Highlights
-
•
A galactose oxidase from Fusarium sambucinum was cloned and expressed in E. coli.
-
•
It could be purified with a one step affinity chromatography step.
-
•
Biochemical characteristics of the enzyme are comparable to galactose oxidases.
Keywords: Galactose oxidase, Fusarium sambucinum, Gene cloning, Heterologous expression, Steady-state kinetics, Alternative electron acceptors
Abstract
A gene encoding a galactose oxidase (GalOx) was isolated from Fusarium sambucinum cultures and overexpressed in Escherichia coli yielding 4.4 mg enzyme per L of growth culture with a specific activity of 159 U mg−1. By adding a C-terminal His-tag the enzyme could be easily purified with a single affinity chromatography step with high recovery rate (90%). The enzyme showed a single band on SDS–PAGE with an apparent molecular mass of 68.5 kDa. The pH optimum for the oxidation of galactose was in the range of pH 6–7.5. Optimum temperature for the enzyme activity was 35 °C, with a half-life of 11.2 min, 5.3 min, and 2.7 min for incubation at 40 °C, 50 °C, and 60 °C, respectively. From all tested substrates, the highest relative activity was found for 1-methyl-β-galactopyranoside (226 U mg−1) and the highest catalytic efficiency (kcat/Km) for melibiose (2700 mM−1 s−1). The enzyme was highly specific for molecular oxygen as an electron acceptor, and showed no appreciable activity with a range of alternative acceptors investigated. Different chemicals were tested for their effect on GalOx activity. The activity was significantly reduced by EDTA, NaN3, and KCN.
Introduction
Galactose oxidase (GalOx1; d-galactose:oxygen 6-oxidoreductase, EC 1.1.3.9) is a monomeric 68-kDa enzyme that contains a single copper ion [1] and an amino acid-derived cofactor [2,3], formed by cross-linking of a Cys and a Tyr residue in the direct vicinity of the copper [4–6]. The thioether bond of the Tyr-Cys cross-link is post-translationally generated [4,7] and has been shown to affect the stability, the reduction potential [8] and the catalytic efficiency of the enzyme [9,10]. It has been classified as a member of the carbohydrate active-enzyme family AA5, subfamily 2 [11]. GalOx catalyzes the two-electron oxidation [3,12] of the C6-hydroxyl group of nonreducing d-galactose residues [13] as well as a range of primary alcohols to the corresponding aldehydes with concomitant reduction of oxygen to hydrogen peroxide [14–17]. During catalysis both the metal ion and the cysteine-modified tyrosine group undergo 1-electron redox interconversions [18]. Despite a wide substrate specificity, GalOx is strictly regioselective and no secondary alcohols are oxidized [19]. However, the enzyme accepts a wide variety of primary alcohols such as benzyl alcohol [20], and glycerol [21] as reducing substrates. GalOx displays remarkable stereospecificity in its reaction with sugars [22], being highly sensitive for the orientation of the C4-OH group, and hence it shows activity with galactose but not with glucose. Because of this specificity, various analytical techniques are based on GalOx, such as the determination of lactose in milk and dairy products [23] or the histochemical examination of mucus-secreting cells [24]. Furthermore, GalOx has been used in biosensors for the measurement of galactose and its derivatives in biological fluids [25], to label galactose residues in glycoconjugates [26], and for the induction of interferon in human lymphocyte culture [27,28]. GalOx is viewed as a competitive and cost-effective catalyst compared to chemical conversion for the manufacturing of fine chemicals for pharmaceutical purposes or in food industry, for example GalOx was used for conversion of sugars like d-galactose to food-grade cross-linking agents [29–32]. Another important application for GalOx is the modification of cell surface carbohydrates and has been used in cell labeling studies and histochemical staining [19]. GalOx is interesting for the use in industrial processes such as derivatization of guar gum and related polymers as well [33,34].
The enzyme is secreted by a number of fungal species, of which Fusarium graminearum (formerly classified as Dactylium dendroides) is the most extensively studied [35–43]. The production and purification of GalOx has been reported from its natural fungal source [26,39,44–49], furthermore, various GalOx genes were cloned and successfully expressed in the filamentous fungi Aspergillus nidulans [50,51], Aspergillus oryzae and Fusarium venenatum [52], which have no endogenous GalOx, in the methylotrophic yeast Pichia pastoris [4,10,33,36,53–56] and in the bacterium Escherichia coli [55,57–59]. Typically, wild-type fungal GalOx is produced as a preproform carrying an N-terminal signal sequence, which is removed upon secretion, yielding the immature proform. The prosequence in this form was suggested to function as an intramolecular chaperone supporting copper binding and cofactor formation [42,50]. The maturation of GalOx requires several successive steps including cleavage of the signal sequence, which directs translocation, metal binding and cofactor processing [12,43]. Subsequently, the prosequence is removed and the Tyr-Cys cofactor is formed by self-processing reactions [2,7].
In the present paper we describe cloning and recombinant expression of a new gao gene without its prepro sequence from Fusarium sambucinum in E. coli. Furthermore, the purification and biochemical characterization of the enzyme are reported. Alternative electron acceptors, and possible activators as well as inhibitors were tested for their effect on GalOx activity.
Materials and methods
Chemicals, strains and vectors
Chemicals for enzyme assays, buffers and media were purchased from Sigma–Aldrich (Steinheim, Germany) and were of the highest purity available. 2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) was purchased from Amresco (Solon, OH, USA). Restriction enzymes, dNTP mix, Rapid DNA Ligation Kit and standard for Agarose gelelectrophoresis (GeneRuler DNA Ladder Mix) were from Fermentas (Vilnius, Lithuania) and the Phusion polymerase was from New England BioLabs (Ipswich, UK). Synthetic oligonucleotides were synthesized by VBC-Biotech (Vienna, Austria). E. coli strain BL21 (DE3) was purchased from Invitrogen (Carlsbad, CA, USA), the cloning vector pJET 1.2 was from Fermentas and the expression vector pET21a was from Novagen (Madison, WI, USA). The HisPrep FF 16/10 column was from GE Healthcare Bioscience AB (Uppsala, Sweden). SDS–PAGE protein standard (Precision Plus Protein prestained standard) was from BioRad (Herts, UK). The electron acceptors ferrocenium (FcPF6), guaiacol, 2,6-dimethoxyphenol, caffeic acid, p-coumaric acid, ferulic acid, sinapic acid, Thioflavin T, 2-(4′-methylaminophenyl)benzothiazole, 1,1′-diethyl-2,2′-carbocyanine iodide, 1,4-benzoquinone, 2,6-dichloro-indophenol, and ferricyanide were purchased from Sigma–Aldrich. F. sambucinum (synonym Gibberella pulicaris) strain MA1886 was kindly provided by Gerhard Adam (Department of Applied Genetics and Cell Biology, BOKU Vienna, Austria).
