Abstract
Genome-wide association studies (GWASs) have already identified at least 22 common susceptibility loci associated with an increased risk of colorectal cancer (CRC). This study examined the relationship between these single nucleotide polymorphisms (SNPs) and the clinical outcomes of patients with colorectal cancer. Seven hundred seventy-six patients with surgically resected colorectal adenocarcinoma were enrolled in the present study. Twenty-two of the GWAS-identified SNPs were genotyped using a Sequenom MassARRAY. Among the 22 SNPs, two (rs1321311G>T in CDKN1A and rs10411210C>T in RHPN2) were significantly associated with the survival outcomes of CRC in a multivariate survival analysis. In a recessive model, the rs1321311 TT genotype (vs. GG + GT) and rs10411210 TT genotype (vs. CC + CT) were associated with a worse prognosis for disease-free survival (adjusted HR = 1.90; 95% confidence interval = 1.00-3.60; P = 0.050, adjusted HR = 1.94; 95% confidence interval = 1.05-3.57; P = 0.034, respectively) and overall survival (adjusted HR = 2.05; 95% confidence interval = 1.00-4.20; P = 0.049, adjusted HR = 2.06; 95% confidence interval = 1.05-4.05; P = 0.036, respectively). None of the other SNPs was significantly associated with any clinicopathologic features or survival. The present results suggest that the genetic variants of the CDKN1A (rs1321311) and RHPN2 (rs10411210) genes can be used as prognostic biomarkers for patients with surgically resected colorectal cancer.
Introduction
Although the benefit of adjuvant chemotherapy in resected colorectal cancer (CRC) has already been established by several studies, 40% of patients experience local or distant recurrences even after curative resection [1]. In particular, the heterogeneity of CRC is considered one of major difficulties for choosing treatments and the effect of chemotherapy seems to vary depending on the tumor biology. Thus, a clinical or biologic biomarker that predicts recurrence, survival, or response patterns to chemotherapy is needed for patients with resected CRC.
Single nucleotide polymorphisms (SNPs) have already been widely implicated in cancer development, prognosis, and treatment response. Recent advances using genome-wide association studies (GWASs) have enabled the identification of multiple CRC-related SNPs [2–8]. Notwithstanding, various studies have also reported that genetic variants generated from candidate gene or pathway-based studies are associated with the clinical outcome of CRC patients, which can be used to categorize patients with different survival or responses to specific treatments. However, no GWAS has yet examined the direct relationship between genetic variations and the survival of CRC patients. Three studies have examined the relationship between GWAS-identified CRC risk variants and the clinical outcomes of CRC, yet two were performed with a small sample size and the third was focused on Caucasian populations, which differ from other ethnic groups [9–11]. Accordingly, the present study examined the relationship between GWAS-identified genetic variants and the clinical outcomes of a relatively large group of Korean CRC patients.
Materials and Methods
Study population
The tissues investigated in this study were obtained from 776 Korean patients who underwent a curative surgical resection between May, 2001 and May, 2008 at Kyungpook National University Hospital (Daegu, Korea). Medical records were retrospectively reviewed to identify the relationship between GWAS-identified genetic variants and the clinical outcomes. In brief, the study included patients with histologically confirmed adenocarcinoma of the colon and rectum who underwent curative surgery but did not receive neoadjuvant therapy before surgery. Each patient was examined every 3 to 6 months for the first three years following the diagnosis of CRC, and every year thereafter, in accordance with the national guidelines. Written informed consent for genetic analysis including gene expression and SNP genotyping was received from all the patients before surgery, and the study was approved by the Institutional Research Board at Kyungpook National University Hospital. The CRC diagnosis and staging were assessed according to the WHO classifications and TMN classifications from the 7th edition of the American Joint Committee on Cancer (AJCC).
