Abstract
Matrix metalloproteinases (MMPs) play a central role in the breakdown of the extracellular matrix and are typically upregulated in cancer cells. The goal of the present study is to develop microwells suitable for capture of cells and detection of cell-secreted proteases. Hydrogel microwells comprised of poly(ethylene glycol) (PEG) were photopatterned on glass and modified with ligands to promote cell adhesion. To sense protease release, peptides cleavable by MMP9 were designed to contain a donor/acceptor FRET pair (FITC and DABCYL). These sensing molecules were incorporated into the walls of the hydrogel wells to enable a detection scheme where cells captured within the wells secreted protease molecules which diffused into the gel, cleaved the peptide, and caused a fluorescence signal to come on. By challenging sensing hydrogel microstructures to known concentrations of recombinant MMP9, the limit of detection was determined to be 0.625 nM with a linear range extending to 40 nM. To enhance sensitivity and to limit cross-talk between adjacent sensing sites, microwell arrays containing small groups (~20 cells/well) of lymphoma cells were integrated into reconfigurable PDMS microfluidic devices. Using this combination of sensing hydrogel microwells and reconfigurable microfluidics, detection of MMP9 release from as few as 11 cells was demonstrated. Smart hydrogel microstructures capable of sequestering small groups of cells and sensing cell function have multiple applications ranging from diagnostics to cell/tissue engineering. Further development of this technology will include single-cell analysis and function-based cell sorting capabilities.
INTRODUCTION
Matrix metalloproteinases (MMPs) play a pivotal role in the modulation of extracellular matrices (ECMs) by degrading almost all ECM components.1–5 Especially, matrix metalloproteinase 9 (MMP9) hydrolyzes type IV collagen, laminin, and fibronectin, which are the major components of the basement membranes.4 The proteolytic activities of MMP9 are closely correlated with morphogenesis, inflammation, tissue remodeling, and various pathological processes including cancer cell invasion and metastasis.1–4 The expression and activation of MMP9 are at much higher levels in almost all human cancers in comparison to normal tissues, leading to facilitation of cancer invasion mainly via degradation of basement membranes.3,4,6 Therefore, the development of sensitive and accurate sensing platforms for MMPs has received significant attention for clinical cancer diagnosis and treatment.
Recently, our laboratory demonstrated an electrochemistry-based sensing platform for detecting protease secretion from cells by combining redox-labeled peptide surfaces with microfluidic devices.7 In this device, the cleavage of redox-labeled peptides by cell-secreted MMP9 resulted in a decrease in electrochemical signal, generating a 3-fold higher electrochemical signal from ~400 activated cells in comparison to quiescent cells.7 Various other methods based on immunoassay,8 surface plasmon resonance (SPR),9 and fluorescence10,11 have been developed for the analysis of MMP expression and secretion. The fluorescence resonance energy transfer (FRET) assay, in particular, is considered to be one of the most sensitive analytical techniques.10,12 By the design of protease-cleavable peptides to contain with donor and acceptor fluorophore pairs, it is possible to adapt FRET-based signal transduction for protease detection.
