Abstract
Over the past two decades, the secreted protein sonic hedgehog (SHH) has emerged as a critical morphogen during embryonic lung development, regulating the interaction between epithelial and mesenchymal cell populations in the airway and alveolar compartments. There is increasing evidence that the SHH pathway is active in adult lung diseases such as pulmonary fibrosis, asthma, chronic obstructive pulmonary disease, and lung cancer, which raises two questions: (1) What role does SHH signaling play in these diseases? and (2) Is it a primary driver of the disease or a response (perhaps beneficial) to the primary disturbance? In this review we aim to fill the gap between the well-studied period of embryonic lung development and the adult diseased lung by reviewing the hedgehog (HH) pathway during the postnatal period and in adult uninjured and injured lungs. We elucidate the similarities and differences in the epithelial–mesenchymal interplay during the fibrosis response to injury in lung compared with other organs and present a critical appraisal of tools and agents available to evaluate HH signaling.
Keywords: hedgehog, lung development, lung fibrosis, fibroblast
Clinical Relevance
New roles for sonic hedgehog signaling, which is essential for embryonic lung development, have emerged during postnatal lung development and in adult lung disease, such as pulmonary fibrosis, chronic obstructive pulmonary disease, and asthma. Our review of the current literature spanning from embryonic period to adulthood highlights the latest findings with particular focus on the regulation of mesenchymal cells due to their importance in the pathogenesis of pulmonary fibrosis and fibrosis of other solid organs.
Developmental signaling pathways orchestrate interactions among endoderm, mesoderm, and ectoderm, resulting in distinct tissue architectures that enable proper organ function and response to injury. Once organ development is completed, many of these pathways are suppressed or restricted to tissue-specific stem cell maintenance. Several pathways regulating embryonic lung development (1) have sparked interest because they are reexpressed in adult disease states. One such system is the hedgehog (HH) pathway, a signaling cascade that regulates morphogenesis of lung and other organs in a concentration-dependent manner (2). HH signaling also maintains adult stem cells in various fully developed tissues (3–5) and is involved in several cancers (6). In this review we focus on the role of HH signaling in the lung during development and in disease. We do not review the extensive work on the role of HH in cancer (6). The online supplement contains an appraisal of tools and agents available to evaluate HH signaling.
The INs and OUTs of the HH Signaling Pathway
The hh gene was first described in Drosophila melanogaster, where it regulates dorsal–ventral differentiation and segment polarity (7). Three vertebrate orthologs were identified: sonic (Shh), indian (Ihh), and desert (Dhh) Hh (8–12). Sonic HH (SHH), the most broadly expressed HH ligand, influences morphogenesis of many organs (13). The other two Hh genes have more restricted developmental roles, Ihh in development of bone (14) and Dhh in development of the gonads and nerve sheaths (15, 16). The molecular mechanisms underlying HH signaling are complex, and the interested reader is directed to more comprehensive reviews (17, 18). Figure 1 depicts the HH signaling pathway and highlights molecules known to be important in the lung.
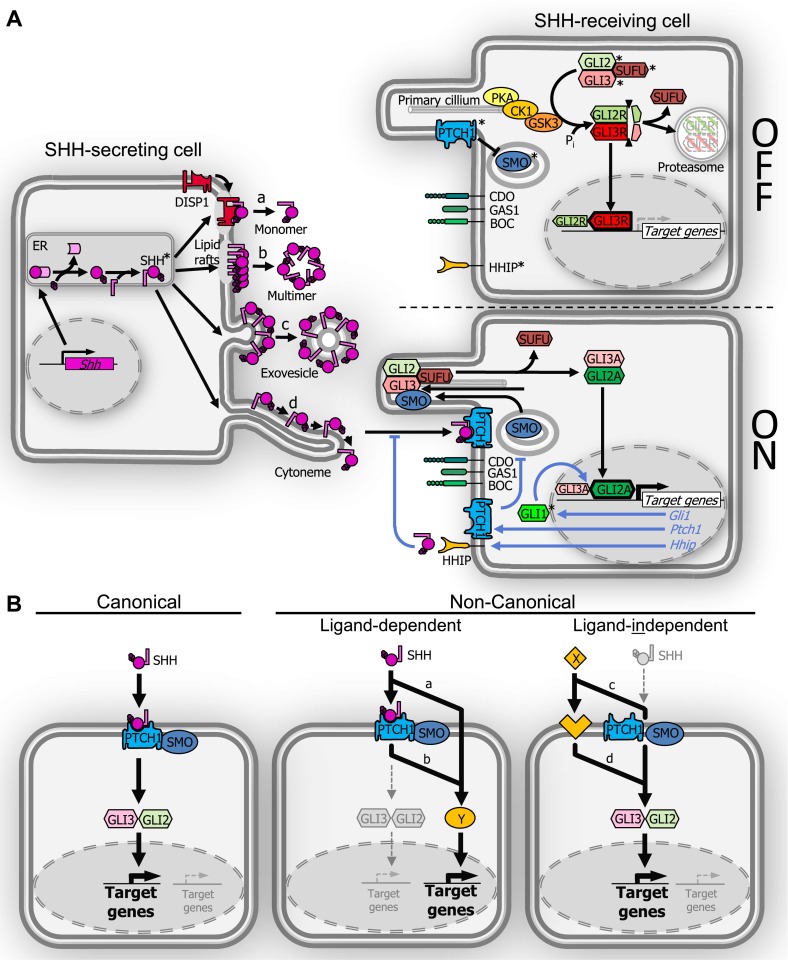
Figure 1.
Schematic depiction of sonic hedgehog (SHH) synthesis, secretion, and signaling. Pathway molecules with described roles in lung development and/or disease are marked (*). (A) In the SHH-secreting cell, SHH-precursor protein undergoes autoproteolytic cleavage and C-terminal addition of a cholesterol moiety followed by N-terminal palmitoylation. Lipidated SHH is able to translocate to lipid rafts in the outer cell membrane. Several modes of secretion have been postulated: (a) a monomeric form requiring DISP1, (b) a multimeric form, (c) in exovesicles or lipoproteins, and (d) along tentacle-like cell cytonemes. The HH signal-receiving cell is shown without (OFF) or with (ON) pathway activation. In the OFF-state, Patched1 (PTCH1) receptor sequesters SMO at the base of the primary cilium. In the absence of SMO activation, GLI2/GLI3 are phosphorylated through the PKA/CK1/GSK3 complex, followed by proteolytic cleavage into their repressor forms GLI2R/GLI3R, which dissociate from Suppressor of Fused (SUFU) and either translocate to the nucleus to repress target gene expression (mainly through GLI3R) or undergo proteasomal degradation. SHH binding to PTCH1 (ON-state) removes sequestration of SMO, which then moves into the primary cilium to induce conversion of GLI2/GLI3 to their activator forms Gli2A/Gli3A. Gli2A/Gli3A can then dissociate from SUFU and translocate to the nucleus to activate target gene transcription (mainly through GLI2A). Direct transcriptional targets (italic blue font and arrows) can provide either a positive (GLI1) or a negative (PTCH1, HHIP) feedback loop for HH signaling. Further modulation of HH signal transduction is provided by cell surface “co-receptors” CAM-regulated by oncogenes (CDO), brother of CDO (BOC), and growth arrest–specific gen1 (GAS1). (B) Canonical and noncanonical HH signaling. Canonical pathway activation (left panel) involves SHH ligand, cell membrane molecules PTCH1/SMO, and GLI transcription factors. In ligand-dependent, noncanonical signaling (middle panel), SHH activates target genes using an alternate pathway Y with (a) or without (b) involvement of PTCH1 and/or SMO. In HH ligand–independent, noncanonical signaling (right panel), alternate X-ligand–mediated pathways activate GLI-mediated target transcription with (c) or without (d) involvement of PTCH1 and/or SMO.
Vertebrate HH ligands are processed and secreted in a manner that is not entirely understood. Synthesis of SHH involves posttranslational modification of its 45-kD precursor by autoproteolytic removal of a C-terminal peptide (19) and C-terminal attachment of cholesterol (20, 21), followed by N-terminal addition of palmitate to the remaining 19-kD peptide (22, 23). Lipid-modified SHH monomers localize to sterol-rich microdomains in the outer plasma membrane (24). Due to its lipid modification, SHH is relatively insoluble, and secretion requires dispatched1 (Disp1) (25–27). This multipass transmembrane protein contains a sterol-sensitive domain like those in the HH receptors Patched1 (PTCH1) and Patched2 (PTCH2) and directs cellular HH ligand release to the cell membrane (25–28). It may also facilitate long-range transport of SHH through tissue after secretion (29). SHH localizes to apical and basolateral regions (27, 30), but how this affects SHH signaling is uncertain.
Several other mechanisms that facilitate extracellular movement of SHH have been postulated: release of soluble SHH monomers (31), formation of multimers (32), assembly into lipoproteins (33) and exovesicles (34), and cytoneme formation for cell-mediated delivery (35, 36) (Figure 1A, a–d). Controversy remains as to how HH ligands reach their target cells and whether a single mechanism or a combination of processes is involved. SHH dispersion is also controlled by its own pathway components, including PTCH1 (37) and HH inhibitory protein (HHIP) (38). Ptch1 and Hhip1 transcription is induced by HH signaling, and the proteins then function in a negative feedback manner by sequestering SHH, thereby modulating its ability to traverse tissues.
