Abstract
A promising application of DNA self-assembly is the fabrication of chromophore-based excitonic devices. DNA brick assembly is a compelling method for creating programmable nanobreadboards on which chromophores may be rapidly and easily repositioned to prototype new excitonic devices, optimize device operation, and induce reversible switching. Using DNA nanobreadboards, we have demonstrated each of these functions through the construction and operation of two different excitonic AND logic gates. The modularity and high chromophore density achievable via this brick-based approach provide a viable path toward developing information processing and storage systems.
Keywords: DNA nanotechnology, DNA bricks, FRET, Boolean logic, nanophotonic devices
Nanophotonics exploits excitonic and plasmonic interactions between light and matter on a scale below the free-space wavelength of light.1−3 Molecular self-assembly organizes matter with atomic precision in two- and three-dimensions. When combined, these technologies enable the construction of high-speed and highly compact devices that employ near-field electromagnetic coupling to achieve high performance.4−6 Recent work by Pistol et al. described chromophores as single-molecule optical devices that can be used for excitonic-based nanoscale computing, sensing, and memory applications.7 For such applications, chromophores must be arranged precisely and proximally to enhance the nonradiative, dipole–dipole coupling between neighboring chromophores. This dipole–dipole coupling facilitates an excitonic transfer process known as Förster resonance energy transfer (FRET).8 The distance over which FRET occurs between chromophores with 50% efficiency, termed the Förster radius, is typically 5 nm or less.8 Thus, a minimum requirement for the fabrication of excitonic circuits is subnanometer resolution control over chromophore position. In addition to the distance between chromophores, additional factors, such as spectral overlap and relative molecular orientation, also play a role in the excitonic transfer efficiency. As molecular orientation can be difficult to predict a priori, a practical approach to excitonic circuit fabrication necessitates a framework for rapid prototyping.
Over the past decade, the simple design rules governing DNA self-assembly have been utilized to create a multitude of templated architectures9−15 expanding well beyond basic duplexes.16 Empowered by these design rules, DNA nanotechnology has been employed to predict and fabricate a variety of nanophotonic devices,2,7,17−44 including those demonstrating excitonic control.7,17−37 However, the majority of multichromophore excitonic circuit studies have synthesized excitonic transmission lines (i.e., wires) on single DNA duplexes, which restricts the complexity and functionality achievable with excitonic circuits.17−23 Notable exceptions are the polyduplex structures recently reported by Buckhout-White et al., which demonstrate complex excitonic circuitry using multiarm DNA junctions.32,33 To enable greater complexity and the assembly of multiple circuit elements, DNA origami, has been used to fabricate excitonic devices, such as transmission lines,24 switches,25 light-harvesting antennae,26 and logic gates.27,28 As a platform technology, DNA origami involves folding a long DNA scaffold into a defined structure via short DNA strands called staples.9 With DNA origami, excitonic circuits can be synthesized by conjugating chromophores to selected staple strands, but half of the structure, the scaffold, plays only a structural role, as chromophore conjugation to the scaffold is impractical. Consequently, excitonic devices synthesized with DNA origami are typically static structures that limit the feasibility of rapidly prototyping excitonic circuitry. Since DNA nanotechnology primarily enables chromophore positioning, and relative chromophore orientation remains difficult to predict or control, rapid prototyping is essential for achieving complex excitonic functionality.31
Recently, the concept of a molecular canvas (i.e., a molecular nanobreadboard) was introduced, whereby single-stranded DNA (ssDNA) oligomers, called DNA bricks, self-assemble to form origami-like structures without the need for long scaffold strands.12−14 Bricks consist of four unique sequence domains that selectively hybridize to four neighboring bricks. As structural building blocks, bricks are particularly modular for two- (2D) and three-dimensional (3D) architectures due to a master DNA library that can be down selected into a multitude of molecularly precise shapes or repeated units during assembly.45 The emergent approach of DNA brick-based nanobreadboards enables the rapid, facile reconfiguration of 2D and 3D architectures to be achieved by simply selecting the desired bricks from the library, whereas DNA origami requires the redesign of all staple strands. Additionally, since a long scaffold strand is not used within the DNA brick approach and any DNA brick can carry covalently attached chromophores, the achievable packing density and position control of chromophores on a DNA brick nanobreadboard is twice that of DNA origami.