While the level of research activity in protease detection is quite high, fairly few reports have described protease detection from specific cells or groups of cells.7,9 The key challenge here is facile integration of cells and sensing elements. In our previous study,7 micropatterned surfaces containing protease sensing electrodes and cell attachment sites were used for cell sensor integration. However, the need to fabricate individual electrodes for each group of cells introduces complexity and limits applications requiring high-throughput screening. The goal of this study is to design an optical protease sensing strategy more amenable to higher throughput detection from cells. To satisfy this goal, we focused on photopatterning of poly(ethylene glycol) (PEG) hydrogels. These are nonfouling hydrogels that have been used extensively for controlling cell attachment on surfaces7,13 and also for encapsulation of biorecognition elements such as enzymes and antibodies.14 In terms of biosensing, the use of hydrogels allows both improvement of the loading capacity and stabilization of sensing molecules. In the past, our laboratory has demonstrated the encapsulation of enzymes and chromophores into hydrogel microstructures for biosensing.15,16 We have also made extensive use of hydrogel micropatterning to create microwells for sequestering cells and forming them into arrays.16,17
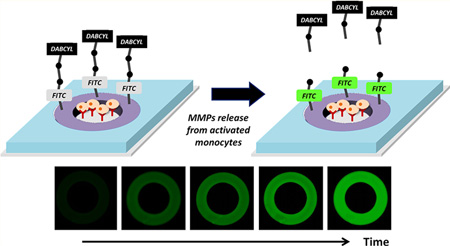
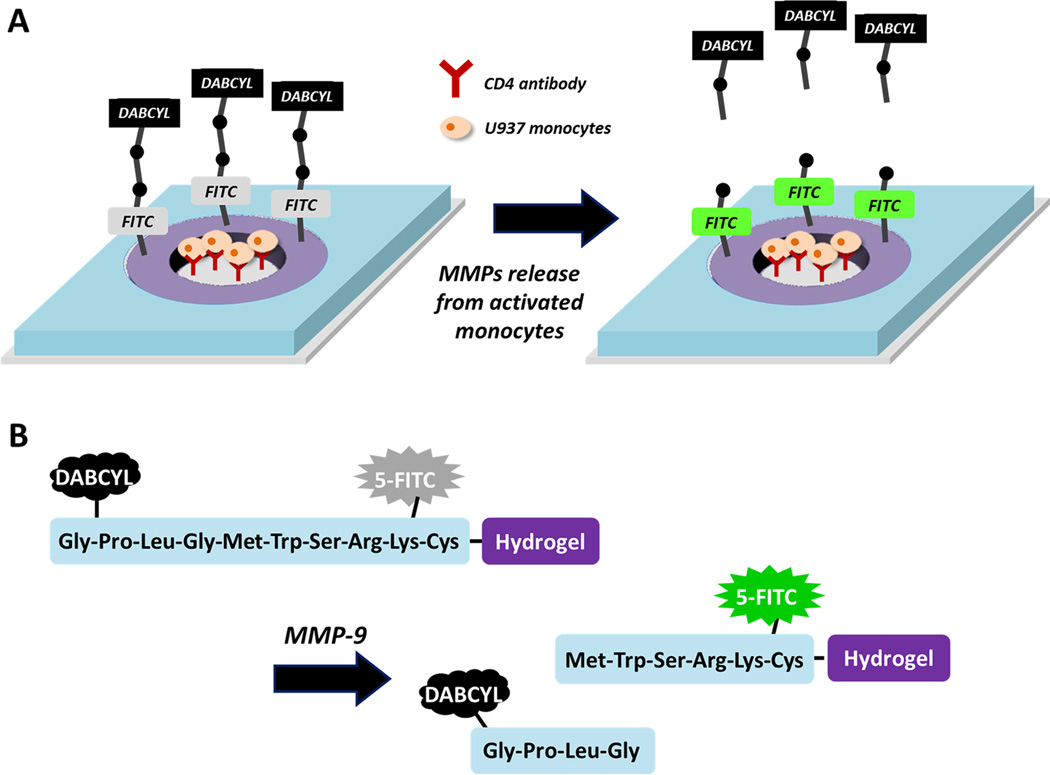
In this work, we sought to create hydrogel microwells sequestering cells and sensing cell-secreted MMP9. MMP9-specific peptides were modified with a donor/acceptor FRET pair (FITC and DABCYL) and covalently linked into the gel. Figure 1A shows the design of sensing surfaces where peptide-carrying hydrogel rings were used to define sites for cell adhesion. Once captured, the cells were stimulated to produce proteases, which in turn diffused into the gel, cleaving the acceptor chromophore DABCYL and causing fluorescence to come on (Figure 1B). To enhance sensitivity and to limit cross-talk between adjacent sensing sites, the microwell arrays were integrated into a reconfigurable microfluidic device. This device contained a microstructured membrane that descended from the roof onto microwells with cells, isolating microwells, and decreasing the volume around the individual well to ~40 nL. Combining sensing hydrogel wells and reconfigurable microfluidics made it possible to detect protease release from as few as 11 lymphoma cells. The microsystem combining hydrogel microwells for cell sequestration/sensing with reconfigurable microfluidics will have applications in cell-based diagnostics and high-throughput screening.
Figure 1.
Sensing MMP9 secretion from cells. (A) Individual protease sensing element consisting of hydrogel rings with incorporated FRET peptides (purple) surrounded by nonsensing PEG gel layer serving as negative control (blue). The glass regions opened in the gel layer were functionalized with antibodies for cell capture. Lymphoma cells bound within the hydrogel rings upon stimulation released MMP9. Protease molecules diffused into the gel and cleaved FRET peptide, causing fluorescence to come on. (B) Design of MMP9 sensing peptide (Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Lys-Cys) with a cleavage site between Gly and Met.