Once HH ligands reach their target cells, several molecules mediate the signaling response. Three core components of the signaling response to HH ligand were identified in D. melanogaster (39): the cell surface receptor patched (Ptc), the heptahelical transmembrane protein smoothened (Smo) that transmits the signal into the cell, and the GLI-family transcription factor cubitus interruptus (Ci) that relays the signal to the nucleus. In vertebrates, ptc and ci are replaced by the Ptch family (Ptch1, Ptch2) and the Gli family of zinc-finger DNA-binding proteins (Gli1, Gli2, Gli3) (40–43).
Reception of the HH signal is mediated by the interaction between the transmembrane proteins PTCH and SMO. In the absence of ligand, PTCH sequesters SMO from the plasma membrane, resulting in HH pathway inhibition. PTCH is inhibited upon binding HH ligand (44), thereby freeing SMO to signal to the GLI transcription factors. The ability of one PTCH molecule to inhibit approximately 50 SMO molecules suggests an indirect interaction involving another signaling molecule (44). Although this putative signaling molecule is unknown, agonistic and antagonistic sterol-like molecules have been identified that bind SMO (45). Oxysterols, for example, are potent activators of HH signaling (46, 47). HH signal transduction is also modulated by several cell surface “co-receptors.” HHIP binds to SHH and IHH (38, 48), thereby decreasing HH pathway activity. Conversely, growth arrest–specific gen1, CAM-regulated by oncogenes (CDO), and brother of CDO promote pathway activity (49–51).
The GLI family of transcription factors relays information to the nucleus about the amount of HH ligand at the cell surface. Similar to Ci in D. melanogaster, GLI proteins are posttranslationally altered in response to HH ligand and therefore can act as transcriptional activators or repressors. GLI2 and GLI3 contain an amino-terminal repressor and a carboxy-terminal activator domain (52). GLI1 lacks the repressor domain and thus acts mainly as an activator. Gli1 is a transcriptional target of HH signaling and reliably reports HH pathway activity (53). GLI2 is the most important pathway activator, whereas GLI3 has primarily repressor function. It is ultimately the balance between Gli2A and GLI3R accumulation and the resulting target gene transcription in the nucleus that influences the pathway output.
In the absence of HH signals, two processes define the pathway status: (1) GLI2 and GLI3 are modified by PKA/CK1/GSK3-dependent phosphorylation to enter the proteasomal pathway for truncation, resulting (for GLI3) in repressor function as GLI3R (and to a lesser extent GLI2R), and also degradation of both proteins (54–56). (2) Suppressor of Fused (SUFU) inhibits GLI transcriptional activity in the absence of ligand (57). Upon HH pathway activation, GLI2 degradation and repressor formation are reduced, allowing nuclear accumulation of GLI2A (to a lesser extent the same occurs with GLI3A) and induction of target gene transcription. A consensus Gli binding site has been defined (58), and many GLI binding sites have been identified, but only a few genes have been shown to be direct transcriptional targets of GLI transcription factors. Among these confirmed targets are the Hh pathway members Ptch1 and Gli1 (59, 60), whose proteins provide negative and positive feedback loops to the pathway, respectively. Other tissue-specific direct targets include FoxA2, FoxF1, Nkx2.2, Myf5, Bcl2, Nmyc (6), and Pdgfra (61).
The primary cilium, a small dynamic tubular structure that transiently forms in interphase and is required for correct cell mitosis (62), is necessary for HH signaling in most cells. Without HH ligand, PTCH1 localizes to the cilium and blocks HH signaling by preventing SMO entry (63). Hh ligand binding drives PTCH1 out of the cilium, permitting ciliary SMO accumulation and downstream pathway activation (64). SMO signals to GLI2 and GLI3, allowing their movement through the primary cilium together with microtubular transport proteins (65, 66). In the HH-OFF state, GLI3 undergoes processing to its repressor state GLI3R, which dissociates from SUFU and translocates to the nucleus (67, 68). In the HH-ON state, GLI2 and GLI3 are enriched in the ciliary tip, where their modification facilitates dissociation from SUFU and nuclear accumulation of primarily the activator form GLI2A (66–68).
Three concepts of HH signaling deserve final mention. First, HH ligand gradients, which control anterior–posterior and dorsal–ventral patterning of the embryo (9, 69, 70), similarly influence secondary lung bud formation (71). An important mechanism for generating gradients of HH activity is likely the expression of the negative regulators PTCH1 and HHIP, whose genes are direct transcriptional targets of SHH signaling. Second, although many developmental processes involving HH ligands follow canonical pathway activation, there is growing evidence of noncanonical HH signaling during development and in adulthood (for review see Reference 72), where SHH signals to a pathway independent of GLI-mediated transcription (73–75) or GLI function is influenced by another signaling pathway (76–78) (Figure 1B). Finally, PTCH1 can act as a dependence receptor (79, 80). Dependence receptors induce apoptosis in the absence of cognate ligands (81). The fact that primary lung fibroblasts from idiopathic pulmonary fibrosis (IPF) lungs are protected from IFN/TNF/Fas-ligand–induced apoptosis by Shh (82) raises the possibility that fibrosis is maintained by epithelial SHH expression, which prevents PTCH1-mediated mesenchymal cell apoptosis that would otherwise occur.
HH Signaling in Embryonic Lung Development: What Do We Know?
Embryonic lung development follows the principle of branching morphogenesis. The endodermal cell layer grows into the surrounding splanchnic mesenchyme, generating a branched tubular structure surrounded by mesenchyme-derived structures such as blood vessels, lymphatics, and nerves. Lung formation is divided into five phases (1). The first four phases (embryonic, pseudoglandular, canalicular, and saccular) result in the typical branching structure ending in alveolar sacs with surrounding stromal scaffold and vascular structures. During the final (postnatal) alveolar phase, the terminal sacs give rise to mature alveolar ducts and alveoli. In humans, the last stage spans almost 10 years, whereas in murine lung development it is completed in 4 weeks (83). An elaborate network of growth factors, transcription factors, and extracellular matrix molecules orchestrates embryonic lung growth (1, 84). Localization of Shh/SHH expression (Figure 2) and knockout of Hh pathway molecules (Table 1), among other results, demonstrate that SHH signaling is a crucial aspect of this network.
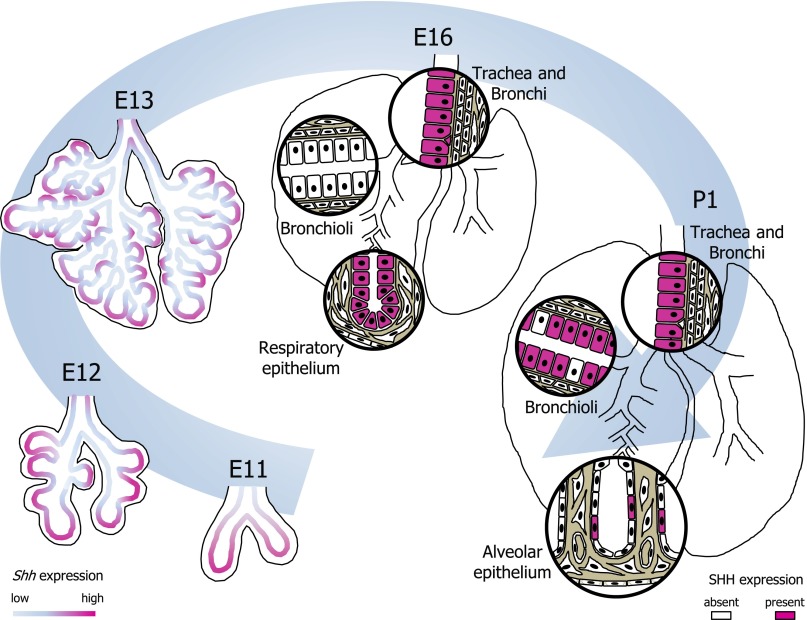
Figure 2.
Schematic of SHH-expressing cells in the developing lung from embryonic day (E)11 to postnatal day (P)1. SHH-expressing cells and cell layers are highlighted in fuchsia. Shh expression is highest at the tips of the primary and secondary lung buds, but also present at lower levels along the developing bronchi. This pattern continues throughout the pseudoglandular stage. Around E16, SHH expression is transiently absent in the distal bronchioli but is vividly present in the respiratory epithelium and along larger bronchi and trachea. Toward P1 the majority of bronchial epithelial cells express SHH, whereas in the saccular-stage alveolar compartment SHH expression is restricted to a subset of epithelial cells.
Table 1.