Isolation and cloning of the GalOx gene
F. sambucinum MA1886 was cultivated in 50 mL Sabouraud medium (5 g L−1 peptone from casein, 5 g L−1 peptone from meat, 10 g L−1 glucose, 10 g L−1 maltose, 5 g L−1 yeast extract) in shaken flasks at 25 °C and 110 rpm for 3 days. Fungal mycelia were collected by centrifugation at 4 °C and 5000×g for 15 min and the pellet was washed in 50 mL saline solution (5 g L−1 NaCl, 0.12 g L−1 MgSO4·7H2O). Genomic DNA was isolated from 100 mg of frozen mycelia ground in liquid nitrogen by the phenol–chloroform-extraction as described by Chomczynski and Sacchi [60]. The gao gene coding for GalOx was amplified by PCR using degenerated primers based on the published sequences from related organisms (Accession Number: FGSG_11032.3/M86819/FOXG_09956.2/FVEG_08555.3): 5′-GCCTCAGCA/TCCC/TA/CTCGG-3′ and 5′-CTGAGTAACGA/CGAAG/TA/CGT-3′, purified by agarose gel electrophoreses and subcloned into the pJET 1.2 cloning vector using the CloneJET PCR Cloning Kit (Fermentas). Restriction sites were introduced using the following forward primers: 5′-TCGCACATATGTACCTTTTGTCACTCGCTC-3′ and 5′-GCTGACATATGGCCTCAGCACCCATTGGA-3′ for gao with and without the prepro sequence, respectively, and 5′-GCTACGCGGCCGCCTGAGTAACGCGAAT-3′ as the reverse primer (restriction sites underlined). Subsequent, the PCR product was digested with NdeI and NotI and cloned in the equally treated expression vector pET21a in frame with the C-terminal His6-tag by the Rapid DNA Ligation Kit. The resulting plasmid was transformed into E. coli BL21 (DE3) by electroporation. DNA sequencing was performed as a commercial service (LGC Genomics; Berlin, Germany). The amino acid sequence derived from the GalOx gene was used to generate a three-dimensional model based on the published structure of GalOx from F. graminearum [5] using SWISS-MODEL [61–63].
Heterologous expression and purification
Cultivation of E. coli BL21 (DE3) for production of the recombinant enzyme was performed in 30 mL of double concentrated LB medium (20 g L−1 peptone from casein, 10 g L−1 yeast extract and 10 g L−1 NaCl) with 50 mg L−1 ampicillin in 125-mL baffled flasks. Cells were grown at 37 °C and 120 rpm until reaching an OD600 of 0.4–0.6. Then recombinant protein expression was induced by addition of 5% lactose and cultivation was continued at 25 °C and 130 rpm overnight. The cell pellet after centrifugation was resuspended in 20 mM potassium phosphate buffer pH 7.0, and an aliquot of 500 μL was homogenized by Precellys24 (PEQLAB, Erlangen, Germany). The cell homogenate was tested for the presence of GalOx activity. Large scale cultivation was done in 1-L baffled flasks containing 300 mL medium [59].
The biomass from these cultivations was harvested by centrifugation at 4000×g for 20 min and 4 °C, and resuspended in phosphate buffer (20 mM, pH7.0). After disruption in a French Press at 100 MPa the crude cell extract was separated from cell debris by centrifugation (30,000×g, 4 °C, 30 min) and used for protein purification by Immobilized Metal Affinity Chromatography (IMAC) with a 20 mL Ni-charged Sepharose 6 Fast Flow column (HisPrep FF 16/10; GE Healthcare). Before loading the sample the column was equilibrated with 10 column volumes (CV) of buffer A (20 mM KH2PO4, 1 M NaCl, 10 mM imidazole, pH 8.0). After the protein sample was applied to the column, it was washed with 3 CV of the same buffer, and eluted in a linear gradient from 0.01 to 1 M imidazole in 10 CV. Fractions containing GalOx activity were pooled and the purity of the purified GalOx was checked by electrophoresis. SDS–PAGE was performed in principle as described by Laemmli [64] using the PerfectBlue vertical electrophoresis apparatus (PEQLAB) and the Precision Plus Protein Dual Color kit as mass standard. Proteins were visualized by Coomassie brilliant blue staining.
Enzyme activity assay
Prior to activity measurement GalOx was activated by incubation with 1 mM CuSO4 for 30 min at 800 rpm and 25 °C. GalOx was measured with the chromogenic ABTS (2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)) assay [65]. The absorbance change at 420 nm (ε420 = 43.2 mM−1 cm−1) was recorded at 30 °C for 180 s. The standard assay mixture (total volume, 1 mL) contained 1 μmol of ABTS in 20 mM potassium phosphate buffer (pH 7.0), 2 U horseradish peroxidase, 100 μmol d-galactose, and a suitable amount of GalOx sample. One Unit of GalOx activity was defined as the amount of enzyme that is necessary for the oxidation of 2 μmol of ABTS per min, which equals the consumption of 1 μmol of O2 per min, under the conditions described above. Protein concentrations were determined at 595 nm by the Bradford assay [66] using the BioRad Protein Assay Kit with BSA as standard.
pH dependence of activity
A pH–activity profile was determined in the range of pH 2.5–10.0 using the buffer systems citric acid (pH 2.5–6.0), potassium phosphate (pH 6.0–8.0) and Tris (pH 8.0–10.0), each at a concentration of 50 mM. Activity measurements were performed otherwise as described for the standard assay.
Temperature optimum and thermal stability
Determination of the temperature optimum of GalOx was achieved by measuring the activity with the standard assay at different temperatures in the range of 30–70 °C. Thermal stability of GalOx was determined by incubating the protein at 30, 40, 50 and 60 °C. Samples were taken at various time points, cooled on ice, and residual GalOx activity was measured using the standard ABTS assay after reactivating the enzyme by incubation with CuSO4.
