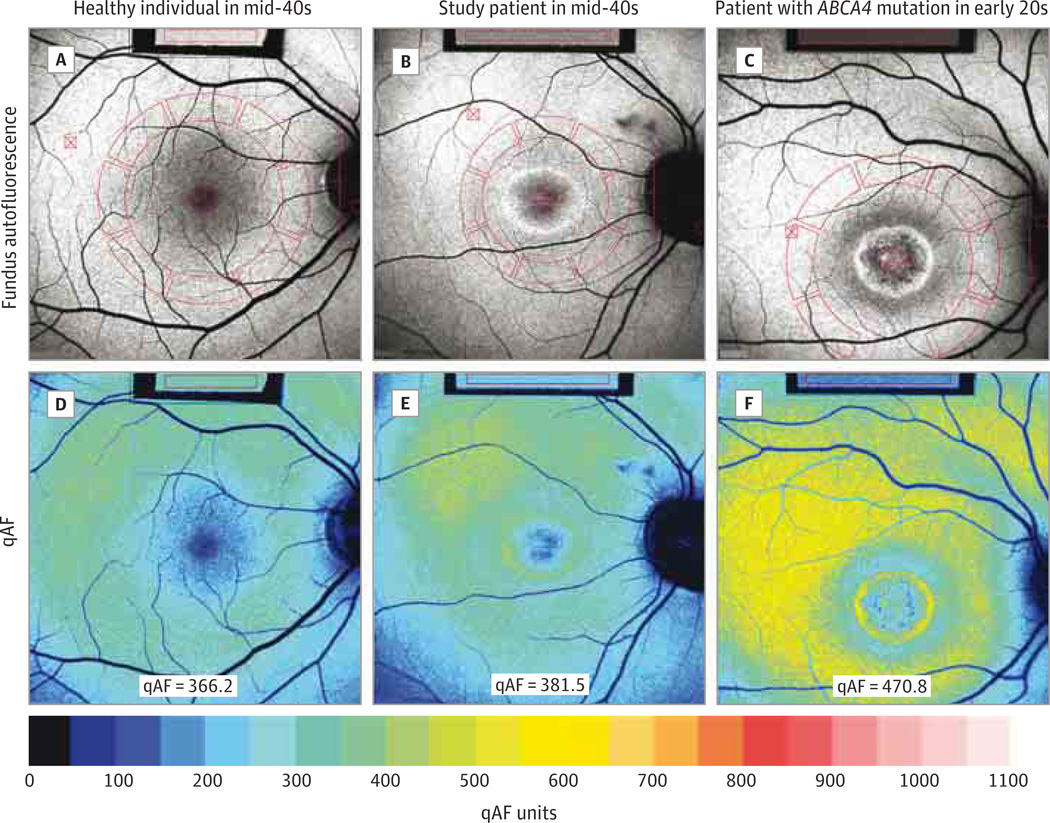
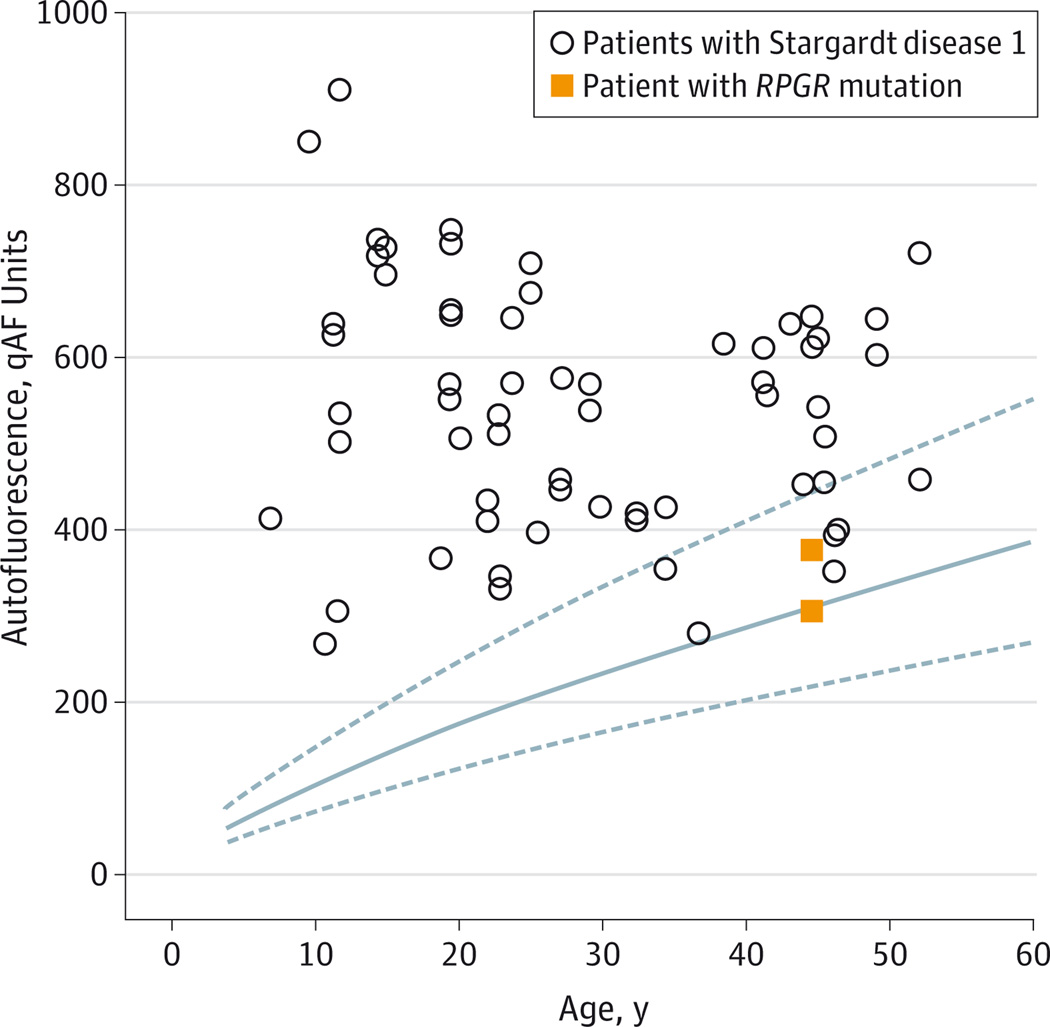
A man in hismid-40s had blurred central vision, photophobia, and nyctalopia since childhood. Examination showed a BEM that was evident in fundus AF images, but there were no flecks, intraretinal pigment migration, retinal vessel attenuation, or optic nerve pallor (Figure 1).He had previously been clinically diagnosed as having STGD1 and genetic screening with an ABCA4 array had revealed a single copy of an ABCA4 gene variant (rs1800548, p.E471K). Sanger sequencing confirmed this change. Retinal disease was reported in his brother and maternal grandfather. As previously described,2,5 the proband’s mother had reported being asymptomatic but was found to exhibit decreased visual acuity, BEM in the right eye, and tapetal reflex in both eyes. Whole-exome sequencing confirmed the ABCA4 variant and detected a novel mutation in the open reading frame 15 domain of RPGR (c.3070G>T, pGlu1024×, OMIM #312610). The mean qAF values of the patient were found to be within the range observed in healthy eyes. The patient’s mean values were also lower than in many patients of the same age with STGD14 (Figure 1 and Figure 2).
Figure 1. Fundus Autofluorescence and Color-Coded Quantitative Fundus Autofluorescence (qAF).
Mean grayscale levels in the circularly arranged segments (red) normalized to the reference (red rectangle) (A–C) provide the qAF values (arbitrary grayscale units),4 which are color coded (D–F).
Figure 2. Quantitative Fundus Autofluorescence (qAF) Values by Age for Study Patient With RPGR Mutation vs Patients With Stargardt Disease 1 and Healthy Individuals.