Hedgehog Pathway Gene Knockout Lung Phenotypes
| Genotype | Lung Phenotype | Comments | Citation |
|---|---|---|---|
| Shh−/− | Single-lobe hypoplastic lungs with decreased epithelium/mesenchyme; malformations of the trachea and trachea-esophageal fistula | Lethal at birth | 93, 94 |
| Shh+/− | No reported abnormalities | Viable | 162 |
| Ptch1−/− | Lethal before lung development begins | Lethal at E8.5–E9.5 | 53, 96 |
| Ptch1+/− | No reported abnormalities | Viable | 96 |
| Gli1−/− | Normal appearing | Viable | 99 |
| Gli2−/− | Hypoplastic lungs with severe patterning defects (single lobe right lung) and diminished epithelium/mesenchyme; mildly hypoplastic trachea and esophagus | Lethal at birth | 91 |
| Gli2+/− | Normal appearing | Viable | 99 |
| Gli3−/− | Hypoplastic lungs of decreased size and abnormal shape of the lobes | Lethal around E14.5 | 92 |
| Gli3+/− | Normal appearing | Viable | 103 |
| Gli1−/−; Gli2+/− | Hypoplastic lungs of decreased size, less severe than Gli1−/−; Gli2−/− | 50% lethal until P21 | 99 |
| Gli1−/−; Gli2−/− | Severely hypoplastic, two lobes | Lethal at birth | 53, 99 |
| Gli2−/−; Gli3+/− | Hypoplastic lungs with abnormal proximal lung development with failure to separate left and right lung and formation of a tracheobronchial fistula; distal lung partially formed | Lethal at birth | 91 |
| Gli2−/−; Gli3−/− | Most severe phenotype; fail to form trachea, lung, and esophagus | Lethal at E10.5; some embryos survive until E13.5 | 91 |
| Hhip−/− | Single-lobe hypoplastic lungs with defective formation of the second generation of lung buds | Lethal at birth | 38 |
| Hhip+/− | No abnormalities reported | Viable | 38 |
| Hhip−/−; Ptch1+/− | More severe lung hypoplasia than Hhip+/− lungs; mesenchyme thickened | Lethal at birth | 38 |
Definition of abbreviations: E, embryonic day; P, postnatal day.
HH Pathway Molecule Expression in the Lung: Where and When
Shh is expressed in respiratory epithelium throughout embryonic lung development in a complex and changing pattern, starting around embryonic day (E)10 (85–88) (Figure 2). Shh expression is high at the tips of the growing bronchial tubules but absent more proximally, suggesting a polarizing role for SHH during branching. This graded SHH expression pattern continues throughout the pseudoglandular and canalicular stages until E16.5 (85). After E16.5, SHH is expressed in proximal and distal airways but only in a subset of the epithelial cells (likely nonciliated) (85). From E17 on, SHH is expressed strongly in the saccular epithelial compartment, only decreasing after birth. Overall expression of Shh and Ptch1 gradually decreases from E15.5 to birth (87). Remarkable similarities were detected when comparing expression of SHH and its pathway molecules in human and murine embryonic lung (89).
Ptch1 is expressed in the lung mesenchyme around E11.5, highest around the distal tips and lower along the bases of the lung buds (mirroring the Shh pattern) (87, 90, 91). Ptch1 expression remains significant during branching morphogenesis but decreases during late gestation (87). SMO is reportedly expressed in epithelium and mesenchyme between E12.5 and E16.5 (89). Gli1, Gli2, and Gli3 are expressed in the mesenchyme during the pseudoglandular stage, and their levels decrease near birth (92). Although all three Glis are expressed strongly in distal mesenchyme, Gli2 is also present in mesenchyme around proximal trachea, and Gli3 is expressed in intermediate areas between lung buds. Hhip is expressed in mesenchyme underlying epithelial regions of high Shh expression, starting around E10.5 and overlapping with Ptch1 and Gli1 expression (38, 48).
Lessons from Functional Inhibition and Overexpression of HH Pathway Molecules
Shh is indispensible for embryonic lung formation, regulating branching morphogenesis and mesenchymal proliferation. Shh−/− mice have single-lobed hypoplastic lungs with decreased epithelium and mesenchyme, malformations of the trachea, and trachea-esophageal fistulae (93, 94). The preservation of proximal and distal epithelial cell phenotypes and the lack of bronchial smooth muscle cells indicate that SHH influences the mesenchymal scaffold rather than epithelial cell differentiation. This idea is supported by investigations using a doxycycline-inducible SP-C promoter–driven Shh conditional knockout (CKO) (95). Shh CKO before E12.5 causes more severe defects in branching morphogenesis, whereas Shh CKO after E12.5 produces mild abrogation of distal bronchial morphogenesis but leaves proximal branching intact. SPC promoter–driven SHH overexpression in Shh−/− mice rescues distal branching but does not affect lung lobulation or the trachea-bronchial cartilage defects (95). The role of Ptch1 in lung development is obscure due to early lethality of null mutants (53, 96).
The effects of SHH on branching are tightly regulated by its downstream targets PTCH1 and HHIP, which are produced in an overlapping pattern and sequester SHH (38, 95). Hhip−/− lungs are hypoplastic due to defective formation of the second generation of lung buds. This phenotype is partially rescued by PTCH1 overexpression and involves failure to localize mesenchymal fibroblast growth factor (FGF)10, a target and antagonist of SHH signaling, to areas of new bud formation (38).
SHH also affects proliferation and differentiation of lung mesenchyme. Embryonic Shh CKO and SHH overexpression result in respiratory failure at birth but with different phenotypes. Shh−/− mice have decreased proliferation of mesenchymal cells (94), whereas SP-C promoter–driven Shh overexpression increases proliferation of lung mesenchyme during late gestation, suggesting that balanced mesenchymal induction by SHH is vital to normal lung formation (87). These observations were accompanied by the expected changes in expression of the SHH transcriptional targets Ptch1 and Gli1 in the mesenchyme (87, 93–95). SHH signaling also regulates lung mesenchymal cell lineages, such as the entry of mesothelial cells into the lung mesenchyme (97).
Although no lung abnormalities have been reported in Ptch1+/− mice (96), the requirement of the GLI transcription factors during lung development is well documented (Table 1). Gli2−/− mice have hypoplastic lungs with severe patterning defects, have diminished mesenchyme, and die at birth (91, 98), supporting a role for SHH in facilitating stromal cell expansion by canonical signaling through GLI2. Gli3−/− mice exhibit decreased lung size and abnormal shape of the lobes (92). In contrast, Gli1−/− mice have normal lungs and viability, as do Gli2+/− mice. However, Gli1−/−;Gli2+/− compound mutants reveal decreased lung size and death soon after birth, indicating overlapping functions for Gli1 and Gli2 and a supportive role for Gli1 in the developing lung (99). The lungs of Gli1−/−;Gli2−/− double mutants have only two lobes and are even smaller than Gli1+/−;Gli2−/− and Gli1−/−;Gli2+/− lungs (53). The most severe lung defects are seen in Gli2−/−;Gli3−/− double mutants, which fail to form trachea, lung, and esophagus (91). Although a single Gli3 allele in Gli2−/−;Gli3+/− mutants partially rescues the distal lung phenotype, proximal lung development is still abnormal, characterized by failure to separate left and right lung and formation of a tracheobronchial fistula (91). The observations that Shh−/− lungs show increased Gli3R levels and, strikingly, that GLI3 loss partially rescues the distal lung phenotype with increased mesenchymal cell proliferation in Shh−/− mice (100) indicate that opposing gradients between SHH and GLI3R contribute to lung development. The fact that the Gli2−/−;Gli3−/− lung phenotype is worse than that of the Shh−/− lung is consistent with the idea that both increased GLI3R and decreased GLI2A contribute to the Shh null phenotype. Finally, the adaptor molecule SUFU, involved in GLI protein processing, affects GLI output during lung development, as Dermo1Cre-dependent CKO of mesenchymal SuFu causes hypoplastic lungs with defective distal branching and absence of myofibroblasts (61).
Morphogens that Are Regulated by HH Signaling
Lung development requires the concerted activity of many morphogens, and the SHH signaling pathway communicates with other key pathways (1). Shh CKO affects not only target gene expression of its own pathway molecules but also affects that of other genes (95). ChIP assays confirmed at least 28 direct GLI1-binding genes in embryonic mouse tissue (59), three of them with important roles in the developing lung.
FGF10 is the only FGF shown to be essential for lung development. Fgf10−/− mice fail to form lungs distal to the trachea (101, 102). FGF10 is maximally expressed in mesenchyme around the growing lung buds (103), and its expression pattern promotes proximal–distal differentiation during branching (104). Shh restricts FGF10 expression to the distal tips of the lung buds and, together with Hhip, inhibits FGF10 expression in the interbud regions, allowing localized new bud outgrowth (38, 93). In Shh−/− lungs, FGF10 expression expands to almost all mesenchyme (93), supporting the model of antagonism between SHH and FGF10.
Bone morphogenetic protein 4, a TGF-β superfamily member, is expressed in proximal mesenchyme and distal epithelium at the tips of the branching lung (105). SHH is able to induce mesenchymal bone morphogenetic protein 4 expression in early (E11.5) (90, 94) but not late lung development (E18.5) (87).
The forkhead transcription factor FoxF1 is expressed at highest levels in the subepithelial lung mesenchyme between the distal bulbous part of the bud and proximal tubular part (106). Decreased FoxF1 expression in Shh−/− lungs is rescued by ectopic SHH treatment. FoxF1−/− lungs share features of Shh−/− lungs, consistent with other data that FoxF1 is a transcriptional target of HH signaling (107).
Morphogens that Regulate HH Signaling
The forkhead transcription factors FOXA1 and FOXA2 are expressed in epithelial cells during embryonic lung development (108, 109). Epithelium-specific FoxA1 and FoxA2 deletion causes defects in branching morphogenesis and respiratory epithelial cell maturation (109). FoxA1−/−;FoxA2−/− mutants have decreased expression of epithelial Shh and mesenchymal Ptch and Hhip as well as decreased expression of transcription factors that control pulmonary smooth muscle differentiation. In light of the bronchial wall defects and absence of cells expressing α-smooth muscle actin in Shh−/− lungs (95), these findings raise the possibility that epithelial FOXA1 and FOXA2 act upstream of Shh, controlling SHH-induced mesenchymal cell expansion and differentiation.
