Microarrays of biomolecules and cells have played important roles in genomics, diagnostics, and drug screening.[1] Live-cell capture on micropatterned surfaces is particularly valuable for diagnostic or biosensing applications,[2] as cell arrays are amenable to rapid characterization and sorting. For example, antibody-coated micropost arrays have been used to detect and isolate rare circulating tumor cells from peripheral blood with high fidelity.[3] Microwell arrays have been used for the capture and analysis of single immune cells.[4] While it is often important to release or sort subsets of cells from these arrays for more thorough downstream analysis (e.g. by PCR or Western blot), methods for the retrieval of specific cells from micropatterned surfaces remain limited. Electrochemical stimulation can be exploited to release cells from particular regions within micropatterns,[5] but these approaches require that cell arrays be registered with electrodes, and they can become somewhat complicated for larger arrays. Arguably, light-activated cell-release strategies provide more flexibility, particularly for retrieval from highly dense arrays. Laser capture microdissection or laser catapulting can be used to remove specific cells from micropatterned surfaces,[4a, 6] but the former approach works with fixed cells while the latter involves harsh conditions, often resulting in cell injury. Another example of light-activated cell sorting is the micropallet array developed by Albritton et al., in which cells are cultured on polymer microstructures (e.g. photoresist patterns) that can later be dislodged from the surface by laser pulses.[7]
The goal of this study was to employ photocleavable hydrogels for capture and release of cells on microbeads. Hydrogels in general may be bioactive or nonfouling depending on their composition and may be microfabricated using photolithography or soft lithography.[8] Photodegradable hydrogels have garnered considerable attention recently for tissue-engineering applications because of new possibilities for modulating matrix properties or delivering signals in 3D scaffolds.[9] However, we are only aware of one report describing the use of a photocleavable gel for cell capture and release. In this report, Yamaguchi et al. describe a hydrogel comprised of poly(ethylene glycol) connected to lipid moieties by photolabile linkers. Cells bound to lipid moieties on the gel were released upon cleavage of lipid groups by UV light.[10] In contrast, we describe the incorporation of photocleavable groups into the bulk of the hydrogel and demonstrate the capture and release of cells and beads based on photoinitiated degradation of the gel.
Micro- and nanobeads are commonly chosen as solid substrates for diagnostic immunoassays,[11] target DNA detection,[12] and cell-sorting applications[13] because of their large surface area-to-volume ratio, broad range of both coding strategies and functional surface chemistries, and ease of manipulation in microfluidic devices (acoustic, magnetic, mechanical, etc.).[14] In one example, Lam and colleagues developed the one-bead-one-compound (OBOC) combinatorial method to synthesize vast peptide libraries on polystyrene microbeads.[15] High-throughput screening of cell binding on OBOC libraries has been used to identify high-affinity peptide ligands against several cancer cell surface receptors.[16] However, sorting cell-containing beads from cell-free beads requires labor-intensive manual picking of single beads.[17] The goal of the present study was to demonstrate the use of photodegradable hydrogels for cell sorting while addressing a specific technical challenge: the screening cell–microbead interactions.
