Abstract
Computational docking is a useful tool for predicting macromolecular complexes, which are often difficult to determine experimentally. Here we present the DOT2 software suite, an updated version of the DOT intermolecular docking program. DOT2 provides straightforward, automated construction of improved biophysical models based on molecular coordinates, offering checkpoints that guide the user to include critical features. DOT has been updated to run more quickly, allow flexibility in grid size and spacing, and generate a complete list of favorable candidate configu-rations. Output can be filtered by experimental data and rescored by the sum of electrostatic and atomic desolvation energies. We show that this rescoring method improves the ranking of correct complexes for a wide range of macromolecular interactions, and demonstrate that biologically relevant models are essential for biologically relevant results. The flexibility and versatility of DOT2 accommodate realistic models of complex biological systems, improving the likelihood of a successful docking outcome.
Keywords: protein-protein interactions, Fourier analysis, atomic solvation parameter, molecular recognition, Poisson-Boltzmann
Introduction
The prediction of interactions between macromolecules has long been a goal of computational chemistry. Efforts have focused primarily on protein-protein interactions, but protein-nucleic acid and protein-carbohydrate interactions are also important targets because of their role in the intrinsic processes of life. When the binding interface between two macromolecules is unknown, a comprehensive search is needed to find the native complex. Unfortunately, a complete search of all possible complexes between two large flexible macromolecules is impossible because the number of configurations is truly vast. The docking problem can be simplified by treating the individual macromolecules as rigid bodies and searching over the three translational and three rotational degrees of freedom. These searches are efficiently performed with convolution techniques in which the properties of each molecule are mapped onto grids and a very rapid translational search is performed for two molecules.1–3 One molecule is rotated, its properties remapped, and the rapid translational search is repeated. With an appropriate set of orientations, a complete, systematic search of over 100 billion configurations can be performed in a few hours.
We developed DOT3 to perform an exhaustive, rigid-body search for two macromolecules. DOT uses convolution methods to calculate the sum of the van der Waals and electrostatic energies in the intermolecular interaction. Many programs that use convolution methods have been developed,4 including Molfit,5 FTDock,6 GRAMM,7 ZDOCK,8 PIPER,9 ASPDock,10 and F2DOCK.11 In two programs, HEX12 and FRODOCK,13 the rotational search is also performed with convolution methods. One advantage of DOT is the use of Poisson-Boltzmann methods to calculate the electrostatic potential of one molecule. We implemented this detailed electrostatic energy model in DOT because of our interest in highly polar intermolecular interactions, such as those in protein-DNA and electron-transfer complexes. This solvent continuum electrostatic model takes into account dielectric, solvation, and ionic strength effects. The electrostatic energy is calculated as the set of partial atomic charges of a second molecule moving in the electrostatic potential field of the first molecule. Applications of DOT include protein-protein,14–21 protein-DNA,22–28 and protein-peptide interactions,29,30 as well as interactions among helices.31 DOT has also been used through CLUSPRO,32–46 but CLUSPRO uses only the DOT van der Waals term in the scoring.
A critical problem in macromolecular docking is effective scoring of the large number of configurations. Ideally, complexes close to the native complex would form a distinguishing cluster among the best ranked configurations. The correct complex can often be identified when coordinates from the known complex are searched, but the imperfect fit of unbound molecules presents a more difficult problem. The rigid-body docking parameters must be sufficiently loose to accommodate some conformational change so that configurations near the correct complex are not excluded. Unfortunately, molecular descriptions that allow imperfect fit often result in highly ranked incorrect configurations. Including flexibility helps this problem,47–49 but localization of likely solutions is necessary before adding flexibility is feasible, and even then it is computationally costly.
Although rigid-body docking of unbound coordinates is often insufficient to distinguish the correct interactions, combining rigid-body computational docking with experimental data can be highly effective. Likely candidates can be selected from the list of docked complexes based on known information or new experiments can be designed to verify the docking results. With this in mind, we have designed the DOT2 macromolecular docking suite for use by experts on the system under study. The DOT procedure has checkpoints that guide the user, utilities to help the user include critical features of each macromolecule, and a filtering mechanism that can be applied to many kinds of data. DOT output is constructed so that the list of favorable configurations remains connected to DOT parameters and reference coordinates, ensuring reliable reproduction of runs and generation of coordinates. The format for the output list is expandable, so that additional evaluations remain tied to the information needed to create full coordinates.
Here, we describe the design aspects of the DOT2 suite that contribute to its usability, versatility, and adaptability to a wide range of macromolecular systems. In addition, we evaluate DOT2, with its improved molecular potentials, against the Benchmark 2.0 of protein-protein complexes50 and compare our results with those from ZDOCK, which also employs a convolution-based rigid-body docking algorithm.
Methods
The DOT2 procedure has three main steps for obtaining a list of configurations between two molecules: preprocessing, docking with DOT, and evaluation. In the preprocessing step, the electrostatic and van der Waals properties of each molecule are calculated and DOT input files are generated. In the docking step, the DOT program maps these properties onto grids and then systematically translates and rotates one molecule (moving) around a stationary molecule. In the efficient translational search, one orientation of the moving molecule is centered at all grid points and interaction energies are calculated by convolving the potential field of the stationary molecule with atom-based properties of the moving molecule. The moving molecule is then rotated, its properties remapped onto the grid, and the translational search repeated. With the standard set of 54,000 orientations (about 6 spacing) applied to the moving molecule and a cubic grid of 128 Å on a side with 1 Å spacing, intermolecular energies are calculated for about 108 billion configurations. DOT outputs a list of ranked configurations and their interaction energies. In the evaluation step, configurations are scored, clustered, and examined for fit to experimental data.
Selection of coordinates and assignment of stationary and moving molecules
Before starting the DOT procedure, the user must select coordinates from the starting PDB files that best represent the biophysical state. Good starting models are crucial for a valid docking calculation and often require system-specific knowledge not present in the PDB file. For example, a metal ion may be an essential cofactor (keep) or an added heavy atom derivative (remove). Small molecules may be cofactors (keep) or derived from the buffer, potentially blocking the binding region (remove). The appropriate oligomerization state for each protein should be built. PDB coordinates often contain some incomplete side chains, which can unfavorably influence the docking calculation. Although the DOT2 suite does not contain residue-building tools, many programs are readily available for this purpose, such as Swiss PDB Viewer.51
PDB files often lack coordinates for flexible loops or N- or C-termini. If experimental evidence indicates that these regions are involved in intermolecular interactions, the user may decide to incorporate a carefully built model. Without such evidence, it is better to exclude these potentially flexible regions from the model because they may, as part of a rigid model, block binding surfaces.
Given coordinates that best describe the biological state, the two molecules must be assigned as stationary or moving based on the following criteria.
-
(1)
The most important criterion is the size of the molecules. The DOT calculation is most accurate and efficient when the larger molecule is stationary and the smaller molecule is moving. Applying a given set of orientations to the smaller molecule provides a finer search over its molecular surface than applying the same rotation set to a larger molecule. The calculation time is dependent on the grid dimensions (NlogN, where N is the total number of grid points) and is linearly dependent on the number of orientations applied to the moving molecule. Grid dimensions are calculated based on the size of the molecules (see below). Assigning the larger molecule as stationary results in a smaller grid, hence a shorter calculation time. To help the user, molecular diameters are calculated and provided in the log file during the preprocessing procedure.
-
(2)
The molecular environment may determine the choice. For example, by assigning a membrane-bound protein as the stationary molecule, the membrane region can be incorporated into the molecular description as an excluded region of low dielectric.
-
(3)
Molecular properties can influence the choice. The molecular properties of the stationary molecule are calculated once, allowing a detailed description of its electrostatic and shape properties. Both the shape potential, which is defined by volumes bounded by molecular surfaces, and the electrostatic potential, which is calculated by Poisson-Boltzmann methods, are computationally intensive. On the other hand, the shape and electrostatic properties of the moving molecule must be rapidly calculated because they are mapped onto the grid for each orientation. In DOT, the moving molecule is represented by its atomic coordinates and partial atomic charges. Therefore, the choice may depend on the need to describe one molecule in more detail than the other.
In protein-DNA systems in which a DNA fragment represents a much longer DNA substrate, the DNA is best assigned as the moving molecule in order to give a uniform charge distribution throughout the DNA.23 If the DNA is the stationary molecule, the electrostatic potential, as calculated by Poisson-Boltzmann methods, is strongly modulated around the ends of the DNA by solvent effects. For example, in a 12-bp dsDNA fragment only the central 4 base pairs show the full negative potential of a long DNA fragment.
Preprocessing: preparation of DOT input files with Prepscript
The DOT2 software suite provides a largely automated procedure, driven by the script Prepscript, that prepares DOT input files starting with two coordinate files in PDB format. Given complete amino-acid residues, nucleotides, and cofactors that are defined in the molecular libraries, Prep-script determines the grid dimensions for the specific molecular system, calculates potentials for the stationary and moving molecules, and creates the DOT parameter file. Prepscript uses the outside programs Reduce,52 which adds hydrogen atoms and corrects residue geometry, APBS,53 which calculates electrostatic potentials by Poisson-Boltzmann methods, and MSMS,54 which calculates molecular surfaces. Prepscript is a heavily commented bash (http://www.gnu.org/software/bash, Free Software Foundation, Inc.) shell script that can be customized by the user for a specific system or new application. The utility programs used by Prepscript can be run independently, further enhancing adaptability. Prepscript generates a detailed log file so that the user can check for expected molecular characteristics, such as the correct total charge. Incorporated into Prepscript are internal checks that detect incomplete molecular descriptions and provide informative error messages. A careful check is essential because flawed molecular descriptions can turn the difficult macromolecular docking problem into an impossible one.
Molecular libraries
Prepscript uses two molecular libraries. The Reduce library contains the connectivity needed to build hydrogen atoms for all standard protein residues, RNA and DNA nucleotides, and cofactors. Reduce follows the residue and atom name conventions used in the remediated PDB (http://www.rcsb.org/pdb). The format of this extensive library is transparent, allowing customization for new functional groups.
The DOT atomic charge library contains molecular radii and partial atomic charges based on the AMBER library of heavy atoms with added polar hydrogens.55 The DOT library contains standard amino acids (including charged N- and C-termini and the multiple protonation states of His and Cys), RNA and DNA nucleotides, and some common cofactors, such as heme. Additional cofactors can be added, but the user must supply reasonable atomic charges. The docking calculation is not highly sensitive to small perturbations in partial atomic charges, but having the correct total charge is important. Atomic charges can be based on similarity with functional groups already in the library, or if necessary on ab initio quantum mechanical calculations.
Grid size
The grid size is chosen to ensure that artifacts from the periodic Fourier calculation are negligible. The grid must be large enough that the moving molecule remains within the grid whenever it is close to the stationary molecule. Further, potentials should be close to zero at the grid boundaries. The grid dimensions selected by Prepscript also permit efficient calculations of the convolutions by Fast Fourier methods and of the electrostatic potential by multi-grid methods. (The multi-grid Partial Differential Equation solver used by APBS prefers that the number of grid points in each direction be a multiple of 32, e.g. 64, 96, 128, 160, 192, 224, or 256.) To select the grid dimension, Prepscript determines the maximum extents (S) of the stationary molecule in the x, y, and z directions and the maximum diameter (M) of the moving molecule. For the default cubic grid, the minimum acceptable grid dimension larger than S + 2M is used. A rectangular grid can also be created, based on the three dimensions of S. Grid spacing less than 1 Å can also be specified. A smaller grid spacing will increase the memory footprint and run time of the computation, but may improve the physical representation of the molecules.