Steady-state kinetic measurements
Steady-state kinetic constants were measured for GalOx for different electron donor substrates. All kinetic measurements were performed at 30 °C in 20 mM phosphate buffer (pH 7.0). Measurement of kinetic constants for various sugar substrates were done with oxygen (air saturation) and the standard ABTS assay. d-Galactose (1–500 mM), 1-methyl-β-galactopyranoside (5–200 mM), melibiose (1–250 mM), raffinose (10–250 mM), and lactose (5–250 mM) were used as substrate. Kinetic constants were calculated by nonlinear least-square regression, fitting the data to the Henri–Michaelis–Menten equation (Sigma Plot 9, Systat; Chicago, IL, USA).
Alternative electron acceptors
The enzyme reactions were carried out in a glove box (Whitley DG250, Don Whitley Scientific, Shipley, UK) and were followed spectrophotometrically using an Agilent 8453 UV–visible spectrophotometer (Agilent Technologies, Santa Clara, CA, USA) at 30 °C, and quantified at wavelengths indicated later in the manuscript (Table 3). To eliminate oxygen the glove box was evacuated and flushed with a nitrogen/hydrogen mixture (99/1) repeatedly. Residual oxygen was removed with the built-in palladium catalyst and the produced water vapor absorbed with silica gel. All buffers and reagents used were flushed with the same gas mixture.
Table 3.
Effect of various alternative electron acceptors on the activity of GalOx.
| Electron acceptor | Conc. stock (mM) | Amax (nm) | Absorption coefficient (mM−1 cm−1) | Relative activity (%) |
|---|---|---|---|---|
| O2 | Air | 420 | 43.2 | 100 |
| ABTS cation radical | 1 | 420 | 43.2 | 0 |
| Ferrocenium ion | 1 | 300 | 4.3 | 0 |
| 1,4-Benzoquinone | 10 | 290 | 2.24 | 0 |
| 2,6-Dichloro-indophenol (DCIP) | 3 | 600 | 11.8 | 0 |
| Ferricyanide | 4 | 420 | 0.98 | 0 |
| Guaiacol radical | 10 | 465 | 12.1 | 0 |
| 2,6-Dimethoxyphenol radical | 10 | 469 | 49.6 | 0 |
| Caffeic acid radical | 10 | 315 | n.d. | 0 |
| p-Coumaric acid radical | 10 | 285 | n.d. | 0 |
| Ferulic acid radical | 10 | 285 | n.d. | 0 |
| Sinapic acid radical | 10 | 305 | n.d. | 0 |
| Thioflavin T | 10 | 220, 400, 490, 950 | n.d. | 0 |
| 2-(4′-Methylaminophenyl)benzothiazole (BTA-1) | 10 | 350 | n.d. | 0 |
| 1,1′-Diethyl-2,2′-carbocyanine iodide | 10 | 420, 614 | n.d. | 0 |
As possible alternative electron acceptors for GalOx, the following compounds were tested: ferrocenium ion (FcPF6), 1,4-benzoquinone, 2,6-dichloro-indophenol (DCIP), and ferricyanide. The activity test was performed with 0.2 U GalOx as with the standard assay, but using the respective electron acceptor instead of oxygen. The reaction stoichiometry is one for the two-electron acceptors (1,4-benzoquinone and DCIP) and two for the one-electron acceptors (ferrocenium ion and ferricyanide). Furthermore, the oxidized, radical forms of the ABTS cation, the phenols guaiacol, 2,6-dimethoxyphenol (DMP), caffeic acid, p-coumaric acid, ferulic acid and sinapic acid, the benzothiazoles thioflavin T and 2-(4′-methylaminophenyl)benzothiazole (BTA-1) and the cyanine dye 1,1′-diethyl-2,2′-carbocyanine iodide were tested as electron acceptors. Due to the short lifetime of these radicals they were produced immediately before the analyses. The oxidation of the corresponding compound to the radical was performed by recombinant laccase from Botrytis aclada expressed in P. pastoris [67] in stirred quartz cuvettes containing 1 mM of the radical builder, 100 mM d-galactose and 1.5 U laccase in 20 mM potassium phosphate buffer pH 6.5 in a final volume of 2.7 mL. The reaction was followed spectrophotometrically. After the oxidation was completed, laccase was inhibited by adding 300 μL 10 mM NaF solution, oxygen was removed by flushing with nitrogen, and the reaction was started immediately by adding 0.2 U GalOx to 1 mL of the radical solution. The negative control was performed without GalOx. For a positive control ascorbate (1.25 mM) was added instead of GalOx.
Effect of various compounds on GalOx activity
The effect of various compounds on the activity of purified GalOx was evaluated by performing the enzyme assay with the addition of 5 mM of each substance (Table 4) except Tween80, which was used in a concentration of 2.5%. Before measuring the activity the enzyme sample was incubated in the assay buffer containing the tested substance and ABTS for 5 min at 30 °C and the reaction was then started by addition of d-galactose.
Table 4.
Effect of various compounds on GalOx activity. GalOx was incubated 5 min at 30 °C with the compound before measuring activity.
| Relative activity (%) | |
|---|---|
| MgCl2 | 104 |
| KCl | 109 |
| NaCl | 98 |
| NH4Cl | 98 |
| MnCl2 | 96 |
| NaF | 103 |
| KCN | <1 |
| NaN3 | <1 |
| EDTA | 43 |
| Tween80 | 110 |
Results and discussion
Isolation and heterologous expression of GalOx-encoding gene
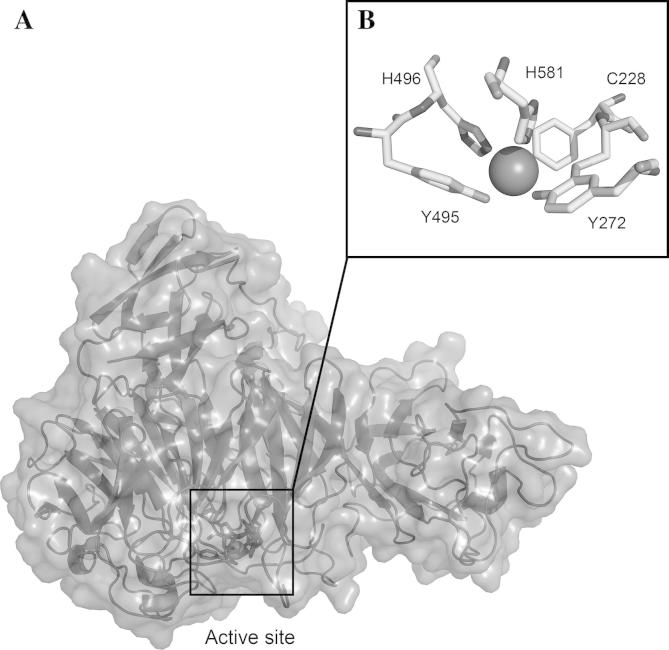
Mycelium of F. sambucinum from a culture grown in liquid medium was harvested and the genomic DNA was isolated. Degenerated primers based on published sequences were used to amplify the gao gene coding for GalOx including its signal sequence. The gene consists of an open reading frame of 2037 bp encoding a polypeptide of 679 amino acids. The sequence (GenBank accession No. KM052576) contains no introns and a 37 amino acid prepro sequence. The similarity to the protein sequences of GalOx from F. graminearum [5] and Fusarium oxysporum[68] are 96% and 81%, respectively. The amino acid sequence derived from the F. sambucinum gao gene was used to generate a three-dimensional homology model based on the published structure of mature GalOx (1gog) from F. graminearum [5] using SWISS-MODEL [61–63] (Fig. 1). The amino acids in the active site (Fig. 1B) as well as in the second shell surrounding it are completely conserved. The architecture of the substrate-binding pocket as well as the residues responsible for copper binding are also identical. The changes in amino acid sequences are mainly found on the surface of the protein.