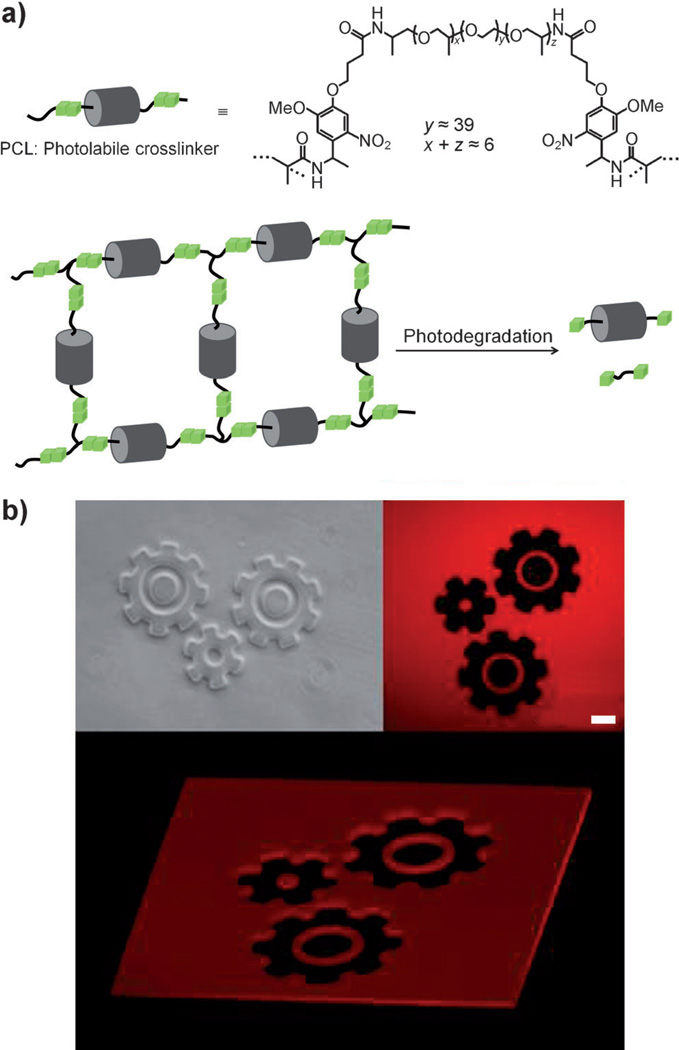
The above-mentioned hydrogel material, herein referred to as photogel, consists of poly(ethylene glycol) (PEG) and a photolabile linker (PLL) containing o-nitrobenzyl groups, which are sensitive to near-UV light (λ = 365 nm). Recently, Anseth and co-workers reported a method for photolabile crosslinker (PCL) synthesis,[18] in which o-nitrobenzyletherbased acrylate monomers were synthesized in an eight-step procedure before coupling to PEG-bis(amine). Here, we developed a novel PCL synthesis route that does not require chromatographic separation and may be accomplished in three steps : 1) Fmoc-PLL coupling to terminal amino groups of O,O’-bis(2-aminopropyl) poly(propylene glycol)-block-poly(ethylene glycol)-block-poly(propylene glycol) 1900 (Jeffamine ED-2001), 2) removal of Fmoc groups, and 3) coupling of methacryl groups. These Jeffamine-containing products can be easily isolated in each step by ether precipitation using a liquid-phase polymer-supported synthesis method.[19] The resulting structure of the PCL is a linear sequence of PLL-methacrylate groups at both ends of Jeffamine connected by amide bonds (see Figure 1a).
Figure 1.
Structure of PCL and photogel polymerization/degradation. a) PCL crosslinking is achieved by radical polymerization using acryl groups. Photogel is degraded by exposure of photolabile o-nitrobenzyl groups to 365 nm UV light. b) Fluorescent confocal microscopy of UV-exposed rhodamine-incorporated photogel shows complete gel degradation with less than 20 µm resolution. Scale bar: 100 µm.
The synthesized PCL was grafted with radical polymer initiators using the methacryl groups and polymerized by exposure to 420 nm UV light in the presence of an appropriate photoinitiator. Subsequent exposure to 365 nm UV light cleaved o-nitrobenzyl groups within the PCL and promoted gel degradation. As highlighted in Figure 1b by the use of rhodamine-containing photogel and standard photolithography, degradation may occur locally, with the feature size determined by the resolution of the chrome photomask.
Figure S1b in the Supporting Information highlights the flexibility of this photopatterning strategy to carry out multistep microfabrication processes whereby microstructures are first fabricated by exposure to 420 nm UV light through a photomask and then augmented by additional photodegradation steps using 365 nm UV light. Scheme 1 illustrates the fabrication of bead arrays and bead-sorting experiments detailed below.
Scheme 1.