Capitalizing on the unique aspects of DNA bricks, we demonstrate the first excitonic wires, switches, and logic devices prototyped on DNA brick-based nanobreadboards. In this work, we show (1) four-chromophore systems with two dynamically controlled inputs permitting bilevel switches that can be coupled to demonstrate fully excitonic AND logic, and (2) dynamic excitonic switching and logic operations controlled by isothermal DNA reactions in stoichiometric quantities. To facilitate prototyping of excitonic logic devices, our current work takes advantage of the increased design flexibility of DNA bricks over DNA origami. Indeed, because of the inability of the scaffold strand to support chromophores, our designs could not have been carried out using standard DNA origami (Figure 1a,b). Prototyping was expedited by the ease with which any selected brick could be functionalized with a chosen chromophore. Prototyping and dynamic behavior were further facilitated by the incorporation of single stranded extensions (tethers) on two of the bricks that served as recognition sites at which chromophore-carrying oligomers were attached or removed. While two designs are described in detail here, multiple chromophore architectures are possible, thereby enabling the exploration of any and all logic gate operations. Additionally, in contrast to duplex-based circuits,17−23 the brick-based nanobreadboard greatly promotes coupling of multiple excitonic devices to achieve greater circuit complexity. Future screening will guide optimum information processing and storage in DNA-based excitonic devices.4−6,31
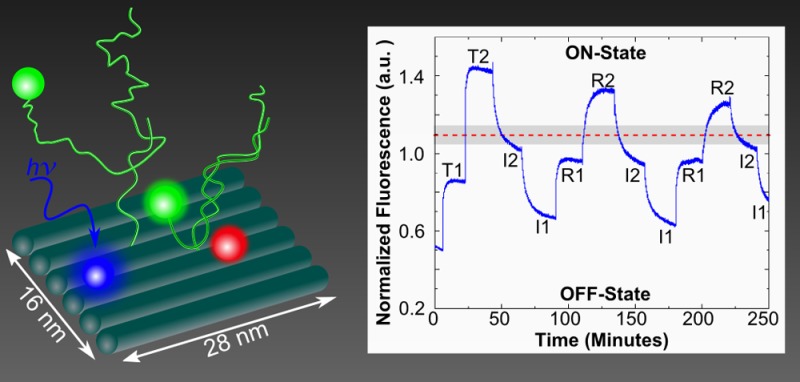
Figure 1.
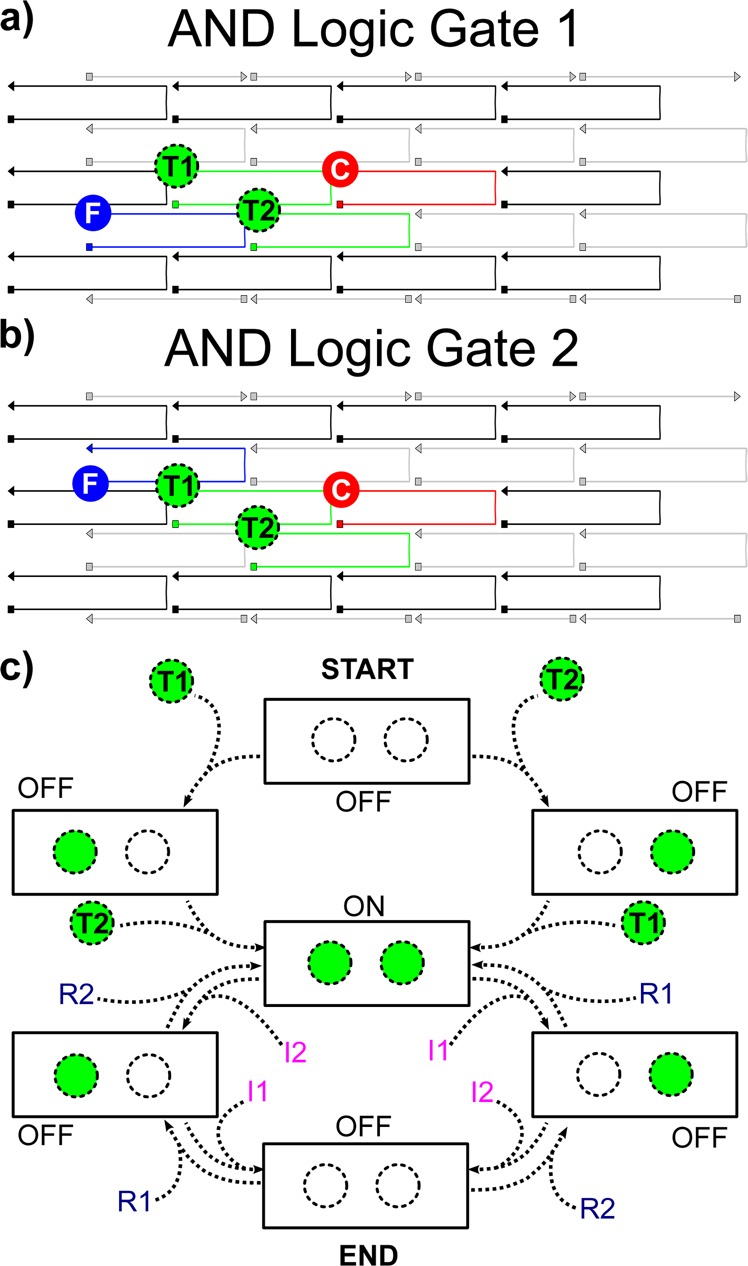
Schematic of (a) AND logic gate 1 and (b) AND logic gate 2 illustrating DNA brick reconfigurability. AND logic gates 1 and 2 only differ by the position of FAM (blue). (c) Schematic of sequentially switchable excitonic AND logic gate operation. Both logic gates are initially fabricated in an OFF-state with only FAM (blue) and Cy5 (red) chromophores attached. When introduced into solution, TAMRA (T1 and T2, green with dashed outline) functionalized ssDNA oligomers hybridize to the corresponding recognition sites (i.e., tethers), switching the gate into an ON-state configuration and generating a fluorescent output signal. Following addition of T1 and T2 to switch the AND gate to the ON-state, invasion strands (I1 and I2) and restoration strands (R1 and R2) are sequentially added to the sample solution to cycle between OFF- (invasion) and ON- (restoration) states.