Fig. 1.
3D structure of GalOx of F. sambucinum. (A) Overall structure showing the predominantly β-structure. (B) The active site of GalOx showing the copper ligands and the thioether cross-link. The structural model was generated by homology modeling based on the published structure of mature GalOx from F. graminearum (PDB 1gog) using SWISS_MODEL.
Based on the determined nucleotide sequence, modified oligonucleotide primers containing restriction sites for NdeI and NotI were constructed. The gene was cloned with and without its prepro sequence into the expression vector pET21a, adding a C-terminal His6 tag to the protein. After transformation of the plasmids into E. coli BL21(DE3) different clones were selected, cultivated on a small scale in double-concentrated LB medium, and 5% of lactose was used as inducer for expression of the gao gene with and without its prepro sequence. No active enzyme was found in the clones containing the full-length gao gene containing its prepro sequence. From the clones containing the gao gene without its prepro sequence the best producer was selected and used for larger scale production of GalOx in 1-L shaking flasks. Routinely, 4.4 mg L−1 of active, soluble GalOx (as calculated from a volumetric activity of 704 U L−1 and a specific activity of the homogenous enzyme of 159 U mg−1) were obtained in shaking flask cultivation after incubation at 25 °C for 16 h. This translates to a space–time yield of 0.28 mg h−1 L−1.
Enzyme purification and molecular properties
Recombinant GalOx was purified 204-fold from the crude cell extract in one single chromatographic step by Immobilized Metal Affinity Chromatography using a HisPrep column as outlined in Table 1. Even strict pooling of only the purest fractions resulted in a high yield of 91% and the final recombinant GalOx preparation had a specific activity of 159 U mg−1. The purification procedure yielded an enzyme preparation that was apparently homogenous as judged by SDS–PAGE (Fig. 2), which shows a single band. As estimated from this SDS PAGE, GalOx has an apparent molecular mass of 68.5 kDa. When compared to the calculated molecular mass of 70.3 kDa derived from the amino acid sequence, SDS–PAGE underestimates the molecular mass by 2.5%. The faster migration indicates that the thioether bond between Cys228 and Tyr272 is formed in the enzyme [7,51].
Table 1.
Purification of recombinant GalOx from F. sambucinum by immobilized metal affinity chromatography (IMAC).
| Purification step | Total protein (mg) | Total activity (U) | Specific activity (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude-extract | 210 | 165 | 0.78 | 1 | 100 |
| IMAC | 0.95 | 150 | 159 | 204 | 91 |
Fig. 2.
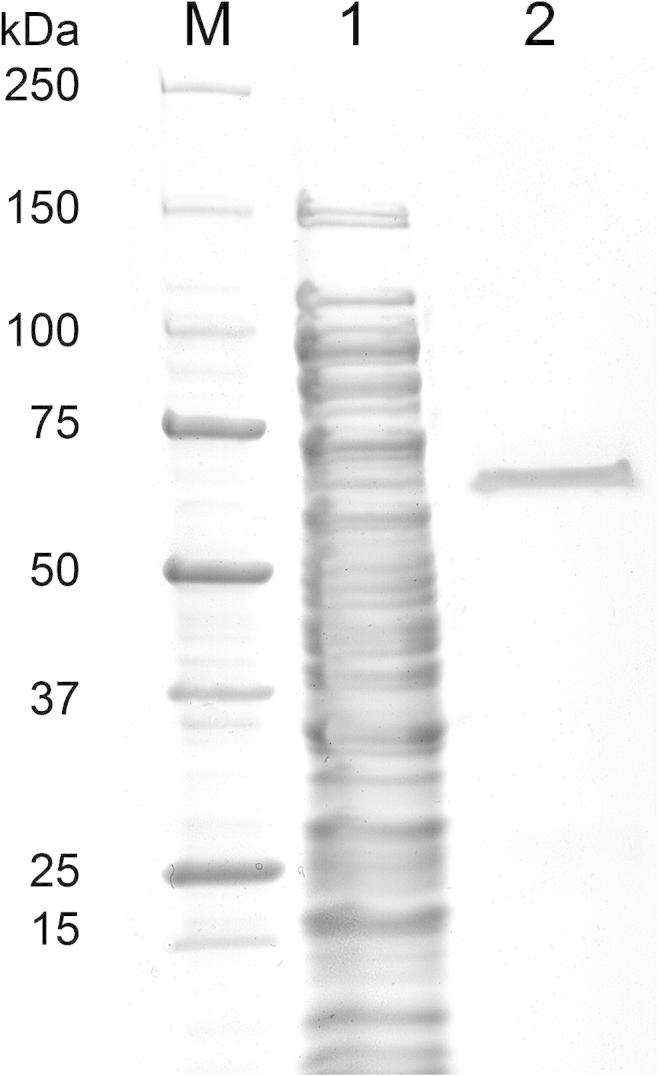
SDS–PAGE analysis of GalOx from F. sambucinum produced in E. coli. Lane M, precision plus protein standard (BioRad); lane 1, crude cell extract; lane 2, purified GalOx after IMAC.