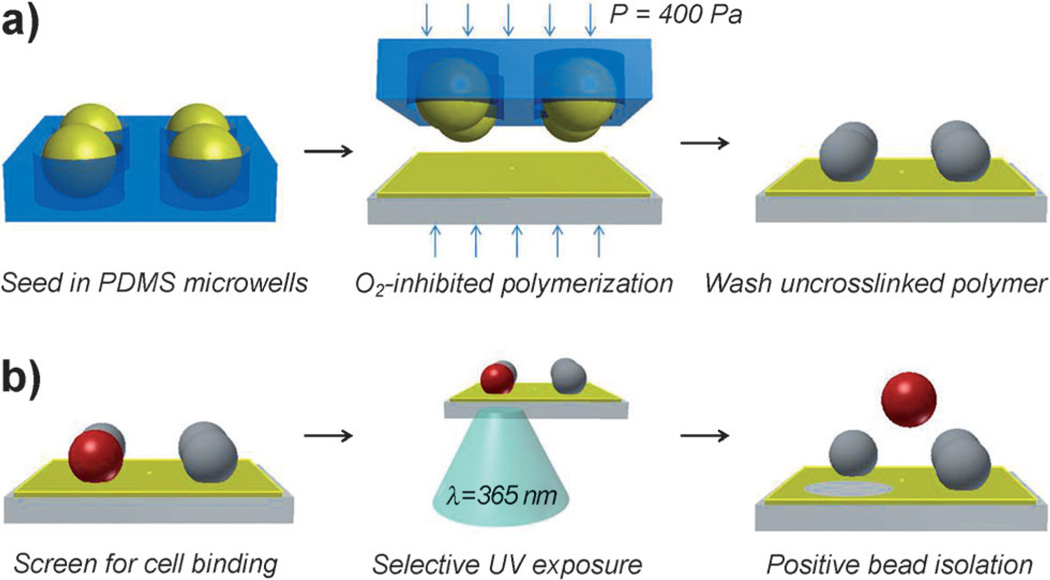
Photogel preparation and process flow for patterning arrays of microbeads for selective bead retrieval. a) PEG-PS beads are swollen with photogel prepolymer, seeded into PDMS microwells, and transferred to a previously formed 15 µm-thick photogel layer on acrylated glass by µTM. Non-uniform polymerization allows uncrosslinked prepolymer to be washed from bead surfaces. b) Microbead arrays are screened for binding activity and positive beads are isolated by degrading photogel with 365 nm UV light.
Our bead patterning technique was inspired by microtransfer molding (µTM), a soft-lithography method whereby a polydimethylsiloxane (PDMS) mold is filled with a liquid precursor, polymerized while in contact with a substrate and then peeled off, leaving molded 3D microstructures on the transfer surface.[20] In our study, this approach was modified to enable transfer of PEG-engrafted polystyrene (PEG-PS) microbead arrays onto glass substrates precoated with photogel. Subsequently, after interaction of the bead arrays with a cell line of interest, the cell-containing beads could be retrieved by simply degrading photogel anchors using 365 nm UV light from a fluorescence microscope (Scheme 1).
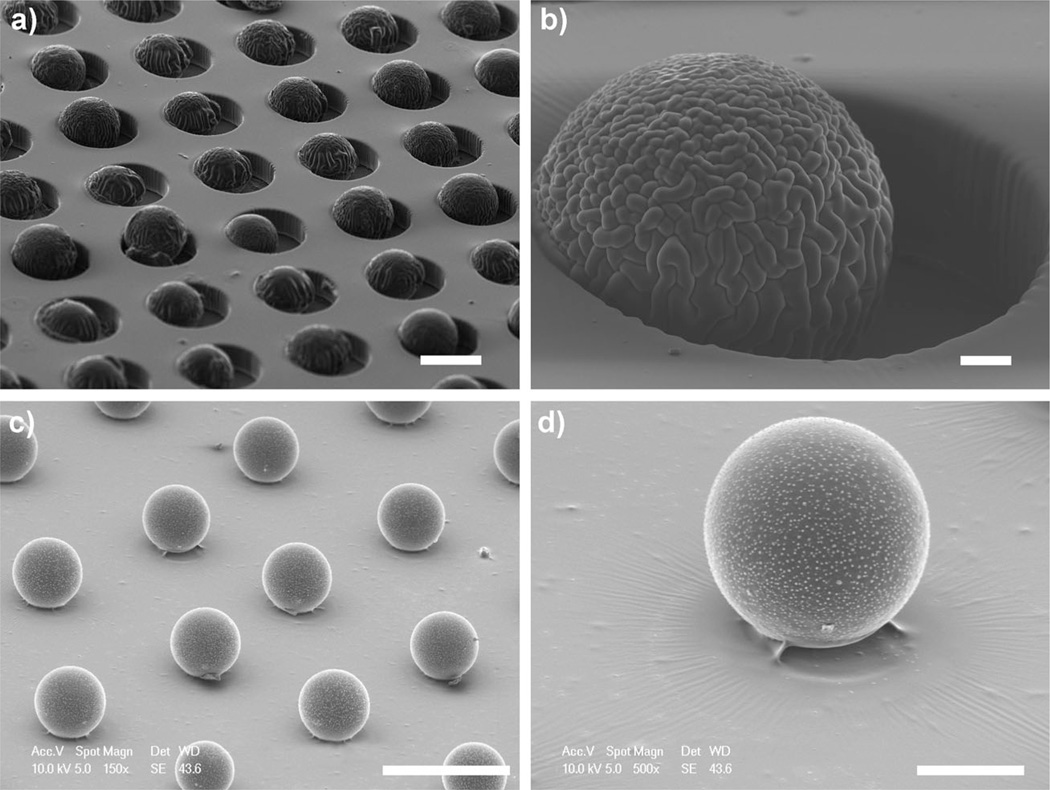
One key design criterion of this system is ensuring that bead surfaces remain available for cell binding while firmly anchored onto the gel substrates. To accomplish this, we took advantage of oxygen-inhibited polymerization in PDMS molds reported by Dendukuri et al.[21] This polymerization strategy leverages the fact that PDMS is permeable to environmental oxygen, which can inhibit free-radical polymerization by forming chain-terminating peroxides. Thus, when carried out inside the PDMS mold, free-radical polymerization occurs most efficiently in regions furthest from PDMS surfaces. This phenomenon has been exploited in continuous-flow lithography to generate polymeric structures in microfluidic devices without channel congestion.