Summary
Classification methods based on machine learning (ML) techniques are becoming widespread analysis tools in neuroimaging studies. They have the potential to enhance the diagnostic power of brain data, by assigning a predictive index, either of pathology or of treatment response, to the single subject’s acquisition. ML techniques are currently finding numerous applications in psychiatric illness, in addition to the widely studied neurodegenerative diseases.
In this review we give a comprehensive account of the use of classification techniques applied to structural magnetic resonance images in autism spectrum disorders (ASDs). Understanding of these highly heterogeneous neurodevelopmental diseases could greatly benefit from additional descriptors of pathology and predictive indices extracted directly from brain data. A perspective is also provided on the future developments necessary to translate ML methods from the field of ASD research into the clinic.
Keywords: autism spectrum disorders, brain alterations, machine learning, magnetic resonance imaging, translational research
Introduction
The autism spectrum disorders (ASDs) are a heterogeneous group of neurodevelopmental pathologies characterized by a strong genetic basis (Jeste and Geschwind, 2014) and early altered neuroanatomical correlates (Hazlett et al., 2011; Wolff et al., 2012). They are considered to be among the most common and most debilitating childhood disorders, whose features include life-long impairments in socio-communicative abilities and narrow interests/repetitive behaviors, which have an impact on most domains of cognitive and adaptive functioning. In the USA, the ASDs have an estimated prevalence of 1:68 (Centers for Disease Control and Prevention, 2014), which is 37-fold greater than that reported in studies published thirty years ago. The true increase in ASD prevalence remains, however, to be confirmed, since it has been suggested that the above prevalence is at least partly a consequence of the inclusion of milder cases in the ASD spectrum, as well as of the improved identification of ASDs (King and Bearman, 2009; Liu et al., 2010).
Although, in the past few decades, studies using structural magnetic resonance imaging (sMRI) have highlighted alterations in the neuroanatomy of ASD patients, a univocal, reliable and consistent pattern of alterations has yet to be identified. Therefore, the diagnosis of ASD currently remains based on behavioral symptoms, i.e. impairment in socio-communicative abilities, as well as on the presence of restricted and stereotyped behaviors (American Psychiatric Association, 2013). Specifically, the assessment of the child is performed by expert clinicians, and it includes the administration of a battery of psychiatric and psychological tests that involve both patient observation, e.g. the Autism Diagnostic Observation Schedule Generic by Lord et al. (2000), and caregiver interview, e.g. the Autism Diagnostic Interview-Revised by Lord et al. (1994). However, these methods could lack reliability, being prone to suffer from a parental recall bias concerning the child’s first ASD symptoms or from relatively low levels of diagnostic specificity in toddlers (Ventola et al., 2006; Chawarska et al., 2007). In particular, it can be hard not only to distinguish ASD from other developmental disabilities (e.g. intellectual disability, language disorders), but also to differentiate ASD behavioral profiles from both normal variations in development and temporary delays. Therefore, there is a pressing need to develop more objective methods of ASD detection and, ultimately, to identify quantitative diagnostic markers for the disorder (Walsh et al., 2011).
Recently, the scientific community has partially revised its opinion on ASD outcome. Researchers and clinicians are starting to regard ASD as a treatable disorder, on the basis of the finding that some patients, thanks to an early ASD diagnosis and a subsequent early and intensive behavioral intervention, may reach normal cognitive function and cease to meet the diagnostic criteria for ASD (Fein et al., 2013). Consequently, research into the early identification of ASDs represents a public health priority. Moreover, an early ASD diagnosis and early intervention could have a positive economic impact, making it possible to reduce the lifetime cost of caring for a person with ASD. Without early intervention, this cost is estimated to stand at $3.2 million (Ganz, 2006).
This review sets out to highlight the role of sMRI in clinical and research applications, focusing in particular on novel analysis methodologies based on machine learning (ML) techniques that have the potential to impact greatly on ASD early diagnosis, treatment monitoring and stratification of subjects for tailored intervention (Ecker and Murphy, 2014). A dedicated effort is still necessary to translate these research instruments into clinically useful biomarkers able to predict autism risk prior to the onset of symptoms and even response to treatment.
The role of sMRI in ASDs: clinical and research applications
Given its non-invasive nature, MRI could be used in clinical practice as part of the comprehensive medical assessment of ASD patients, in order to exclude brain alterations. However, this role of sMRI is a subject of debate. While the guidelines of the American Academy of Neurology and Child Neurology Society (Filipek et al., 2000) judged MRI scanning to be a procedure that is not recommended in the standard clinical evaluation of ASD, more recently some authors have highlighted its possible utility for revealing brain abnormalities in these patients (Boddaert et al., 2009; Erbetta et al., 2013). Moreover, quantitative analyses based on sMRI have indicated the existence of widespread brain abnormalities in ASDs, reporting, for example, differences in total brain, frontoparietotemporal cortex, corpus callosum and cerebellar volume between ASD patients and controls (for a review see Chen et al., 2011). In particular, sMRI studies in the first years of life of ASD subjects have identified an abnormal pattern of brain growth (Courchesne et al., 2001; Sparks et al., 2002), characterized by accelerated postnatal growth followed by a progressive attenuation of the difference between patients and controls with increasing age. While some researchers argue that abnormal brain enlargement is mainly explained by an excessive increase in white matter (WM) (Courchesne et al., 2001; Herbert et al., 2003), others think that the gray matter (GM) is involved, alone or in association with WM involvement (Palmen et al., 2005; Schumann et al., 2010). As regards the regions involved in ASD brain enlargement, converging findings suggest an anteroposterior gradient of severity, with the most consistent increase reported in the frontotemporal regions (Carper et al., 2002; Schumann et al., 2010).