Kinetic properties
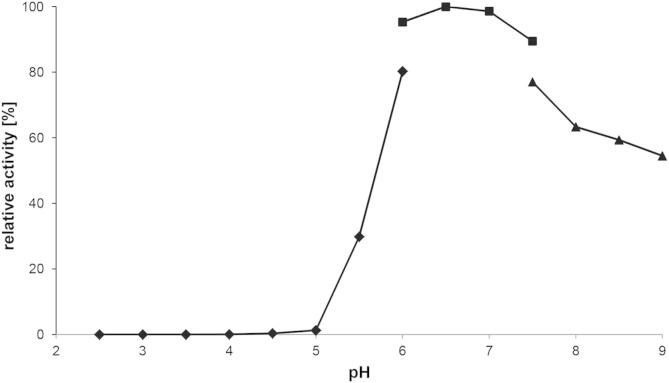
The pH and temperature optima were determined using d-galactose as electron donor and oxygen as electron acceptor. The pH-profile (Fig. 3) for recombinant GalOx is rather broad with a bell-shaped curve, showing more than 95% activity in the range of pH 6–7.5. Below pH 5 GalOx shows no activity. This is in good agreement with data reported previously for native GalOx from different sources [44,48,49].
Fig. 3.
Effect of the pH on the activity of GalOx from F. sambucinum produced in E. coli. The buffers used were 50 mM citrate (●), 50 mM phosphate (■) and 50 mM Tris (▴).
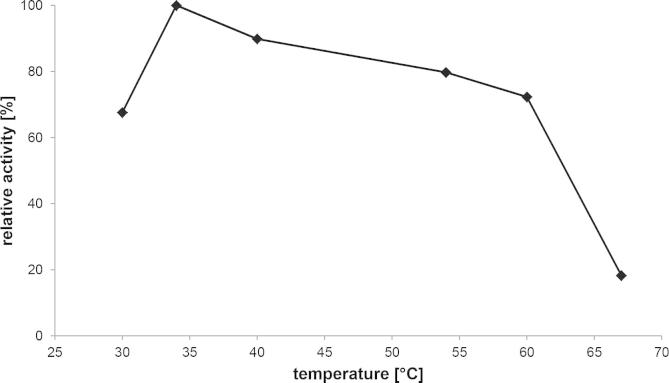
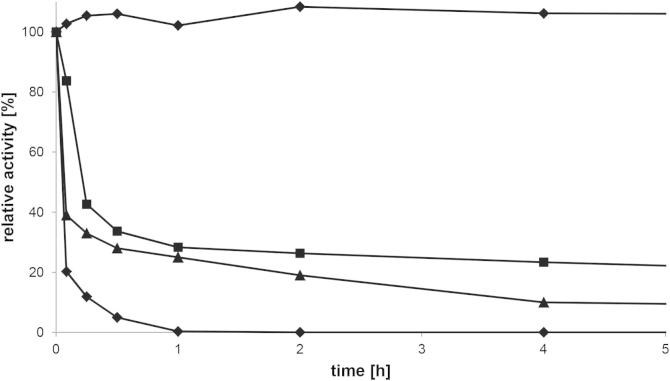
A temperature optimum of 35 °C for purified GalOx was estimated by measuring the activity at various temperatures (Fig. 4). The enzyme was active up to 70 °C during the 3-min assay. The thermal stability was determined by incubating purified GalOx at various temperatures (30 °C, 40 °C, 50 °C, and 60 °C) and measuring the residual activity of aliquots taken at the times indicated. The results (Fig. 5) show that the enzyme is stable at 30 °C for at least 24 h of incubation. The calculated half-life was 11.2 min, 5.3 min, and 2.7 min for incubation at 40 °C, 50 °C, and 60 °C, respectively. The thermostability of GalOx from F. sambucinum is therefore significantly lower than reported for GalOx from other Fusarium strains [49,57].
Fig. 4.
Temperature optimum of GalOx from F. sambucinum produced in E. coli.
Fig. 5.
Thermal stability of GalOx from F. sambucinum produced in E. coli. Pre-incubation of GalOx at 30 °C (●), 40 °C (■), 50 °C (▴) and 60 °C (♦), respectively, for various time points.
GalOx has a broad substrate specificity, which is one of the most interesting characteristics of the enzyme [19]. Steady-state kinetic constants for various substrates were determined using oxygen (air) as electron acceptor. The initial rates of substrate turnover were recorded using different substrate concentrations in the standard ABTS assay at 30 °C and pH 7.0. Kinetic data are summarized in Table 2. The enzyme showed the highest relative activity with 1-methyl-β-galactopyranoside (142% relative to galactose) and approx. 80% relative activity with melibiose, raffinose, and lactose. The highest catalytic efficiency (kcat/Km) was found for melibiose (2700 M−1 s−1) followed by raffinose (2500 M−1 s−1) whereas the corresponding value for d-galactose was 3-fold lower. The lowest catalytic efficiency was measured for lactose as a result of an unfavorably high Michaelis constant of 683 mM. The lowest measured Km value, 16 mM for melibiose, is still rather high when compared to other carbohydrate-active enzymes, which seems to be due to the broad substrate specificity for GalOx [33].
Table 2.
Apparent kinetic constants of GalOx from F. sambucinum produced in E. coli for several electron donors.
| Substrate | Vmax (μmol min−1 mg−1) | Km (mM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| d-Galactose | 47 ± 1.1 | 61 ± 4.2 | 54 | 890 |
| 1-Methyl-β-galactopyranoside | 67 ± 1.0 | 39 ± 1.7 | 77 | 2000 |
| Lactose | 38 ± 4.2 | 683 ± 95.5 | 44 | 64 |
| Melibiose | 37 ± 0.8 | 16 ± 1.5 | 42 | 2700 |
| Raffinose | 43 ± 1.2 | 20 ± 2.3 | 49 | 2500 |
It is of interest to know whether GalOx can transfer electrons to other acceptors than oxygen. To answer this question we tested a range of alternative electron acceptors, some of which are used by other copper-containing oxidoreductases. The one-electron acceptors included also different organic radicals. Due to the short life span of these radicals they were produced directly prior to their use by oxidation with laccase, which was inhibited with fluoride when the radical-forming reaction was completed. GalOx did not show significant activity with any of the tested electron acceptors (Table 3). These results are similar to those published by Aisaka et al., who tested different possible electron acceptors but also failed to find an alternative to oxygen [69]. GalOx is therefore a true oxidase without detectable dehydrogenase activity.
The effect of various compounds, mainly various metal ions, on GalOx activity was determined (Table 4). Monovalent and divalent cations like Mg2+, K+, Na+, NH4+, and Mn2+ showed no significant effect on the enzyme activity. The nonionic detergent Tween80 and fluoride had also no effect on GalOx activity. The enzyme activity was reduced to less than 50% by EDTA. This result is different to published data [44,46,49] for GalOx from Fusarium acuminatum, Gibberella fujikuroi and Polyporus circinatus, respectively, where EDTA did not inhibit activity significantly. As expected other metallo-enzyme inhibitor such as azide and cyanide completely inactivated GalOx.