SNP selection and genotyping
Thirty-five polymorphisms associated with CRC susceptibility were selected from the US National Human Genome Research Institute (NHGRI) Catalog of Published Genome-Wide Association Studies (as accessed in March 2013). Among these 35 polymorphisms, 22 polymorphisms were applied in this study, while the others were excluded for the following reasons: the minor allele frequency was ≤ 5% in the HapMap JPT data of the public SNP database (http://www.ncbi.nlm.nih.gov/SNP) or the SNPs could not be applied to the SEQUENOM’s MassARRAY platform. All the obtained tissues were immediately snapped frozen in liquid nitrogen and stored -80°C until the relevant experiments were conducted. The genomic DNA was extracted from fresh colorectal mucosal tissue at the time of surgery using a QIAamp genomic DNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The genotype analysis was performed using SEQUENOM’s MassARRAY iPLEX assay according to the instructions of the manufacturer. To validate the genotyping, approximately 5% of samples were randomly selected for re-genotyping using a sequencing method or restriction fragment length polymorphism (RFLP) assay by a different investigator and the results were 100% concordant.
Statistical analysis
The Hardy-Weinberg equilibrium was tested using a goodness-of-fit χ2 test with 1 df. The genotypes for each SNP were analyzed as a three-group categorical variable (referent model), and also grouped according to a dominant and recessive model. Overall survival (OS) was measured from the day of surgery to the date of the last follow-up or date of death. Disease-free survival (DFS) was calculated from the day of surgery until recurrence or death from any cause. The survival estimates were calculated using the Kaplan-Meier method. The differences in OS or DFS according to the SNPs were compared using log-rank tests. Cox’s proportional hazard regression model was used for the multivariate survival analyses, which were always adjusted for age (< 64 versus ≥ 64 years), the preoperative CEA level (normal versus elevated), differentiation (well and moderate versus poorly), sex (male versus female), primary site (colon versus rectum), and stage (I to III). The proportional hazards assumption was checked and all graphs for the log-log survivor functions of the two SNP aid groups (rs10411210 and rs1321311) were parallel except for DFS graph of rs10411210. The hazard ratio (HR) and 95% confidence interval (CI) were also estimated. A cut-off P value of 0.05 (two-sided) was adopted for all the statistical analyses. The statistical data were obtained using an SPSS software package (SPSS 11.5 Inc. Chicago, IL, USA) or SAS Genetic software (SAS Institute, Cary, NC).
Results
Patient and disease characteristics
The clinical and pathologic characteristics of the patients are listed in Table 1. The median age was 64 years (range 21–89), 444 (57.2%) patients were diagnosed with colon cancer, and 327 (42.1%) patients had rectal cancer. The pathologic stages after surgical resection were as follows: stage I (N = 141, 18.2%), stage II (N = 318, 41.0%), and stage III (N = 317, 40.9%). Among the 635 patients with stage II or III diseases, 570 patients (89.8%) received adjuvant chemotherapy based on 6 cycles of 5-fluorouracil/leucovorin ± radiotherapy (390), 12 cycles of 5-fluorouracil/leucovorin/oxaliplatin (n = 67), 8 cycles of capecitabine (n = 41), or doxifluridine for 1 year (n = 72).
Table 1. Patient characteristics (N = 776).
| Variables | N | % |
|---|---|---|
| Age, median (years) | 64 | |
| <64 | 378 | 48.7 |
| ≥64 | 398 | 51.3 |
| Sex | ||
| Male | 432 | 55.7 |
| Female | 344 | 44.3 |
| Primary site | ||
| Colon | 444 | 57.2 |
| Rectum | 327 | 42.1 |
| Colon+ Rectum | 5 | 0.6 |
| Histological differentiation | ||
| Well | 94 | 12.1 |
| Moderate | 656 | 84.6 |
| Poor or signet ring | 26 | 3.4 |
| CEA | ||
| Normal | 642 | 82.7 |
| Elevated | 134 | 17.3 |
| CA19–9 | ||
| Normal | 675 | 87.0 |
| Elevated | 101 | 13.0 |
| Pathologic stage a | ||
| I | 141 | 18.2 |
| II | 318 | 41.0 |
| III | 317 | 40.9 |
| Relapse | ||
| No | 545 | 70.2 |
| Yes | 231 | 29.8 |
| Death | ||
| No | 593 | 76.4 |
| Yes | 183 | 23.6 |
a the 7th AJCC staging system
Overall treatment outcomes
At the last overall analysis (December 2012), 231 patients had experienced a disease relapse, 163 patients had died as a result of CRC, and 20 patients had died from causes unrelated to CRC. At the median follow-up duration of 78.5 months, the estimated 5-year OS and DFS for all the patients was 83.1% and 76.0% respectively, plus the survival differed according to the stage (P<0.001).