The two excitonic AND logic gates assembled on DNA brick nanobreadboards, with differing chromophore arrangements were designed to (1) demonstrate reconfigurability for rapid prototyping and (2) investigate the effects of chromophore placement on AND logic gate performance. The designs, labeled AND logic gates 1 and 2, are illustrated in Figure 1a and b, respectively (strand sequences and chromophore details are provided in the Supporting Information, S1). Each nanobreadboard is a two-dimensional (6 helices × 94 base pair) structure12 with four chromophores positioned in zigzag (AND logic gate 1) or quasi-linear (AND logic gate 2) arrays to form excitonic transmission lines. The nanobreadboard is composed of three types of DNA bricks: (i) eight 21 nucleotide (nt) strands, (ii) 18 42 nt strands, and (iii) two 58 nt strands containing a 16 nt tether. Of the four chromophores that constitute the AND logic gates, two are permanently attached to the nanobreadboard (F and C, Figure 1a,b), while the other two (T1 and T2, Figure 1a,b) are independently added to or removed from DNA tethers via DNA hybridization and toehold-mediated strand displacement.39 The use of tethers allows independent control over chromophore spacing and the numbers of base pairs involved in DNA hybridization and strand displacement. This innovation is a significant performance advantage over previous designs in which binding to an origami scaffold strand restricted the number of base pairs involved in the switching reactions and necessitated the use of excessive fuel concentrations to drive switching reactions.25
Carboxyfluorescein, commonly referred to as FAM (F), and a cyanine derivative, Cy5 (C), served as the exciton donor (i.e., optical input) and acceptor (i.e., optical output) chromophores, respectively. F (blue circle, Figure 1a) and C (red circle, Figure 1a) were covalently attached to 42 nt staple strands and were embedded permanently into the nanobreadboard. Two dynamic tetramethylrhodamine (TAMRA) chromophores (excitonic logic inputs) were covalently attached to 26 nt single-stranded (ssDNA) oligomers and acted to position the logic inputs for AND switching. The two oligomers were distinguishable by sequence and labeled as TAMRA 1 (i.e., T1, logic input 1) or TAMRA 2 (i.e., T2, logic input 2) in Figure 1a (green circles with dashed outline). T1 and T2 were designed with a 16 nt complementary region that attached to the nanobreadboard tethers, leaving 10 nt toeholds for subsequent removal by invasion strands.
Exploiting toehold-mediated strand displacement,39 ssDNA invasion and restoration strands were designed to either remove or restore T1 and T2 from or onto the nanobreadboards, respectively, to enable isothermal reversible switching. The isothermal nature of the switching processes facilitates practical device operation and complex multielement circuit design, both of which would be difficult to achieve if input switching required thermal cycling. Invasion strand 1 (I1) hybridizes with the 10 nt toehold on T1 and invasion strand 2 (I2) hybridizes with the 10 nt toehold on T2, thereby specifically removing the corresponding chromophore from the nanobreadboard through a three-way branch migration process. Similar to T1 and T2, I1 and I2 are designed with a 10 nt toehold for the attachment of restoration strand 1 (R1) and restoration strand 2 (R2), respectively. R1 and R2 completely hybridize with the respective invasion strands, thus releasing T1 or T2 back into solution. Once released, T1 and T2 rehybridize to the nanobreadboard tethers. It should be noted that no additional T1 or T2 are added to the solution, which validates the use of these logic devices in systems where inputs are controlled by external processes (e.g., biological processes). The R1 and R2 sequences have been specifically designed to minimize unwanted complementary regions with the nanobreadboard tethers, thereby reducing competition with the T1 and T2 strands. The restoration strands have only a 6 nt overlap with the nanobreadboard tethers and a 25 nt overlap with the invasion strands, and the free-energy is minimized by formation of the desired complementary pairs (see Supporting Information, S1 for further details). This design work was accomplished using UNIQUIMER45 to generate random sequences for all strands and NUPACK46 to analyze thermodynamically favorable strand hybridizations via free-energy minimization.
The excitonic AND logic gates are in the ON-state when both T1 and T2 are attached concurrently onto the nanobreadboard, thereby completing a FRET transmission line that enables exciton flow from F (excited at 450 nm) to C (monitored at 668 nm). To maximize nearest neighbor FRET and minimize non-nearest neighbor FRET, the positions of chromophore attachment sites have been chosen such that only neighboring chromophores have separation distances that lie within the Förster radius. Exciton transmission is interrupted when either or both logic input chromophores (i.e., T1 and T2) are removed from the nanobreadboard, resulting in an OFF-state logic gate configuration (Figure 1c). Once T1 and T2 have been introduced into solution, the gate can be reversibly switched between the ON- or OFF- states via the successive addition of invasion (OFF) and restoration (ON) strands.