Structural MRI applied to ASD diagnosis: machine learning implementations
In the last few years several post-processing methods for application to MRI brain data have been developed and implemented to obtain diagnostic models for ASD with the aim of improving the behavioral assessment of the disorder.
Traditional neuroimaging analysis approaches were designed mainly to reveal brain abnormalities by statistically comparing data from two groups of subjects (usually affected subjects and a control group) either matched or controlled for possible confounding variables (e.g. age, gender, head size). The most widely used methods include manually or automatically traced regions of interest (ROIs) analysis, where, in general, a prior hypothesis about the involvement of specific ROIs is tested through a statistical comparison between the two groups of subjects. Manual ROI tracing requires the intervention of experts in brain anatomy and raises the issues of inter-rater and intra-rater reliability. Automated tools can help in ROI tracing, even though they often demand computationally intensive sMRI data pre-processing to co-register and harmonize brain images of different subjects.
The availability of powerful computing resources has allowed the implementation of whole-brain methods, e.g. those involving the investigation of cortical thickness in multiple ROIs into which, through an automated procedure, the brain is parcellated. The most widely used method for deriving the brain surface (Dale et al., 1999) and extracting the ROI values of certain features (e.g. cortical thickness average and standard deviation) is implemented in the FreeSurfer analysis suite1 (Fischl, 2012).
Whole-brain investigation at the voxel level can also be performed with voxel-based morphometry (VBM) approaches (Ashburner and Friston, 2000). In a computationally demanding pre-processing step the sMRI of each subject is segmented into the GM, WM and cerebrospinal fluid components; then, all the subjects’ images are co-registered to a common template image, for example through the iterative DARTEL algorithm developed by Ashburner (2007); finally, the GM (or WM) local concentrations or volumes are statistically compared between the two groups of subjects at each single voxel location (mass univariate statistics). Two of the most widely used software tools for performing VBM analysis are SPM2 (Wellcome Trust Centre for Neuroimaging) and FSL3 (Jenkinson et al., 2012).
All these analysis techniques, in addition to their undoubted ability to assess brain involvement in a pathological condition at group level, are also able to provide information at single subject level (e.g. ROI- or voxel-wise tissue characterization and computation of volumetric changes). However, these kinds of analyses have so far had a limited impact on clinical practice, which would benefit mostly from methods specifically developed to allow the drawing of inferences on the outcome of each individual (Orrù et al., 2012).
More recently, ML techniques, e.g. those based on support vector machines (SVMs) (Vapnik., 1995), have been shown to be valuable instruments both for conducting group studies in a multivariate fashion and for making predictive diagnoses in single subjects. For a large variety of diseases, two-class classification problems, either for diagnosis prediction or for transition and treatment prognosis, have been solved with ML techniques.
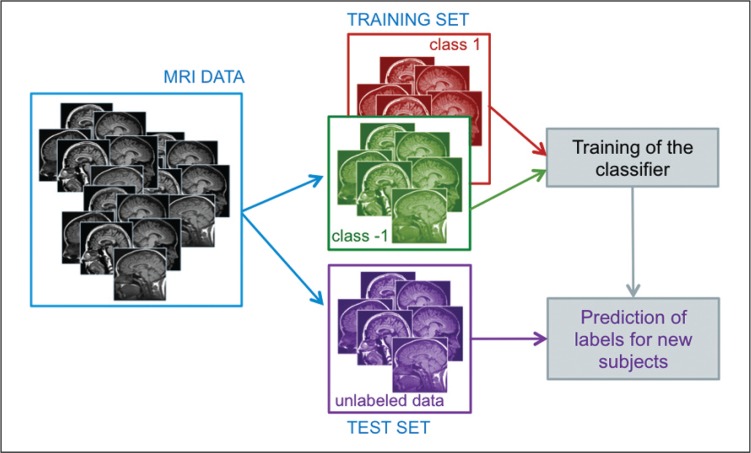
Machine learning refers to all procedures in which the learning by example paradigm is implemented. They require a training set, from which to learn the differences between the groups of subjects (e.g. patients and controls), and a validation set in order to make an unbiased evaluation of the classifier performance on previously unseen data, i.e. data not entered in the training phase (Fig. 1).
Figure 1.
Machine learning approach: scheme of the training and validation phases of a classifier that uses subject’s information extracted from structural MRI data during the classifier training to predict the class membership of cases not involved in the training procedure.