FGF9 is expressed in epithelial and mesenchymal cells during lung development (110). FGF9−/− lungs have deficient distal branching (less severe than FGF10−/−) but normal Shh expression (111). In vitro, FGF9 inhibits mesenchymal cell differentiation without affecting SHH-induced proliferation of mesenchymal cells (90). However, because FGF9 partially rescues the distal vascular phenotype in Shh−/− lungs, but not vice versa, it is possible that FGF9 modulates SHH signaling in vivo during formation of the lung capillary network (112).
WNT5A, one of the ligands of the WNT signaling cascade, is expressed in lung epithelium and mesenchyme during embryonic lung development (113). Wnt5a−/− lungs show defective distal lung morphogenesis with increased mesenchyme and overexpression of Shh and Ptch1 (113), whereas SP-C promoter–driven epithelial Wnt5A overexpression decreases lung mesenchyme and Shh/Ptch1 expression (114), suggesting a regulatory role for WNT5A in SHH-induced mesenchymal proliferation.
Knockout of transforming growth factor β receptor II, a receptor for TGF-β ligands that is involved in epithelial–mesenchymal communication during development, causes cystic lung with defective branching (115). The Hh signaling transcriptional targets Ptch1 and Gli1 were increased without altered Shh expression, suggesting the possibility of noncanonical pathway regulation.
Eyes absent1 (EYA1) and sine oculis1 (SIX1), two homeobox transcriptions factors involved in eye development and expressed in lung epithelium and mesenchyme, were also found to affect lung development (116–118). Six1−/− and Eya1−/− lungs show a hypoplastic phenotype with branching defects, which is exaggerated in Six1−/−;Eya1−/− mutants. Lung mesenchyme is increased, as seen after SHH overexpression (87), and increased SHH signaling is observed. Cyclopamine, a Smo antagonist (details are provided in the online supplement), partially rescues lung structure, confirming the importance of increased SHH signaling in these mutant mice. Similar findings were reported for adrenoceptor ARα2b−/− mice (119).
From these studies it is clear that (1) during embryonic lung development, expression of SHH and its signaling molecules are highly regulated in location and time and (2) SHH expression exerts its effect on different cellular compartments because it is essential for branching events in the bronchial compartment and is critical to the respiratory epithelial compartment at the canalicular and saccular stages, where it regulates balanced mesenchymal expansion. These studies prompt a fundamental question: Does SHH-mediated regulation of lung mesenchyme play a role after birth and in adult disease?
HH Signaling during Postnatal Lung Development
Postnatally, mature lung is generated in two phases: an early phase of alveolar septum formation (alveolarization) followed by maturation of alveolar walls and the microvasculature (120). In mice, the second phase commences at about postnatal day (P)10. An important part of septal maturation is a decrease of mesenchymal cells that accompanies the fusion of capillaries as growing, matrix-producing septa transform into mature, thin-walled structures (121, 122). The significance of alveolarization is illustrated by bronchopulmonary dysplasia, which develops in infants secondary to perinatal lung injury and results in abnormal alveolar wall morphology (123, 124). Numerous factors promote or disrupt alveolarization (125, 126), but until recently no role for SHH had been identified.
Murine models established the first evidence of HH signaling in the postnatal lung. Although Shh and Ptch1 expression decreases during late gestation, it is still present at birth (87). At P1, SHH expression is observed in almost all epithelial cells lining the conducting airways, whereas in the alveolar sacs SHH is restricted to a subgroup of epithelial cells (85). In this study, SHH was detected until P15 but was below the detection threshold at P24. Hyperoxia-induced lung injury, a model of bronchopulmonary dysplasia, causes up-regulation of Shh and Ptch1 in epithelium and mesenchyme, respectively, and thickening of alveolar walls at P7 and P14 (127), raising the possibility that SHH signaling plays a role in postnatal lung pathology.
Using Gli1nlacZ/+ reporter mice (53), we detected HH-responding cells (Gli1-positive cells) throughout the postnatal period and into adulthood (128). Two observations are important: (1) Gli1-positive cells, mostly fibroblasts, are found in alveolar and peribronchial/perivascular zones. To further illustrate HH-responding cell location, we used the Gli1creERT2-Rosa26mT/mG lineage tracer (129). Gli1 lineage, labeled at P3, is detectable at P10 in three functionally distinct locations: in and around the peribronchial smooth muscle layer, scattered in alveolar septa, and in mesothelial cells of the visceral pleura (Figure 3). (2) Gli1-positive cells (Gli1nlacZ/+ reporter) in the alveolar zones are abundant at P7 in septal walls and tips but are increasingly less numerous at P14 and P21 while remaining constant around airways and vessels. The decreased septal expression coincides with the end of the alveolarization phase. Between P14 and P21, the central region of septa is reduced to a fibrous meshwork interwoven with capillaries, and the number of fibroblasts decreases by 10 to 20% (122). The coincidence of fewer HH-responding cells with the transition to septal maturation suggests a functional connection. Analysis of primary lung fibroblasts from P4 to P12 mice revealed significant Ptch1 and Gli1 expression until P8 (130), supporting the presence of HH signaling until the end of alveolarization. In this study, SHH stimulated chemotactic fibroblast migration, presumably an important feature of septal elongation.
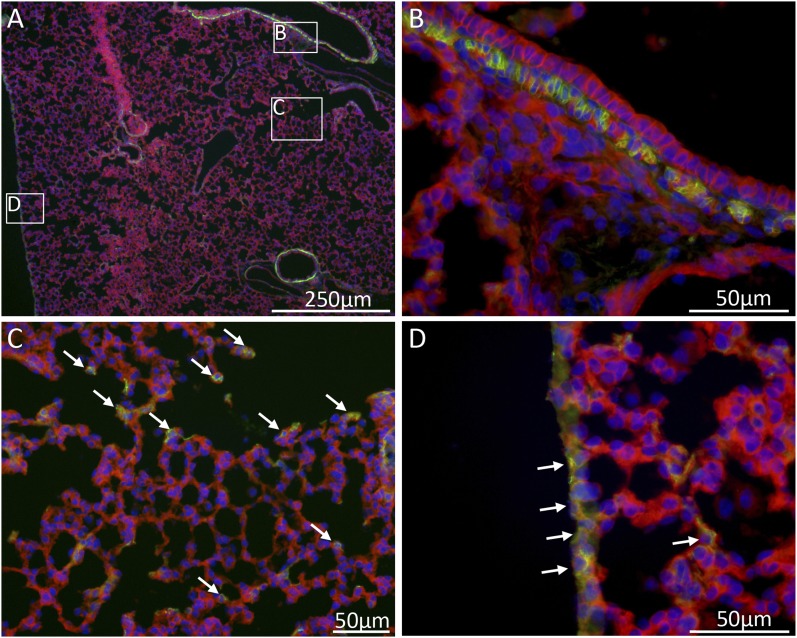
Figure 3.
Hh-responding cells in different functional areas of the lung. Gli1-expressing cells were marked at P3 with a single dose of tamoxifen using the Gli1creERT2-Rosa26mT/mG lineage tracer mouse. Immunofluorescence images of lung sections at P12 illustrate that cells in the Gli1-lineage (green) are present in three locations: in the peribronchial smooth muscle layer (B), scattered in the alveolar septal walls (C, arrows), and in many mesothelial cells of the visceral pleura (D, arrows). Unmarked cells appear red. DAPI-stained cell nuclei appear blue.
A role for postnatal HH signaling has recently been confirmed. When HH signaling is reduced in vivo beginning at P3, before its normal decline around P8, enlarged airspaces develop, without a decreased number of septal tips or grossly abnormal elastin formation, suggesting accelerated lung maturation due to premature reduction of mesenchyme (128). Conversely, treatment with a SMO agonist from P1 to P7 causes “lung hyperplasia” with preserved epithelial differentiation at P9 and P21 (131), hinting that enhancing HH signaling affects lung maturation by preventing fibroblast loss.
HH Signaling in Pulmonary Fibrosis
Given the mitogenic effect of SHH on mesenchyme during development, it is intriguing that increased HH signaling is associated with fibrosis in lung (Table 2) and other organs (132–134). IPF is a progressive fibrosing interstitial pneumonia of unknown etiology, characterized by fibroblastic foci and deposition of extracellular matrix leading to destruction of alveolar structures (135, 136). The current paradigm of IPF pathogenesis is that epithelial injury leads to aberrant epithelial–mesenchymal communication in a manner that prevents epithelial repair, promotes expansion and activation of mesenchymal cells, and stimulates angiogenesis (136, 137). In human IPF lungs, microarray data showed increased expression of several developmental pathway genes, including PTCH1 (138). SHH expression is high in epithelial cells lining fibrotic areas in samples of IPF and other interstitial pneumonias but is undetectable in normal lung (139–141). Abnormal expression of PTCH1, SMO, and GLI1 was also found in IPF lungs (82, 142). The localization of PTCH1 and GLI1, which are expressed only in mesenchymal cells during development, to epithelial and mesenchymal cells in fibrotic lungs suggests that the strict separation of SHH-expressing and SHH-responsive cells is lost during the fibrotic process. However, these opposing observations of the expression pattern during development and in disease also raise the more general issue of technical difficulties in analyzing tissues that contain different cell types and gene expression levels.
Table 2.