EXPERIMENTAL SECTION
Materials
Matrix metalloproteinase 9 (MMP9, human, recombinant), poly(ethylene glycol) diacrylate (PEG-DA, MW 700), poly(ethylene glycol) (PEG, MW 200), and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma-Aldrich. Acrylic poly(ethylene glycol) maleimide (Acryl-PEG-Mal, MW3400) was obtained from Laysan Bio (Arab, AL). 1-[4-(2-Hydroxyethoxy)phenyl]-2-hydroxy-2-methyl-1-propan-1-one (Irgacure 2959) was purchased from Ciba Specialty Chemicals (Basel, Switzerland). Phosphate-buffered saline (PBS) was purchased from TEKnova (Hollister, CA). Mouse antihuman CD4 antibodies (anti-CD4 Ab) were obtained from Beckman Coulter (Miami, FL). Glass slides (75 × 25 mm2) and cover glasses (24 × 30 × 0.13 mm) were purchased from Fisher Scientific (Pittsburgh, PA). (3-Acryloxypropyl)trichlorosilane was purchased from Gelest (Morrisville, PA). The peptide Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Lys-Cys (GPLGMWSRKC) was synthesized by GL Biochem (Shanghai, People’s Republic of China). All other chemicals were purchased from Sigma (St. Louis, MO) or Aldrich Chemicals (Milwaukee, WI). MATLAB (MathWorks Inc., Natick, MA) and COMSOL Multiphysics (COMSOL, Inc., Burlington, MA) were used for modeling of peptide cleavage and calculating the diffusion coefficient of hydrogel and MMP9 release rate for cells.
Lymphoma cells (U-937 cells) were purchased from the American Type Culture Collection (ATCC) and cultured in 10% (v/v) fetal bovine serum (FBS, Invitrogen, Carlsbad, CA), 100 U/mL of penicillin, and 100 µg/mL of streptomycin in RPMI-1640 media (VWR, West Chester, PA) at 37 °C under a humidified 5% CO2 atmosphere.
Micropatterning of Composite (Functional/Nonfunctional) Hydrogel Microwells
The glass substrates were cleaned in an oxygen plasma chamber (YES-R3, San Jose, CA) at 300 W for 15 min and then incubated in 0.1% (v/v) (3-acryloxypropyl)trichlorosilane in anhydrous toluene for 1 h under a nitrogen purge. The slides were rinsed with fresh toluene, dried under nitrogen, and then cured at 100 °C for 2 h. Acryl silane modified substrates were used immediately or placed in a desiccator until future use. Sensing hydrogel rings (inner diameter 100 µm, outer diameter 200 µm) were prepared by UV-initiated polymerization of PEG-DA and Acryl-PEG-Mal. Monomer solutions for functional hydrogel were prepared by dissolving 20% (v/v) PEG-DA (MW 700), 40% (v/v) PEG (MW 200), 10% (v/v) of 20 mM Acryl-PEG-Mal, and 1% Irgacure 2959 in PBS. Monomer solutions were loaded onto the silane-treated glass and then covered with a cover glass (24 × 30 × 0.13 mm) to create a uniform layer of monomer solutions. The slides were irradiated through the photomask for 1.5 s by a 365 nm UV light source (80 mW/cm2; OmniCure Series 1000, Lumen Dynamics Group, Mississauga, Ontario, Canada), washed with DI water, and dried using nitrogen gas.
To define the site for cell attachment, the silanized glass substrates containing sensing hydrogel rings were further patterned with nonfunctional PEG hydrogel microwells (diameter 200 µm). A solution of the monomer mixture (20% PEG-DA, 40% PEG, 1% Irgacure 2959) was pipetted onto substrates, registered with existing patterns using a photomask with fiduciary marks, and exposed using a mask aligner (ABM, Inc., San Jose, CA, USA).