[22] In our micropatterning strategy, microbeads were swollen in photogel precursor solution and then seeded into PDMS molds (shown in Figure 2a,b). Subsequently, the bead laden microwell arrays were brought into contact with photogel-coated glass substrates and polymerized with radical polymer initiators (see Supporting Information for polymerization details). As illustrated by the micrographs in Figure 2c,d, only a small region of each bead was anchored to the substrate, while over 90% of the bead surface area, corresponding to those regions more proximal to the gas-permeable PDMS, was available for cell binding after washing uncrosslinked oligomer.
Figure 2.
Scanning electron microscopy (SEM) images of µTM process for patterning microbeads. a,b) 75 µm beads swollen in photogel precursor solution are seeded into “soft” PDMS microwells. c,d) A bead array is transferred and anchored to a photogel substrate by µTM. Oxygen-inhibited polymerization near PDMS surfaces allows washing of uncrosslinked oligomer at bead surfaces after patterning. Scale bars: a) 100 µm, b) 10 µm, c) 200 µm, d) 50 µm.
We observed a tradeoff between transfer yield (i.e. photogel anchor strength) and exposed surface area of the bead, each a function of tunable fabrication conditions. For 75 µm beads with approximately 90% surface exposure (see Figure S2), the combined efficiency of PDMS microwell seeding, µTM, and washing steps yielded (68 ± 11)% bead transfer from the initial 1225 PDMS microwells on a 1 cm2 mold. The transfer yield of beads could be increased further, but at the cost of decreased surface exposure. In addition, several bead sizes (from 20 to 110 µm) were patterned with similar yield by modifying PDMS mold geometry (see the Supporting Information for detailed fabrication methods).