Molecular properties may be a factor in choosing the size of the grid. For example, the stationary molecule may create a strong electrostatic potential that extends far out into solvent, as found with some electron-transfer proteins. In these cases, the user can specify larger grid dimensions in Prepscript.
Processing PDB files
Prepscript first creates the heavy (nonhydrogen) and polar hydrogen atom coordinates needed to calculate the molecular properties. Hydrogen atoms are added with the program Reduce. We chose the Reduce program because it detects and corrects problems with protein geometry and is extendable to new functional groups. Reduce determines the best orientation of the side-chain amide groups of Asn and Gln and the imidazole ring of His based on the local environment, selecting either the initial coordinates or performing a 180 rotation to best match hydrogen-bonding patterns. The local environment is also used to determine the protonation state of each His residue. Reduce also corrects internal clashes of side chains. Prepscript then removes nonpolar hydrogen atoms, leaving the polar hydrogen atoms present in the DOT charge library. Partial atomic charges and molecular radii are then assigned.
Prepscript verifies that the total charge on each molecular system is an integer, giving a specific error message if this is not true. A nonintegral charge can be due to incomplete protonation, missing side-chain atoms, or cofactors that are not defined in the Reduce or DOT charge libraries. These checks are key for helping the user to build coordinates that represent the system. The user can override the check, as we did for specific Benchmark 2.0 systems (described below). Prepscript also checks that the total molecular charge is within −20 to +20, a range that can be adjusted for highly charged molecules.
Molecular properties for the van der Waals energy term
In DOT, the van der Waals energy is proportional to the number of atoms in the moving molecule that lie within a favorable interaction layer surrounding the stationary molecule.56 The shape of the moving molecule is represented by the positions of its heavy atoms. Previously,3,23 we included polar hydrogen atoms as part of the moving molecule shape, but this overemphasized polar groups, resulting in false positives with highly polar interfaces.
The shape potential of the stationary molecule consists of of an excluded volume surrounded by a 3.0 Å favorable layer. Prepscript applies the program MSMS54 to the heavy atoms to create molecular surfaces bounding these volumes. The standard molecular surface (based on van der Waals radii and a 1.4Å probe sphere) defines the boundary of the excluded volume. A second surface, calculated using van der Waals radii expanded by 3 Å, defines the outer boundary of the favorable layer. A parity-fill algorithm then classifies grid points as being inside or outside these surfaces. This automated method replaces our initial procedure for defining these regions.23
The file containing the shape potential of the stationary molecule consists of a list of spheres, radii, and their fill values. This is a convenient and versatile format for properties that are best described by a sparse set. Each grid point in the excluded and favorable volumes is the center of a small sphere (radius 0.1 Å). Fill values for the spheres are forbidden (F, corresponding to a value of 1000) for the excluded interior of the molecule and attractive (A, corresponding to a value of 1) for grid points in the 3 Å favorable region surrounding the stationary molecule. The user can add spheres to customize the molecular system. For example, if the stationary molecule is one monomer of a dimer, the user could append large (5 Å) spheres with value 0 centered at the positions of dimer interface atoms. When DOT processes the shape potential file, the values within these spheres are mapped onto the shape potential grid, overwriting the favorable layer at the dimer interface. Grid points assigned as forbidden override other values, maintaining the excluded volume. Values within spheres can also be summed (S), allowing the value at a grid point to reflect overlapping spheres. This may be useful for emphasizing specific regions on a protein surface, such as deep pockets or regions identified experimentally as being in the molecular interface.
Molecular properties for the electrostatic energy term
The electrostatic energy term is calculated as the set of atomic point charges of the moving molecule placed in the electrostatic potential of the stationary molecule. The electrostatic potential of the stationary molecule is calculated with APBS, which uses a continuum treatment of dielectric and salt effects and solves the linearized Poisson-Boltzmann equation by finite difference methods. This approach takes into account dielectric, solvation, and ionic strength effects on the stationary molecule, but neglects these effects on the moving molecule. A protein dielectric of 3, a solvent dielectric of 80, and an ionic strength of 150 mM are typically used, but the user can adjust these parameters. The electrostatic potential file gives a value for all grid points, a format that is convenient for describing a potential that is nonzero for most of the grid.
Within DOT, the electrostatic potential is modified to be compatible with the shape potential.23,57 The lenient point-based shape potential allows moving molecule atoms to approach as close as the stationary molecule surface, but, physically, the closest possible approach is approximately 1.4 Å out from the molecular surface; a moving molecule atom inside this region can see large, unrealistic electrostatic potential values, up to 15 kcal/mol/e. To eliminate this artifact, Prepscript determines electrostatic clamping values that are then passed to DOT as parameters. To calculate electrostatic clamping values, a surface is created with MSMS54 using van der Waals radii expanded by 1.4 Å. The maximum and minimum electrostatic potentials outside this surface are then determined. These clamping values are usually within the the range of −5 to +5 kcal/mol/e. Larger values often indicate a problem with the setup of the potentials of the stationary molecule.
Create DOT Parameter File
The parameter file contains the information needed to run DOT, including the grid size, the molecular property files, the rotation set file (default is 54,000 orientation, about 6.0° spacing), the number of moving molecule atoms that can penetrate the excluded volume of the stationary molecule, electrostatic clamping values, the number of configurations to be output (default is 2,000), and the molecular coordinates needed to regenerate the configurations output by DOT. The file is an easily modified text file. For example, the user may specify a different rotational set (sets range from 64 to 232,022 orientations) or more output configurations, such as the 200,000 used for the Benchmark 2.0 systems (see below).
Docking: the DOT Core
DOT uses convolution functions to rapidly compute electrostatic and van der Waals energies for the complex. The DOT2 core has increased speed and accuracy, a smaller memory footprint, and improved merging of results. The convolution functions used within the DOT core have been previously described.3 Briefly, DOT does two convolutions, one of the electrostatic potential of the stationary molecule with the atomic point charges of the moving molecule and one of the shape potential of the stationary molecule with the positions of the heavy atoms of the moving molecule. The moving molecule is then rotated, and the two convolutions repeated for the new orientation.
DOT first maps the molecular properties onto grids. For the shape of the moving molecule, each heavy atom coordinate is mapped to the nearest grid point. The charge distribution of the moving molecule is placed onto the grid using trilinear interpolation relative to the atomic centers. The shape potential of the stationary molecule is mapped onto the grid according to the radii and fill values to give the excluded volume and the surrounding favorable layer. DOT reads the electrostatic potential file of the stationary molecule in either APBS53 or UHBD58 format and then modifies the electrostatic potential grid. First, all grid points within the excluded volume are set to zero. This modification is particularly important when atoms of the moving molecule are allowed to penetrate the excluded volume, a region where the electrostatic potential varies rapidly due to proximity to atom centers. Second, the electrostatic clamping values are applied, preventing moving molecule atoms close to the stationary molecule from seeing artificially large potentials. The application of both modifications can be controlled within the DOT parameter file. For example, a user can specify that the interior of the electrostatic grid is not zeroed or can multiply it by a scaling factor. This allows versatility if the electrostatic potential is replaced by a different property.
The most time-consuming part of the DOT calculation is the correlations, computed by the Convolution Theorem using Fast Fourier Transforms (FFTs). DOT uses Hermitian symmetric transforms to obtain a factor of two improvement in both speed and memory usage. These transforms are implemented in the highly portable and efficient open-source FFTW3 library.59 A typical DOT calculation on two medium proteins can be run in under an hour using six processors of a modern multicore machine.
Another improvement in the DOT core is reliable counting of intermolecular collisions. The number of moving molecule atoms allowed to penetrate the excluded volume of the stationary molecule (‘bumps’) is specified in the parameter file. DOT discards all solutions with more than the allowed number of bumps. We have found that allowing up to 10 bumps is sufficient to compensate for the imperfect fit of two unbound protein structures in many systems.
Previously, DOT allowed only integral grid spacing and cubic grids. DOT2 allows nonintegral grid spacing and the use of rectangular grids, which we have found convenient for molecules with one very long dimension.27 Iteration through the grids is now more efficient, improving the performance of the evaluation routine.
To achieve efficient retention of all favorable configurations, the collection of the output configurations was improved. In DOT 1.0, only the most favorable configuration at each grid point was retained as the results from all rotations were merged. This merge was very efficient but multiple favorable configurations at a specific grid point were represented only by the best ranked one. In DOT2, results are stored in a heap-based priority queue. The merge of each rotation into a master queue is still very efficient. The total time required for queue insertions is less than NlogM, where N is the number of grid points and is the number of results requested by the user. Results with a score worse than the worst of the current M results are trivially discarded, so there is no heap operation for most configurations. The speed and memory footprint of DOT2 depends little on the number of results specified. The 200,000 configurations retained in the Benchmark 2.0 runs (see below) gave a small memory footprint and good performance.
Evaluation
The output list of favorable-energy configurations from DOT is designed to facilitate analysis by keeping further evaluations tied to the original configuration in a compact format. The list, termed the E6D file, contains the DOT score, the three translations and three rotations needed to generate the position of the moving molecule relative to the stationary molecule, the individual energy components, and the ranking of each configuration. Additional evaluations, such as alternate scoring or the RMSD fit of the moving molecule to a reference position, are added to the E6D file as new user-labeled columns. Scripts in the DOT2 suite assist tasks such as sorting the E6D file by any column of data or creating PDB files for a user-specified number of the top-ranked configurations.
We have examined alternate scoring methods, focusing on the van der Waals term. The DOT electrostatic term, with the modifications described above, is well behaved, but the DOT van der Waals term, which approximates the surface area buried in the complex, does not take into account differences among atom types. For example, there is no penalty for moving polar side chains out of solvent and into a nonpolar environment. To provide a better estimate of this term, we have implemented two scoring methods to re-rank the configurations output from DOT. The first re-ranking method uses the atomic contact energies (ACE) potential developed by Zhang et al.,60 based on the approach by Miyazawa and Jernigan.61 ACE is a pairwise potential consisting of 18 protein atom types that is based on statistical analysis of atom pairing frequency observed in the interior of known protein structures. We used a cutoff distance of 6 Å, linearly scaled from 5 Å to 7 Å, to select the atom pairs across the intermolecular interface. The DOT log file lists atoms for which there are no ACE parameters, such as those of cofactors. An advantage of ACE is its computational efficiency. The ACE term lacks solvent-screened electrostatic interactions,60 so we used the sum of ACE and the DOT electrostatic energy (ACE+Elec) to re-rank the list of DOT configurations. The speed of ACE allows it to be applied to large lists from DOT, for example, the 200,000 configurations in the Benchmark 2.0 systems (see below).