These techniques can be applied to analyze image features extracted either at the voxel or at the ROI level. In neuroimaging studies the image features can generally be reported in a N × P matrix of numbers, where N is the number of subjects (typically less than 100) and P the total number of available neuroimaging measures for each subject (typically about either 106 voxel features or 103 ROI features). Since, in general, N« P, particular caution should be applied to the classifier training phase and to the checks necessary to establish its generalization ability. Among the ML techniques, the SVMs have shown themselves to be particularly suitable for handling high dimensional patterns (large P) in the presence of a limited number of cases (small N).
The vector of features x extracted from the sMRI of each subject i can be expressed as {xi1, …, xiP} ∈ ℝN×P, where i=1, … N. In the simplest case, where there are only two different classes, a classifier can be formalized as a decision function f : ℝN×P → {−1, +1}, which assigns an observation xi to one of the classes denoted with the +1 and −1 labels, respectively. The decision function f, chosen among a class of parameterized functions, has to be learned from data during the training phase. If the class of linear functions is chosen, as in linear-kernel SVM applications, the decision function f corresponds to a separating hyperplane parameterized in terms of its normal vector w and a bias term b. The class label l of a new subject characterized by the feature vector x is thus predicted as l = f(x; w,b) = sgn(wT·x + b). By learning the input (x)–output (l) relationship on known examples, the classifier is trained. To find the optimal separating hyperplane, suitable loss functions have to be implemented and particular care should be taken to avoid overfitting the data (because in that case, the satisfactory performance obtained during the training phase would not be generalized in the validation phase on new data). The classifier performance is usually provided in terms of both the sensitivity (percentage of subjects with disease correctly identified, i.e. true-positive rate) and the specificity (percentage of control subjects correctly identified, i.e. true-negative rate), or of the accuracy (overall percentage of subjects correctly classified). However, by varying the classifier decision threshold, the trade-off between the sensitivity and the rate of false-positive detection can be shown as the receiver operating characteristic (ROC) curve (Metz, 2006), and the area under the ROC curve (AUC) represents a quantitative index for comparing the ROC curves of different classifiers (Hanley and McNeil, 1982). In particular, the AUC represents the accuracy of the instrument in predicting subjects who will or will not have the disease: an AUC ≥ 0.9 suggests ‘high’ diagnostic accuracy (Swets and Pickett, 1982). Moreover, to ensure an unbiased estimate of the classifier performance it is appropriate to implement a cross-validation scheme, both in the model selection phase (e.g. the estimate of free model parameters) and in the final evaluation of the classification accuracy, possibly implementing nested cross-validation loops.
The application of a linear classifier in neuroimaging studies, referring in particular to linear-kernel SVMs, has an additional advantage. The weight vector w can be represented as a map with the same dimensions as the original data (discrimination map), where the map intensity is proportional the discriminant power of each voxel (Mourão-Miranda et al., 2005). The possibility to localize the most discriminant areas of the brain in a case-control study allows the single subject predictive information provided by the SVM classifiers to be complemented with an anatomical description of the brain involvement in that particular pathology.
Most applications of ML to neuroimaging data were originally developed within the Alzheimer’s disease research community, starting with the first automated classification efforts of Fan et al. (2008) and Klöppel et al. (2008). More recently, they have been implemented in large-scale analyses of the multicenter MRI data collected within the prospective Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Weiner et al., 2012). The main contribution the ML technique can make in this field of research is to favor early diagnosis, by predicting the outcome of subjects with mild cognitive impairment (Chincarini et al., 2011; Hinrichs et al., 2011; Cho et al., 2012; Retico et al., 2014).
Even though some authors have highlighted critical aspects of ML techniques as applied to brain data (Lemm et al., 2011; Orrù et al., 2012), these methods are becoming widespread analysis instruments also in psychiatric disease investigations. Machine learning has already been applied to sMRI data of patients with schizophrenia (Castellani et al., 2012), major depression (Costafreda et al., 2009) and autism (as reported in Table I, and detailed below).
Table I.
Implementation details and classification performance achieved by MRI-based autism detection procedures.