Genotype frequencies and effects on survival
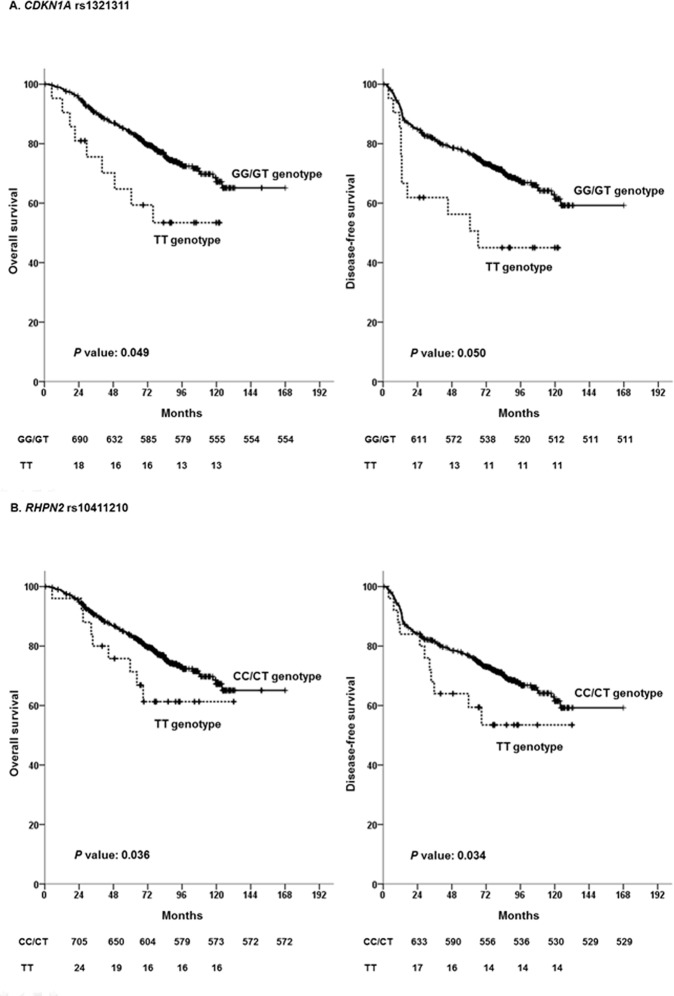
The frequencies of each genotype are shown in Table 2 and were confirmed to be in Hardy-Weinberg equilibrium (P>0.05). Among the 22 SNPs (S1 Table and S2 Table), two SNPs (rs1321311G>T in Cyclin-dependent kinase inhibitor 1A (CDKN1A) and rs10411210C>T in Rho GTPase binding protein 2 (RHPN2) were found to be significantly associated with the survival outcomes of CRC in the multivariate survival analysis. In the recessive model, the rs1321311 TT genotype (vs. GG + GT) and rs10411210 TT genotype (vs. CC + CT) were associated with a worse prognosis for DFS (adjusted HR = 1.90; 95% confidence interval = 1.00–3.60; P = 0.050, adjusted HR = 1.94; 95% confidence interval = 1.05–3.57; P = 0.034, respectively) and OS (adjusted HR = 2.05; 95% confidence interval = 1.00–4.20; P = 0.049, adjusted HR = 2.06; 95% confidence interval = 1.05–4.05; P = 0.036, respectively) (Table 3 and Fig. 1). None of the other SNPs was significantly associated with survival. With regard to clinicopathologic parameters, age (adjusted HR for DFS = 1.77; P<0.001, adjusted HR for OS = 2.56; P<0.001), preoperative CEA level (adjusted HR for DFS = 1.81; P<0.001, adjusted HR for OS = 1.78; P = 0.002), and pathologic stage (adjusted HR for DFS = 2.75; P<0.001, adjusted HR for OS = 2.49; P<0.001) were identified as significantly associated with DFS and OS (Table 3).