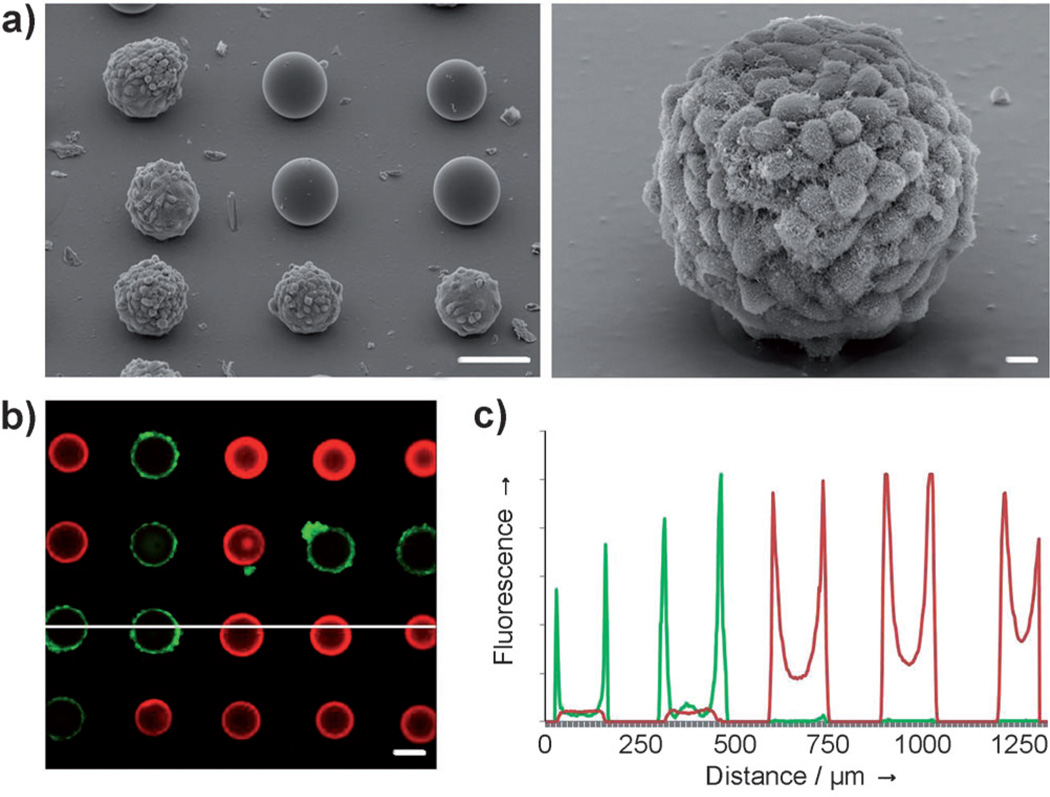
As a proof-of-principle demonstration of screening cell– bead interactions, we mixed blank PEG-PS beads with beads containing arginine–glycine–aspartic acid (RGD) sequences prior to patterning. RGD is a primary integrin-binding motif found in extracellular matrix proteins, and numerous RGD-modified biomaterials have been developed for integrinmediated cell adhesion.[23] Cysteine-terminated, RGD-containing peptides were assembled by a thiol-reactive maleimide-PEG-NHS linker onto amine-functionalized bead surfaces, then mixed randomly with blank acetylated beads (see the Supporting Information for details). Micropatterned arrays with a 1:1 ratio of blank to RGD+ beads were incubated with NIH 3T3 cells, a mouse fibroblast line that expresses RGD-binding surface receptors. These cells are notorious for binding nonspecifically to a variety of substrates and were selected for our study to emphasize nonfouling properties of the photogel and highlight specificity of the screen. 24–48 h after seeding, RGD+ beads were coated with a cell monolayer, while blank beads and photogel were virtually cell free (Figure 3). Over 99% of the RGD+ beads contained cells two days after capture. No fibroblast attachment was observed on beads labeled with nonsense (RGE) peptides (data not shown). When challenged with Hep G2 cells that do not bind to RGD, neither peptide-labeled nor blank beads contained cells (data not shown).
Figure 3.
Specific cell attachment and growth on RGD-labeled beads patterned on a nonfouling photogel surface. a) SEM images of the formation of a 3T3 cell monolayer on functionalized beads adjacent to blank beads (scale bars: left image 100 µm, right image 10 µm). b) Confocal microscopy of calcein-stained (green) 3T3 cells seeded onto a mixed array of “blank” rhodamine-labeled (red) and RGD-labeled beads; after washing, cell attachment and spreading only occurs on RGD-labeled beads (scale bar: 100 µm). c) Fluorescence profile corresponding to the white line in (b).