Evidence for Sonic Hedgehog Signaling in Human Lung Disease
| Disease | Method | Findings | Citation |
|---|---|---|---|
| Parenchymal lung diseases | |||
| IPF | Microarray | PTCH1 gene expression altered in IPF lungs | 138 |
| ISH | SHH highly expressed in epithelium of fibrotic areas | 139, 142 | |
| qRT-PCR | SHH, PTCH1, and GLI1 up-regulated in IPF lungs | 82 | |
| IHC | SHH expressed in hyperplastic alveolar type II cells in fibrotic areas | 140, 141 | |
| IHC | PTCH1, SMO, and GLI1 expressed in fibroblastic foci of IPF lungs | 82 | |
| ELISA | SHH elevated in BALF from IPF lungs | 142 | |
| NSIP | ISH | SHH weakly expressed in epithelium, but higher than in normal lung | 139 |
| IHC | SHH expressed in epithelial cells of thickened alveolar walls | 140, 141 | |
| COP | IHC | SHH expressed in buds of organizing exudate | 141 |
| Airway diseases | |||
| COPD | GWAS | SNPs in region close HHIP gene on 4q31 linked to decreased lung function (FEV1/FVC ratio) and COPD-related phenotypes | 152, 153 |
| qRT-PCR | HHIP decreased in COPD lungs | 153 | |
| WB | HHIP decreased in COPD lungs | 153 | |
| Asthma | GWAS | SNPs in regions of HHIP on 4q31 and the PTCH1 gene on 9q22-31 linked to decreased lung function (FEV1/FVC ratio) and asthma-related phenotypes | 154 |
Definition of abbreviations: BALF, bronchoalveolar lavage fluid; COP, cryptogenic organizing pneumonia; COPD, chronic obstructive pulmonary disease; GWAS, gene-wide association study; IHC, immunohistochemistry; IPF, idiopathic pulmonary fibrosis; ISH, in situ hybridization; NSIP, nonspecific interstitial pneumonia; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SNP, single-nucleotide polymorphism; WB, Western blot.
Experimental lung fibrosis models also show abnormalities in the HH pathway. FITC-induced lung fibrosis (143) results in SHH overexpression in airway and alveolar epithelial cells (140, 144). Endotracheal bleomycin administration, the most studied animal fibrosis model (145), results in up-regulated expression of Shh and Gli1 in fibrotic lesions (146). In Gli1nlacZ/+ mice, the number of Gli1-positive cells, which are mostly fibroblasts and myofibroblasts, is increased in fibrotic lesions when compared with uninjured lung (128). Numbers of Gli1-positive cells in morphologically normal lung are also increased, indicating either de novo activation of a resident pool of potentially HH-responsive cells and/or production of additional HH-responding cells (e.g., by local fibroblast proliferation).
Inhibition of HH signaling at the level of SHH or SMO in the bleomycin model fails to prevent fibrosis (128, 146). Enhancement of HH signaling by Shh overexpression during the fibrotic phase worsens lung fibrosis (128), pointing toward a potential role for HH signaling in controlling fibroblast expansion and/or survival. In vitro data support this hypothesis: in primary lung fibroblasts, SHH up-regulates Ptch1 and Gli1 expression, increases proliferation, and protects from TNF-α/IFN-γ/FasL–induced apoptosis (82). We also reported increased survival of SHH-stimulated primary lung fibroblasts in vitro (128). These findings suggest that HH signaling may sustain fibrosis and prevent resolution. GANT61, an inhibitor of GLI2/3, decreases fibrosis (146). Although caution is warranted because of possible off-target effects and unknown effects on GLI3R, this result raises the possibility of ligand-independent HH pathway activation in fibrosis.
The hypothesis that epithelium-derived SHH drives the mesenchymal cell response in fibrotic lung is supported by evidence from other organs. Skin lesions of patients with systemic sclerosis reveal SHH overexpression and abnormal HH signaling (132). SHH stimulation of human primary skin fibroblasts induces a myofibroblastic phenotype and increases collagen expression. Dermal Shh overexpression and bleomycin treatment cause skin fibrosis in mice, and the latter is blocked by the SMO inhibitor LDE223 and Smo siRNA. Ptch1+/− mice, which likely have elevated HH signaling, are more susceptible to bleomycin-induced skin fibrosis (132, 147). In the liver, chronic cholestasis and nonalcoholic steatohepatitis are characterized by enhanced HH signaling, which promotes activation of hepatic stellate cells to a myofibroblastic phenotype (133, 148). In the kidney, ureteral obstruction–induced fibrosis is accompanied by myofibroblast transformation of fibroblasts with active HH signaling and is blocked in Gli1−/− mice and by SMO antagonists (134, 149).
HH Signaling in COPD and Asthma
Obstructive airway diseases such as COPD and asthma are also associated with altered HH signaling (Table 2). Both diseases manifest peribronchial fibrosis as a result of chronic inflammation and tissue remodeling (150, 151), processes that involve fibroblast expansion and matrix deposition, similar to lung fibrosis. Gene-wide association studies linked a locus near HHIP on 4q31 to decreased lung function and COPD-related phenotypes (152, 153). A SNP in this region is located in a potential enhancer region of the HHIP promoter and thus could alter HHIP gene expression. HHIP protein is reduced in COPD lungs, implying a role for HH signaling (153). Because HHIP is a direct transcriptional target of Hh signaling, this observation could indicate decreased Hh pathway activation in COPD; however, because HHIP is a negative feedback molecule in the HH pathway, a direct effect of low HHIP levels would be enhanced HH signaling.
Another gene-wide association study revealed linkages of the HHIP and PTCH1 regions to decreased lung function and asthma-related phenotypes (154); however, it is unclear how HH signaling might contribute to asthma pathophysiology. One possibility is that HH signals from airway epithelial cells are received by CD4 T cells, which play an important role in airway inflammation and require HH signaling for normal differentiation in the thymus (155, 156). Although SHH expressed by airway epithelium promotes TH2 differentiation of CD4 T cells, thereby exacerbating the allergic response in a dust mite–induced asthma model, CD4-specific gene deletion of Ptch1 did not alter allergic response in an ovalbumin-induced asthma model, raising the question of noncanonical HH pathway activation in CD4 T cells. It must also be noted that lymphocytes do not have primary cilia, but alternate modes of HH signaling may exist (157). Another possibility is that epithelial SHH signals to peribronchial stromal cells, including fibroblasts and pericytes, with subsequent implications for tissue remodeling. At least in adult uninjured murine lung, the vast majority of HH-responding cells are located in the interstitial spaces around airways and vessels (128). Given the use of inhaled glucocorticoids in asthma and COPD, it is thought provoking that several clinically used glucocorticoid derivatives, such as budenoside, can modulate SMO localization and HH pathway activity, thereby increasing sensitivity to HH ligand input (158).
Conclusions and Future Directions
In this review we highlight the general principles of HH signaling and apply the knowledge available from embryonic lung development to review the latest insights into the functions of HH signaling during postnatal lung development and in adult disease, both occurring during time periods when HH signaling changes. One newly emerging concept of HH signaling is that it influences the alveolar phase during postnatal lung development and, by extension, adult lung morphology and function. A second concept in need of further testing involves regulation of the stromal compartment in fibrotic lung diseases.
An element connecting the embryonic period, postnatal lung development, and adult fibrotic diseases appears to be a fibroblast population capable of responding to HH signals. Up- or down-regulation of HH might push HH-responsive fibroblasts down different paths. An appropriately timed reduction in HH signaling might be necessary to decrease the number of fibroblasts and thereby allow a physiologic process such as alveolar wall thinning and maturation to occur. In contrast, the continuation and/or enhancement of HH signaling in response to epithelial cell injury might sustain or expand fibroblasts in areas of tissue remodeling, such as fibroblastic lesions in IPF, thereby preventing fibrosis resolution.
The lung and other organs manifest mesenchymal HH pathway activation during fibrosis, and in some examples experimental HH inhibition is antifibrotic, suggesting a more general mechanism involving HH pathway activation. To what extent HH signaling contributes to lung fibrosis is a critical question because HH pathway inhibitors are becoming available for clinical use. The ability of tumor-derived HH to cause a stromal response supporting tumor growth further substantiates the importance of the HH pathway in regulating pathological mesenchymal cell behavior (159–161). The physiologic role of HH signaling in uninjured adult lung, if there is one, remains unclear. However, the presence of HH-responding cells in the perimeter of the bronchovascular bundles indicates there could be a pool of mesenchymal cells that have progenitor characteristics and that normally replenish the lung stroma but expand abnormally during lung disease and injury repair.
Acknowledgments
Acknowledgments
The authors thank Edward Laufer from Columbia University for providing the lungs of the Gli1creERT2-Rosa26mT/mG lineage tracer mouse and William Rom for his ongoing generous support of our work.