| Study | Number of subjects | Age range (years) | Classification method | Features | Subjects in training/test set | Validation scheme | Performance |
|---|---|---|---|---|---|---|---|
| Akshoomoff et al., 2004 | 52 male ASD subjects and 15 age-and gender-matched controls | 1.7–5.2 | Discriminant function analysis | GM and WM volumes; area of the anterior and cerebellar vermis; cerebellar GM and WM volumes | All | None | Sensitivity=95.8% Specificity=92.3% |
| Neeley et al., 2007 | 33 male ASD subjects; 24 age- and gender-matched TD controls; 24 age- and gender-matched RD subjects (all high functioning) | only the average 14±6 is provided | Classification and regression tree (CART) and logistic regression | Volumes of 26 structures (either GM or WM) of the temporal lobe | 56/1 | Leave-one-out cross validation | Best performance in ASD vs TD separation (obtained in the classification of LFG-GM, RTS, RITG-GM): Sensitivity=85% Specificity=83% |
| Singh et al., 2008 | 16 ASD and 11 TD controls | not provided | Linear Programming Boosting (LPBoost) | Cortical thickness computed at each vertex of the cortica surface | 24/3 | 10 iterations of 9-fold cross validation | Accuracy ≈90% |
| Ecker et al., 2010a | 20 male ASD subjects and 20 age-, gender- and IQ-matched controls; 19 ADHD matched to the ASD group (validation set) | 20–68 | SVM (linear kernel) | Five morphometric features for each vertex of the cortical surface | 38/2 | Leave-pair-out cross validation | Best performance (obtained in classification of cortical thickness of the right hemisphere): Sensitivity=90% Specificity=90% Accuracy=90% |
| Ecker et al., 2010b | 22 male ASD subjects and 22 age- and gender matched controls (all high functioning) | 18–42 | SVM (linear kernel) | Voxel-wise GM and WM maps | 43/1 | Leave-one-out cross validation GM | Best performance obtained in classification: Sensitivity=77% Specificity=86% Accuracy=81% |
| Jiao et al., 2010 | 22 ASD subjects and 16 age- and gender-matched controls (all high functioning) | 6–15 | Four ML techniques: SVM, MLPs, FTs, LMTs | Regional cortical thickness and volumes for 66 brain structures | 90%/10% | 10 iterations of 10-fold cross validation | Best classification performance obtained by LMT on cortical thickness: AUC=93% Sensitivity=95% Specificity=75% Accuracy=87% |
| Uddin et al., 2011 | 24 ASD subjects (22 males and 2 females) and 24 age-, gender- and IQ-matched controls (all high functioning) | 8–18 | SVM (with radial basis function kernel) | Voxel-wise GM and WM maps | 90%/10% | 10-fold cross validation | Best performance obtained in classification of GM in the posterior cingulate cortex: Accuracy ≈90% |
| Calderoni et al., 2012 | 38 female ASD subjects and 38 age-, gender- and IQ-matched controls | 2–7 | SVM (linear kernel) | Voxel-wise GM maps | 74/2 | Leave-pair-out cross validation | AUC=80% |
| Sato et al., 2013 | 82 male subjects and 84 age matched controls (all high functioning) | 18–42 | support vector regression with radial basis function | Inter-regional cortical thickness correlation | 165/1 | Leave-one-out cross validation | r=0.362 (p<0.001) correlation between predicted and observed ADOS score |
Abbreviations: ADHD=attention deficit hyperactivity disorder; ASD=autism spectrum disorder; ADOS=Autism Diagnostic Observation Schedule; AUC=area under the receiver operating characteristic curve; FTs=functional trees; GM=gray matter; IQ=intelligence quotient; LFG=left fusiform gyrus; LMTs=logistic model trees; ML=machine learning; MLPs= multilayer perceptrons; RD=reading deficit; RITG=right inferior temporal gyrus; RTS=right temporal stem; SVMs=support vector machines; TD=typical development; WM=white matter.
In their pioneering work, Akshoomoff et al. (2004) performed manual tracing anatomical parcellation of six pre-selected brain regions and analyzed volume-based features to discriminate ASD toddlers from controls. Neeley et al. (2007) performed a multivariate analysis of 26 temporal lobe structures and used classification and regression tree analysis and discriminant analysis to identify structures with possible discriminatory power. They found that the volumes of the GM and WM of the left fusiform gyrus, of the GM of the right inferior temporal gyrus, and the volume of the right temporal stem discriminated ASD subjects from controls with 85% sensitivity and 83% specificity.
Surface-based morphometry methods based on cortical thickness revealed 90% classification accuracy in distinguishing ASD subjects from controls, evaluated in cross validation (Singh et al., 2008). Multiparameter classification approaches on cortical thickness measures also provided very good discrimination performances: accuracy of 87% (Jiao et al., 2010) and of up to 90% in the left hemisphere (Ecker et al., 2010a).
Among the voxel-wise volume-based classification methods, the SVM classifiers were used to discriminate ASD from control female children on the basis of the altered GM volumes, achieving an AUC of 80% and identifying a highly discriminant brain circuit involving bilaterally the superior frontal gyrus and the right temporoparietal junction (Calderoni et al., 2012). Both GM and WM have been analyzed in a study of adult subjects, obtaining a best classification accuracy of 81% when GM images only were utilized (Ecker et al., 2010b), and accuracies of up to approximately 90% when regions within the default mode network only were considered in a group of younger subjects (Uddin et al., 2011).
In the work of Sato et al. (2013), the support vector regression (SVR) method with radial basis function was applied to inter-regional cortical thickness correlations with the aim not of distinguishing ASD from control subjects, but of predicting the Autism Diagnostic Observation Schedule (ADOS) autistic symptom severity score. They obtained a correlation between predicted and observed ADOS scores of r=0.362 (p<0.001).