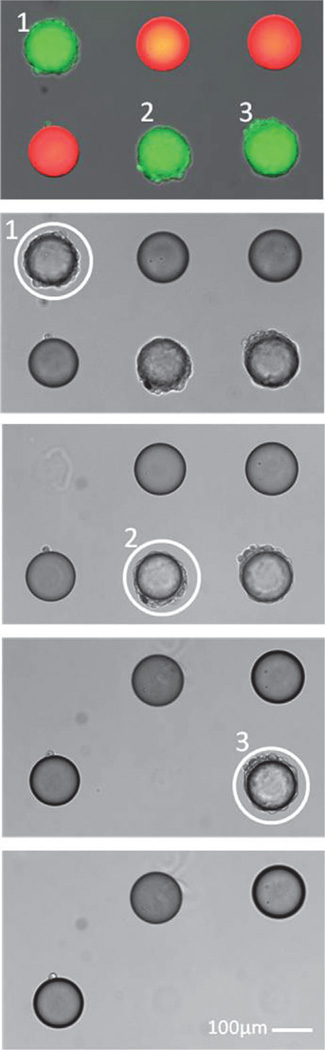
Experiments aimed at optimizing gel degradation (see Figure S3 in the Supporting Information for details) showed that exposure to 600W UV light (6 J cm−2) for 10 s was sufficient to initiate degradation of gel anchors. Using this optimized exposure, individual beads were released from the array after screening for cell binding. Figure 4 shows an example of selective, sequential release of cell-containing beads from an array (a video of bead release is provided in the Supporting Information).
Figure 4.
Simultaneous screening and selective release of beads. Three positive hits are identified by calcein fluorescence imaging, then released sequentially by exposure to 365 nm UV light (indicated by white circles) without disturbing blank (rhodamine labeled, red) beads.
A simple strategy for bead sorting was developed. After incubation with cells, beads were labeled with cell-tracker dye such that cell-containing beads could be easily identified using an upright fluorescence microscope under photogel-compatible light (λ = 488 nm; Figure 4, top panel, green beads). Collection was accomplished by placing glass substrates with bead arrays face down into a collection chamber fabricated from PDMS (see Figure S4 in the Supporting Information for apparatus scheme) and then illuminating specific bead anchors with 365 nm UV light from the microscope. Released beads then settled to the bottom of the collection chamber and could be retrieved from there. To prevent functional damage, it was important to limit cell irradiation. We have previously observed high cell viability after up to 10 min UV irradiation at the intensity used for gel degradation.[24] Further, direct exposure of cells to UV light was minimized by illuminating through the glass slide and at the focal plane of photogel anchors. Figure S5 illustrates how focusing through a microscope aperture corresponds to locally degraded regions of photogel for selective microbead release adjacent to undisturbed neighboring beads. It should be noted that with the present photogel formulation, bead release occurred approximately 1 h after exposure, likely because of disentanglement and diffusion of polymer chains after cleavage. The time to release will be further reduced in the future by decreasing the molecular weight of the photogel precursor polymer and by introducing agitation or thermal energy.
In summary, we have developed a cell- and bead-sorting approach by combining photopatternable hydrogels and microbead arrays. We described a new method for the synthesis of photogel precursors and demonstrated that the photogel acts in a manner similar to photoresist. Interestingly, the same photogel may be polymerized or degraded depending on the wavelength of the UV light and therefore combines features of both positive- and negative-tone resists. We highlighted one application of such a photogel for screening cell binding to microbead libraries. When incubated with arrays of microbeads that are partially embedded in the photogel, cells were found to attach on RGD-containing beads but not on blank beads or beads that carry nonsense (control) peptide. Minimal cell attachment occurred on a nonfouling PEG-based photogel. Beads that contained cells could then be sorted from the array simply by short exposure of the anchoring photogel to UV light through a microscope objective. Microbeads offer an advantageous reagent delivery vehicle, and our patterning technique is theoretically not limited to specific bead sizes or surface chemistries common to bead functionalization. Consequently, we anticipate that microbead patterning in photolabile hydrogels will offer a flexible platform for bead-based biological studies. More broadly, we envision micropatterned photogels described here as having many applications related to cell capture and release for screening, diagnostics, or tissue engineering.
Supplementary Material
Footnotes
The authors gratefully acknowledge Sven Hausner and Kevin Lau for their assistance with experimental design. This work was supported by an NIH grant (C4EB012836-01) to J.S. and an NSF EFRI grant (0937997) to A.R.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201303965.