Footnotes
This work was supported by National Institutes of Health/National Heart, Lung and Blood Institute grants T32-ES7267-20 (W.N.R.) and 5R21-HL104455 (J.S.M.), NRSA Fellowship grant F32-HL120637from the National Institutes of Health/National Heart, Lung and Blood Institute (M.C.K.), a Grant-in-Aid from the Stony Wold-Herbert Foundation NYC (M.C.K.), an award of the NYU Physician Scientist Training Program (M.C.K.), NCI grant R01CA128158 (A.L.J.), an Irma T. Hirschl Scholar Award from the Irma T. Hirschl/Monique Weill-Caulier Trusts (J.S.M.), and a grant for fibrosis research from the Will Rogers Foundation (M.C.K., J.S.M.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0132TR on July 28, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Maeda Y, Davé V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 3.Brownell I, Guevara E, Bai CB, Loomis CA, Joyner AL. Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell. 2011;8:552–565. doi: 10.1016/j.stem.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng YC, Levine CM, Zahid S, Wilson EL, Joyner AL. Sonic hedgehog signals to multiple prostate stromal stem cells that replenish distinct stromal subtypes during regeneration. Proc Natl Acad Sci USA. 2013;110:20611–20616. doi: 10.1073/pnas.1315729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrova R, Garcia AD, Joyner AL. Titration of GLI3 repressor activity by sonic hedgehog signaling is critical for maintaining multiple adult neural stem cell and astrocyte functions. J Neurosci. 2013;33:17490–17505. doi: 10.1523/JNEUROSCI.2042-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teglund S, Toftgård R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 8.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 9.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 10.Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 11.Chang DT, López A, von Kessler DP, Chiang C, Simandl BK, Zhao R, Seldin MF, Fallon JF, Beachy PA. Products, genetic linkage and limb patterning activity of a murine hedgehog gene. Development. 1994;120:3339–3353. doi: 10.1242/dev.120.11.3339. [DOI] [PubMed] [Google Scholar]
- 12.Roelink H, Augsburger A, Heemskerk J, Korzh V, Norlin S, Ruiz i Altaba A, Tanabe Y, Placzek M, Edlund T, Jessell TM, et al. Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell. 1994;76:761–775. doi: 10.1016/0092-8674(94)90514-2. [DOI] [PubMed] [Google Scholar]
- 13.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 14.Lai LP, Mitchell J. Indian hedgehog: its roles and regulation in endochondral bone development. J Cell Biochem. 2005;96:1163–1173. doi: 10.1002/jcb.20635. [DOI] [PubMed] [Google Scholar]
- 15.Bitgood MJ, Shen L, McMahon AP. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 16.Parmantier E, Lynn B, Lawson D, Turmaine M, Namini SS, Chakrabarti L, McMahon AP, Jessen KR, Mirsky R. Schwann cell-derived Desert hedgehog controls the development of peripheral nerve sheaths. Neuron. 1999;23:713–724. doi: 10.1016/s0896-6273(01)80030-1. [DOI] [PubMed] [Google Scholar]
- 17.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 18.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 19.Maity T, Fuse N, Beachy PA. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc Natl Acad Sci USA. 2005;102:17026–17031. doi: 10.1073/pnas.0507848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters C, Wolf A, Wagner M, Kuhlmann J, Waldmann H. The cholesterol membrane anchor of the Hedgehog protein confers stable membrane association to lipid-modified proteins. Proc Natl Acad Sci USA. 2004;101:8531–8536. doi: 10.1073/pnas.0308449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tukachinsky H, Kuzmickas RP, Jao CY, Liu J, Salic A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Reports. 2012;2:308–320. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor FR, Wen D, Garber EA, Carmillo AN, Baker DP, Arduini RM, Williams KP, Weinreb PH, Rayhorn P, Hronowski X, et al. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry. 2001;40:4359–4371. doi: 10.1021/bi002487u. [DOI] [PubMed] [Google Scholar]
- 23.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004;18:641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 25.Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, Beachy PA. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111:63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- 26.Tian H, Jeong J, Harfe BD, Tabin CJ, McMahon AP. Mouse Disp1 is required in sonic hedgehog-expressing cells for paracrine activity of the cholesterol-modified ligand. Development. 2005;132:133–142. doi: 10.1242/dev.01563. [DOI] [PubMed] [Google Scholar]
- 27.Zavros Y, Orr MA, Xiao C, Malinowska DH. Sonic hedgehog is associated with H+-K+-ATPase-containing membranes in gastric parietal cells and secreted with histamine stimulation. Am J Physiol Gastrointest Liver Physiol. 2008;295:G99–G111. doi: 10.1152/ajpgi.00389.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- 29.Etheridge LA, Crawford TQ, Zhang S, Roelink H. Evidence for a role of vertebrate Disp1 in long-range Shh signaling. Development. 2010;137:133–140. doi: 10.1242/dev.043547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vincent S, Thomas A, Brasher B, Benson JD. Targeting of proteins to membranes through hedgehog auto-processing. Nat Biotechnol. 2003;21:936–940. doi: 10.1038/nbt844. [DOI] [PubMed] [Google Scholar]
- 31.Creanga A, Glenn TD, Mann RK, Saunders AM, Talbot WS, Beachy PA. Scube/You activity mediates release of dually lipid-modified Hedgehog signal in soluble form. Genes Dev. 2012;26:1312–1325. doi: 10.1101/gad.191866.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng X, Goetz JA, Suber LM, Scott WJ, Jr, Schreiner CM, Robbins DJ. A freely diffusible form of Sonic Hedgehog mediates long-range signalling. Nature. 2001;411:716–720. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- 33.Eugster C, Panáková D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev Cell. 2007;13:57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435:172–177. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 35.Sanders TA, Llagostera E, Barna M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 2013;497:628–632. doi: 10.1038/nature12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rojas-Ríos P, Guerrero I, González-Reyes A. Cytoneme-mediated delivery of hedgehog regulates the expression of bone morphogenetic proteins to maintain germline stem cells in Drosophila. PLoS Biol. 2012;10:e1001298. doi: 10.1371/journal.pbio.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briscoe J, Chen Y, Jessell TM, Struhl G. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol Cell. 2001;7:1279–1291. doi: 10.1016/s1097-2765(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 38.Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17:342–347. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forbes AJ, Nakano Y, Taylor AM, Ingham PW. Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev Suppl. 1993:115–124. [PubMed] [Google Scholar]
- 40.Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O’Brien SJ, Wong AJ, Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- 41.Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao FT, Law ML, Seuanez HN, O’Brien SJ, Vogelstein B. The GLI-Kruppel family of human genes. Mol Cell Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 43.Zaphiropoulos PG, Undén AB, Rahnama F, Hollingsworth RE, Toftgård R. PTCH2, a novel human patched gene, undergoing alternative splicing and up-regulated in basal cell carcinomas. Cancer Res. 1999;59:787–792. [PubMed] [Google Scholar]
- 44.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 45.Mas C, Ruiz i Altaba A. Small molecule modulation of HH-GLI signaling: current leads, trials and tribulations. Biochem Pharmacol. 2010;80:712–723. doi: 10.1016/j.bcp.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 46.Corcoran RB, Scott MP. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc Natl Acad Sci USA. 2006;103:8408–8413. doi: 10.1073/pnas.0602852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nachtergaele S, Mydock LK, Krishnan K, Rammohan J, Schlesinger PH, Covey DF, Rohatgi R. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat Chem Biol. 2012;8:211–220. doi: 10.1038/nchembio.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 49.Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev Cell. 2011;20:775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 2007;21:1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10:647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 53.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 54.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 55.Wang B, Li Y. Evidence for the direct involvement of betaTrCP in Gli3 protein processing. Proc Natl Acad Sci USA. 2006;103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svärd J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergström A, Ericson J, Toftgård R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 58.Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, Wong WH, McMahon AP. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- 60.Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin C, Chen MH, Yao E, Song H, Gacayan R, Hui CC, Chuang PT. Differential regulation of Gli proteins by Sufu in the lung affects PDGF signaling and myofibroblast development. Dev Biol. 2014;392:324–333. doi: 10.1016/j.ydbio.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quarmby LM, Parker JD. Cilia and the cell cycle? J Cell Biol. 2005;169:707–710. doi: 10.1083/jcb.200503053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 64.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 65.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ericson J, Briscoe J, Rashbass P, van Heyningen V, Jessell TM. Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harb Symp Quant Biol. 1997;62:451–466. [PubMed] [Google Scholar]