Figure 2 demonstrates the efficacy of the DNA brick-based nanobreadboard for prototyping excitonic transmission lines, switches, and logic gates. Nanobreadboards with chromophore arrangements corresponding to each logic state were synthesized, annealed, and filtered with Amicon centrifugal filter units prior to obtaining spectroscopic measurements (Supporting Information, S2.2). Panels b and c plot the emission intensity of the logic output and illustrate the equilibrium logic states as defined by Figure 2a for AND gates 1 and 2, respectively, when all strands are in stoichiometry. Clear differences in the overall excitonic transmission efficiency are observed for different states of the logic gates. The nonzero intensities measured for states i–iii reflect excitonic cross-talk and bleed-through between the chromophores present on the devices and represent the intrinsic behaviors that vary based on the logic gate designs. The threshold window for each gate is indicated by a shaded gray area that delineates the region between the minimum fluorescence obtained with the attachment of both logic input chromophores (T1 and T2) and the maximum fluorescence obtained with the attachment of only one logic input chromophore (T1 or T2) for a given logic gate design. The logic threshold (i.e., the fluorescence level at which the gate output switches between OFF and ON) is taken to be the median fluorescence value within the threshold window range and is indicated by the red dashed line in Figure 2b,c. As can be seen from the data, simply shifting the location of F by one double helix reduced the cross-talk and overall transmission efficiency of AND logic gate 2 relative to gate 1 but resulted in a slightly larger threshold window. Logic gate performance is further analyzed using the equilibrium threshold tolerance (Table 1), which is given as a dimensionless value and is described in further detail in Supporting Information, S5. A larger equilibrium threshold tolerance (i.e., threshold range relative to overall fluorescence intensity) indicates superior logic gate performance arising from greater tolerance to errors within the system that may arise from malformed nanobreadboards and deviations from stoichiometry during synthesis and operation. While AND logic gate 2 exhibited a slightly larger threshold window range, it showed a greater equilibrium threshold tolerance by approximately a factor of 2 in its equilibrium states, which highlights the impact that small design changes can have on device performance based on multiple factors involved in the excitonic energy transfer.
Figure 2.
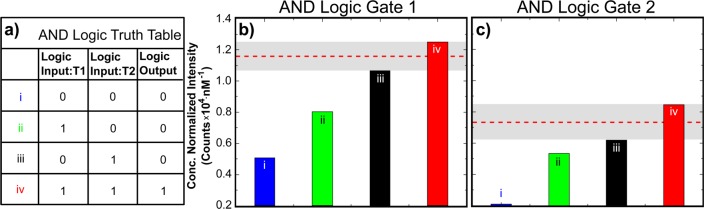
Excitonic AND logic truth tables showing the logic values associated with the ON- (Logic Output: 1) or OFF- (Logic Output: 0) states for the attachment (Logic Input: 1) or absence (Logic Input: 0) of T1 or T2 onto/from the nanobreadboard. Adjacent to the truth table are bar plots representing the concentration normalized fluorescence emission at 668 nm (Logic Output) corresponding to each logic state for AND logic gate designs 1 (b) and 2 (c). The threshold windows are indicated by the shaded gray areas. The logic threshold defines the midpoint of the logic threshold window and is indicated by a red dashed line in each bar chart. All spectra were collected by exciting F at 450 nm. Spectra were normalized by nanobreadboard concentration (∼25 nM).
Table 1. AND Logic Gate Operational Parameters.
| operational parameters | AND logic gate 1 | AND logic gate 2 |
|---|---|---|
| equilibrium threshold tolerance | 0.16 | 0.31 |
| dynamic threshold tolerancea | 0.06 | 0.11 |
| threshold window loss/cycleb | 40.5% loss | 29.9% loss |
Average threshold tolerance values for two separate reaction kinetics data sets.
Averages calculated using reaction kinetics data from three separate switching cycles.
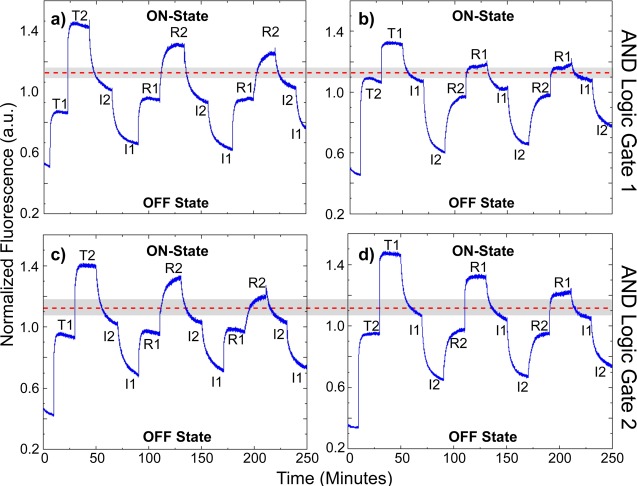
In contrast to the static equilibrium logic gate data shown in Figure 2, Figure 3 summarizes the isothermal dynamic switching results (i.e., fluorescence emission changes as a function of time) showing the dynamics of sequentially switchable AND logic operations for both gate designs. The data were obtained by exciting F at 450 nm and monitoring C emission at 668 nm. Figure 3 shows sequential switching for both AND logic gate 1 (Figure 3a,b) and logic gate 2 (Figure 3c,d) through three switching cycles. A switching cycle is defined as the transition of logic from OFF (Logic Output: 0) to ON (Logic Output: 1) and back to OFF resulting from the successive addition of restoration (R) and invasion (I) strands in stoichiometric amounts at ∼20 min intervals. Reflective of dynamic changes in overall excitonic transmission efficiencies, the data show clearly distinguishable output intensities for the various logic states, even after three full switching cycles. State transition rates were consistent throughout switching and were highest for the hybridization of T1 to the nanobreadboard, both initially and upon restoration. To enable analysis of intrinsic gate dynamics and account for extrinsic effects of dilution and photobleaching, each data set was subjected to a normalization procedure that is outlined in the Supporting Information, S3. The analysis and tabulation of switching rates for each transition are provided in Supporting Information, S4.