Contributor Information
Christian Siltanen, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
Dong-Sik Shin, Email: dshin@ucdavis.edu, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
Julie Sutcliffe, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA); Division of Hematology/Oncology, Department of Internal Medicine, Center for Molecular and Genomic Imaging, University of California Davis, Davis, CA 95616 (USA).
Alexander Revzin, Email: arevzin@ucdavis.edu, Department of Biomedical Engineering, University of California Davis, One Shields Ave, Davis, CA 95616 (USA).
References
- 1.Michelini E, Cevenini L, Mezzanotte L, Coppa A, Roda A. Anal. Bioanal. Chem. 2010;398:227–238. doi: 10.1007/s00216-010-3933-z. [DOI] [PubMed] [Google Scholar]
- 2.a) Revzin A, Maverakis E, Chang HC. Biomicrofluidics. 2012;6:021301–02130113. doi: 10.1063/1.4706845. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu Y, Matharu Z, Howland MC, Revzin A, Simonian AL. Anal. Bioanal. Chem. 2012;404:1181–1196. doi: 10.1007/s00216-012-6149-6. [DOI] [PubMed] [Google Scholar]; c) Nakanishi J, Takarada T, Yamaguchi K, Maeda M. Anal. Sci. JACS. 2008;24:67–72. doi: 10.2116/analsci.24.67. [DOI] [PubMed] [Google Scholar]
- 3.Nagrath S, Sequist L, Maheswaran S, Bell D, Irimia D, Ulkus L, Smith M, Kwak E, Digumarthy S, Muzikansky A, Ryan P, Balis U, Tompkins R, Haber D, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Revzin A, Sekine K, Sin A, Tompkins R, Toner M. Lab Chip. 2005;5:30–37. doi: 10.1039/b405557h. [DOI] [PubMed] [Google Scholar]; b) Zhu H, Stybayeva G, Silangcruz J, Yan J, Ramanculov E, Dandekar S, George MD, Revzin A. Anal. Chem. 2009;81:8150–8156. doi: 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Varadarajan N, Julg B, Yamanaka YJ, Chen H, Ogunniyi AO, McAndrew E, Porter LC, Piechocka-Trocha A, Hill BJ, Douek DC, Pereyra F, Walker BD, Love JC. J. Clin. Invest. 2011;121:4322–4331. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Story CM, Papa E, Hu CC, Ronan JL, Herlihy K, Ploegh HL, Love JC. Proc. Natl. Acad. Sci. USA. 2008;105:17902–17907. doi: 10.1073/pnas.0805470105. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab Chip. 2010;10:1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A, Heath JR. Nat. Med. 2011;17:738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Yousaf M, Houseman B, Mrksich M. Proc. Natl. Acad. Sci. USA. 2001;98:5992–5996. doi: 10.1073/pnas.101112898. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhu H, Yan J, Revzin A. Colloids Surf. B. 2008;64:260–268. doi: 10.1016/j.colsurfb.2008.02.010. [DOI] [PubMed] [Google Scholar]; c) Shah SS, Kim M, Foster E, Vu T, Patel D, Chen LJ, Verkhoturov SV, Schweikert E, Tae G, Revzin A. Anal. Bioanal. Chem. 2012;402:1847–1856. doi: 10.1007/s00216-011-5613-z. [DOI] [PubMed] [Google Scholar]; d) Li Y, Yuan B, Ji H, Han D, Chen SQ, Tian F, Jiang XY. Angew. Chem. 2007;119:1112–1114. doi: 10.1002/anie.200603844. Angew. Chem. Int. Ed.2007, 46, 1094 – 1096; [DOI] [PubMed] [Google Scholar]; e) Inaba R, Khademhosseini A, Suzuki H, Fukuda J. Biomaterials. 2009;30:3573–3579. doi: 10.1016/j.biomaterials.2009.03.045. [DOI] [PubMed] [Google Scholar]
- 6.Lee JY, Jones C, Zern MA, Revzin A. Anal. Chem. 2006;78:8305–8312. doi: 10.1021/ac0613333. [DOI] [PubMed] [Google Scholar]