The second re-ranking method uses atomic solvation parameters (ASP) to approximate desolvation energies. ASP values, based on octanol/water transfer energies per unit surface area for 10 atom types, have been optimized for protein-protein binding.62 Like ACE, ASP does not include direct electrostatic interactions, so configurations were re-ranked by the sum of ASP and the DOT electrostatic energy (ASP+Elec). In our implementation of ASP, the external, exposed surface area of each atom was determined both in the isolated molecules and in each docked complex using the program MSMS. The desolvation energy for each complex was calculated as the sum of the change in area for each atom upon going from the complex to the isolated molecule multipled by its ASP value. To explore the best estimate for the exposed surface area of each atom, we evaluated both the solvent-excluded surface (typical molecular surface) and the solvent-accessible surface (surface created by the center of a probe sphere) using probe spheres with radii of 0.8, 1.4, and 3.0 Å. These 6 area estimates were applied to the top 2,000 DOT and top 2,000 ACE+Elec configurations for all docked Benchmark 2.0 systems (see below). We found that the solvent-excluded surface created with a 1.4 Å probe, which approximates the atomic surface contacted by the surface of a water molecule, gave the best results. The ASP calculation requires creating the molecular surface for each docked complex, a computationally expensive step. In our current implementation, ASP is about 3,000-fold slower than ACE, making it feasible for thousands of configurations but not for hundreds of thousands. ACE and ASP are implemented as scripts that use a table-based lookup, so new scoring methods can easily be implemented based on these scripts.
Filtering with experimentally determined constraints
Experimental constraints that give specific information about contacts or interactions under biological conditions are an invaluable aid in filtering out false positive solutions. This information must be translated into interatomic distance constraints to be applied to the list of configurations output by DOT. Some experimental data, however, can be difficult to interpret as specific distance constraints. We developed the program Dotxyzfilter for greater versatility in defining distance constraints. With Dotxyzfilter, the user can identify configurations that satisfy a subset of a list of constraints. We have found this useful in hydrogen/deuterium exchange mass spectrometry experiments that show that a certain number of amides have become protected within a protein region, but cannot distinguish the specific amides. For example, with Dotxyzfilter, the user can search for all configurations that have at least 6 backbone nitrogen atoms within a 12-residue sequence of one molecule that lie within 7 Å of any atom of the other molecule. Input and output files for Dotxyzfilter are E6D files, allowing the effect of each constraint to be examined, multiple constraints to be combined, and constraints to be applied consecutively.
Applying DOT to Benchmark 2.0
Preprocessing, docking, and evaluation were applied to the protein-protein docking Benchmark 2.063 using the strategy shown in Figure 1. Coordinates of bound complexes and superposed, unbound proteins were downloaded from zlab.bu.edu/benchmark2. The molecule labeled ‘receptor’ was assigned as the stationary molecule and the molecule labeled ‘ligand’ was assigned as the moving molecule. All systems were run through the default mode of Prepscript, which includes the key checks for the integrity of the coordinates. In this mode, the coordinates were adjusted by Reduce to correct the orientation of His, Asn, and Gln side chains, determine the protonation state of His residues, and resolve internal steric clashes. The total charge of each molecule (allowed range of −40 to +40) and the presence of all molecular components in the Reduce and DOT charge libraries were tested. The check for a nonintegral total charge identified coordinate sets that contained incomplete side chains. If these problems occurred in either the bound or unbound coordinates (Supporting Table S1), we usually did not carry the system further. In a few cases with incomplete side chains, indicated in Table 1, we overrode the Prepscript checks to investigate the effect on the docking outcome.
Figure 1.
The strategy for docking the Benchmark 2.0 systems using the automated preparation and evaluation procedures of DOT2.
Table 1.
Application of DOT to Benchmark 2.0
| PDB | Total in DOT 200,000 (best) | DOT-> ASP+Eleca Number in top | ACE+Elec -> ASP+Elecb Number in top | ZDOCK Number in top | ||||
|---|---|---|---|---|---|---|---|---|
| 2,000 | 100 | 30 (best) | 2,000 | 100 | 30 (best) | 2,000 (best) | ||
| 1ACB_b | 379 (213) | 16 | 12 | 8 (1) | 376 | 36 | 15 (1) | |
| 1ACB_u (0 bumps) | 15 (9480) | 0 | 0 | 0 (–) | 1 | 0 | 0 (1392) | 0 (–)h |
| 1ACB_u (10 bumps) | 3 (103,429) | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 0 (–) |
| 1AHW_b | 1561 (2) | 219 | 74 | 20 ( 3) | 0 | 0 | 0 (–) | |
| 1AHW_u (0 bumps) | 898 (33) | 61 | 6 | 1 (19) | 0 | 0 | 0 (–) | CDRsc 24 (8) |
| 1AHW_u (10 bumps) | 965 (342) | 16 | 6 | 0 ( 55) | 0 | 0 | 0 (–) | |
| 1AK4_b | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | |
| 1AK4_u | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 0 (–) |
| 1AKJ_bd | 789 (1) | 169 | 9 | 0 ( 33) | 0 | 0 | 0 (–) | |
| 1AKJ_u | 853 (34) | 29 | 0 | 0 ( 178) | 0 | 0 | 0 (–) | 6 (40) |
| 1AY7_b | 2427 (1) | 400 | 89 | 27 ( 1) | 0 | 0 | 0 (–) | |
| 1AY7_u | 492 (405) | 5 | 3 | 2 ( 14) | 0 | 0 | 0 (–) | 24(111) |
| 1B6C_b | 339 (20) | 39 | 39 | 30 ( 1) | 139 | 100 | 30 ( 1) | |
| 1B6C_u | 300 (572) | 2 | 2 | 2 ( 3) | 193 | 60 | 16 (4) | 18(1) |
| 1BJl_b | 67 (4177) | 0 | 0 | 0 (–) | 61 | 55 | 15 (3) | |
| 1BJl_uUBBf (0 bumps) | 4 (68,905) | 0 | 0 | 0 (–) | 3 | 3 | 3 ( 4) | CDRs 61 (2) |
| 1BVK_b | 53 (15,955) | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | |
| 1BVK_u (0 bumps) | 30 (24,205) | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | |
| 1BVK_u (10 bumps) | 2 (162,117) | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | CDRs 0 (3970) |
| 1BVN_b | 1208 (1) | 222 | 96 | 28 ( 1) | 91 | 80 | 29 ( 1) | |
| 1BVN_u | 1551 (107) | 48 | 40 | 24 ( 1) | 53 | 19 | 2 ( 17) | 48 (14) |
| 1CGI_b | 466 (1) | 129 | 100 | 30 ( 1) | 455 | 100 | 30 ( 1) | |
| 1CGI_u | 175 (174) | 6 | 3 | 2(5) | 4 | 4 | 4(2) | 11 (23) |
| 1D6R_be | 1064 (96) | 59 | 0 | 0 ( 542) | 0 | 0 | 0 (–) | |
| 1D6R_ue | 3 (108,109) | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 2 (984) |
| 1DFJ_b | 200 (1) | 175 | 98 | 30 ( 1) | 0 | 0 | 0 (–) | |
| 1DFJ_u | 832 (2) | 289 | 93 | 29 ( 1) | 0 | 0 | 0 (–) | 73 (1) |
| 1DQJ_b | 113 (1428) | 3 | 3 | 3 ( 10) | 0 | 0 | 0 (–) | |
| 1DQJ_u | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | CDRs 0 (2287) |
| 1E6J_b | 66 (30,300) | 0 | 0 | 0 (–) | 64 | 56 | 6 ( 23) | |
| 1E6J_u (0 bumps) | 133 (6070) | 0 | 0 | 0 (–) | 86 | 26 | 1 ( 14) | CDRs 121 (1) |
| 1E6J_u (10 bumps) | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | |
| 1E96_be | 177 (67) | 3 | 0 | 0 ( 229) | 17 | 0 | 0 ( 979) | |
| 1E96_ue | 242 (39) | 6 | 1 | 1 ( 21) | 54 | 2 | 0 ( 42) | 16 (16) |
| 1EAW_bd | 728 (1) | 158 | 1 | 0 ( 100) | 5 | 1 | 0 ( 90) | |
| 1EAW_ud | 2820 (12) | 125 | 0 | 0 ( 393) | 6 | 0 | 0 ( 1516) | 62 (3) |
| 1F34_b | 323 (1) | 136 | 81 | 30 ( 1) | 145 | 85 | 30 ( 1) | |
| 1F34_u | 393 (63) | 34 | 0 | 0 ( 117) | 117 | 3 | 0 ( 74) | 13 (5) |
| 1FQl_b | 402 (4) | 44 | 40 | 25 (1) | 19 | 12 | 5 (11) | |
| 1FQ1_u | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | Not run |
| 1FQJ_bd | 747 (1) | 130 | 0 | 0 ( 293) | 0 | 0 | 0 (–) | |
| 1FQJ_u | 42 (16,731) | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 0 (5551) |
| 1FSK_b | 293 (119) | 13 | 13 | 13(1) | 4 | 4 | 3(11) | |
| 1FSK_u UBB (0 bumps) | 171 (399) | 2 | 2 | 2 ( 4) | 0 | 0 | 0 (–) | CDRs 163 (1) |
| 1GCQ_bd | 319 (13) | 29 | 1 | 0 ( 80) | 0 | 0 | 0 (–) | |
| 1GCQ_u | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 1 (429) |
| 1GP2_b | 376 (1) | 67 | 61 | 30 ( 1) | 102 | 69 | 29 ( 1) | |
| 1GP2_ue | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 0 (–) |
| 1GRN_bd,e | 563 (1) | 254 | 43 | 16(1) | 30 | 16 | 8 ( 2) | |
| 1GRN_ue (0 bumps) | 1118 (291) | 32 | 0 | 0 ( 500) | 0 | 0 | 0 (–) | 6 (807) |
| 1GRN_ue (10 bumps) | 1244 (1425) | 3 | 0 | 0 ( 1560) | 0 | 0 | 0 (–) | |
| 1HEl_b | 443 (1) | 76 | 70 | 30 ( 1) | 38 | 38 | 22 ( 1) | |
| 1HEl_ue | 211 (735) | 3 | 0 | 0 ( 158) | 0 | 0 | 0 (–) | 2 (258) |
| 1HIA_b | 1254 (22) | 199 | 47 | 14 ( 4) | 95 | 14 | 4 (2) | |
| 1HIA_ue | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 0 (–) | |
| H2M_b | 697 (1) | 171 | 58 | 20 ( 1) | 0 | 0 | 0 (–) | |
| H2M_ue (0 bumps) | 51 (694) | 3 | 1 | 0 ( 37) | 0 | 0 | 0 (–) | 0 (34,162) |
| H2M_ue (10 bumps) | 113 (11,749) | 0 | 0 | 0 ( 0) | 0 | 0 | 0 (–) | |
| H9R_b | 438 (113) | 25 | 0 | 0 ( 106) | 0 | 0 | 0 (–) | |
| H9R_u UBB (0 bumps) | 185 (151) | 3 | 0 | 0 ( 166) | 0 | 0 | 0 (–) | CDRs 16 (20) |
| UPS_b | 1738 (3) | 308 | 73 | 16(1) | 0 | 0 | 0 (–) | |
| UPS_u (0 bumps) | 1080 (40) | 50 | 4 | 1 ( 7) | 0 | 0 | 0 (–) | CDRs 18 (53) |
| UPS_u (10 bumps) | 880 (462) | 6 | 0 | 0 ( 107) | 0 | 0 | 0 (–) | |
| 1K5D_bd | 394 (1) | 105 | 0 | 0 ( 102) | 43 | 0 | 0 ( 156) | |
| 1K5D_u (0 bumps) | 47 (463) | 2 | 0 | 0 ( 1668) | 0 | 0 | 0 (–) | 8 (134) |
| 1K5D_u (10 bumps) | 143 (10,317) | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | |
| 1KAC_b | 864 (5) | 59 | 10 | 3 ( 18) | 0 | 0 | 0 (–) | |
| 1KAC_ud | 409 (80) | 9 | 0 | 0 ( 812) | 0 | 0 | 0 (–) | 3 (72) |
| 1KTZ_b | 1332(508) | 18 | 0 | 0 ( 171) | 64 | 20 | 2 ( 26) | |
| 1KTZ_u (0 bumps) | 267 (4403) | 0 | 0 | 0 (–) | 76 | 1 | 0 ( 83) | 7 (804) |
| 1KTZ_u (10 bumps) | 10 (118,002) | 0 | 0 | 0 (–) | 1 | 0 | 0 ( 513) | |