Figure 3.
Dynamic switching data demonstrating sequentially switchable excitonic AND logic functionality by examining fluorescent emission changes resulting from injecting TAMRA (T1 or T2), invasion (I1 or I2), or restoration (R1 or R2) strands. The data were obtained by exciting F at 450 nm and monitoring the fluorescence of C at 668 nm. Repeated AND logic operation by introducing (a) T1 to AND logic gate 1 prior to T2; (b) T2 to AND logic gate 1 prior to T1; (c) T1 to AND logic gate 2 prior to T2; (d) T2 to AND logic gate 2 prior to T1. All data were normalized by concentration and corrected for photobleaching (Supporting Information, S3). The logic threshold (red dashed line) defines the midpoint of the logic threshold window. The dynamic threshold window (gray area) defines the region between the minimum fluorescence obtained with the attachment of both logic input chromophores (T1 and T2) and the maximum fluorescence obtained with the attachment of one logic input chromophore (T1 or T2) for a given logic gate design operated over three complete OFF–ON–OFF switching cycles.
For the dynamic switching data, a dynamic threshold tolerance was used to assess the performance of each logic gate. The dynamic threshold tolerance for each logic gate design exhibited a reduction resulting from the threshold window loss that occurred with each successive switching cycle (Table 1). The threshold window loss per cycle is a qualitative parameter used to assess the reduction in threshold window with successive switching cycles. The threshold window loss values for each logic gate design were calculated using the stepwise ratios of C emission fluorescence intensities for each cycle, adjusted for dilution (Table 1). AND logic gate 2 was found to exhibit ∼25% lower threshold window loss over three switching cycles than AND logic gate 1. This smaller threshold window loss for AND logic gate 2 results in a higher average dynamic threshold tolerance for the total dynamic switching data, as shown in Table 1, where AND logic gate 2 has a dynamic threshold tolerance almost twice that of AND logic gate 1.
The results obtained from AND logic gates 1 and 2 illustrate the advantage of utilizing a reconfigurable nanobreadboard to rapidly prototype, evaluate, and optimize excitonic logic gate designs. Although both designs exhibit successful excitonic AND logic gate operations, significant differences were observed in device performance. A number of factors may influence the performance of the logic gates including: (i) the Förster radius, (ii) chromophore orientation, (iii) photobleaching, (iv) incomplete hybridization for DNA-controlled dynamic processes, and (v) base-pair quenching. The magnitude and relative contributions of each of these factors are quite difficult to predict a priori and can be complicated to decouple or correct for. Examples of these complicating factors can be observed by assessing the fluorescence of the equilibrium logic states for each AND logic gate design (Figure 2), where one can qualitatively compare the relative FRET efficiencies between the two designs. According to Förster theory, FRET is dependent on the relative distances and orientations between two chromophores.8 For a chromophore separation that is large compared to the Förster radius, FRET efficiency decreases with distance as a function of 1/r6.8 The relative orientation between chromophores is characterized by the coupling factor, κ2, and is very often approximated to be 2/3 due to the rapid randomization of the chromophore dipoles.8,47−51 Because the excitation wavelength of 450 nm was chosen to lie outside of the C absorption range (Figure S2.4, Supporting Information) and all spectra were normalized by concentration, the differences in emission at 668 nm (Figure 2) most likely result from altering the attachment point of F. For AND logic gates 1 and 2, in the initial OFF-state (i.e., without T1 and T2), the distances between the attachment points of F and C are calculated as 10.72 and 10.54 nm, respectively (Figure 1a,b). If the change of attachment point translates only into a corresponding change of distance between F and C, one would expect ∼10% greater direct FRET for AND logic gate 2 than AND logic gate 1 in the OFF-state. However, in actuality AND logic gate 1 shows ∼40% greater emission at 668 nm than does AND logic gate 2. This difference between the observed fluorescence and that calculated based on the change in distance between attachment sites may arise from the flexible ethylene spacers that link the chromophore to the DNA causing the position of the chromophores to not coincide precisely with the attachment point of the DNA. Additionally, since chromophores tend to intercalate between bases or find preferential orientations, κ2 need not be the same for the two gates.50,51 Finally, it has been shown in previous studies that specific nucleotide bases may contribute to the fluorescence-quenching of certain chromophores via photoinduced electron transfer, which may be a contributing factor within this case.52 Since these potential complicating factors are not easily predictable a priori nor discernible from the static equilibrium state data, the prototyping capability of DNA brick nanobreadboards is crucial for quickly assessing device performance.