- 70.Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chuang PT, McMahon AP. Branching morphogenesis of the lung: new molecular insights into an old problem. Trends Cell Biol. 2003;13:86–91. doi: 10.1016/s0962-8924(02)00031-4. [DOI] [PubMed] [Google Scholar]
- 72.Jenkins D. Hedgehog signalling: emerging evidence for non-canonical pathways. Cell Signal. 2009;21:1023–1034. doi: 10.1016/j.cellsig.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 73.Yam PT, Langlois SD, Morin S, Charron F. Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron. 2009;62:349–362. doi: 10.1016/j.neuron.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 74.Polizio AH, Chinchilla P, Chen X, Manning DR, Riobo NA. Sonic Hedgehog activates the GTPases Rac1 and RhoA in a Gli-independent manner through coupling of smoothened to Gi proteins. Sci Signal. 2011;4:pt7. doi: 10.1126/scisignal.2002396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bijlsma MF, Borensztajn KS, Roelink H, Peppelenbosch MP, Spek CA. Sonic hedgehog induces transcription-independent cytoskeletal rearrangement and migration regulated by arachidonate metabolites. Cell Signal. 2007;19:2596–2604. doi: 10.1016/j.cellsig.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 76.Dennler S, André J, Alexaki I, Li A, Magnaldo T, ten Dijke P, Wang XJ, Verrecchia F, Mauviel A. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, Li CW, Hsu JL, Miller SA, Wang X, et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell. 2012;21:374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 79.Mille F, Thibert C, Fombonne J, Rama N, Guix C, Hayashi H, Corset V, Reed JC, Mehlen P. The Patched dependence receptor triggers apoptosis through a DRAL-caspase-9 complex. Nat Cell Biol. 2009;11:739–746. doi: 10.1038/ncb1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thibert C, Teillet MA, Lapointe F, Mazelin L, Le Douarin NM, Mehlen P. Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science. 2003;301:843–846. doi: 10.1126/science.1085405. [DOI] [PubMed] [Google Scholar]
- 81.Bredesen DE, Mehlen P, Rabizadeh S. Receptors that mediate cellular dependence. Cell Death Differ. 2005;12:1031–1043. doi: 10.1038/sj.cdd.4401680. [DOI] [PubMed] [Google Scholar]
- 82.Lozano Bolanos A, Mendoza Milla C, Cisneros Lira J, Ramirez R, Checa M, Barrera L, Garcia-Alvarez J, Carbajal V, Becerril C, Gaxiola M, et al. Role of sonic hedgehog in idiopathic pulmonary fibrosis Am J Physiol Lung Cell Mol PhysiolIn press [DOI] [PubMed] [Google Scholar]
- 83.Shi W, Bellusci S, Warburton D. Lung development and adult lung diseases. Chest. 2007;132:651–656. doi: 10.1378/chest.06-2663. [DOI] [PubMed] [Google Scholar]
- 84.Warburton D, Schwarz M, Tefft D, Flores-Delgado G, Anderson KD, Cardoso WV. The molecular basis of lung morphogenesis. Mech Dev. 2000;92:55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- 85.Miller LA, Wert SE, Whitsett JA. Immunolocalization of sonic hedgehog (Shh) in developing mouse lung. J Histochem Cytochem. 2001;49:1593–1604. doi: 10.1177/002215540104901213. [DOI] [PubMed] [Google Scholar]
- 86.Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 87.Bellusci S, Furuta Y, Rush MG, Henderson R, Winnier G, Hogan BL. Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1997;124:53–63. doi: 10.1242/dev.124.1.53. [DOI] [PubMed] [Google Scholar]
- 88.Sato H, Murphy P, Giles S, Bannigan J, Takayasu H, Puri P. Visualizing expression patterns of Shh and Foxf1 genes in the foregut and lung buds by optical projection tomography. Pediatr Surg Int. 2008;24:3–11. doi: 10.1007/s00383-007-2036-1. [DOI] [PubMed] [Google Scholar]
- 89.Zhang M, Wang H, Teng H, Shi J, Zhang Y. Expression of SHH signaling pathway components in the developing human lung. Histochem Cell Biol. 2010;134:327–335. doi: 10.1007/s00418-010-0738-2. [DOI] [PubMed] [Google Scholar]
- 90.Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Dev Biol. 2003;258:169–184. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 91.Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- 92.Grindley JC, Bellusci S, Perkins D, Hogan BL. Evidence for the involvement of the Gli gene family in embryonic mouse lung development. Dev Biol. 1997;188:337–348. doi: 10.1006/dbio.1997.8644. [DOI] [PubMed] [Google Scholar]
- 93.Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- 94.Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- 95.Miller LA, Wert SE, Clark JC, Xu Y, Perl AK, Whitsett JA. Role of Sonic hedgehog in patterning of tracheal-bronchial cartilage and the peripheral lung. Dev Dyn. 2004;231:57–71. doi: 10.1002/dvdy.20105. [DOI] [PubMed] [Google Scholar]
- 96.Goodrich LV, Milenković L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 97.Dixit R, Ai X, Fine A. Derivation of lung mesenchymal lineages from the fetal mesothelium requires hedgehog signaling for mesothelial cell entry. Development. 2013;140:4398–4406. doi: 10.1242/dev.098079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rutter M, Wang J, Huang Z, Kuliszewski M, Post M. Gli2 influences proliferation in the developing lung through regulation of cyclin expression. Am J Respir Cell Mol Biol. 2010;42:615–625. doi: 10.1165/rcmb.2008-0390OC. [DOI] [PubMed] [Google Scholar]
- 99.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 100.Li Y, Zhang H, Choi SC, Litingtung Y, Chiang C. Sonic hedgehog signaling regulates Gli3 processing, mesenchymal proliferation, and differentiation during mouse lung organogenesis. Dev Biol. 2004;270:214–231. doi: 10.1016/j.ydbio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 101.Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, et al. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 103.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 104.Volckaert T, Campbell A, Dill E, Li C, Minoo P, De Langhe S. Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development. 2013;140:3731–3742. doi: 10.1242/dev.096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL. Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development. 1999;126:4005–4015. doi: 10.1242/dev.126.18.4005. [DOI] [PubMed] [Google Scholar]
- 106.Mahlapuu M, Enerbäck S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development. 2001;128:2397–2406. doi: 10.1242/dev.128.12.2397. [DOI] [PubMed] [Google Scholar]
- 107.Astorga J, Carlsson P. Hedgehog induction of murine vasculogenesis is mediated by Foxf1 and Bmp4. Development. 2007;134:3753–3761. doi: 10.1242/dev.004432. [DOI] [PubMed] [Google Scholar]
- 108.Wan H, Kaestner KH, Ang SL, Ikegami M, Finkelman FD, Stahlman MT, Fulkerson PC, Rothenberg ME, Whitsett JA. Foxa2 regulates alveolarization and goblet cell hyperplasia. Development. 2004;131:953–964. doi: 10.1242/dev.00966. [DOI] [PubMed] [Google Scholar]
- 109.Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- 110.Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 111.Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- 112.White AC, Lavine KJ, Ornitz DM. FGF9 and SHH regulate mesenchymal Vegfa expression and development of the pulmonary capillary network. Development. 2007;134:3743–3752. doi: 10.1242/dev.004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li C, Xiao J, Hormi K, Borok Z, Minoo P. Wnt5a participates in distal lung morphogenesis. Dev Biol. 2002;248:68–81. doi: 10.1006/dbio.2002.0729. [DOI] [PubMed] [Google Scholar]
- 114.Li C, Hu L, Xiao J, Chen H, Li JT, Bellusci S, Delanghe S, Minoo P. Wnt5a regulates Shh and Fgf10 signaling during lung development. Dev Biol. 2005;287:86–97. doi: 10.1016/j.ydbio.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 115.Li M, Li C, Liu YH, Xing Y, Hu L, Borok Z, Kwong KY, Minoo P. Mesodermal deletion of transforming growth factor-beta receptor II disrupts lung epithelial morphogenesis: cross-talk between TGF-beta and Sonic hedgehog pathways. J Biol Chem. 2008;283:36257–36264. doi: 10.1074/jbc.M806786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.El-Hashash AH, Al Alam D, Turcatel G, Bellusci S, Warburton D. Eyes absent 1 (Eya1) is a critical coordinator of epithelial, mesenchymal and vascular morphogenesis in the mammalian lung. Dev Biol. 2011;350:112–126. doi: 10.1016/j.ydbio.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.El-Hashash AH, Al Alam D, Turcatel G, Rogers O, Li X, Bellusci S, Warburton D. Six1 transcription factor is critical for coordination of epithelial, mesenchymal and vascular morphogenesis in the mammalian lung. Dev Biol. 2011;353:242–258. doi: 10.1016/j.ydbio.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu K, Reddy R, Berika M, Warburton D, El-Hashash AH. Abrogation of Eya1/Six1 disrupts the saccular phase of lung morphogenesis and causes remodeling. Dev Biol. 2013;382:110–123. doi: 10.1016/j.ydbio.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 119.Haubold M, Gilsbach R, Hein L. Alpha2B-adrenoceptor deficiency leads to postnatal respiratory failure in mice. J Biol Chem. 2010;285:34213–34219. doi: 10.1074/jbc.M110.129205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Burri PH. Structural aspects of postnatal lung development: alveolar formation and growth. Biol Neonate. 2006;89:313–322. doi: 10.1159/000092868. [DOI] [PubMed] [Google Scholar]
- 121.Burri PH. The postnatal growth of the rat lung: 3. Morphology. Anat Rec. 1974;180:77–98. doi: 10.1002/ar.1091800109. [DOI] [PubMed] [Google Scholar]
- 122.Kauffman SL, Burri PH, Weibel ER. The postnatal growth of the rat lung: II. Autoradiography. Anat Rec. 1974;180:63–76. doi: 10.1002/ar.1091800108. [DOI] [PubMed] [Google Scholar]