Table 2. Information and genotypes of 22 polymorphisms.
| rsID | Base change | Chromosome | Gene | Position | 1 | 2 | 3 | MAF | HWE-P |
|---|---|---|---|---|---|---|---|---|---|
| rs10411210 | C/T | 19 | RHPN2 | 19q13.11 | 527 (68.8) | 214 (27.9) | 25 (3.3) | 0.172 | 0.568 |
| rs10795668 | G/A | 10 | KRT8P16—TCEB1P3 | 10p14 | 348 (45.1) | 336 (43.5) | 88 (11.4) | 0.332 | 0.614 |
| rs10936599 | T/C | 3 | MYNN | 3q26.2 | 266 (34.3) | 372 (48) | 137 (17.7) | 0.417 | 0.725 |
| rs11169552 | C/T | 12 | DIP2B, ATF1 | 12q13.12 | 334 (44.7) | 334 (44.7) | 79 (10.6) | 0.329 | 0.739 |
| rs1321311 | G/T | 6 | SRSF3—CDKN1A | 6p21.2 | 534 (71.7) | 189 (25.4) | 22 (3) | 0.156 | 0.294 |
| rs3802842 | A/C | 11 | C11orf93 | 11q23.1 | 237 (30.7) | 397 (51.4) | 139 (18) | 0.437 | 0.222 |
| rs3824999 | A/C | 11 | POLD3 | 11q13.4 | 284 (36.8) | 365 (47.3) | 122 (15.8) | 0.395 | 0.793 |
| rs4444235 | C/T | 14 | RPS3AP46—BMP4 | 14q22.2 | 249 (32.2) | 367 (47.5) | 157 (20.3) | 0.440 | 0.306 |
| rs4779584 | T/C | 15 | SCG5—GREM1 | 15q13.3 | 551 (73) | 182 (24.1) | 22 (2.9) | 0.150 | 0.146 |
| rs4939827 | C/T | 18 | SMAD7 | 18q21.1 | 448 (57.7) | 279 (36) | 49 (6.3) | 0.243 | 0.531 |
| rs5934683 | T/C | X | GPR143—SHROOM2 | Xp22.2 | 642 (83.4) | 79 (10.3) | 49 (6.4) | 0.115 | 0.123 |
| rs6687758 | A/G | 1 | DUSP10—CICP13 | 1q41 | 384 (49.7) | 321 (41.5) | 68 (8.8) | 0.296 | 0.937 |
| rs6983267 | T/G | 8 | SRRM1P1—POU5F1B | 8q24.21 | 205 (27.7) | 394 (53.2) | 141 (19.1) | 0.457 | 0.047 |
| rs7014346 | G/A | 8 | SRRM1P1—POU5F1B | 8q24.21 | 355 (46.2) | 348 (45.3) | 65 (8.5) | 0.311 | 0.114 |
| rs7758229 | G/T | 6 | SLC22A3 | 6q25.3 | 451 (58.5) | 277 (35.9) | 43 (5.6) | 0.235 | 0.956 |
| rs9929218 | G/A | 16 | CDH1 | 16q22.1 | 570 (73.5) | 193 (24.9) | 13 (1.7) | 0.141 | 0.468 |
| rs10505477 | T/C | 8 | SRRM1P1—POU5F1B | 8q24.21 | 149 (19.6) | 385 (50.7) | 226 (29.7) | 0.449 | 0.514 |
| rs11903757 | T/C | 2 | NABP1—SDPR | 2q32.3 | 682 (91.2) | 65 (8.7) | 1 (0.1) | 0.045 | 0.669 |
| rs2057314 | T/C | 6 | DCBLD1 | 6q22.1 | 228 (30) | 378 (49.7) | 155 (20.4) | 0.452 | 0.942 |
| rs7136702 | T/C | 12 | N/A | 12q13 | 223 (29.3) | 365 (47.9) | 174 (22.8) | 0.468 | 0.294 |
| rs7315438 | C/T | 12 | TBX3—UBA52P7 | 12q24.21 | 260 (34.6) | 364 (48.5) | 127 (16.9) | 0.411 | 0.983 |
| rs961253 | C/A | 20 | TARDBPP1—BMP2 | 20p12.3 | 605 (80) | 143 (18.9) | 8 (1.1) | 0.105 | 0.889 |
Table 3. Multivariate analysis for survival.