Limitations of existing studies
In the last few years, as reported in table 1, several studies have implemented sMRI-based predictive models in ASD patients, but none of these focused on very young ASD children. Current understanding of the cerebral alterations in ASDs is indeed based on brain imaging studies performed mostly in adolescent and adult subjects. Therefore, the distinctive brain profile of ASD when symptoms start constitutes a neglected area of research.
On the other hand, previous studies performed using ML techniques applied to sMRI in ASD patients are often found to present several limitations. First, the wide age range of previously investigated patient samples [e.g. 8–18 years in the study by Uddin et al. (2011); 20–68 years in Ecker et al. (2010a); and 18–42 years in Sato et al. (2013)] could represent a major weakness as regards the analysis of ASD patients, in whom the atypical brain development is an age-dependent effect. Second, including only high-functioning ASD patients in the study sample (e.g. Neeley et al., 2007; Jiao et al., 2010; Ecker et al., 2010b; Uddin et al., 2011; Sato et al., 2013) makes it impossible to extend the conclusions to ASD subjects with intellectual disability, who represent approximately one third of this population (Centers for Disease Control and Prevention, 2014). Third, previous studies have mainly included samples made up only of male subjects (e.g. Akshoomoff et al., 2004; Neeley et al., 2007; Ecker et al., 2010a,b), or of males and females together, but without separate gender analyses (Uddin et al., 2011). In this way, gender differences in the brain anatomy of ASD patients [see, for example, Schumann et al. (2010) and Nordahl et al. (2011)] could not be detected. A first study fully dedicated to the investigation of brain alterations in female ASD subjects was conducted by Calderoni et al. (2012). Fourth, little is currently known about the influence of behavioral and pharmacological treatments on brain structure and function in ASD patients. Comparing pre- and post-therapy data from the MRI scans of ASD subjects can help to reveal how therapy affects cerebral plasticity. Two reports dealing with this approach have recently been published (Pardini et al., 2012; Voos et al., 2013).
In view of these considerations, there is a need to perform ML studies of sMRI data in large samples of clinically well characterized ASD subjects falling within narrow age ranges (both males and females, high-functioning and low-functioning) who should be scanned longitudinally in order to highlight neuroanatomical differences, similarities and distinctive features of each ASD subgroup.
Among the algorithms for the analysis of sMRI data, the surface-based methods (e.g. Singh et al., 2008; Jiao et al., 2010; Ecker et al., 2010a; Sato et al., 2013) are not optimal instruments for ASD studies, despite their ability to investigate expected abnormalities in the cortical thickness of ASD subjects. They are unable to analyze subcortical structures (e.g. amygdala, basal ganglia), which instead constitute crucial neuroanatomical substrates of the ASD symptomatology. Machine learning studies focused on specific ROIs require an a priori hypothesis of the brain involvement in ASD, and they are particularly time consuming, especially when the ROI tracing is performed manually. By contrast, whole-brain approaches risk failing to detect subtle ASD signs due to confounding effects within the data.
To overcome the limitations that differently affect all analysis methods, a more general framework could be implemented, wherein the ML classifications of both whole brain and ROI-based features are performed at the same time, and the results of the two classifiers are then combined to enhance the classification performance. In general, there is no reason to assume that any single classification scheme might be optimal for the detection of pathological subjects. It is more likely that different methods have complementary strengths, and the availability, in the same subject, of the outputs of multiple systems will allow a direct comparison between different classification schemes (e.g. voxel-wise vs ROI-based approaches) and possibly improve the overall performance. In addition, when the combination of the two classifiers is carried out, this novel strategy would allow simultaneous microscopic-scale (voxel level) and macroscopic-scale (ROI level) analysis of the brain.
Even though this review focuses on the diagnostic power of sMRI of the brain, ML techniques can easily be extended to accommodate data acquired with different modalities, e.g. diffusion tensor imaging, functional MRI (fMRI), electroencephalography and near-infrared spectroscopy. Data fusion approaches, i.e. encoding information extracted from fractional anisotropy (FA) and mean diffusivity maps, or from FA and fMRI connectivity, in a single vector of features to be classified, have been attempted in the field of ASD research too (Ingalhalikar et al., 2011; Deshpande et al., 2013). Such within-modality approaches can obviously be extended to accommodate image features extracted by other modalities, e.g. sMRI, fMRI and non-imaging features. When dealing with large data samples and multiple data sources for each subject a powerful e-infrastructure could be useful to efficiently handle the computational effort. A publicly accessible grid-/cloud-based infrastructure has been developed within the neuGRID initiative4. It provides researchers with suitable computing resources and the software packages most widely used within the neuroimaging community (Redolfi et al., 2009; Redolfi et al., 2013). In conclusion, the combination of complete clinical information with innovative and powerful data analysis techniques would provide a deeper knowledge of the neurobiological basis of ASD.