The dynamic switching data shown in Figure 3 reveals a highly dynamic system consisting of multiple competing time-dependent effects. The extrinsic factor that seems the most detrimental to the logic performance is the photobleaching of F, which leads to a time-dependent decrease in output fluorescence intensity (see Supporting Information, S3). The rate of photobleaching of F is proportional to the intensity of the excitation light. Conversely, the rate of photobleaching is reduced when exciton transfer from the initially excited donor chromophore F occurs between chromophores along the FRET transmission line (Supporting Information, S3). Because FRET is an emission process that shortens the excited state lifetime of F, it acts as a protecting agent against photobleaching, which is the result of an excited state chemical reaction process.53−55 Accordingly, because the act of dynamically attaching chromophores onto the nanobreadboard and completing the transmission line increases the FRET efficiency between the chromophores, the effect of photobleaching on the system is reduced. A first principle account of this effect is impractical because of the high degree of difficulty to determine factors such as how the concentration of reactive oxygen species varies with time. In lieu of this, we have opted to correct for photobleaching by normalizing for an average photobleaching effect that consequently may correct for other effects, including the incomplete hybridization of strands. It should also be noted that, as an extrinsic factor, photobleaching is observed primarily as an artifact to our efforts to measure the kinetics of state changes by continually monitoring the C output emission, and does not constitute an intrinsic defect in the logic gate design.
Ideally, the threshold window loss per cycle would be a measure of the percentage of strands that incompletely hybridize to form the desired product. Because the injection strands are identical between logic gate designs and were shown to give similar rate constants (Supporting Information, S4) that are comparable to previous work,25 it is assumed that they do not contribute to threshold window loss. Rather, since the injection strands are added in equimolar amounts (in contrast to our previous work),25 it is likely that concentration and volumetric errors result in one or more of the following three processes involving incomplete hybridization between (i) T1, T2, and the nanobreadboard tethers, (ii) the invasion strands (i.e., I1 and I2) with T1 and T2, or (iii) the restoration strands (i.e., R1 and R2) with the invasion strands. All three of these processes would result in a threshold window loss by either decreasing the ON-state fluorescence (i.e., processes i and iii) or increasing the OFF-state fluorescence (i.e., process ii). Decoupling the incomplete hybridization process from the effects of photobleaching is beyond the scope of this report. However, this does show DNA brick-based nanobreadboards and DNA hybridization/strand-displacement can both expedite the trial and error process, as well as give some qualitative insight into the factors that contribute to differences in device performance. Further improvements may be achieved by (i) using chromophore types that are less prone to photobleaching, (ii) modifying the chromophore arrangements to increase FRET along the transmission line (e.g., decreased chromophore separation distance and/or shorter tethers), or (iii) adding injection strands in excess to decrease the occurrence of incomplete hybridization, thereby improving AND logic gate performance by decreasing the threshold window loss and resultant threshold tolerance reduction.
In this study, we report the synthesis and isothermal dynamic operation of two excitonic AND logic gates formed from four-chromophore transmission lines on DNA brick-based nanobreadboards. We establish that DNA brick-based nanobreadboards can be employed as a convenient means by which to rapidly prototype and evaluate the relative performance of excitonic-based devices. This is accomplished by two means: (1) through the modular nature of the DNA brick-based design in which only a small subset of the ssDNA oligomers requires redesign in order to change chromophore attachment points and (2) by DNA hybridization and toehold-mediated strand displacement to repeatedly attach and remove chromophore-functionalized strands to and from nanobreadboard tethers to allow dynamic operation. Two excitonic AND logic gate designs were investigated to demonstrate (1) reconfigurable excitonic-based device operability on DNA brick architectures and (2) isothermal sequentially switchable AND logic gate functionality. Both designs showed successful AND logic gate switching that was followed for three switching cycles. Additionally, it was found that the AND logic gate 2 design yielded better equilibrium and dynamic logic gate performance. Further optimization of chromophore types, chromophore arrangements, and amount of injection strand excess may lead to even better device performance. Because DNA brick nanobreadboards are composed entirely of synthetic DNA oligomers, chromophores can be covalently attached at any base position, thereby permitting a chromophore density twice that of DNA origami. The higher chromophore density, modularity, and greater circuit complexity that can be achieved by this approach may offer the keys to constructing excitonic-based information processing and storage systems.
Acknowledgments
The authors thank the students and staff within the Nanoscale Materials and Device group for valuable assistance with this work, Dr. Bryan Wei (Tsinghua University) for descriptive information on UNIQUIMER, and Prof. Andrew Turberfield (Oxford University) for helpful discussions on photobleaching. The research was supported in part by (1) NSF IDR No. 1014922, (2) NIH Grant No. P20 RR016454, (3) NIH Grant No. K25GM093233, (4) the W. M. Keck Foundation Award, and (5) the Micron Technology MSE Ph.D. Fellowship.
Supporting Information Available
Strand sequences, dye information, experimental methods, AFM images and methods, CanDo images, photobleaching correction processing, reaction rate calculations, and threshold window calculations. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Saikin S. K.; Eisfeld A.; Valleau S.; Aspuru-Guzik A. Photonics meets excitonics: Natural and artificial molecular aggregates. Nanophotonics 2013, 2, 1–18. [Google Scholar]
- Naruse M.; Tate N.; Aono M.; Ohtsu M. Analysis of optical near-field energy transfer by stochastic model unifying architectural dependencies. Rep. Prog. Phys. 2013, 76, 1–50. [Google Scholar]
- Mathur N. Beyond the silicon roadmap. Nat. Nanotechnol. 2002, 419, 573–574. [DOI] [PubMed] [Google Scholar]
- Xia Y.; Whitesides G. M. Soft lithography. Annu. Rev. Mater. Sci. 1998, 28, 153–184. [Google Scholar]
- 2012 International Technology Roadmap for Semiconductors (ITRS), http://www.itrs.net/Links/2012ITRS/2012Chapters/2012Overview.pdf, accessed August, 2014.