| DFS | OS | ||||||
|---|---|---|---|---|---|---|---|
| P-value | HR | 95% CI | P-value | HR | 95% CI | ||
| Age, ≥64 years | 4.6×10 –5 | 1.77 | 1.35–2.34 | 1.2×10 –8 | 2.56 | 1.85–3.53 | |
| Male sex | 0.336 | 1.23 | 0.95–1.61 | 0.294 | 1.46 | 0.81–1.97 | |
| CEA, elevated | 2.0×10 –4 | 1.81 | 1.32–2.48 | 0.002 | 1.78 | 1.25–2.55 | |
| Stage, II/III | 6.6×10 –13 | 2.75 | 2.09–3.63 | 7.8×10 –9 | 2.49 | 1.82–3.39 | |
| Differentiation, well/moderate | 0.451 | 0.71 | 0.29–1.73 | 0.569 | 0.75 | 0.28–2.03 | |
| Primary site, colon | 0.142 | 0.89 | 0.48–1.21 | 0.111 | 0.77 | 0.51–1.92 | |
| CDKN1A (rs1321311), TT | vs.CC/CT | 0.050 | 1.90 | 1.00–3.60 | 0.049 | 2.05 | 1.00–4.20 |
| RHPN2(rs10411210), TT | vs.GG/GT | 0.034 | 1.94 | 1.05–3.57 | 0.036 | 2.06 | 1.05–4.05 |
Fig 1. Survival curves according to genotype in combined analysis: CDKN1A rs1321311 (A) and RHPN2 rs10411210 (B).
P values correspond to multivariate Cox model adjusted for age, CEA level, differentiation, sex, primary site, and pathologic stage.
Discussion
The prognostic impact of GWAS-identified genetic variants was investigated in CRC patients who had been surgically treated with curative intent. As a result, the current study demonstrated that the CDKN1A rs1321311 and RHPN2 rs10411210 polymorphisms among the 22 selected variants were significantly associated with survival after adjusting for clinical and pathologic factors.
A GWAS is an investigation of many common genetic variants in different individuals to discover if any variant is associated with a specific trait. GWASs have already been actively applied to explore disease susceptibility, prognosis, and drug response prediction [12]. To date, three recent studies have examined the relationship between GWAS-identified CRC risk variants and the clinical outcomes of CRC [9–11]. In Western countries, Phipps et al. examined the relationship between GWAS-identified variants and survival of the 2611 CRC patients, and found a strong association between a SNP (rs4939827) in the SMAD 7 gene and reduced disease-specific survival and OS [10]. Another study based on Western populations evaluated the survival results of 285 stage II or III CRC patients receiving fluorouracil-based chemotherapy, and found an association between one SNP (rs10318) and the recurrence of stage II and between three SNPs (rs10749971, rs961253, and rs355527) and the recurrence of stage III [9]. However, the influence of ethnic variations should also be considered when interpreting the prognostic impact of GWAS-identified variants [13]. Thus, a recent study by Xing et al. reported an association between two variants (rs4779584 and rs10795668) and a reduced risk of survival and a marginal effect between rs10795668 and chemotherapy in 380 Chinese CRC patients [11]. Notwithstanding, there are several important differences between the data in the present study and that in previous CRC prognostic studies. First, given the homogeneous ethnic background of Korean patients, any potential confounding effect due to ethnicity is likely to be small in the current study. Moreover, this study included a relatively large number of patients. Furthermore, the adjuvant treatment application was consistent and the loss to follow-up was very low. However, even though the present data identified certain gene variants as a statistically significant prognostic factor for survival in operated CRC, these results should be interpreted cautiously. Considering multiple comparison issue, we cannot rule out the possibility of a type I error in the analysis of individual SNP. Therefore, additional studies with larger sample sizes are required.