- 7.a) Quinto-Su P, To’a Salazar G, Sims C, Allbritton N, Venugopalan V. Anal. Chem. 2008;80:4675–4679. doi: 10.1021/ac800129a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shadpour H, Sims CE, Thresher RJ, Allbritton NL. Cytometry Part A. 2009;75:121–129. doi: 10.1002/cyto.a.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang Y, To’a Salazar G, Pai J-H, Shadpour H, Sims CE, Allbritton NL. Lab Chip. 2008;8:734–740. doi: 10.1039/b800286j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Suh KY, Seong J, Khademhosseini A, Laibinis PE, Langer R. Biomaterials. 2004;25:557–563. doi: 10.1016/s0142-9612(03)00543-x. [DOI] [PubMed] [Google Scholar]; b) Revzin A, Russell RJ, Yadavalli VK, Koh W-G, Deister C, Hile DD, Mellott MB, Pishko MV. Langmuir. 2001;17:5440–5447. doi: 10.1021/la010075w. [DOI] [PubMed] [Google Scholar]
- 9.a) DeForest C, Anseth K. Annu. Rev. Chem. Biomol. Eng. 2012;3:421–444. doi: 10.1146/annurev-chembioeng-062011-080945. [DOI] [PubMed] [Google Scholar]; b) Griffin DR, Kasko AM. J. Am. Chem. Soc. 2012;134:13103–13107. doi: 10.1021/ja305280w. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi S, Yamahira S, Kikuchi K, Sumaru K, Kanamori T, Nagamune T. Angew. Chem. 2012;124:132–135. doi: 10.1002/anie.201106106. Angew. Chem. Int. Ed.2012, 51, 128 – 131. [DOI] [PubMed] [Google Scholar]
- 11.Khan I, Ravindran R, Yee J, Ziman M, Lewinsohn D, Gennaro M, Flynn J, Goulding C, DeRiemer K, Lerche N, Luciw P. Clin. Vaccine Immunol. 2008;15:433–438. doi: 10.1128/CVI.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Jin R, Mirkin C. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Huang N-T, Oh B, Lam R, Fan R, Cornell T, Shanley T, Kurabayashi K, Fu J. Adv. Healthcare Mater. 2013;2:965–975. doi: 10.1002/adhm.201200378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Sivagnanam V, Song B, Vandevyver C, Gijs MAM. Anal. Chem. 2009;81:6509–6515. doi: 10.1021/ac9009319. [DOI] [PubMed] [Google Scholar]; b) Andersson H, Jö;nsson C, Moberg C, Stemme G. Electrophoresis. 2001;22:3876–3882. doi: 10.1002/1522-2683(200110)22:18<3876::AID-ELPS3876>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 15.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 16.a) Peng L, Liu R, Marik J, Wang X, Takada Y, Lam KS. Nat. Chem. Biol. 2006;2:381–389. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]; b) Gagnon MKJ, Hausner SH, Marik J, Abbey CK, Marshall JF, Sutcliffe JL. Proc. Natl. Acad. Sci. USA. 2009;106:17904–17909. doi: 10.1073/pnas.0906925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan C-X, Lam KS. Mol. Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 18.Kloxin A, Tibbitt M, Anseth K. Nat. Protoc. 2010;5:1867–1887. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Han H, Wolfe MM, Brenner S, Janda KD. Proc. Natl. Acad. Sci. USA. 1995;92:6419–6423. doi: 10.1073/pnas.92.14.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) He L, Fullenkamp DE, Rivera JG, Messersmith PB. Chem. Commun. 2011;47:7497–7499. doi: 10.1039/c1cc11928a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X-M, Xia Y, Whitesides GM. Adv. Mater. 1996;8:837–840. [Google Scholar]
- 21.Dendukuri D, Panda P, Haghgooie R, Kim JM, Hatton TA, Doyle PS. Macromolecules. 2008;41:8547–8556. [Google Scholar]
- 22.Dendukuri D, Pregibon DC, Collins J, Hatton TA, Doyle PS. Nat. Mater. 2006;5:365–369. doi: 10.1038/nmat1617. [DOI] [PubMed] [Google Scholar]
- 23.a) Ruoslahti E, Pierschbacher MD. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]; b) Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 24.Shin DS, Seo JH, Sutcliffe JL, Revzin A. Chem. Commun. 2011;47:11942–11944. doi: 10.1039/c1cc15046d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.