| 1KXP_b | 277 (1) | 142 | 93 | 30 ( 1) | 20 | 18 | 13(1) | |
| 1KXP_ud,e | 1635 (1) | 316 | 39 | 10 (5) | 17 | 1 | 0 ( 62) | 20 (1) |
| 1M10_b | 844 (1) | 144 | 39 | 6 ( 12) | 94 | 1 | 1 ( 15) | |
| 1M10_ue | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 0 (–) |
| 1MAH_be | 1038 (1) | 198 | 100 | 30 ( 1) | 186 | 93 | 30 ( 1) | |
| 1MAH_ue | 1151 (1957) | 1 | 1 | 1 ( 1) | 344 | 68 | 22 ( 1) | 85 (1) |
| 1MLC_b | 223 (1668) | 1 | 1 | 1 ( 4) | 1 | 1 | 1 ( 10) | |
| 1MLC_u (0 bumps) | 22 (26816) | 0 | 0 | 0 (–) | 3 | 2 | 0 ( 91) | CDRs 39 (5) |
| 1MLC_u (10 bumps) | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | |
| 1NCA_b | 455 (8) | 47 | 44 | 14 ( 10) | 0 | 0 | 0 (–) | |
| INCA_u UBB (0 bumps) | 113 (616) | 5 | 5 | 3 ( 9) | 0 | 0 | 0 (–) | CDRs 50 (4) |
| 1PPE_b | 1124 (3) | 235 | 61 | 25 ( 1) | 173 | 32 | 7(5) | |
| 1PPE_u | 2327 (223) | 37 | 33 | 8 ( 2) | 508 | 94 | 29 ( 1) | 324 (1) |
| 1QA9_bd | 1181 (1) | 118 | 0 | 0 ( 174) | 0 | 0 | 0 (–) | |
| 1QA9_u | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 2 (850) |
| 1UDI_b | 1583 (1) | 317 | 96 | 29 ( 1) | 71 | 11 | 5(1) | |
| 1UDI_u | 1013 (8) | 16 | 15 | 7(5) | 6 | 1 | 1 ( 19) | 7 (13) |
| 1WEJ_b | 723 (31) | 31 | 31 | 27 ( 1) | 0 | 0 | 0 (–) | |
| 1WEJ_u (0 bumps) | 627 (114) | 15 | 15 | 15(1) | 0 | 0 | 0 (–) | CDRs 55 (1) |
| 1WEJ_u (10 bumps) | 83 (25,317) | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | |
| 2BTF_b | 672 (1) | 141 | 91 | 28 ( 2) | 0 | 0 | 0 (–) | |
| 2BTF_u (0 bumps) | 260 (18) | 12 | 7 | 4(1) | 0 | 0 | 0 (–) | 7 (96) |
| 2BTF_u (10 bumps) | 738 (299) | 9 | 8 | 6 ( 7) | 0 | 0 | 0 (–) | |
| 2MTA_b ETg | 240 (489) | 5 | 5 | 5(1) | 136 | 56 | 23 ( 1) | |
| 2MTA_u ET (0 bumps) | 11 (76,179) | 0 | 0 | 0 (–) | 1 | 0 | 0 ( 507) | 1 (1722) |
| 2PCC_b ET | 1956 (9) | 62 | 13 | 7 ( 7) | 225 | 23 | 8 ( 7) | |
| 2PCC_u ETe (0 bumps) | 476 (337) | 2 | 0 | 0 ( 842) | 0 | 0 | 0 (–) | 1 (1037) |
| 2SIC_b | 98 (1440) | 1 | 1 | 1 ( 26) | 79 | 47 | 7 ( 7) | |
| 2SIC_u (0 bumps) | 19 (8114) | 0 | 0 | 0 (–) | 14 | 5 | 4(5) | 69 (1) |
| 2SIC_u (10 bumps) | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | |
| 2SNI_b | 262 (348) | 12 | 12 | 12(1) | 241 | 100 | 30 ( 1) | |
| 2SNI_u (10 bumps) | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) | 6 (300) |
| 7CEI_bd | 2151 (2) | 166 | 4 | 0 ( 33) | 0 | 0 | 0 (–) | |
| 7CEI_u | 1652 (1) | 81 | 13 | 5(2) | 0 | 0 | 0(–) | 186 (1) |
The top 2,000 DOT configurations rescored by ASP+Elec. Note that the number in the top 2,000 corresponds to that found by DOT scoring.
The top 2,000 ACE+Elec configurations rescored by ASP+Elec.
CDRs: ZDOCK was restrained to the CDRs of the antibody and therefore ZDOCK results are not comparable to the unrestrained DOT runs.
The DOT score gave the best results for bound complexes [top 2,000, top 100, top 30 (best)] 1AKJ: 169, 46, 23 (1); lEAW_b: 158, 39, 18 (1); 1FQJ: 130, 27, 10 (1); 1GCQ: 29, 4, 3 (13); 1GRN: 254, 86, 30 (1); 1K5D: 105, 42, 25 (1); 1M10: 144, 31, 16 (1); 1QA9: 118, 28, 12 (1); 7CEI: 166, 28, 13 (2), and for unbound complexes 1EAW: 125, 10, 2 (12); 1KAC: 9, 1, 0 (80); 1KXP: 316, 43, 17(1).
Noninteger charge, multiple side chains incomplete.
UBB: Coordinates are of an unbound/bound system.
ET: An electron-transfer complex.
ZDOCK found no hits.
Molecules were docked with default parameters, with the grid size determined by the size of each system, as described above. The 200,000 most favorable DOT configurations were retained. Bound and unbound coordinates were docked in three ways. First, as a control, only the single correct orientation of the moving molecule was tested. The best fit of the moving molecule can be translated up to 1 Å from its position in the complex, due to grid effects, but the correct complex was usually clearly distinguished among the best-ranked configurations, particularly when using bound coordinates. In the second docking, a full search was done, in which 54,000 orientations were applied to the moving molecule. No atom of the moving molecule was allowed to penetrate the excluded volume of the stationary molecule (‘0 bumps’). The set of 54,000 orientations (6° search) did not include the crystallographic orientation; the closest orientation differs by about 3°. In the third docking, a full search was also performed, but up to 10 atoms of the moving molecule were allowed to penetrate the excluded volume of the stationary molecule (‘10 bumps’). Allowing 0 bumps usually worked well for the bound complexes. The lenient shape description used by DOT accommodates the good fit, despite the approximations caused by mapping molecular properties onto a grid. For the unbound coordinates, allowing up to 10 bumps generally gave better results, but in some few cases the 0 bumps run was more successful. We have found that 0 bumps works better for protein-DNA complexes23–26 and for protein-protein electron-transfer complexes,3,57 two of which (2PCC, 2MTA) are in Table 1. Also, using 0 bumps often works better for bound/unbound combinations (1BJI, 1FSK, 1I9R, 1NCA in Table 1). For unbound coordinates, results for both the 0 bumps and 10 bumps runs are included in Table 1 when 0 bumps gave significantly better results.
To determine hits among the 200,000 configurations output by DOT, the RMSD between the docked and reference positions of the moving molecule was calculated for interface Cα atoms, given a fixed position for the stationary molecule. The reference position of the moving molecule either corresponded to its crystallographic position in the complex (bound) or the superposed position from Benchmark 2.0 (unbound). Interface residues were defined as those with any atoms within 10 Å of the stationary molecule.8 An RMSD of 5 Å or less was considered a hit. This criterion is equivalent to the RMSD cutoff of 2.5 Å used in the evaluation of ZDOCK,64 in which the interface alpha carbon atoms of both molecules were fit.
All DOT output configurations were re-ranked by ACE+Elec. The top-ranked 2,000 configurations from both DOT and ACE+Elec were then re-ranked by ASP+Elec (Figure 1).
Results
To make all the features of DOT easily accessible, DOT2 now provides automated procedures for preparing DOT input files and for evaluating DOT output. Previously these steps involved running multiple computer programs, which required considerable computational expertise. The automated procedure for preparing input files includes 1) checking and correcting the geometry of input coordinates, 2) checking for integral charge and the presence of residues and cofactors in the Reduce and charge libraries, 3) calculating the appropriate grid size, 4) preparing properly protonated coordinates for electrostatic calculations, 5) calculating the electrostatic potential of the stationary molecule using Poisson-Boltzmann methods with either APBS or UHBD and reasonable parameters, 6) preparing molecular surfaces with different sized radii, 7) filling volumes with appropriate values based on these surfaces, 8) determining electrostatic clamping values, which make the electrostatic potential of the stationary molecule compatible with the shape potential, and 9) ensuring a consistent coordinate frame for all calculations. The DOT output is a compact format containing the user-specified number of the most favorable configurations determined by the DOT score. This list of configurations is designed for automated evaluation, which includes 1) comparing configurations against a reference position to determine quality of fit, 2) calculating ACE and ASP scores for docked configurations, and 3) re-ranking by ACE+Elec. The evaluation routine can be customized to add additional evaluations, such as ASP+Elec re-ranking of a specified number of configurations.
Application of DOT to Benchmark 2.0
We applied DOT to the Benchmark 2.0 set of 84 protein-protein complexes64 for direct comparison with the docking program ZDOCK. We took advantage of the DOT2 automated procedures to customize coordinate preparation, docking, and evaluation (Figure 1). The majority of the systems did not pass the preprocessing checks of Prepscript (Supporting Table S1). The major problem was incomplete side chains (43 systems). The 46 systems that were carried on to the DOT docking step included seven with incomplete side chains in unbound coordinates (1GP2, 1HE1, 1HIA, 1I2M, 1KXP, 1M10, 2PCC) and four with incomplete side chains in both bound and unbound coordinates (1D6R, 1E96, 1GRN, 1MAH). Of the 46 systems, 11 contained antibody fragments (1AHW, 1BJ1, 1BVK, 1DQJ, 1E6J, 1FSK, 1I9R, 1JPS, 1MLC, 1NCA, 1WEJ), eight contained proteases (1ACB, 1CGI, 1D6R, 1F34, 1HIA, 1PPE, 2SIC, 2SNI), and six contained Rho-related small GTP-binding proteins (1E96, 1GCQ, 1GRN, 1HE1, 1I2M, 1K5D). In the docking step, a 6° set of rotations (54,000) was applied to the moving molecule. For molecules of 100-150 residues, this provides a search over the moving molecule surface that is approximately as fine as the 1 Å translational search. Cubic grid sizes varied from 128 Å3 to 256 Å3, depending on the system, resulting in a search of 100 to 800 billion total configurations. For the bound complexes, the moving molecule was not allowed to penetrate the excluded volume of the stationary molecule (‘0 bumps’). For unbound coordinates, two runs were done: one allowing no penetrations (‘0 bumps’) and one allowing up to 10 atoms of the moving molecule to penetrate the excluded volume of the stationary molecule (‘10 bumps’). In general, the 10 bumps runs gave better results for unbound structures. Electron-transfer proteins, which typically have a small interface and a significant electrostatic component to the intermolecular energy, gave better results with 0 bumps. In Benchmark 2.0 systems in which only the bound structure is available for one molecule (1BJI, 1FSK, 1I9R, and 1NCA, labeled UBB in Table 1), only the 0 bumps run was done.