Preparation of Sensing Hydrogel Microwells
The MMP9-specific peptide Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Lys-Cys was conjugated to free maleimide groups in functional hydrogels using thiols on cysteine by incubating micropatterned hydrogel substrates with a solution of peptide (2 mM peptide and 4 mM diisopropylethylamine (DIPEA) in DMF). The immobilization of peptide molecules was first confirmed using MMP9-specific peptides labeled with a simple fluorophore (5-TAMRA). More specifically, a 5-TAMRA-labeled peptide solution containing 2 mM peptide and 4 mM DIPEA in DMF was added to sensing hydrogel microwell substrates at room temperature for 2 h, followed by rinsing with DI water. The thiol–maleimide coupling reaction between peptides and the sensing hydrogel could be verified by measuring the fluorescence of 5-TAMRA-labeled peptides immobilized on sensing hydrogel surfaces. By incorporating FRET peptides instead of 5-TAMRA-labeled peptides into functional hydrogels in the same way, protease-sensing microwells were prepared, containing a sensing hydrogel region and a barrier hydrogel region. In the final step, surfaces were incubated at room temperature for 1 h with anti-CD4 Ab (100 µg/mL) in 1 × PBS, followed by washing with DI water and drying using nitrogen gas.
Fluorescence Signal Detection of Proteolytic Cleavage
FRET-peptide-containing sensing hydrogel surfaces were exposed to recombinant MMP9 (0.625–40 nM) dissolved in working buffer (50 mM Tris-HCl, 1 mM CaCl2, and 0.05% Triton X-100; pH 7.5). The proteolytic cleavage reaction of peptide immobilized on hydrogel surfaces was monitored using a Zeiss 200 M epi-fluorescence microscope (Carl Zeiss Micro-Imaging, Inc. Thornwood, NY) equipped with an AxioCam MRm (CCD monochrome, 1300 pixels × 1030 pixels). In this paper, fluorescence associated with 5-TAMRA- and FRET-labeled peptides was monitored using 550±25 nm/605 ±70 nm TAMRA excitation/emission filters and 470 ± 40 nm/525 ± 50 nm excitation/emission filter for FITC. Acquisition of fluorescence images and their analysis were carried out using AxioVision software (Carl Zeiss MicroImaging, Inc. Thornwood, NY).
Fabrication of Reconfigurable Microfluidic Device
Reconfigurable microfluidic devices with a reversibly collapsible PDMS membrane were fabricated through a multilayer soft lithography, as described in previous reports.18 In brief, the prepolymer solution of PDMS (Sylgard 184, Dow Corning), mixed at a 10:1 curing ratio, was poured onto a photoresist-patterned substrate and placed under vacuum to remove air bubbles. The two-layer microfluidic device is composed of a bottom flow layer (400 µm thickness) and a top control layer (1 cm thicnessk). The control layer was baked first for 25 min at 70 °C, and the flow layer was baked for 20 min while holes were punched for the control layer. Then, the two layers were carefully aligned by eye and adhered together. Finally, the devices were baked for 1 h to become fully cured. Afterward, the devices were cut out and used for experiments. The vacuum-actuated reconfigurable microfluidic device was aligned over the sensing hydrogel microwells to facilitate cell seeding in the open configuration (vacuum applied to the control layer). Upon capturing cells in the microwells and stimulating them, the vacuum was released and the PDMS features descended around the cells/sensing gel microwells in order to monitor MMP secretions from cells in real time.
Monitoring of MMP9 Secretion by Lymphoma cells
A reconfigurable double-channel microfluidic device was integrated to the anti-CD4 Ab immobilized sensing hydrogel substrates for real-time monitoring of MMP9 release from lymphoma cells. U-937 cells suspended in serum-free and Phenol Red free RPMI-1640 media were infused into the channel at 1 µL/min for 20 min. After removing unbound cells by washing with PBS at 10 µL/min for 20 min, a mitogenic solution containing 100 ng/mL of PMA in serum-free and Phenol Red free RPMI-1640 media was introduced into the channel at 10 µL/min for 10 min. PMA stimulation promotes monocyte to macrophage differentiation and upregulates MMP production.2,5 Subsequently, the microfluidic device was actuated to descend the roof onto microwells. Fluorescence signals from sensing hydrogel microwells (n = 4) were monitored at room temperature 20 min after addition of the mitogenic solution. Time-lapse images of sensing hydrogels were acquired at 10 min intervals for a total of 2 h.
RESULTS AND DISCUSSION
This paper describes novel protease-sensing hydrogel microwells integrated with reconfigurable microfluidic devices to enable sensitive detection of cell-secreted MMP9. Peptides carrying a FRET pair were integrated into hydrogel microwells to enable time-resolved detection of protease release from as few as 11 cells.