Conclusions
GalOx is of interest for a number of biotechnological applications. Because of this interest, more detailed knowledge about GalOx from different sources is important since this might reveal novel or improved areas of application. The gao gene coding for GalOx from F. sambucinum can be easily expressed in the bacterial expression host E. coli even without codon optimization. A simple one step affinity purification is sufficient to purify the protein with a yield of 90%. GalOx from F. sambucinum is very well comparable in its biochemical and catalytic properties to other fungal GalOx, which is not surprising when considering the well-conserved geometry of the active site and the substrate-binding site in these enzymes. Because of its similar biochemical properties and its simple and efficient purification protocol this new enzyme could be an alternative for GalOx from other sources.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We thank Gerhard Adam (Institute of Applied Genetics and Cell Biology, University of Natural Resources and Life Sciences, Vienna, Austria) for providing the fungal strain and Roman Kittl for providing the laccase.
This project was supported by a grant from the Austrian Science Foundation (FWF project L 504-B11).
Footnotes
Abbreviations used: GalOx, galactose oxidase; IMAC, immobilized metal affinity chromatography; SDS–PAGE, one dimensional sodium dodecylsulfate polyacrylamide gel electrophoresis; ABTS, 2,2′-azinobis (3-ethylbenzthiazolinesulfonic acid); DCIP, 2,6-dichloro-indophenol; FcPF6, ferrocenium; DMP, 2,6-dimethoxyphenol; BTA-1, 2-(4′-methylaminophenyl)benzothiazole.
Appendix A. Supplementary data
GalOx_F_sambucinum_model.
References
- 1.Cleveland L., Coffman R.E., Coon P., Davis L. An investigation of the role of the copper in galactose oxidase. Biochemistry. 1975;14:1108–1115. doi: 10.1021/bi00677a003. [DOI] [PubMed] [Google Scholar]
- 2.Firbank S.J., Rogers M., Hurtado-Guerrero R., Dooley D.M., Halcrow M.A., Phillips S.E., Knowles P.F., McPherson M.J. Cofactor processing in galactose oxidase. Biochem. Soc. Trans. 2003;31:506–509. doi: 10.1042/bst10.1042/. [DOI] [PubMed] [Google Scholar]
- 3.Knowles P.F., Brown R.D., Koenig S.H., Wang S., Scott R.A., McGuirl M.A., Brown D.E., Dooley D.M. Spectroscopic studies of the active site of galactose oxidase. Inorg. Chem. 1995;34:3895–3902. [Google Scholar]
- 4.Whittaker M.M., Whittaker J.W. Cu(I)-dependent biogenesis of the galactose oxidase redox cofactor. J. Biol. Chem. 2003;278:22090–22101. doi: 10.1074/jbc.M300112200. [DOI] [PubMed] [Google Scholar]
- 5.Ito N., Phillips S.E., Stevens C., Ogel Z.B., McPherson M.J., Keen J.N., Yadav K.D., Knowles P.F. Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature. 1991;350:87–90. doi: 10.1038/350087a0. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker J.W. The radical chemistry of galactose oxidase. Arch. Biochem. Biophys. 2005;433:227–239. doi: 10.1016/j.abb.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 7.Rogers M.S., Baron A.J., McPherson M.J., Knowles P.F., Dooley D.M. Galactose oxidase pro-sequence cleavage and cofactor assembly are self-processing reactions. J. Am. Chem. Soc. 2000;122:990–991. [Google Scholar]
- 8.Itoh S., Taki M., Fukuzumi S. Active site models for galactose oxidase and related enzymes. Coord. Chem. Rev. 2000;198:3–20. [Google Scholar]
- 9.Rokhsana D., Howells A.E., Dooley D.M., Szilagyi R.K. Role of the Tyr-Cys cross-link to the active site properties of galactose oxidase. Inorg. Chem. 2012;51:3513–3524. doi: 10.1021/ic2022769. [DOI] [PubMed] [Google Scholar]
- 10.Rogers M.S., Hurtado-Guerrero R., Firbank S.J., Halcrow M.A., Dooley D.M., Phillips S.E., Knowles P.F., McPherson M.J. Cross-link formation of the cysteine 228-tyrosine 272 catalytic cofactor of galactose oxidase does not require dioxygen. Biochemistry. 2008;47:10428–10439. doi: 10.1021/bi8010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levasseur A., Drula E., Lombard V., Coutinho P.M., Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels. 2013;6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas F. Ten years of a biomimetic approach to the copper(II) radical site of galactose oxidase. Eur. J. Inorg. Chem. 2007;2007(17):2379–2404. [Google Scholar]
- 13.Kupper C.E., Rosencrantz R.R., Henßen B., Pelantová H., Thönes S., Drozdová A., Křen V., Elling L. Chemo-enzymatic modification of poly-N-acetyllactosamine (LacNAc) oligomers and N, N-diacetyllactosamine (LacDiNAc) based on galactose oxidase treatment. Beilstein J. Org. Chem. 2012;8:712–725. doi: 10.3762/bjoc.8.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verma P., Pratt R.C., Storr T., Wasinger E.C., Stack T.D. Sulfanyl stabilization of copper-bonded phenoxyls in model complexes and galactose oxidase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18600–18605. doi: 10.1073/pnas.1109931108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittaker J.W. Free radical catalysis by galactose oxidase. Chem. Rev. 2003;103:2347–2363. doi: 10.1021/cr020425z. [DOI] [PubMed] [Google Scholar]
- 16.Rokhsana D., Dooley D.M., Szilagyi R.K. Structure of the oxidized active site of galactose oxidase from realistic in silico models. J. Am. Chem. Soc. 2006;128:15550–15551. doi: 10.1021/ja062702f. [DOI] [PubMed] [Google Scholar]
- 17.Borman C.D., Saysell C.G., Sokolowski A., Twitchett M.B., Wright C., Sykes A.G. Reactivity of galactose oxidase. Coord. Chem. Rev. 1999;190–192:771–779. [Google Scholar]
- 18.Jazdzewski B.A., Tolman W.B. Understanding the copper–phenoxyl radical array in galactose oxidase: contributions from synthetic modeling studies. Coord. Chem. Rev. 2000;200–202:633–685. [Google Scholar]