The 200,000 most favorable configurations retained by DOT for each system were re-ranked by ACE+Elec. We then selected the 2,000 top-ranked configurations from DOT and ACE+Elec for further analysis. These two lists of configurations often had little overlap. For example, bound coordinates of 1ACB (1ACB b, Table 1) gave 16 hits among the top 2,000 from DOT, but 376 hits among the top 2,000 from ACE+Elec. In contrast, bound coordinates from 1AHW (1AHW b) gave 219 hits among the top 2,000 from DOT, but no hits among the top 2,000 from ACE+Elec.
We re-ranked the two lists of 2,000 top configurations, one from DOT and one from ACE+Elec, by ASP+Elec (Figure 1) and then analyzed the number of hits in the top 30, 100, and 2,000 configurations, along with the total hits in the 200,000 configurations retained by DOT and the best-ranked hit from DOT scoring (Table 1). In general, ASP+Elec put more hits in the top 30 and 100 configu-rations than DOT or ACE+Elec. ACE+Elec did not give significantly better scoring than ASP+Elec in any system. DOT scoring, however, gave significantly better results than either ACE+Elec or ASP+Elec in nine bound cases and three unbound cases (Table 1).
Bound systems
We assessed the docking results by three scoring methods: the initial DOT score, the top 2,000 DOT configurations re-ranked by ASP+Elec, and the top 2,000 ACE+Elec configurations re-ranked by ASP+Elec (Table 1). For the bound coordinates, the correct complex was clearly distinguished in the majority of systems (27/46), with at least 50% hits within the top-ranked 30 by at least one scoring method. These systems included four antibody complexes (1AHW, 1BJI, 1JPS, and 1WEJ) in which the full, two-domain Fab was used. Fourteen additional systems had at least one hit within the top-ranked 30, including four Fab complexes (1DQJ, 1E6J, 1FSK, and 1MLC). The bound coordinates of five systems had no hits in the top-ranked 30: 1D6R and 1E96, which have incomplete side chains, the Fab complex 1I9R, and 1AK4 and 1BVK (discussed below).
Unbound systems: comparison with ZDOCK
Results from application of ZDOCK to unbound systems in Benchmark 2.064 allowed direct comparison with DOT. ZDOCK was not applied to the eight Benchmark 2.0 systems ranked difficult, but only one of these systems contained complete side chains and was run through DOT (1FQ1). DOT and ZDOCK both use convolution methods, calculated by fast Fourier transforms, to perform a systematic grid-based search over all space for two rigid macromolecules. Both programs applied 54,000 orientations to the Benchmark 2.0 targets. There are, however, distinct differences between the two programs. In DOT, the grid spacing was 1 Å and the grid size was customized for each system, whereas ZDOCK used a grid spacing of 1.2 Å with cubic grids of either 100 or 128 points on each side.65 The two programs use different protocols for retaining configurations. ZDOCK keeps the best-energy configuration for each orientation of the moving molecule, resulting in 54,000 total configurations. In contrast, DOT retains a user-specified number of the most favorably ranked configurations (200,000 for Benchmark 2.0 systems) regardless of the orientation. We have found this approach useful for identifying clusters, where one orientation can give similar energy configurations among nearby grid points. The two programs use different scoring methods. With DOT, we used three scoring methods: the DOT score based on shape fit and electrostatic energy, ASP+Elec re-ranking of the top 2,000 DOT-scored configurations, and ASP+Elec re-ranking of the top 2,000 configurations scored by ACE+Elec. These were compared with four methods of scoring by ZDOCK.64 Two versions of ZDOCK were run on each system: ZDOCK 2.1 (shape complementarity) and ZDOCK 2.3 (added electrostatics and desolvation terms). These two sets of 54,000 configurations were then re-ranked with ZRANK (versions ZDOCK 2.1 ZR and ZDOCK 2.3 ZR), which uses seven scoring terms that have been optimally weighted using a training set of 10 Benchmark 1.0 systems50 before applying them to Benchmark 2.0 systems. The ZDOCK analysis gives the number of hits in the top-ranked 2,000 and the best ranked hit.64 In Table 1, we provide the ZDOCK scoring method with the highest ranked hit; this method usually had the largest number of hits in the top-ranked 2,000.
DOT results on systems containing antibody fragments (Fab and Fv) are not comparable with ZDOCK results. In ZDOCK, the search was restrained to the CDRs by modification of the antibody shape potential (indicated in Table 1 by ‘CDRs’ in the ZDOCK column). In contrast, DOT searched over the full Fv and two-domain Fab coordinates provided in Benchmark 2.0 for each antibody in Table 1. Since CDRs make up a small fraction of the total surface area of a Fab, it is unsurprising that the restrained ZDOCK search found more hits among the top 2,000 for most Fab antibody systems (1BJI, 1E6J, 1FSK, 1MLC, 1NCA, and 1WEJ). However, the DOT search found more hits among the top 2,000 in two Fab systems (1AHW and 1JPS), as well as finding 15 hits among the top 30 for Fab 1WEJ. In general, disallowing penetrations of the moving ligand with the antibody (0 bumps run) gave better results. 1BVK, which contains an Fv, gave no solutions in DOT. This system is a particularly difficult target; even with restraints to the CDR regions, ZDOCK found no hits in the top 2,000 by any ranking method.
Among other systems, ASP+Elec scoring found a favorable energy cluster making up 50% or more of the top 30 in 1B6C, 1BVN, 1DFJ, 1MAH, and 1PPE. The DOT score found a favorable energy cluster for 1KXP. In general, ZDOCK also appears to do well with these systems, although details on the distribution of good hits in the top 2,000 were not provided. 1ACB, which gave no hits among the 54,000 ZDOCK configurations, gave only a few, poorly scored hits with DOT. 1AK4, which contains a single domain of the HIV capsid protein, gave no hits with either DOT or ZDOCK. The three other unbound systems with no ZDOCK hits (1GP2, 1HIA, and 1M10) also gave no hits with DOT; all three systems have several incomplete side chains. Most other systems with incomplete side chains (1D6R, 1GRN, 1HEI, 1I2M, and 2PCC) gave poor results with both programs. Of the three remaining systems with incomplete side chains, two were among those with the best scores (1MAH and 1KXP) and one (1E96) gave at least one hit in the top 30 with both DOT and ZDOCK. Overall, incomplete side chains, which could modify the interface surface or create artificial surfaces, gave equivalent results with both programs.
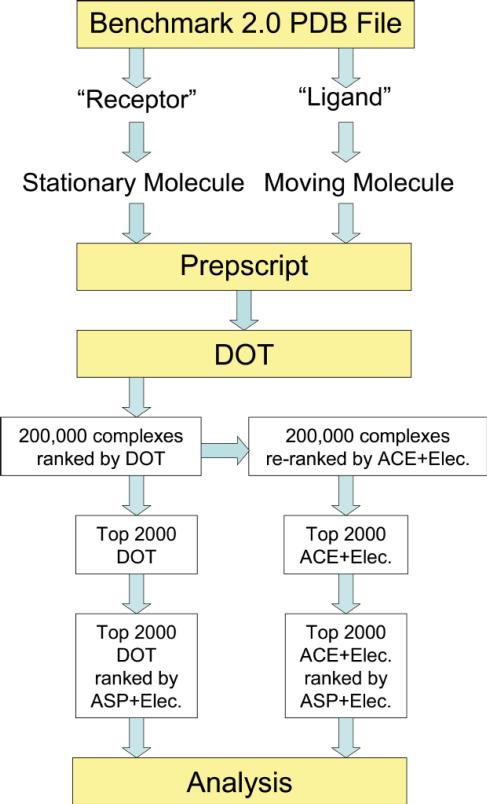
2SIC (subtilisin bound to an inhibitor) showed the greatest difference between DOT and ZDOCK. DOT found no hits in the 10 bumps run used for unbound systems whereas ZDOCK found 69 hits within the top-ranked 2,000 configurations, including rank 1. We investigated this complex further to determine the source of this difference. The assumed biological state of the inhibitor is a dimer in the PDB coordinates, but the Benchmark 2.0 coordinates contain only the monomer present in the asymmetric unit. In the 10 bumps run reported in Table 1, subtilisin, as the stationary molecule, contacts the exposed dimer interface of the inhibitor in most favorably ranked configurations (Figure 2a). To see if reversing the assignment of the molecules influenced results, we docked subtilisin, as the moving molecule, to the stationary inhibitor monomer. The outcome improved significantly (Table 2). With the bound coordinates, the majority of the 30 top-ranked configurations were hits. With the unbound coordinates, the 10 bumps provided more hits in the top 2,000 than the 0 bumps run. We then added the two calcium ions that are present in the subtilisin PDB coordinates, but absent in the Benchmark 2.0 coordinates. The 10 bumps run with unbound coordinates showed significant improvement, with three hits in the top 30 by ASP+Elec re-ranking of the ACE+Elec list (Table 2), but contacts with the exposed dimer interface predominated (Figure 2b). Finally, subtilisin was docked to the inhibitor dimer, which was built by applying the appropriate symmetry transformations. The two equivalent subtilisin-binding sites of the dimer were identified (Figure 2c), with hits making up the majority of the 30 top-ranked configurations for both bound and unbound coordinates (Table 2). Thus, using the biological dimer and including structural calcium ions greatly improved the docking outcome for unbound coordinates; the unsuccessful run with an incomplete system (0/200,000 hits in the 10 bumps run) became definitive, with identification of the correct complex as the largest, favorable energy cluster.
Figure 2.
Improved docking outcome through the use of the biological dimer of an inhibitor. Dockings of unbound coordinates that allowed up to 10 bumps are compared with the crystallo-graphic complex of subtilisin (gray Cα backbone tubes) and the inhibitor monomer (cyan ribbon). The distribution of subtilisin around the inhibitor is shown by the geometric centers (spheres, with the largest red spheres being the most favorable) of the 300 top-ranked subtilisin placements from ASP+Elec re-ranking of the top 2,000 ACE+Elec configurations. One sphere may represent multiple placements with the same center, but different orientations. (a) Docking the moving inhibitor monomer to stationary subtilisin. No matches with the crystallographic complex were found. In most, the exposed dimer interface of the inhibitor interacts with subtilisin. (b) Docking the moving subtilisin with calcium ions to the stationary inhibitor monomer. A small cluster lies at the crystallographic position, but most contact the exposed dimer interface of the inhibitor. (c) Docking moving subtilisin to the stationary inhibitor dimer (second monomer shown as blue ribbons). Two clusters match the crystallographic positions of subtilisin, one centered within the subtilisin structure (upper right) and one (circled, left, bottom) that corresponds to the second subtilisin binding site. These two clusters include 17 of the 30 top-ranked configurations, identifying the correct complex.