- 123.Jobe AH, Bancalari E. Bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2001;163:1723–1729. doi: 10.1164/ajrccm.163.7.2011060. [DOI] [PubMed] [Google Scholar]
- 124.Lemons JA, Bauer CR, Oh W, Korones SB, Papile LA, Stoll BJ, Verter J, Temprosa M, Wright LL, Ehrenkranz RA, et al. NICHD Neonatal Research Network. Very low birth weight outcomes of the National Institute of Child Health And Human Development Neonatal Research Network, January 1995 through December 1996. Pediatrics. 2001;107:E1. doi: 10.1542/peds.107.1.e1. [DOI] [PubMed] [Google Scholar]
- 125.Madurga A, Mizíková I, Ruiz-Camp J, Morty RE. Recent advances in late lung development and the pathogenesis of bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol. 2013;305:L893–L905. doi: 10.1152/ajplung.00267.2013. [DOI] [PubMed] [Google Scholar]
- 126.Ahlfeld SK, Conway SJ. Aberrant signaling pathways of the lung mesenchyme and their contributions to the pathogenesis of bronchopulmonary dysplasia. Birth Defects Res A Clin Mol Teratol. 2012;94:3–15. doi: 10.1002/bdra.22869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dang H, Wang S, Yang L, Fang F, Xu F. Upregulation of shh and ptc1 in hyperoxia-induced acute lung injury in neonatal rats. Mol Med Report. 2012;6:297–302. doi: 10.3892/mmr.2012.929. [DOI] [PubMed] [Google Scholar]
- 128.Liu L, Kugler MC, Loomis CA, Samdani R, Zhao Z, Chen GJ, Brandt JP, Brownell I, Joyner AL, Rom WN, et al. Hedgehog signaling in neonatal and adult lung. Am J Respir Cell Mol Biol. 2013;48:703–710. doi: 10.1165/rcmb.2012-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.King P, Paul A, Laufer E. Shh signaling regulates adrenocortical development and identifies progenitors of steroidogenic lineages. Proc Natl Acad Sci USA. 2009;106:21185–21190. doi: 10.1073/pnas.0909471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McGowan SE, McCoy DM. Platelet-derived growth factor-A and sonic hedgehog signaling direct lung fibroblast precursors during alveolar septal formation. Am J Physiol Lung Cell Mol Physiol. 2013;305:L229–L239. doi: 10.1152/ajplung.00011.2013. [DOI] [PubMed] [Google Scholar]
- 131.Heine VM, Griveau A, Chapin C, Ballard PL, Chen JK, Rowitch DH. A small-molecule smoothened agonist prevents glucocorticoid-induced neonatal cerebellar injury. Sci Transl Med. 2011;3:105ra104. doi: 10.1126/scitranslmed.3002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Horn A, Palumbo K, Cordazzo C, Dees C, Akhmetshina A, Tomcik M, Zerr P, Avouac J, Gusinde J, Zwerina J, et al. Hedgehog signaling controls fibroblast activation and tissue fibrosis in systemic sclerosis. Arthritis Rheum. 2012;64:2724–2733. doi: 10.1002/art.34444. [DOI] [PubMed] [Google Scholar]
- 133.Omenetti A, Porrello A, Jung Y, Yang L, Popov Y, Choi SS, Witek RP, Alpini G, Venter J, Vandongen HM, et al. Hedgehog signaling regulates epithelial-mesenchymal transition during biliary fibrosis in rodents and humans. J Clin Invest. 2008;118:3331–3342. doi: 10.1172/JCI35875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fabian SL, Penchev RR, St-Jacques B, Rao AN, Sipilä P, West KA, McMahon AP, Humphreys BD. Hedgehog-Gli pathway activation during kidney fibrosis. Am J Pathol. 2012;180:1441–1453. doi: 10.1016/j.ajpath.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fernandez IE, Eickelberg O. New cellular and molecular mechanisms of lung injury and fibrosis in idiopathic pulmonary fibrosis. Lancet. 2012;380:680–688. doi: 10.1016/S0140-6736(12)61144-1. [DOI] [PubMed] [Google Scholar]
- 137.Chapman HA. Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu Rev Physiol. 2011;73:413–435. doi: 10.1146/annurev-physiol-012110-142225. [DOI] [PubMed] [Google Scholar]
- 138.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med. 2008;5:e62. doi: 10.1371/journal.pmed.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Coon DR, Roberts DJ, Loscertales M, Kradin R. Differential epithelial expression of SHH and FOXF1 in usual and nonspecific interstitial pneumonia. Exp Mol Pathol. 2006;80:119–123. doi: 10.1016/j.yexmp.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 140.Stewart GA, Hoyne GF, Ahmad SA, Jarman E, Wallace WA, Harrison DJ, Haslett C, Lamb JR, Howie SE. Expression of the developmental Sonic hedgehog (Shh) signalling pathway is up-regulated in chronic lung fibrosis and the Shh receptor patched 1 is present in circulating T lymphocytes. J Pathol. 2003;199:488–495. doi: 10.1002/path.1295. [DOI] [PubMed] [Google Scholar]
- 141.Fitch PM, Howie SE, Wallace WA. Oxidative damage and TGF-β differentially induce lung epithelial cell sonic hedgehog and tenascin-C expression: implications for the regulation of lung remodelling in idiopathic interstitial lung disease. Int J Exp Pathol. 2011;92:8–17. doi: 10.1111/j.1365-2613.2010.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cigna N, Farrokhi Moshai E, Brayer S, Marchal-Somme J, Wémeau-Stervinou L, Fabre A, Mal H, Lesèche G, Dehoux M, Soler P, et al. The hedgehog system machinery controls transforming growth factor-β-dependent myofibroblastic differentiation in humans: involvement in idiopathic pulmonary fibrosis. Am J Pathol. 2012;181:2126–2137. doi: 10.1016/j.ajpath.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 143.Roberts SN, Howie SE, Wallace WA, Brown DM, Lamb D, Ramage EA, Donaldson K. A novel model for human interstitial lung disease: hapten-driven lung fibrosis in rodents. J Pathol. 1995;176:309–318. doi: 10.1002/path.1711760313. [DOI] [PubMed] [Google Scholar]
- 144.Fisher CE, Ahmad SA, Fitch PM, Lamb JR, Howie SE. FITC-induced murine pulmonary inflammation: CC10 up-regulation and concurrent Shh expression. Cell Biol Int. 2005;29:868–876. doi: 10.1016/j.cellbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 145.Moeller A, Ask K, Warburton D, Gauldie J, Kolb M. The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol. 2008;40:362–382. doi: 10.1016/j.biocel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Farrokhi Moshai E, Wemeau-Stervinou L, Cigna N, Brayer S, Marchal Somme J, Crestani B, Mailleux AA. Targeting the hedgehog-gli pathway inhibits bleomycin-induced lung fibrosis in mice. Am J Respir Cell Mol Biol. 2014;51:11–25. doi: 10.1165/rcmb.2013-0154OC. [DOI] [PubMed] [Google Scholar]
- 147.Horn A, Kireva T, Palumbo-Zerr K, Dees C, Tomcik M, Cordazzo C, Zerr P, Akhmetshina A, Ruat M, Distler O, et al. Inhibition of hedgehog signalling prevents experimental fibrosis and induces regression of established fibrosis. Ann Rheum Dis. 2012;71:785–789. doi: 10.1136/annrheumdis-2011-200883. [DOI] [PubMed] [Google Scholar]
- 148.Guy CD, Suzuki A, Zdanowicz M, Abdelmalek MF, Burchette J, Unalp A, Diehl AM NASH CRN. Hedgehog pathway activation parallels histologic severity of injury and fibrosis in human nonalcoholic fatty liver disease. Hepatology. 2012;55:1711–1721. doi: 10.1002/hep.25559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ding H, Zhou D, Hao S, Zhou L, He W, Nie J, Hou FF, Liu Y. Sonic hedgehog signaling mediates epithelial-mesenchymal communication and promotes renal fibrosis. J Am Soc Nephrol. 2012;23:801–813. doi: 10.1681/ASN.2011060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Elias JA, Lee CG, Zheng T, Ma B, Homer RJ, Zhu Z. New insights into the pathogenesis of asthma. J Clin Invest. 2003;111:291–297. doi: 10.1172/JCI17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hogg JC, Timens W. The pathology of chronic obstructive pulmonary disease. Annu Rev Pathol. 2009;4:435–459. doi: 10.1146/annurev.pathol.4.110807.092145. [DOI] [PubMed] [Google Scholar]
- 152.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhou X, Baron RM, Hardin M, Cho MH, Zielinski J, Hawrylkiewicz I, Sliwinski P, Hersh CP, Mancini JD, Lu K, et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum Mol Genet. 2012;21:1325–1335. doi: 10.1093/hmg/ddr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li X, Howard TD, Moore WC, Ampleford EJ, Li H, Busse WW, Calhoun WJ, Castro M, Chung KF, Erzurum SC, et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127:1457–1465. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Furmanski AL, Saldana JI, Ono M, Sahni H, Paschalidis N, D’Acquisto F, Crompton T. Tissue-derived hedgehog proteins modulate Th differentiation and disease. J Immunol. 2013;190:2641–2649. doi: 10.4049/jimmunol.1202541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Michel KD, Uhmann A, Dressel R, van den Brandt J, Hahn H, Reichardt HM. The hedgehog receptor patched1 in T cells is dispensable for adaptive immunity in mice. PLoS ONE. 2013;8:e61034. doi: 10.1371/journal.pone.0061034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de la Roche M, Ritter AT, Angus KL, Dinsmore C, Earnshaw CH, Reiter JF, Griffiths GM. Hedgehog signaling controls T cell killing at the immunological synapse. Science. 2013;342:1247–1250. doi: 10.1126/science.1244689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Y, Davidow L, Arvanites AC, Blanchard J, Lam K, Xu K, Oza V, Yoo JW, Ng JM, Curran T, et al. Glucocorticoid compounds modify smoothened localization and hedgehog pathway activity. Chem Biol. 2012;19:972–982. doi: 10.1016/j.chembiol.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bailey JM, Swanson BJ, Hamada T, Eggers JP, Singh PK, Caffery T, Ouellette MM, Hollingsworth MA. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res. 2008;14:5995–6004. doi: 10.1158/1078-0432.CCR-08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 161.Tian H, Callahan CA, DuPree KJ, Darbonne WC, Ahn CP, Scales SJ, de Sauvage FJ. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci USA. 2009;106:4254–4259. doi: 10.1073/pnas.0813203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]