- Arden W.; Brillouët M.; Cogez P.; Graef M.; Huizing B.; Mahnkopf R.. More than Moore White Paper. http://www.itrs.net/Links/2010ITRS/IRC-ITRS-MtM-v2%203.pdf, accessed August, 2014.
- Pistol C.; Dwyer C.; Lebeck A. R. Nanoscale optical computing using resonance energy transfer logic. IEEE Micro 2008, 28, 7–19. [Google Scholar]
- Förster T. Intermolecular energy migration and fluorescence. Ann. Phys. 1948, 437, 55–75. [Google Scholar]
- Rothemund P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [DOI] [PubMed] [Google Scholar]
- Lin C.; Liu Y.; Rinker S.; Yan H. DNA tile based self-assembly: building complex nanoarchitectures. ChemPhysChem 2006, 7, 1641–1647. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Dietz H.; Liedl T.; Hogberg B.; Graf F.; Shih W. M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B.; Dai M.; Yin P. Complex shapes self-assembled from single-stranded tiles. Nature 2012, 485, 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y.; Ong L. L.; Shih W. M.; Yin P. Three-dimensional structures self-assembled from DNA bricks. Science 2012, 338, 1177–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y.; Ong L. L.; Sun W.; Song J.; Shih W. M.; Yin P. DNA brick crystals with prescribed depths. Nature 2014, 6, 994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Song J.; Besenbacher F.; Dong M.; Gothelf K. V. Self-assembly of DNA origami and single-stranded tile structures at room temperature. Angew. Chem., Int. Ed. 2013, 52, 1–6. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Marblestone A. H.; Teerapittayanon S.; Vasquez A.; Church G. M.; Shih W. M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller M. J.; Tullis R. H. Self-organizing molecular photonic structures based on functionalized synthetic nucleic acid (DNA) polymers. Nanotechnology 1991, 2, 165. [Google Scholar]
- Heilemann M.; Tinnefeld P.; Mosteiro G. S.; Parajo M. G.; Van Hulst N. F.; Sauer M. Multistep energy transfer in single molecular photonic wire. J. Am. Chem. Soc. 2004, 126, 6514–6515. [DOI] [PubMed] [Google Scholar]
- Hannestad J. K.; Sandin P.; Albinsson B. Self-assembled DNA photonic wire for long-range energy transfer. J. Am. Chem. Soc. 2008, 130, 15889–15895. [DOI] [PubMed] [Google Scholar]
- Vyawahare S.; Eyal S.; Mathews K. D.; Quake S. R. Nanometer-scale fluorescence resonance optical waveguides. Nano Lett. 2004, 4, 1035–1039. [Google Scholar]
- Tinnefeld P.; Heilemann M.; Sauer M. Design of molecular photonic wires based on multistep electronic excitonic transfer. ChemPhysChem 2005, 6, 217–222. [DOI] [PubMed] [Google Scholar]
- Ohya Y.; Yabuki K.; Komatsu M.; Ouchi T. Sequential arrangement of chromophores and energy transfer behavior on oligonucleiotides assemblies. Polym. Adv. Technol. 2000, 11, 845–855. [Google Scholar]
- Spillmann C. M.; Buckhout-White S.; Oh E.; Goldmann E. R.; Anicona M. G.; Medintz I. L. Extending FRET cascades on linear DNA photonic wires. Chem. Commun. 2014, 50, 7246–7249. [DOI] [PubMed] [Google Scholar]
- Stein I. H.; Steinhauer C.; Tinnefeld P. Single-molecule four-color FRET visualizes energy-transfer paths on DNA origami. J. Am. Chem. Soc. 2011, 133, 4193–4195. [DOI] [PubMed] [Google Scholar]
- Graugnard E.; Kellis D. L.; Bui H.; Barnes S.; Kuang W.; Lee J.; Hughes W. L.; Knowlton W. B.; Yurke B. DNA-controlled excitonic switches. Nano Lett. 2012, 12, 2117–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta P. K.; Varghese R.; Nangreave J.; Lin S.; Yan H.; Liu Y. DNA-directed artificial light-harvesting antenna. J. Am. Chem. Soc. 2011, 133, 11985–11993. [DOI] [PubMed] [Google Scholar]
- Zadegan R. M.; Jepsen M. D. E.; Thomsen K. E.; Okholm A. H.; Schaffert D. H.; Andersen E. S.; Birkedal V.; Kjems J. Construction of a 4 zeptoliters switchable 3D DNA box origami. ACS Nano 2012, 6, 10050–10053. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [DOI] [PubMed] [Google Scholar]
- Yurke B.; Kuang W. Passive linear nanoscale optical and molecular electronics device synthesis from nanoparticles. Phys. Rev. A 2010, 81, 033814. [Google Scholar]
- LaBoda C.; Duschl H.; Dwyer C. L. DNA-enabled integrated molecular systems for computation and sensing. Acc. Chem. Res. 2014, 47, 1816–1824. [DOI] [PubMed] [Google Scholar]
- Mottaghi M. D.; Rallapalli A.; Dwyer C. RETLab: A fast design automation framework for arbitrary RET networks. Proc. DATE 2014, 1–6. [Google Scholar]