MRI classification methods as future diagnostic biomarkers? Potential benefits and drawbacks
Machine learning methods applied to sMRI could help to improve the efficiency of an early ASD diagnosis through the identification of a peculiar pattern of brain alterations. Early ASD identification allows inclusion in intervention programs that, in turn, impact on outcome, improving socio-communicative skills, reducing mal-adaptive behaviors (Vismara and Rogers, 2010), and normalizing patterns of brain activity (Dawson et al., 2012). In fact, greater knowledge of the brain profile characteristics of very young children with ASDs could help not only in ASD early identification, but also in the implementation of tailored intervention protocols.
On the other hand, several critical aspects of MRI-based ML classification techniques must be taken into account. First of all, in order to suggest the complementary use of ML classification approaches in the ASD diagnostic process, a high accuracy (over 90%) is required. This should derive from high degrees of both sensitivity and specificity of the ML algorithms. As far as sensitivity is concerned, the high heterogeneity of ASD clinical expression necessarily hinders the capability of a single biomarker to detect a high percentage of ASD patients (Grzadzinski et al., 2013). However, despite showing a wide variation in phenotype severity and expression, ASD patients share strong and consistent commonalities, possibly underpinned by alterations in the same brain regions and thus potentially detectable using ML-based approaches. On the other hand, the degree of specificity of distinctive ASD brain patterns could suffer because of the overlap between ASD and other neuropsychiatric pathologies (e.g. intellectual disability, language disorders, attention deficit hyperactivity disorder, regulatory disorders). Even though the brain MRI examination is performed with a clinical indication in ASD subjects (to complete the assessment pathway, with the aim of excluding brain alterations), a major limitation to the wider use of MRI is the fact that it frequently needs to be performed under general anesthesia. In fact, some ASD patients, especially young children and lower functioning individuals, are not able to stay still in the scanner without anesthesia. In these cases, brain MRI acquisition becomes a more complicated, uncomfortable and not completely risk-free procedure. In order to overcome this critical aspect, a possible option is to perform MRI without sedation, during natural sleep. The “sleep MRI” method has been validated and successfully used to examine brain function in toddlers with ASD (Pierce, 2011; Redcay et al., 2007; Nordahl et al., 2008; Dinstein et al., 2011).
In addition, even though encouraging results are provided by ML-based approaches, caution is required in the interpretation and dissemination of their findings, in order not to raise false promises for a technology that is still far from able to substitute the traditional diagnostic approach (Walsh et al., 2011).
In conclusion, the application of ML classification techniques to sMRI data of ASD subjects is still in its infancy and will not soon be able to replace the tailored information provided by a multidisciplinary assessment. Several steps still have to be completed before an ML tool able to support the diagnosis of ASD, even one showing optimal performance, can make the transition from the laboratory to the clinic. However, as ML techniques are valuable tools for uncovering relationships hidden within the complexity of imaging and non-imaging data, dedicated efforts to refine and standardize the analysis strategies should allow them, in the future, to provide significant insights able to contribute to the early diagnosis of ASDs and the stratification of ASD individuals.
Acknowledgments
This work was supported by the Italian Ministry of Health and by the Tuscany Region with the grant GR-2010-2317873 and by the Istituto Nazionale di Fisica Nucleare (INFN), Italy.
Footnotes
References
- Akshoomoff N, Lord C, Lincoln AJ, et al. Outcome classification of preschool children with autism spectrum disorders using MRI brain measures. J Am Acad Child Adolesc Psychiatry. 2004;43:349–357. doi: 10.1097/00004583-200403000-00018. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5) 5th ed. Washington DC: American Psychiatric Publishing; 2013. [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Boddaert N, Zilbovicius M, Philipe A, et al. MRI findings in 77 children with non-syndromic autistic disorder. PLoS One. 2009;4:e4415. doi: 10.1371/journal.pone.0004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderoni S, Retico A, Biagi L, et al. Female children with autism spectrum disorder: an insight from mass-univariate and pattern classification analyses. Neuroimage. 2012;59:1013–1022. doi: 10.1016/j.neuroimage.2011.08.070. [DOI] [PubMed] [Google Scholar]
- Carper RA, Moses P, Tigue ZD, et al. Cerebral lobes in autism: early hyperplasia and abnormal age effects. Neuroimage. 2002;16:1038–1051. doi: 10.1006/nimg.2002.1099. [DOI] [PubMed] [Google Scholar]
- Castellani U, Rossato E, Murino V, et al. Classification of schizophrenia using feature-based morphometry. J Neural Transm. 2012;119:395–404. doi: 10.1007/s00702-011-0693-7. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Prevalence of autism spectrum disorder among children aged 8 years - Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2010. Morbidity and Mortality Weekly Report (MMWR) Surveillance Summaries. 2014;63:1–22. [PubMed] [Google Scholar]