Table 2.
Analysis of 2SIC docking
| Run | Total in DOT 200,000 (best) | DOT -> ASP_Elec Number in top |
ACE_Elec -> ASP_Elec Number in top |
||||
|---|---|---|---|---|---|---|---|
| 2,000 | 100 | 30 (best) | 2,000 | 100 | 30 (best) | ||
| Stationary = Enzyme (no Ca ions); Moving = Inhibitor Monomer | |||||||
| 2SIC_b | 98 (1440) | 1 | 1 | 1 ( 26) | 79 | 47 | 7 ( 7) |
| 2SIC_u (0 bumps) | 19(8114) | 0 | 0 | 0 (–) | 14 | 5 | 4(5) |
| 2SIC_u (10 bumps) | 0 | 0 | 0 | 0 (–) | 0 | 0 | 0 (–) |
| Stationary = Inhibitor Monomer; Moving = Enzyme (no Ca ions) | |||||||
| 2SIC_b | 418 (1) | 116 | 77 | 23 ( 1) | 252 | 68 | 25(2) |
| 2SIC_u (0 bumps) | 61 (438) | 6 | 6 | 1 ( 28) | 33 | 13 | 1 ( 28) |
| 2SIC_u (10 bumps) | 246(1711) | 1 | 1 | 0 ( 90) | 79 | 3 | 0 ( 40) |
| Stationary = Inhibitor Monomer; Moving = Enzyme with 2 Ca ions | |||||||
| 2SIC_b | 392 (2) | 99 | 78 | 25 ( 1) | 249 | 68 | 26 ( 2) |
| 2SIC_u (0 bumps) | 59 (487) | 5 | 5 | 2(19) | 33 | 13 | 0 ( 38) |
| 2SIC_u (10 bumps) | 230 (1741) | 1 | 1 | 0 ( 66) | 97 | 5 | 3 ( 7) |
| Stationary = Inhibitor dimer; Moving = Enzyme with 2 Ca ions | |||||||
| 2SIC_b | 561 (18) | 95 | 81 | 24(1) | 402 | 74 | 24 ( 2) |
| 2SIC_u (0 bumps) | 93 (384) | 4 | 4 | 4(3) | 49 | 14 | 0 ( 33) |
| 2SIC_u (10 bumps) | 193 (2202) | 0 | 0 | 0 (–) | 176 | 38 | 17 (1) |
ASP+Elec
Because of the success of the ASP+Elec scoring, we applied ASP+Elec to the full 200,000 configurations output by DOT for five diverse unbound systems (10 bumps runs). These rankings were compared with results from ZDOCK and the best results from ASP+Elec re-ranking of the top 2,000 from the DOT and ACE+Elec lists (Table 3). In all cases, the full ASP+Elec analysis put significantly more hits in the top 2,000, though not necessarily giving the highest ranked hit. For the antibody/antigen complex 1WEJ, full ASP+Elec ranking hits make up the majority of the 30 top-ranked configurations, making the 10 bumps run now successful. Further, the full 1WEJ search from DOT gave results comparable to those from ZDOCK, which restricted the search to the CDRs. This demonstrates that, in this case, ASP+Elec effectively distinguished the CDR region as the antigen-docking site. 1UDI and 1CGI contain about 20% hits within the top-ranked 100 configurations. Although 1AY7 and 2BTF did not have hits in the top-ranked 30, the full ASP+Elec re-ranking greatly increased the number of hits in the top 2,000.
Table 3.
ASP+Elec scoring of all 200,000 configurations for unbound, 10 bumps runs
| PDB ID | Proteins | Total in DOT 200,000 (best) | Best 2,000 ASP+Elec Number in top |
200,000 ASP+Elec Number in top |
ZDOCK Number in top 2,000 (best) | ||||
|---|---|---|---|---|---|---|---|---|---|
| 2,000 | 100 | 30 (best) | 2,000 | 100 | 30 (best) | ||||
| 1AY7 | Barnase/barstar | 492 (405) | 5 | 3 | 2 (14) | 151 | 1 | 0 (40) | 24(111) |
| 1CGI | Protease/inhibitor | 175 (174) | 4 | 4 | 4 (2) | 81 | 20 | 8 (9) | 11 (23) |
| 1UDI | Glycosylase/inhibitor | 1013 (8) | 16 | 15 | 7 (5) | 378 | 19 | 7 (7) | 7 (13) |
| 1WEJ | Antibody/antigen | 83 (25,317) | 0 | 0 | 0 (–) | 82 | 43 | 18 (1) | 55 (1) |
| 2BTF | Actin/profilin | 738 (299) | 9 | 8 | 6 (7) | 142 | 0 | 0 (105) | 7 (96) |
Discussion
Biology is full of surprises; often the biological interaction of two macromolecules has been found to be very different from the ‘obvious’ one. Given two unbound structures, an ideal docking program would produce one ‘correct’ answer. But even such a program must start with a good model of the biologically relevant problem. Given these considerations, we had three central goals for DOT. The first goal was to comprehensively sample the full set of possible configurations. The second goal was to score these samples sufficiently realistically to map the most likely interactions. The third goal was to make DOT accessible to experts focused on a specific system, who therefore know the critical structural elements and are familiar with experimental data that can help distinguish correct complexes, but who are not necessarily computational specialists.
Comprehensive sampling
Our first goal was achieved with the development of DOT,3,56 which uses correlation functions to perform an exhaustive search. DOT2 uses faster FFTs to compute the convolutions and efficiently collects a large number of the best-ranked configurations. The convolution method, while giving a very fast translational search, does impose some limitations. First, the translational search is over the full grid. Specific positions can be discarded by design of the molecular properties, but all are tested. Second, the stationary and moving molecules are treated differently so docking results can vary depending on the assignment of the two starting molecules. Given a specified rotational set for the moving molecule, a larger molecule would be more coarsely sampled than a smaller molecule. Further, the representations of the moving and stationary molecules are different. In DOT, we use detailed electrostatic and shape potentials for the stationary molecule, taking advantage of the need to calculate these potentials only once. In contrast, the representation of the moving molecule is limited because its properties must be recalculated for every orientation. In DOT, the moving molecule is simply represented by point charges at atomic positions; solvation effects and the lower dielectric of the protein interior are not taken into account. Third, implementation of pair-wise potentials is computationally costly. A separate convolution would be required for each distinct atom type, and the time of the calculation increases linearly with the number of convolutions.
Scoring
To achieve our second goal of reasonable scoring, two key improvements in potentials that we previously prototyped are now automated in DOT2: electrostatic clamping57 and the use of molecular surfaces (rather than van der Waals spheres) to describe the excluded volume of the stationary molecule.23 The DOT2 scoring method ranked a hit as the most favorable configuration in many of the bound Benchmark 2.0 systems and found hits within the top 200,000 for most of the unbound systems. The DOT2 score provides a good estimate of the electrostatic energy, but the van der Waals term, which approximates the surface area buried in the interface, does not take desolvation differences among different atom types into account. To improve the van der Waals term, we examined two methods - ACE and ASP - for rescoring the list of configurations generated by DOT. Our goal was to determine their generality over a wide variety of protein-protein complexes.
ACE is based on a statistical analysis of atom pairing frequency observed in the interior of known protein structures60. Given a list of complexes, ACE can be rapidly calculated. Therefore we applied the ACE+Elec score to the full list of 200,000 DOT configurations output for the Benchmark 2.0 systems and compared the top-ranked 2,000 from each scoring method. Unfortunately neither the DOT nor the ACE+Elec score showed a clear advantage across all systems. Instead, the scoring method preference appeared highly dependent on the system, with the same method usually giving the most hits in the top 2,000 for both bound and unbound coordinates.
ACE has several disadvantages for scoring macromolecular interfaces. Charged and hydrogen-bonding side chains, which are rare in protein interiors but common in macromolecular interfaces, are poorly represented. In addition, the preference of protein backbone atoms for hydrophobic side chains may be overestimated. Statistical potentials based on atom pairing frequency in protein-protein interfaces have been developed and implemented in ZDOCK.66 This improved the docking outcome overall, but at the cost of computing 12 convolutions for this energy term.1 A drawback of these statistical pair-wise potentials is that they are specific to protein residues, and therefore cannot be extended to cofactors or to interactions such as protein-DNA complexes.
The second desolvation model that we investigated was ASP62, which is based on atomic solvation parameters and the change in exposed atomic surface area upon going from the isolated molecules to the complex. ASP is computationally much slower than ACE because a molecular surface must be calculated for each docked complex. Results from the Benchmark 2.0 systems show that the ASP calculation is well worth the computational time and may be generally useful. ASP+Elec rescoring of the top 2,000 configurations from ACE+Elec consistently improved the ranking of hits. ASP+Elec usually improved the ranking of hits within the DOT top 2,000, although there were a few systems where DOT scoring gave the best results (Table 1). ASP+Elec rescoring of all 200,000 DOT configurations (Table 3) significantly enriched the number of hits in the top-ranked 2,000 compared with the scoring methods used in Table 1 or results from ZDOCK. We are currently investigating algorithms that improve the speed of ASP so that ASP+Elec scoring can be routinely applied to the full DOT output, thereby eliminating the need for ACE.
ASP has additional advantages over ACE. The atomic types used in ASP are general, and therefore may be extendable to molecules other than proteins, so that the desolvation energy of exposed cofactors or other kinds of molecules could be estimated. Unlike ACE, ASP does not use pair-wise potentials. Instead, each isolated protein structure determines the basis for calculating ASP. Therefore, ASP has the potential to be implemented as a molecular description that can be used by DOT; a recent approach using an approximate ASP potential is the program ASPDock.10
User accessibility
Although other convolution methods also provide comprehensive sampling and reasonable scoring approaches, DOT2 is unique in its goal of user accessibility and control. Rather than using a ‘black-box’ approach, DOT2 is designed for transparency, versatility, and adaptability in the three separate phases of the computation – preprocessing, docking with the convolution engine (the DOT core), and evaluation. DOT2 is supported by a detailed User Guide that includes the rationale for each step. Our versatile design of DOT allows it to be extended to macromolecules other than proteins or to problems other than macromolecular docking. For example, DOT has recently been used to assemble multidomain proteins using data from atomic force microscopy.67
The DOT2 preprocessing step provides molecular checks and a detailed log file with information to guide the user. Tools for modification and customization of molecular potentials are provided in an extensive library of scripts. The molecular property files are transparent, allowing the user to verify that the final, processed coordinates passed along to the docking step are those that the user intended. Essential groups such as modified amino acids, cofactors, and metal ions are retained, with the user alerted if they are not present in the Reduce and charge libraries. The effectiveness of the preprocessing step is demonstrated by its ability to detect problems in Benchmark 2.0 systems (Supplementary Table S1), which have not been previously considered or identified.
Unlike the programs PIPER9 and ZDOCK,64 DOT maintains a complete list of top-ranked configurations, with the user controlling how many should be reported. The resulting output is a compact list that permits retention of an arbitrarily large number of configurations without loss of computational efficiency. Individual energy components, as well as the total energy, are included in the list so that the user can evaluate the significance of each energy term. Analysis steps add further information to each configuration record. By using the same list format for further analyses, rather than molecular coordinates, the connection between the original configuration and new information is maintained. This design feature greatly facilitated the ACE and ASP evaluations of the Benchmark 2.0 systems.