- 19.Whittaker J.W. Galactose oxidase. Adv. Protein Chem. 2002;60:1–49. doi: 10.1016/s0065-3233(02)60050-6. [DOI] [PubMed] [Google Scholar]
- 20.Whittaker M.M., Whittaker J.W. Catalytic reaction profile for alcohol oxidation by galactose oxidase. Biochemistry. 2001;40:7140–7148. doi: 10.1021/bi010303l. [DOI] [PubMed] [Google Scholar]
- 21.Klibanov A.M., Alberti B.N., Marletta M.A. Stereospecific oxidation of aliphatic alcohols catalyzed by galactose oxidase. Biochem. Biophys. Res. Commun. 1982;108:804–808. doi: 10.1016/0006-291x(82)90900-7. [DOI] [PubMed] [Google Scholar]
- 22.Ito N., Phillips S.E., Yadav K.D., Knowles P.F. Crystal structure of a free radical enzyme, galactose oxidase. J. Mol. Biol. 1994;238:794–814. doi: 10.1006/jmbi.1994.1335. [DOI] [PubMed] [Google Scholar]
- 23.Adányi N., Szabó E.E., Váradi M. Multi-enzyme biosensors with amperometric detection for determination of lactose in milk and dairy products. Eur. Food Res. Technol. 1999;209:220–226. [Google Scholar]
- 24.Roberts G.P., Gupta S.K. Use of galactose oxidase in the histochemical examination of mucus-secreting cells. Nature. 1965;207:425–426. doi: 10.1038/207425a0. [DOI] [PubMed] [Google Scholar]
- 25.Kanyong P., Pemberton R.M., Jackson S.K., Hart J.P. Development of an amperometric screen-printed galactose biosensor for serum analysis. Anal. Biochem. 2013;435:114–119. doi: 10.1016/j.ab.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Mazur Adam W. Galactose oxidase, enzymes in carbohydrate synthesis. Am. Chem. Soc. 1991:99–110. [Google Scholar]
- 27.Dianzani F., Monahan T.M., Scupham A., Zucca M. Enzymatic induction of interferon production by galactose oxidase treatment of human lymphoid cells. Infect. Immun. 1979;26:879–882. doi: 10.1128/iai.26.3.879-882.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson H.M., Dianzani F., Georgiades J.A. Large-scale induction and production of human and mouse immune interferons. In: Sidney P., editor. Methods in Enzymology. Academic Press; 1981. pp. 158–162. [DOI] [PubMed] [Google Scholar]
- 29.Schoevaart R., Kieboom T. Galactose dialdehyde: the forgotten candidate for a protein cross-linker? Carbohydr. Res. 2001;334:1–6. doi: 10.1016/s0008-6215(01)00166-5. [DOI] [PubMed] [Google Scholar]
- 30.Schoevaart R., Kieboom T. Galactose dialdehyde as potential protein cross-linker: proof of principle. Carbohydr. Res. 2002;337:899–904. doi: 10.1016/s0008-6215(02)00051-4. [DOI] [PubMed] [Google Scholar]
- 31.Bonnet V., Duval R., Rabiller C. Oxidation of galactose and derivatives catalysed by galactose oxidase: structure and complete assignments of the NMR spectra of the main product. J. Mol. Catal. B Enzym. 2003;24–25:9–16. [Google Scholar]
- 32.van Wijk A., Siebum A., Schoevaart R., Kieboom T. Enzymatically oxidized lactose and derivatives thereof as potential protein cross-linkers. Carbohydr. Res. 2006;341:2921–2926. doi: 10.1016/j.carres.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson D., Akumanyi N., Hurtado-Guerrero R., Dawkes H., Knowles P.F., Phillips S.E.V., McPherson M.J. Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution. Protein Eng. Des. Sel. 2004;17:141–148. doi: 10.1093/protein/gzh018. [DOI] [PubMed] [Google Scholar]
- 34.Hall L.D., Yalpani M. A high-yielding, specific method for the chemical derivatization of d-galactose-containing polysaccharides: oxidation with d-galactose oxidase followed by reductive amination. Carbohydr. Res. 1980;81:C10–C12. [Google Scholar]
- 35.Barbosa-Tessmann I.P., da Silva D.A., Peralta R.M., Kemmelmeier C. A new species of Fusarium producer of galactose oxidase. J. Basic Microbiol. 2001;41:143–148. doi: 10.1002/1521-4028(200107)41:3/4<143::aid-jobm143>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 36.Deacon S.E., Mahmoud K., Spooner R.K., Firbank S.J., Knowles P.F., Phillips S.E., McPherson M.J. Enhanced fructose oxidase activity in a galactose oxidase variant. ChemBioChem. 2004;5:972–979. doi: 10.1002/cbic.200300810. [DOI] [PubMed] [Google Scholar]
- 37.Ögel Z.B., Brayford D., McPherson M.J. Cellulose-triggered sporulation in the galactose oxidase-producing fungus Cladobotryum (Dactylium) dendroides NRRL 2903 and its re-identification as a species of Fusarium. Mycol. Res. 1994;98:474–480. [Google Scholar]
- 38.Avigad G., Amaral D., Asensio C., Horecker B.L. The d-galactose oxidase of Polyporus circinatus. J. Biol. Chem. 1962;237:2736–2743. [PubMed] [Google Scholar]
- 39.Aisaka K., Terada O. Production of galactose oxidase by Gibberella fujikuroi. Agric. Biol. Chem. 1981;45:2311–2316. [Google Scholar]
- 40.Shatzman A.R., Kosman D.J. Regulation of galactose oxidase synthesis and secretion in Dactylium dendroides: effects of pH and culture density. J. Bacteriol. 1977;130:455–463. doi: 10.1128/jb.130.1.455-463.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koroleva O.V., Rabinovich M.L., Buglova T.T., Iaropolov A.I. Properties of Fusarium graminearum galactose oxidase. Prikl. Biokhim. Mikrobiol. 1983;19:632–637. [PubMed] [Google Scholar]
- 42.McPherson M.J., Ogel Z.B., Stevens C., Yadav K.D., Keen J.N., Knowles P.F. Galactose oxidase of Dactylium dendroides. Gene cloning and sequence analysis. J. Biol. Chem. 1992;267:8146–8152. [PubMed] [Google Scholar]
- 43.Aparecido Cordeiro F., Bertechini Faria C., Barbosa-Tessmann I. Identification of new galactose oxidase genes in Fusarium spp. J. Basic Microbiol. 2010;50:527–537. doi: 10.1002/jobm.201000078. [DOI] [PubMed] [Google Scholar]