- Chawarska K, Klin A, Paul R, et al. Autism spectrum disorder in the second year: stability and change in syndrome expression. J Child Psychol Psychiatry. 2007;48:128–138. doi: 10.1111/j.1469-7610.2006.01685.x. [DOI] [PubMed] [Google Scholar]
- Chen R, Jiao Y, Herskovits EH. Structural MRI in autism spectrum disorder. Pediatr Res. 2011;69:63R–68R. doi: 10.1203/PDR.0b013e318212c2b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chincarini A, Bosco P, Calvini P, et al. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. Neuroimage. 2011;58:469–480. doi: 10.1016/j.neuroimage.2011.05.083. [DOI] [PubMed] [Google Scholar]
- Cho Y, Seong JK, Jeong Y, et al. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage. 2012;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costafreda SG, Chu C, Ashburner J, et al. Prognostic and diagnostic potential of the structural neuroanatomy of depression. PLoS One. 2009;4:e6353. doi: 10.1371/journal.pone.0006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Karns CM, Davis HR, et al. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57:245–254. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dawson G, Jones EJ, Merkle K, et al. Early behavioral intervention is associated with normalized brain activity in young children with autism. J Am Acad Child Adolesc Psychiatry. 2012;51:1150–1159. doi: 10.1016/j.jaac.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Libero LE, Sreenivasan KR, et al. Identification of neural connectivity signatures of autism using machine learning. Front Hum Neurosci. 2013;7:670. doi: 10.3389/fnhum.2013.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinstein I, Pierce K, Eyler L, et al. Disrupted neural synchronization in toddlers with autism. Neuron. 2011;70:1218–1225. doi: 10.1016/j.neuron.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Murphy D. Neuroimaging in autism-from basic science to translational research. Nat Rev Neurol. 2014;10:82–91. doi: 10.1038/nrneurol.2013.276. [DOI] [PubMed] [Google Scholar]
- Ecker C, Marquand A, Mourão-Miranda J, et al. Describing the brain in autism in five dimensions—magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J Neurosci. 2010a;30:10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Rocha-Rego V, Johnston P, et al. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. Neuroimage. 2010b;49:44–56. doi: 10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Erbetta A, Bulgheroni S, Contarino V, et al. Neuroimaging findings in 41 low-functioning children with autism spectrum disorder: a single-center experience. J Child Neurol. 2013 doi: 10.1177/0883073813511856. [DOI] [PubMed] [Google Scholar]
- Fan Y, Batmanghelich N, Clark CM, et al. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fein D, Barton M, Eigsti IM, et al. Optimal outcome in individuals with a history of autism. J Child Psychol Psychiatry. 2013;54:195–205. doi: 10.1111/jcpp.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek PA, Accardo PJ, Ashwal S, et al. Practice parameter: screening and diagnosis of autism: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Child Neurology Society. Neurology. 2000;55:468–479. doi: 10.1212/wnl.55.4.468. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz ML. The costs of autism. In: Moldin SO, Rubenstein JLR, editors. Understanding Autism: From Basic Neuroscience to Treatment. Boca Raton, FL: Taylor and Francis Group; 2006. [Google Scholar]
- Grzadzinski R, Huerta M, Lord C. DSM-5 and autism spectrum disorders (ASDs): an opportunity for identifying ASD subtypes. Mol Autism. 2013;4:12. doi: 10.1186/2040-2392-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Poe MD, Gerig G, et al. Early brain over-growth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry. 2011;68:467–476. doi: 10.1001/archgenpsychiatry.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MR, Ziegler DA, Deutsch CK, et al. Dissociations of cerebral cortex, subcortical and cerebral white matter volumes in autistic boys. Brain. 2003;126:1182–1192. doi: 10.1093/brain/awg110. [DOI] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Xu G, et al. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M, Parker D, Bloy L, et al. Diffusion based abnormality markers of pathology: toward learned diagnostic prediction of ASD. Neuroimage. 2011;57:918–927. doi: 10.1016/j.neuroimage.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, et al. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Chen R, Ke X, et al. Predictive models of autism spectrum disorder based on brain regional cortical thickness. Neuroimage. 2010;50:589–599. doi: 10.1016/j.neuroimage.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M, Bearman P. Diagnostic change and the increased prevalence of autism. Int J Epidemiol. 2009;38:1224–1234. doi: 10.1093/ije/dyp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, et al. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm S, Blankertz B, Dickhaus T, et al. Introduction to machine learning for brain imaging. Neuroimage. 2011;56:387–399. doi: 10.1016/j.neuroimage.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Liu KY, King M, Bearman PS. Social influence and the autism epidemic. AJS. 2010;115:1387–1434. doi: 10.1086/651448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Metz CE. Receiver operating characteristic analysis: a tool for the quantitative evaluation of observer performance and imaging systems. J Am Coll Radiol. 2006;3:413–422. doi: 10.1016/j.jacr.