The need for appropriate coordinate sets
The need for good starting models for computational docking was brought home by our results on the Benchmark 2.0 systems. We had hoped that the protein-protein complexes in Benchmark 2.0 would provide a useful comparison of DOT with ZDOCK. To be useful for the development and evaluation of docking and scoring methods, the benchmark systems should be as close as possible to the biologically active state. Instead, we found numerous problems with the Benchmark 2.0 systems. Investigators focusing on a specific system would certainly address the problems found in the Benchmark 2.0 coordinates by completing side chains, constructing the biological oligomerization state, and including essential cofactors.
The first problem that we identified was incomplete side chains in many protein coordinate sets. This is common in PDB files, but the absence of side chains can create artificial pockets, hydrophobic patches, or altered binding surfaces, making the already difficult docking problem even more difficult. Two bound Benchmark 2.0 systems with incomplete side chains in Table 1 were among the three bound systems that gave the poorest results. The three unbound systems (1GP2, 1HIA, and 1M10) that gave no hits with either ZDOCK or DOT all had incomplete side chains. Many of the other unbound systems with incomplete side chains gave only a few hits among the top 2,000 by both docking methods. Thus, building complete side chains is an important step in the preparation of coordinates for docking calculations.
A second problem was incorrect oligomerization states. The PDB coordinates alone may not represent the complete state. For example, two molecules in a PDB file may represent two distinct monomers in the asymmetric unit, the biological dimer, or part of a larger biological oligomer that must be generated by adding symmetry-related molecules. Unfortunately, not all PDB files contain the correct information needed to generate the biological state. Further, the oligomerization state can change upon going from isolated proteins to the bound complex. Thus, system-specific biochemical knowledge beyond the PDB coordinates may be required to ascertain the oligomerization state of the individual components and whether those states persist in the complex.
Although coordinates for some multimers are present in Benchmark 2.0 (1AKJ and 1I9R, Table 1), many systems are represented by the coordinates in the asymmetric unit instead of the appropriate biological multimer. Examples include the inhibitor in 2SIC and methylamine dehydrogenase (MADH) in 2MTA. In both cases, the PDB files provide the symmetry operations needed to create the full biological complex from the asymmetric unit. In other systems, it is unclear if the PDB files provide the correct oligomerization state. For example, 2SNI defines the assumed biological complex as a 1:1 complex of enzyme:inhibitor, but 2CI2 defines the isolated inhibitor as a hexamer. The oligomerization state of the enzyme:inhibitor complex in 7CEI is even less clear. In 7CEI, the assumed biological state is a 1:1 complex, but that of the isolated inhibitor is a tetramer in 1UNK. Further, the isolated enzyme structure in 1M08, which was determined after 7CEI, specifies that the enzyme is a dimer and supports this with additional experimental evidence.68
Our in-depth docking study of 2SIC (Table 2) demonstrates that using the correct oligomeric state of a protein can have a dramatic effect on the docking outcome. Bound coordinates gave reasonable results with both the monomer and dimer of the inhibitor, presumably because of the excellent fit of the interface. With the unbound inhibitor monomer, interactions at the exposed dimer interface dominated over the imperfect fit at the correct interface (Figure 2a). This is not surprising, given the strong protein-protein interactions often seen for dimer interfaces. With the complete inhibitor dimer, the correct complex was decisively identified as the major cluster in the top 30 configurations using the standard 10 bumps protocol for unbound systems (Figure 2c).
A third problem in Benchmark 2.0 is missing cofactors, particularly metal ions. Additional system-specific knowledge beyond the PDB files is needed to differentiate essential catalytic and structural metal ions from metal ions added to the crystallographic solution, such as heavy-metal derivatives used to aid structure determination. Examples in Benchmark 2.0 include omission of the catalytic copper ion in amicyanin (2MTA), magnesium ions the 11 systems containing ATP or GDP (including 1E96, 1FQ1, 1FQJ, 1GP2, 1K5D, and 1KXP in Table 1), and calcium ions in 2SIC and 2SNI. Essential metal ions contribute to the overall charge of the molecule and local charge distribution near the metal site. Further, the metal ion may have multiple ionization states, only one of which may be compatible with protein binding.
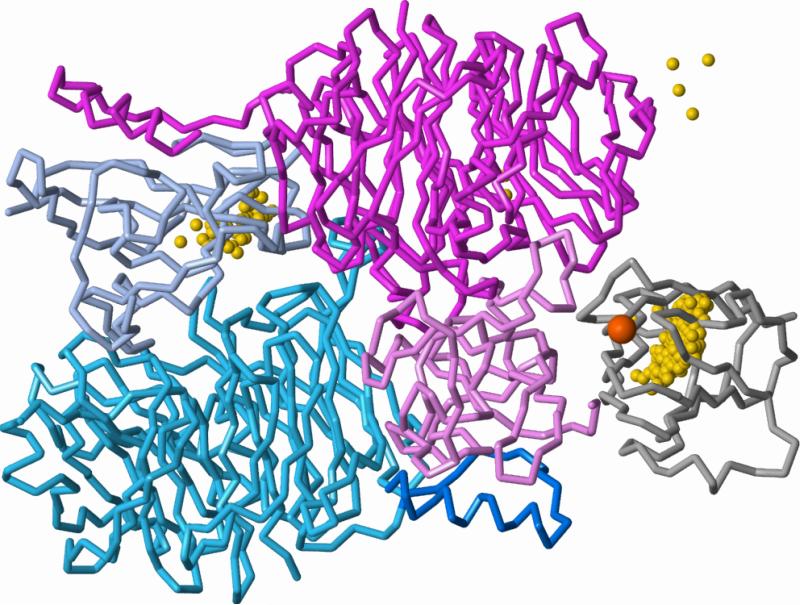
2MTA, which contains methylamine dehydrogenase (MADH) and amicyanin, is an example where knowledge of the system is required to build a reasonable model. MADH is a dimer of heterodimers, but the Benchmark 2.0 coordinates contain only the single heterodimer present in the asymmetric unit (Figure 3). The N-terminal region of one chain in the heterodimer forms part of the domain created by the second heterodimer, and comes within 4 Å of the bound amicyanin. Therefore, representing MADH solely as a single heterodimer creates several artificial features: an exposed dimer interface, a U-shaped surface groove that should be occupied by the N-terminus of the other heterodimer, an N-terminal region extending into solvent, and an incomplete binding surface for amicyanin (Figure 3). The Benchmark 2.0 coordinates also lack the amicyanin copper ion, which is bound to a surface histidine that contacts MADH. With the precisely fit bound coordinates, the correct complex was unambiguously identified, but only after re-ranking by ASP+Elec. With unbound coordinates, the best ZDOCK and DOT scoring methods gave only one hit in the top 2,000. Although a fully representative model of the 2MTA system does not guarantee successful docking results, the cluster of favorably ranked configurations at the artificially exposed dimer surface certainly makes this electron-transfer complex a more difficult target.
Figure 3.
Poor representation of the MADH tetramer as an isolated heterodimer. In Benchmark 2.0, MADH coordinates consist of the single heterodimer (magenta and pink) from 2MTA, artificially exposing the large interface with the second heterodimer (blue). The N-terminal region of the second heterodimer (dark blue) contributes to part of the binding site of amicyanin (right, gray with orange Cu atom) on the first heterodimer. The 600 top-ranked configurations from docking amicyanin to the single heterodimer (geometric centers shown as gold spheres, ASP+Elec re-ranking of the ACE+Elec list) form two major clusters. One cluster (right) lies at the amicyanin-binding site (rank 507 is the only hit). The second cluster (left) contacts the exposed surfaces of the dimer interface and N-terminal region that would be buried in the biological tetramer.
While DOT preprocessing is useful for detecting simple problems such as incomplete side chains or undefined cofactors, constructing a biologically relevant model often requires more extensive structural analysis and biological knowledge. In our investigations of complex biological systems, we found that most have idiosyncrasies, some revealed by preliminary docking studies, that required careful selection and adjustment of coordinates from the starting PDB files. Our studies on the cytochrome c:cytochrome oxidase57 and the linker histone:nucleosome25 interactions showed some of the complexities of model building. Partial models of the multi-chain assemblies of cytochrome oxidase and the nucleosome had to be constructed that were small enough to be computationally feasible, yet still retain essential features. Docking was used to check that artificial surfaces created in these partial models did not contain favorable binding sites. Assigning the correct oxidation states for metal sites in cytochrome c and cytochrome oxidase was important because of their close approach in the electron transfer complex. Analysis of the full crystal environment of the nucleosome identified structured regions involved in crystal contacts that were likely to be disordered in solution, and therefore needed to be removed from the model. The nucleosomal DNA had to be extended to create the full region known to interact with the linker histone. Comparison of the two linker histone molecules in the asymmetric unit revealed that the structure of the first molecule in the PDB file was significantly perturbed by crystal packing interactions and therefore potentially a poor model of the biological structure. With DOT, we were able to create and customize the molecular properties needed for these complex systems.
Conclusions
The transparent, versatile, and modular design of DOT2 provides the flexibility needed to construct detailed models, allowing DOT to be used as an exploratory tool on complex systems and to be extended to problems beyond macromolecular docking. Key to the design is the automated procedure for building the molecular representations, ensuring that these are properly calculated, all in the same coordinate frame, and compatible with each other. This allows the user to focus on what may be the most crucial step for docking, preparation of realistic input models of the macromolecules.
The lenient-fit, full search of DOT compensated for the conformational changes induced upon binding in many Benchmark 2.0 systems. Despite the limitations imposed by the convolution methodology, correct complexes were found for most unbound systems within the most favorable 200,000 configurations as scored by DOT. Rescoring with ASP+Elec, which includes atomic desolvation in the energy, appeared to be effective over more systems than scoring by either DOT or the pair-wise, protein-specific ACE. To the degree to which they could be compared, DOT and ZDOCK gave similar results on Benchmark 2.0 systems. Unfortunately, the presence of incompletely described systems in Benchmark 2.0 significantly compromises the utility of the Benchmark as a reliable tool for evaluating docking programs or optimizing potentials.
Further, the complexes in the Benchmark are limited to those that can be successfully crystallized. It is unclear how well this limited set represents the vast array of biologically important protein-protein interactions, which range from the very fast interactions of electron-transfer complexes to irreversibly bound ones. Recently, docking protocols and molecular potentials10,66 have been evaluated based their overall performance on Benchmark 2.0 or Benchmark 3.0 (which includes Benchmark 2.0). The absence of analysis of the failures encountered within the Benchmark systems makes it impossible to determine if failures are due to incorrect starting models, poor biophysical representations used in the docking, or large conformational changes that make the system intractable to rigid-body docking methods. Improved performance may be due to better results on systems that dominate the Benchmark, such as protease/inhibitor and antibody complexes, rather than better modeling of the forces that underlie all protein-protein interactions.
Supplementary Material
Acknowledgments
We thank Susan Lindsey for creation of the DOT2 web site and assistance with software development and infrastructure.
Funding
This work was supported by the National Science Foundation (DBI 99-04559 and MCB 1020765), the National Institutes of Health (GM070996 and GM046312), and the University of California, San Diego, Center for AIDS Research (CFAR, National Institutes of Health grant P30 AI036214).