- 44.Cooper J.A.D., Smith W., Bacila M., Medina H. Galactose oxidase from Polyporus circinatus, Fr. J. Biol. Chem. 1959;234:445–448. [PubMed] [Google Scholar]
- 45.Markus Z., Miller G., Avigad G. Effect of culture conditions on the production of d-galactose oxidase by Dactylium dendroides. Appl. Microbiol. 1965;13:686–693. doi: 10.1128/am.13.5.686-693.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aisaka K., Terada O. Purification and properties of galactose oxidase from Gibberella fujikuroi. Agric. Biol. Chem. 1982;46:1191–1197. [Google Scholar]
- 47.Whittaker M.M., Whittaker J.W. The active site of galactose oxidase. J. Biol. Chem. 1988;263:6074–6080. [PubMed] [Google Scholar]
- 48.Gasparotto E.P.L., Abrao S.C.C., Inagaki S.Y., Tessmann D.J., Kemmelmeier C., Tessmann I.P.B. Production and characterization of galactose oxidase produced by four isolates of Fusarium graminearum. Braz. Arch. Biol. Technol. 2006;49 [Google Scholar]
- 49.Alberton D., Silva de Oliveira L., Peralta R.M., Barbosa-Tessmann I.P. Production, purification, and characterization of a novel galactose oxidase from Fusarium acuminatum. J. Basic Microbiol. 2007;47:203–212. doi: 10.1002/jobm.200610290. [DOI] [PubMed] [Google Scholar]
- 50.McPherson M.J., Stevens C., Baron A.J., Ogel Z.B., Seneviratne K., Wilmot C., Ito N., Brocklebank I., Phillips S.E., Knowles P.F. Galactose oxidase: molecular analysis and mutagenesis studies. Biochem. Soc. Trans. 1993;21(Pt 3):752–756. doi: 10.1042/bst0210752. [DOI] [PubMed] [Google Scholar]
- 51.Baron A.J., Stevens C., Wilmot C., Seneviratne K.D., Blakeley V., Dooley D.M., Phillips S.E., Knowles P.F., McPherson M.J. Structure and mechanism of galactose oxidase. The free radical site. J. Biol. Chem. 1994;269:25095–25105. [PubMed] [Google Scholar]
- 52.Xu F., Golightly E., Schneider P., Berka R., Brown K., Johnstone J., Baker D., Fuglsang C., Brown S., Svendsen A., Klotz A. Expression and characterization of a recombinant Fusarium spp. galactose oxidase. Appl. Biochem. Biotechnol. 2000;88:23–32. [Google Scholar]
- 53.Whittaker M.M., Whittaker J.W. Expression of recombinant galactose oxidase by Pichia pastoris. Protein Expr. Purif. 2000;20:105–111. doi: 10.1006/prep.2000.1287. [DOI] [PubMed] [Google Scholar]
- 54.Dietzsch C., Spadiut O., Herwig C. A fast approach to determine a fed batch feeding profile for recombinant Pichia pastoris strains. Microb. Cell Fact. 2011;10:85–95. doi: 10.1186/1475-2859-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spadiut O., Olsson L., Brumer H. A comparative summary of expression systems for the recombinant production of galactose oxidase. Microb. Cell Fact. 2010;9:68–81. doi: 10.1186/1475-2859-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paukner R., Staudigl P., Choosri W., Sygmund C., Halada P., Haltrich D., Leitner C. Galactose oxidase from Fusarium oxysporum – expression in E. coli and P. pastoris and biochemical characterization. PLoS One. 2014;9:e100116. doi: 10.1371/journal.pone.0100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun L., Petrounia I.P., Yagasaki M., Bandara G., Arnold F.H. Expression and stabilization of galactose oxidase in Escherichia coli by directed evolution. Protein Eng. 2001;14 doi: 10.1093/protein/14.9.699. [DOI] [PubMed] [Google Scholar]
- 58.Deacon S.E., McPherson M.J. Enhanced expression and purification of fungal galactose oxidase in Escherichia coli and use for analysis of a saturation mutagenesis library. ChemBioChem. 2011;12:593–601. doi: 10.1002/cbic.201000634. [DOI] [PubMed] [Google Scholar]
- 59.Choosri W., Paukner R., Wührer P., Haltrich D., Leitner C. Enhanced production of recombinant galactose oxidase from Fusarium graminearum in E. coli. World J. Microbiol. Biotechnol. 2010;27:1349–1353. doi: 10.1007/s11274-010-0585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 61.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 62.Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peitsch M.C. Protein modeling by E-mail. Nat. Biotechnol. 1995;13:658–660. [Google Scholar]
- 64.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 65.Leitner C., Volc J., Haltrich D. Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor. Appl. Environ. Microbiol. 2001;67:3636–3644. doi: 10.1128/AEM.67.8.3636-3644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 67.Kittl R., Mueangtoom K., Gonaus C., Khazaneh S.T., Sygmund C., Haltrich D., Ludwig R. A chloride tolerant laccase from the plant pathogen ascomycete Botrytis aclada expressed at high levels in Pichia pastoris. J. Biotechnol. 2012;157:304–314. doi: 10.1016/j.jbiotec.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 68.Ma L.J., van der Does H.C., Borkovich K.A., Coleman J.J., Daboussi M.J., Di Pietro A., Dufresne M., Freitag M., Grabherr M., Henrissat B., Houterman P.M., Kang S., Shim W.B., Woloshuk C., Xie X., Xu J.R., Antoniw J., Baker S.E., Bluhm B.H., Breakspear A., Brown D.W., Butchko R.A., Chapman S., Coulson R., Coutinho P.M., Danchin E.G., Diener A., Gale L.R., Gardiner D.M., Goff S., Hammond-Kosack K.E., Hilburn K., Hua-Van A., Jonkers W., Kazan K., Kodira C.D., Koehrsen M., Kumar L., Lee Y.H., Li L., Manners J.M., Miranda-Saavedra D., Mukherjee M., Park G., Park J., Park S.Y., Proctor R.H., Regev A., Ruiz-Roldan M.C., Sain D., Sakthikumar S., Sykes S., Schwartz D.C., Turgeon B.G., Wapinski I., Yoder O., Young S., Zeng Q., Zhou S., Galagan J., Cuomo C.A., Kistler H.C., Rep M. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kazuo A., Takayuki U., Osamu T. Kinetic properties of galactose oxidase from Gibberella fujikuroi. Agric. Biol. Chem. 1984;48:1425–1431. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GalOx_F_sambucinum_model.