For the current study, the TT genotype of CDKN1A rs1321311 was significantly associated with worse survival outcomes. CDKN1A is a conserved protein belong to the CDK inhibitor Cip/Kip family, that has been implicated in cell cycle regulation [14]. In particular, this molecule is known to be involved in the regulation of fundamental cellular programs, such as cell proliferation, differentiation, migration, senescence, and apoptosis [15]. Although its mechanism and effect on cancer are still unknown, several reports have indicated that CDKN1A may be a biologic predictor and beneficial target for cancer treatment using cell cycle alteration [16]. Interestingly, several previous studies have demonstrated an association between a decreased expression of CDKN1A and the recurrence of stage II CRC, along with tumor suppression function in the colon of CDKN1A-deficient mice [17,18]. Therefore, the above findings suggest that CDKN1A can be a possible biomarker for CRC. Dunlop et al. also performed a GWAS meta-analysis to identify common variants influencing CRC risk, including 29778 cases and 29204 controls, and found a strong association between the CDKN1A (rs1321311) polymorphism and an increased CRC risk [19]. Similarly, the current study also found an association between the TT genotype and an association with a worse survival. However, there is no previous data on the prognostic role of CDKN1A (rs1321311) polymorphisms in CRC. In patients with head and neck cancer, the CT/TT genotype has been associated with a worse second primary malignancy risk than the CC genotype [20].
Another finding from the present study was a significant association between the RHPN2 rs10411210 TT genotype and poor survival outcomes in patients with surgically resected CRC. RHPN2 is a master regulator of actin cytoskeleton rearrangement and plays a role in various actin-modulating protein targets [21]. Yet, despite increasing evidence that RHPN2 plays important roles in several aspects of tumorigenesis and cancer progression, it is still unclear whether the polymorphism itself alters the function of the protein expression [22]. Indeed, several GWASs have already established a significant association between the RHPN2 polymorphism and an increased risk of CRC [3,23,24]. Thus, the association between CRC risk and the TT genotype is consistent across these studies. However, such previous studies have only focused on the risk of CRC, and not the impact on the survival outcomes of CRC. Consequently, the present results show that the role of the RHPN2 polymorphism still requires further clarification for predicting the prognosis of CRC.
In conclusion, the current findings indicated that genetic variations of CDKN1A and RHPN2 may influence the prognosis for CRC. However, since the exact mechanism and function of these gene variants have not yet been fully defined, the present findings need to be confirmed in further studies with other populations in order to clarify the association between these polymorphisms and the prognosis of CRC.
Supporting Information
.
(DOCX)
.
(DOCX)
Acknowledgments
This work was supported in part by a National Research Foundation of Korea Grant funded by the Korean Government (NRF-2011–0015862), and in part by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korean Ministry of Education, Science and Technology (NRF-2013R1A1A4A01008144).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by a National Research Foundation of Korea Grant funded by the Korean Government (NRF-2011-0015862), and in part by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korean Ministry of Education, Science and Technology (NRF-2013R1A1A4A01008144).