The qAF values (arbitrary grayscale units) acquired from each eye of our patient with an RPGR mutation and qAF values from patients with Stargardt disease 1 are plotted by age.4 Solid line indicates mean qAF values from 277 healthy individuals; dashed lines, 95% confidence interval.4
Discussion
X-linked retinitis pigmentosa accounts for up to 20% of families with retinitis pigmentosa, and RPGR is the most frequently mutated gene in X-linked retinitis pigmentosa.6 In our proband, the inheritance pattern was typical of X-linked disease and the report of a tapetal reflex in the mother was consistent with X-linked retinitis pigmentosa. It was ultimately apparent that the BEM phenotype had also been conferred by the mutation in RPGR. Nevertheless, given that disease caused by mutations in ABCA4 can manifest as BEM, that disease-causing variants inABCA4 are copious (approximately 800 sequence variants), and that a second mutation is frequently not detected in the presence of clinical disease, it was initially reasonable to consider ABCA4 pathogenicity on the basis of a single allele and the clinical presentation. Here was a case in which an additional clinical tool, qAF, could guide genetic screening so as to potentially avoid the pitfalls of phenotypic and genetic heterogeneity. Subsequent to the identification of the ABCA4 variant p.E471K by array screening, data available from the Exome Variant Server7 indicated that this allele is unlikely to be disease causing.
Acknowledgments
Funding/Support: This work was supported by grants EY024091, EY019861, and P30EY019007 from the National Eye Institute and a grant from Research to Prevent Blindness to the Department of Ophthalmology, Columbia University.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Dr Sparrow had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Marsiglia, Tsang, Sparrow.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Marsiglia, Mahajan, Zernant, Sparrow.
Critical revision of the manuscript for important intellectual content: Marsiglia, Lee, Delori, Tsang, Sparrow.
Statistical analysis: Delori, Sparrow.
Obtained funding: Sparrow.
Administrative, technical, or material support: Marsiglia, Lee, Mahajan, Zernant, Sparrow.
Study supervision: Tsang, Sparrow.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest and none were reported.
Contributor Information
Marcela Marsiglia, Department of Ophthalmology, Columbia University, New York, New York.
Winston Lee, Department of Ophthalmology, Columbia University, New York, New York.
Vinit B. Mahajan, Department of Ophthalmology and Visual Sciences, University of Iowa, Iowa City.
Jana Zernant, Department of Ophthalmology, Columbia University, New York, New York.
François C. Delori, Schepens Eye Research Institute, Harvard Medical School, Boston, Massachusetts; Department of Ophthalmology, Harvard Medical School, Boston, Massachusetts.
Stephen H. Tsang, Department of Ophthalmology, Columbia University, New York, New York.
Janet R. Sparrow, Department of Ophthalmology, Columbia University, New York, New York.
References
- 1.Thiadens AA, Phan TM, Zekveld-Vroon RC, et al. Writing Committee for the Cone Disorders Study Group Consortium. Clinical course, genetic etiology, and visual outcome in cone and cone-rod dystrophy. Ophthalmology. 2012;119(4):819–826. doi: 10.1016/j.ophtha.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Bassuk AG, Sujirakul T, Tsang SH, Mahajan VB. A novel RPGR mutation masquerading as Stargardt disease. Br J Ophthalmol. 2014;98(5):709–711. doi: 10.1136/bjophthalmol-2013-304822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zernant J, Schubert C, Im KM, et al. Analysis of the ABCA4 gene by next-generation sequencing. Invest Ophthalmol Vis Sci. 2011;52(11):8479–8487. doi: 10.1167/iovs.11-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke TR, Duncker T, Woods RL, et al. Quantitative fundus autofluorescence in recessive Stargardt disease. Invest Ophthalmol Vis Sci. 2014;55(5):2841–2852. doi: 10.1167/iovs.13-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acton JH, Greenberg JP, Greenstein VC, et al. Evaluation of multimodal imaging in carriers of X-linked retinitis pigmentosa. Exp Eye Res. 2013;113:41–48. doi: 10.1016/j.exer.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson DA, Khan NW, Othman MI, et al. Rd9 is a naturally occurring mouse model of a common form of retinitis pigmentosa caused by mutations in RPGR-ORF15. PLoS One. 2012;7(5):e35865. doi: 10.1371/journal.pone.0035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project. [Accessed June 5, 2014];Exome Variant Server. http://evs.gs.washington.edu/EVS/. [Google Scholar]