- Buckhout-White S.; Claussen J. C.; Melinger J. S.; Dunningham Z.; Ancona M. G.; Goldman E. R.; Medintz I. L. A triangular three-way dye DNA switch capable of reconfigurable molecular logic. RSC Adv. 2014, 4, 48860–48871. [Google Scholar]
- Buckhout-White S.; Spillmann C. M.; Algar W. R.; Khachatrian A.; Melinger J. S.; Goldman E. R.; Ancona M. G.; Medintz I. L. Assembling programmable FRET-based photonic networks using designer DNA scaffolds. Nat. Commun. 2014, 5, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T.; Ogura Y.; Tanida J. A nanoscale set-reset flip-flop in fluorescence resonance energy transfer-based circuits. Appl. Phys. Express 2013, 6, 015201. [Google Scholar]
- Genot A. J.; Bath J.; Turberfield A. J. Reversible logic circuits made of DNA. J. Am. Chem. Soc. 2011, 133, 20080–20083. [DOI] [PubMed] [Google Scholar]
- Pei H.; Liang L.; Yao G.; Li J.; Huang Q.; Fan C. Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew. Chem., Int. Ed. 2012, 51, 9020–9024. [DOI] [PubMed] [Google Scholar]
- Bui H.; Onodera C.; Tan Y.; Graugnard E.; Kuang W.; Lee J.; Knowlton W. B.; Yurke B.; Hughes W. B. Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 2010, 10, 3367–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Luo D.; Niu Y.; Wang Y.; Cui G. Fabrication of logic circuits based on DNA origami. J. Comput. Theor. Nanosci. 2012, 9, 1680–1685. [Google Scholar]
- Simmel F. C.; Yurke B. A DNA-based device switchable between three distinct mechanical states. Appl. Phys. Lett. 2002, 80, 883–885. [Google Scholar]
- Saghatelian A.; Völcker N. H.; Guckian K. M.; Lin V. S.; Ghadiri M. R. DNA-based photonic logic gates: AND, NAND, and INHIBIT. J. Am. Chem. Soc. 2003, 125, 346–347. [DOI] [PubMed] [Google Scholar]
- Okamoto A.; Tanaka K.; Saito I. DNA logic gates. J. Am. Chem. Soc. 2004, 126, 9458–9463. [DOI] [PubMed] [Google Scholar]
- Mitchell G. P.; Mirkin C. A.; Letsinger R. L. Programmed assembly of DNA functionalized quantum dots. J. Am. Chem. Soc. 1999, 121, 8122–8123. [Google Scholar]
- Li H.; Park S. H.; Reif J. H.; LaBean T. H.; Yan H. DNA-templated self-assembly of protein and nanoparticle linear arrays. J. Am. Chem. Soc. 2004, 126, 418–419. [DOI] [PubMed] [Google Scholar]
- Sharma J.; Ke Y.; Lin C.; Chhabra R.; Wang Q.; Nangreave J.; Liu Y.; Yan H. DNA-tile-directed self-assembly of quantum dots into two-dimensional nanopatterns. Angew. Chem., Int. Ed. 2008, 47, 5157–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund P. W. K.; Andersen E. S. Nanotechnology: The importance of being modular. Nat. Nanotechnol. 2012, 485, 584–585. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Wei B.; Yuan Y.; Mi Y. UNIQUIMER 3D, a software system for structural DNA nanotechnology design, analysis and evaluation. Nucleic Acids Res. 2009, 37, 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadeh J. N.; Steenberg C. D.; Bois J. S.; Wolfe B. R.; Pierce M. B.; Khan A. R.; Dirks R. M.; Pierce N. A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [DOI] [PubMed] [Google Scholar]
- Lewis D. F.; Zhang L.; Zuo X. DNA as a helical ruler: Exciton-coupled circular dichroism in DNA conjugates. J. Am. Chem. Soc. 2005, 127, 10002–10003. [DOI] [PubMed] [Google Scholar]
- Khan Y. R.; Dykstra T. E.; Scholes G. D. Exploring the Förster limit in a small FRET pair. Chem. Phys. Lett. 2008, 461, 305–309. [Google Scholar]
- Kato T.; Kashida H.; Kishida H.; Yada H.; Okamoto H.; Asanuma H. Development of a robust model system of FRET using base surrogates tethering fluorophores for strict control of their position and orientation within DNA duplex. J. Am. Chem. Soc. 2013, 135, 741–750. [DOI] [PubMed] [Google Scholar]
- Van der Meer B. W. Kappa-squared: From nuisance to new sense. Rev. Mol. Biotechnol. 2002, 82, 181–196. [DOI] [PubMed] [Google Scholar]
- Van der Meer B. W.FRET–Förster Resonance Energy Transfer: From theory to applications, 1st ed.; Wiley-VCH: Weinheim, 2013. [Google Scholar]
- Torimura M.; Kurata S.; Yamada K.; Yokomaku T.; Kamagata Y.; Kanagawa T.; Kurane R. Fluorescence-quenching phenomenon by photoinduced electron transfer between fluorescent dye and a nucleotide base. Anal. Sci. 2001, 17, 155–160. [DOI] [PubMed] [Google Scholar]
- Song L.; Hennink E. J.; Young T.; Tanke H. J. Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy. Biophys. J. 1995, 68, 2588–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal T.; Gascoigne R. J. Photobleaching-corrected FRET efficiency imaging of live cells. Biophys. J. 2004, 26, 3923–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.