References
- 1. Ku G, Tan IB, Yau T, Boku N, Laohavinij S, Cheng AL, et al. (2012) Management of colon cancer: resource-stratified guidelines from the Asian Oncology Summit 2012. Lancet Oncol 13: e470–481. 10.1016/S1470-2045(12)70424-2 [DOI] [PubMed] [Google Scholar]
- 2. Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, et al. (2007) A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet 39: 1315–1317. [DOI] [PubMed] [Google Scholar]
- 3. Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, et al. (2010) Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet 42: 973–977. 10.1038/ng.670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, et al. (2008) Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet 40: 26–28. [DOI] [PubMed] [Google Scholar]
- 5. Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, et al. (2013) Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 144: 799–807.e724. 10.1053/j.gastro.2012.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. (2008) Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 40: 631–637. 10.1038/ng.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. (2007) A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 39: 984–988. [DOI] [PubMed] [Google Scholar]
- 8. Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, et al. (2008) A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 40: 623–630. 10.1038/ng.111 [DOI] [PubMed] [Google Scholar]
- 9. Dai J, Gu J, Huang M, Eng C, Kopetz ES, Ellis LM, et al. (2012) GWAS-identified colorectal cancer susceptibility loci associated with clinical outcomes. Carcinogenesis 33: 1327–1331. 10.1093/carcin/bgs147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Phipps AI, Newcomb PA, Garcia-Albeniz X, Hutter CM, White E, Fuchs CS, et al. (2012) Association between colorectal cancer susceptibility loci and survival time after diagnosis with colorectal cancer. Gastroenterology 143: 51–54.e54. 10.1053/j.gastro.2012.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xing J, Myers RE, He X, Qu F, Zhou F, Ma X, et al. (2011) GWAS-identified colorectal cancer susceptibility locus associates with disease prognosis. Eur J Cancer 47: 1699–1707. 10.1016/j.ejca.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 12. Patel JN, McLeod HL, Innocenti F (2013) Implications of genome-wide association studies in cancer therapeutics. Br J Clin Pharmacol 76: 370–380. 10.1111/bcp.12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He J, Wilkens LR, Stram DO, Kolonel LN, Henderson BE, Wu AH, et al. (2011) Generalizability and epidemiologic characterization of eleven colorectal cancer GWAS hits in multiple populations. Cancer Epidemiol Biomarkers Prev 20: 70–81. 10.1158/1055-9965.EPI-10-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsihlias J, Kapusta L, Slingerland J (1999) The prognostic significance of altered cyclin-dependent kinase inhibitors in human cancer. Annu Rev Med 50: 401–423. [DOI] [PubMed] [Google Scholar]
- 15. Romanov VS, Pospelov VA, Pospelova TV (2012) Cyclin-dependent kinase inhibitor p21(Waf1): contemporary view on its role in senescence and oncogenesis. Biochemistry (Mosc) 77: 575–584. 10.1134/S000629791206003X [DOI] [PubMed] [Google Scholar]
- 16. Stivala LA, Cazzalini O, Prosperi E (2012) The cyclin-dependent kinase inhibitor p21CDKN1A as a target of anti-cancer drugs. Curr Cancer Drug Targets 12: 85–96. [DOI] [PubMed] [Google Scholar]
- 17. Belt EJ, Brosens RP, Delis-van Diemen PM, Bril H, Tijssen M, van Essen DF, et al. (2012) Cell cycle proteins predict recurrence in stage II and III colon cancer. Ann Surg Oncol 19 Suppl 3: S682–692. 10.1245/s10434-012-2216-7 [DOI] [PubMed] [Google Scholar]
- 18. Poole AJ, Heap D, Carroll RE, Tyner AL (2004) Tumor suppressor functions for the Cdk inhibitor p21 in the mouse colon. Oncogene 23: 8128–8134. [DOI] [PubMed] [Google Scholar]
- 19. Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, Whiffin N, et al. (2012) Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet 44: 770–776. 10.1038/ng.2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z, Sturgis EM, Zhang F, Lei D, Liu Z, Xu L, et al. (2012) Genetic variants of p27 and p21 as predictors for risk of second primary malignancy in patients with index squamous cell carcinoma of head and neck. Mol Cancer 11: 17 10.1186/1476-4598-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peck JW, Oberst M, Bouker KB, Bowden E, Burbelo PD (2002) The RhoA-binding protein, rhophilin-2, regulates actin cytoskeleton organization. J Biol Chem 277: 43924–43932. [DOI] [PubMed] [Google Scholar]
- 22. Wilson KF, Erickson JW, Antonyak MA, Cerione RA (2013) Rho GTPases and their roles in cancer metabolism. Trends Mol Med 19: 74–82. 10.1016/j.molmed.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carvajal-Carmona LG, Cazier JB, Jones AM, Howarth K, Broderick P, Pittman A, et al. (2011) Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum Mol Genet 20: 2879–2888. 10.1093/hmg/ddr190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hutter CM, Chang-Claude J, Slattery ML, Pflugeisen BM, Lin Y, Duggan D, et al. (2012) Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res 72: 2036–2044. 10.1158/0008-5472.CAN-11-4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.
(DOCX)
.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.