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Bokde AL, Born C, et al. Classifying brain states and determining the discriminating activation patterns: Support Vector Machine on functional MRI data. Neuroimage. 2005;28:980–995. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Neeley ES, Bigler ED, Krasny L, et al. Quantitative temporal lobe differences: autism distinguished from controls using classification and regression tree analysis. Brain Dev. 2007;29:389–399. doi: 10.1016/j.braindev.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordahl CW, Lange N, Li DD, et al. Brain enlargement is associated with regression in preschool-age boys with autism spectrum disorders. Proc Natl Acad Sci U S A. 2011;108:20195–20200. doi: 10.1073/pnas.1107560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordahl CW, Simon TJ, Zierhut C, et al. Brief report: methods for acquiring structural MRI data in very young children with autism without the use of sedation. J Autism Dev Disord. 2008;38:1581–1590. doi: 10.1007/s10803-007-0514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrù G, Pettersson-Yeo W, Marquand AF, et al. Using Support Vector Machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci Biobehav Rev. 2012;36:1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Palmen SJ, Hulshoff Pol HE, Kemner C, et al. Increased gray-matter volume in medication-naive high-functioning children with autism spectrum disorder. Psychol Med. 2005;35:561–570. doi: 10.1017/s0033291704003496. [DOI] [PubMed] [Google Scholar]
- Pardini M, Elia M, Garaci FG, et al. Long-term cognitive and behavioral therapies, combined with augmentative communication, are related to uncinate fasciculus integrity in autism. J Autism Dev Disord. 2012;42:585–592. doi: 10.1007/s10803-011-1281-2. [DOI] [PubMed] [Google Scholar]
- Pierce K. Early functional brain development in autism and the promise of sleep fMRI. Brain Res. 2011;1380:162–174. doi: 10.1016/j.brainres.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redcay E, Kennedy DP, Courchesne E. fMRI during natural sleep as a method to study brain function during early childhood. Neuroimage. 2007;38:696–707. doi: 10.1016/j.neuroimage.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Redolfi A, Bosco P, Manset D, et al. Brain investigation and brain conceptualization. Funct Neurol. 2013;28:175–190. [PMC free article] [PubMed] [Google Scholar]
- Redolfi A, Mc Clatchey R, Anjum A, et al. Grid infrastructures for computational neuroscience: the neuGRID example. Future Neurology. 2009;4:703–722. [Google Scholar]
- Retico A, Bosco P, Cerello P, et al. Predictive models based on Support Vector Machines: whole-brain versus regional analysis of structural MRI in the Alzheimer’s disease. J Neuroimaging. 2014 doi: 10.1111/jon.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JR, Hoexter MQ, Oliveira PP, Jr, et al. Inter-regional cortical thickness correlations are associated with autistic symptoms: a machine-learning approach. J Psychiatr Res. 2013;47:453–459. doi: 10.1016/j.jpsychires.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Schumann CM, Bloss CS, Barnes CC, et al. Longitudinal magnetic resonance imaging study of cortical development through early childhood in autism. J Neurosci. 2010;30:4419–4427. doi: 10.1523/JNEUROSCI.5714-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Mukherjee L, Chung MK. Cortical surface thickness as a classifier: boosting for autism classification. Med Image Comput Comput Assist Interv. 2008;11:999–1007. doi: 10.1007/978-3-540-85988-8_119. [DOI] [PubMed] [Google Scholar]
- Sparks BF, Friedman SD, Friedman SD, et al. Brain structural abnormalities in young children with autism spectrum disorder. Neurology. 2002;59:184–192. doi: 10.1212/wnl.59.2.184. [DOI] [PubMed] [Google Scholar]
- Swets JA, Pickett RM. Evaluation of Diagnostic systems: Methods from Signal Detection Theory. New York: Academic Press; 1982. [Google Scholar]
- Uddin LQ, Menon V, Young CB, et al. Multivariate searchlight classification of structural magnetic resonance imaging in children and adolescents with autism. Biol Psychiatry. 2011;70:833–841. doi: 10.1016/j.biopsych.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik V. The Nature of Statistical Learning Theory. Berlin: Springer-Verlag; 1995. [Google Scholar]
- Ventola PE, Kleinman J, Pandey J, et al. Agreement among four diagnostic instruments for autism spectrum disorders in toddlers. J Autism Dev Disord. 2006;36:839–847. doi: 10.1007/s10803-006-0128-8. [DOI] [PubMed] [Google Scholar]
- Vismara LA, Rogers SJ. Behavioral treatments in autism spectrum disorder: what do we know? Annu Rev Clin Psychol. 2010;6:447–468. doi: 10.1146/annurev.clinpsy.121208.131151. [DOI] [PubMed] [Google Scholar]
- Voos AC, Pelphrey KA, Tirrell J, et al. Neural mechanisms of improvements in social motivation after pivotal response treatment: two case studies. J Autism Dev Disord. 2013;43:1–10. doi: 10.1007/s10803-012-1683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P, Elsabbagh M, Bolton P, et al. In search of bio-markers for autism: scientific, social and ethical challenges. Nat Rev Neurosci. 2011;12:603–612. doi: 10.1038/nrn3113. [DOI] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2012;8(1 Suppl):S1–68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JJ, Gu H, Gerig G, et al. Differences in white matter fiber tract development present from 6 to 24 months in infants with autism. Am J Psychiatry. 2012;169:589–600. doi: 10.1176/appi.ajp.2011.11091447. [DOI] [PMC free article] [PubMed] [Google Scholar]