Footnotes
ZDOCK performs 6 complex convolutions, which is slightly more expensive than 12 of the real convolutions computed by DOT. More recent versions of ZDOCK use an improved convolution engine, which reduces the cost of each convolution, but still requires multiple convolutions.
DOT2 is available at http://www.sdsc.edu/CCMS/DOT for free download under an open source license. The DOT 2.0 User Guide is at http://www.sdsc.edu/CCMS/DOT/dotuser.pdf
References
- 1.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Proc. Natl. Acad. Sci. USA. 1992;89:2195. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vakser IA, Aflalo C. Proteins. 1994;20:320. doi: 10.1002/prot.340200405. [DOI] [PubMed] [Google Scholar]
- 3.Mandell JG, Roberts VA, Pique ME, Kotlovyi V, Mitchell JC, Nelson E, Tsilgeny I, Ten Eyck LF. Prot. Eng. 2001;14:105. doi: 10.1093/protein/14.2.105. [DOI] [PubMed] [Google Scholar]
- 4.Moreira IS, Fernandes PA, Ramos MJ. J. Comput. Chem. 2010;31:317. doi: 10.1002/jcc.21276. [DOI] [PubMed] [Google Scholar]
- 5.Heifetz A, Katchalski-katzir E, Eisenstein M. Protein Sci. 2002;11:571. doi: 10.1110/ps.26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabb HA, Jackson RM, Sternberg MJE. J. Mol. Biol. 1997;272:106. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 7.Vakser IA. Proteins. 1997;1:226. [PubMed] [Google Scholar]
- 8.Chen R, Li L, Weng Z. Proteins. 2003;52:80. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 9.Kozakov D, Brenke R, Comeau SR, Vajda S. Proteins. 2006;65:392. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Guo D, Huang Y, Liu S, Xiao Y. BMC Bioinformatics. 2011;12:36. doi: 10.1186/1471-2105-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajaj C, Chowdhury R, Siddavanahalli V. IEEE/ACM Trans Comput Biol Bioinform. 2011;8:45. doi: 10.1109/TCBB.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie DW, Kemp GJL. Proteins. 2000;39:178. [PubMed] [Google Scholar]
- 13.Garzon JI, Lopéz-Blanco JR, Pons C, Kovacs J, Abagyan R, Fernandez-Recio J, Chacon P. Bioinformatics. 2009;25:2544. doi: 10.1093/bioinformatics/btp447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen SWW, Pellequer JL, Schved JF, Giansily-Blaizot M. Throm. Haemost. 2002;88:74. [PubMed] [Google Scholar]
- 15.Giorgione J, Hysell M, Harvey DF, Newton AC. Biochemistry. 2003;42:11194. doi: 10.1021/bi0350046. [DOI] [PubMed] [Google Scholar]
- 16.Zheng D, Kurenova E, Ucar D, Golubovskaya V, Magis A, Ostrov D, Cance WG, Hochwald SN. Biochem. Biophys. Res. Commun. 2009;388:301. doi: 10.1016/j.bbrc.2009.07.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolmos E, Nowak M, Werner M, Fischer K, Schwarz G, Mathews S, Schoof H, Nagy F, Bujnicki JM, Davis SJ. HFSP J. 2009;3:350. doi: 10.2976/1.3218766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo M, Shapiro R, Morris GM, Yang X-L, Schimmel P. J. Phys. Chem. B. 2010;114:16273. doi: 10.1021/jp1082517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatachari NJ, Walker LA, Tastan O, Le T, Dempsey TM, Li Y, Yanamala N, Srinivasan A, Klein-Seetharaman J, Montelaro RC, et al. Virol. J. 2010;7:119. doi: 10.1186/1743-422X-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Lee Y, Lazar P, Son M, Baek A, Thangapandian S, Jeong NY, Yoo YH, Lee KW. J. Mol. Graph. Model. 2011;29:996. doi: 10.1016/j.jmgm.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Rosano C. Mitochondrion. 2011;11:513. doi: 10.1016/j.mito.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Hopfner K-P, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Cell. 2001;105:473. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 23.Roberts VA, Case DA, Tsui V. Proteins. 2004;57:172. doi: 10.1002/prot.20193. [DOI] [PubMed] [Google Scholar]
- 24.Adesokan AA, Roberts VA, Lee KW, Lins RD, Briggs JM. J. Med. Chem. 2004;47:821. doi: 10.1021/jm0301890. [DOI] [PubMed] [Google Scholar]
- 25.Fan L, Roberts VA. Proc. Natl. Acad. Sci. USA. 2006;103:8384. doi: 10.1073/pnas.0508951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. Cell. 2008;133:789. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BI, Weinfeld M, et al. J. Biol. Chem. 2011;286:32638. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts VA, Pique ME, Hsu S, Li S, Slupphaug G, Rambo RP, Jamison J, Liu T, Lee JH, Tainer JA, et al. Nucl. Acids Res. 2012;40:6070. doi: 10.1093/nar/gks291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietra F. J Chem Inf Model. 2009;49:972. doi: 10.1021/ci800463h. [DOI] [PubMed] [Google Scholar]
- 30.Pietra F. Chem. Biodivers. 2011;8:816. doi: 10.1002/cbdv.201100060. [DOI] [PubMed] [Google Scholar]
- 31.Patargias G, Zitzmann N, Dwek R, Fischer WB. J. Med. Chem. 2006;49:648. doi: 10.1021/jm050721e. [DOI] [PubMed] [Google Scholar]
- 32.Crowley PB, Hunter DM, Sato K, McFarlane W, Dennison C. Biochem. J. 2004;378:45. doi: 10.1042/BJ20031423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruschus JM, Greene LE, Eisenberg E, Ferretti JA. Protein Sci. 2004;13:2029. doi: 10.1110/ps.03390504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sondermann H, Nagar B, Bar-Sagi D, Kuriyan J. Proc. Natl. Acad. Sci. USA. 2005;102:16632. doi: 10.1073/pnas.0508315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagchi A, Ghosh TC. Biochem. Biophys. Res. Commun. 2005;335:609. doi: 10.1016/j.bbrc.2005.07.115. [DOI] [PubMed] [Google Scholar]
- 36.Bagchi A, Roy P. Biochem. Biophys. Res. Commun. 2005;331:1107. doi: 10.1016/j.bbrc.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 37.Bagchi A, Ghosh TC. Theochem. 2006;758:113. [Google Scholar]
- 38.Bagchi A, Ghosh TC. Biophys. Chem. 2006;119:7. doi: 10.1016/j.bpc.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 39.de Armas HN, Dewilde M, Verbeke K, Maeyer MD, Declerck PJ. Structure. 2007;15:1105. doi: 10.1016/j.str.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Buttani V, Gaertner W, Losi A. Eur. Biophys. J. 2007;36:831. doi: 10.1007/s00249-007-0155-1. [DOI] [PubMed] [Google Scholar]
- 41.Rumpel S, Becker S, Zweckstetter M. J. Biomol. NMR. 2008;40:1. doi: 10.1007/s10858-007-9204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann WA, Arduini A, Nicol SM, Camacho CJ, Lessard JL, Fuller-Pace FV, de Lanerolle P. J. Cell Biol. 2009;186:193. doi: 10.1083/jcb.200905016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson TA, Qiu J, Plaut AG, Holyoak T. J. Mol. Biol. 2009;389:559. doi: 10.1016/j.jmb.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mccoy JG, Johnson HD, Singh S, Bingman CA, Lei IK, Thorson JS, G. N. P. Proteins. 2009;74:50. doi: 10.1002/prot.22131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esser J, Rakonjac M, Hofmann B, Fischer L, Provost P, Schneider G, Steinhilber D, Samuelsson B, Radmark O. Biochem. J. 2010;425:265. doi: 10.1042/BJ20090856. [DOI] [PubMed] [Google Scholar]
- 46.Tettamanti G, Cattaneo AG, Gornati R, de Eguileor M, Bernardini G, Binelli G. Gene. 2010;450:85. doi: 10.1016/j.gene.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 47.Mashiach E, Schneidman-Duhovny D, Peri A, Shavit Y, Nussinov R, Wolfson HJ. Proteins. 2010;78:3197. doi: 10.1002/prot.22790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mashiach E, Nussinov R, Wolfson HJ. Nucl. Acids Res. 2010;38:W457. doi: 10.1093/nar/gkq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demerdash ONA, Buyan A, Mitchell JC. Proteins. 2010;78:3156. doi: 10.1002/prot.22811. [DOI] [PubMed] [Google Scholar]
- 50.Chen R, Mintseris J, Janin J, Weng Z. Proteins. 2003;52:88. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 51.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 52.Word JM, Lovell SC, Richardson JS, Richardson DC. J. Mol. Biol. 1999;285:1735. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 53.Baker NA, Sept D, Joseph S, Holst JJ, McCammon JA. Proc. Natl. Acad. Sci. USA. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanner MF, Olson AJ, Spehner J-C. Biopolymers. 1996;38:305. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 55.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Jr., Weiner P. J. Am. Chem. Soc. 1984;106:765. [Google Scholar]
- 56.Ten Eyck LF, Mandell JG, Roberts VA, Pique ME. In: Hayes A, Simmons M, editors. Proceedings of the 1995 ACM/IEEE Supercomputing Conference; San Diego. Los Alamitos, CA: IEEE Computer Society Press; 1995. p. 22. www.sdsc.edu/CCMS/Papers/DOT_sc95.html. [Google Scholar]
- 57.Roberts VA, Pique ME. J. Biol. Chem. 1999;274:38051. doi: 10.1074/jbc.274.53.38051. [DOI] [PubMed] [Google Scholar]
- 58.Gilson MK, Davis ME, Luty BA, McCammon JA. J. Phys. Chem. 1993;97:3591. [Google Scholar]
- 59.Frigo M, Johnson SG. Proc.IEEE. 2005;93:216. [Google Scholar]
- 60.Zhang C, Vasmatzis G, Cornette JL, DeLisi C. J. Mol. Biol. 1997;267:707. doi: 10.1006/jmbi.1996.0859. [DOI] [PubMed] [Google Scholar]
- 61.Miyazawa S, Jernigan RL. Macromolecules. 1985;18:534. [Google Scholar]
- 62.Fernandez-Recio J, Totrov M, Skorodumov C, Abagyan R. Proteins. 2005;58:134. doi: 10.1002/prot.20285. [DOI] [PubMed] [Google Scholar]
- 63.Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, Weng Z. Proteins. 2005;60:214. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- 64.Pierce B, Weng Z. Proteins. 2007;67:1078. doi: 10.1002/prot.21373. [DOI] [PubMed] [Google Scholar]
- 65.Chen R, Weng Z. Proteins. 2002;47:281. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 66.Mintseris J, Pierce B, Wiehe K, Anderson R, Chen R, Weng Z. Proteins. 2007;69:511. doi: 10.1002/prot.21502. [DOI] [PubMed] [Google Scholar]
- 67.Trinh MH, Odorico M, Pique ME, Teulon J-M, Roberts VA, Ten Eyck LF, Getzoff ED, Parot P, Chen SW, Pellequer J-L. Structure. 2012;20:113. doi: 10.1016/j.str.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng Y-S, Hsia K-C, Doudeva LG, Chak K-F, Yuan HS. J. Mol. Biol. 2002;324:227. doi: 10.1016/s0022-2836(02)01092-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.