Optimizing Peptide Loading into Hydrogel Microstructures
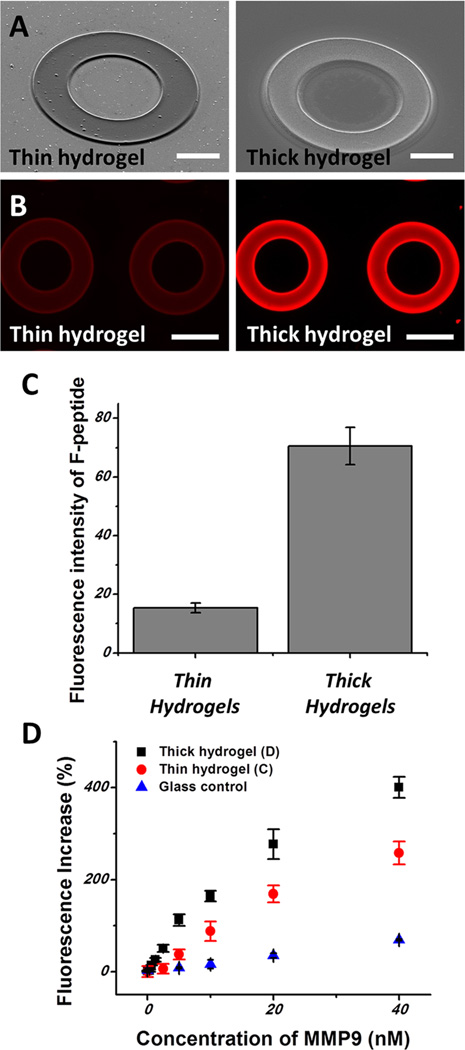
Three variants of protease sensing surfaces were prepared and compared: peptide-carrying hydrogel microstructures of two thicknesses, 5 and 15 µm (Figure 2A), and also planar glass substrates. The planar glass substrates were also prepared on acryl silane modified surfaces in a way similar to that used for hydrogel substrates. A solution containing 10% of 20 mM Acryl-PEG-Mal linkers and 1% Irgacure 2959 was loaded onto surfaces and exposed to 365 nm UV light for 1.5 s. The thickness of the hydrogel was tuned by changing the volume of prepolymer during the patterning process. Cysteine-terminated peptides were incorporated into the gel using maleimide–thiol chemistry and bound to glass substrates via Acryl-PEG-Mal linkers.
Figure 2.
(A) Scanning electronic microscope images of thin hydrogel and thick hydrogel (scale bar 50 µm). (B) Fluorescence images of thin and thick hydrogels after immobilization of TAMRA-peptides (scale bar 100 µm). (C) Fluorescence intensities of TAMRA-peptide containing thin and thick hydrogel patterns, respectively. (D) Fluorescence signals after incubating peptide-containing hydrogels and glass control with different concentrations of MMP9 for 90 min.
To check the loading capacity of various substrates, peptides were functionalized with a fluorophore (TAMRA). The peptides were integrated onto maleimide-containing substrates via thiols on the terminal cysteine of the peptide. Figure 2B,C shows results comparing fluorescence signals from thin vs thick hydrogel rings. As seen from these data, a ~4-fold higher signal was observed from thicker (15 µm) gel rings.
In the next set of experiments, surfaces/gels were functionalized with FRET peptides and challenged with different concentrations of recombinant MMP9 to determine calibration curves for three types of sensing surfaces. Figure 2D demonstrates that the weakest responses and lowest sensitivity were observed from planar glass substrates, whereas the highest sensitivity responses were seen with thick (15 µm) gel rings. Defining sensitivity as the slope of the calibration curves in Figure 2D (change in fluorescence signal per change in analyte concentration), the sensitivities of thick hydrogels, thin hydrogels, and glass surfaces were 9.98, 6.44, and 1.66 nM−1, respectively. On the basis of these studies, 15 µm hydrogel rings were selected for subsequent cell detection experiments.
While certain proteases, for example MMP2 and MMP9, share cleavable peptide domains and may not be distinguished on the basis of enzymatic activity, other types of proteases have limited cross-reactivity. In a previous study employing electrical detection, we demonstrated that MMP9 cleavable peptide was not responsive to proteases such as tumor necrosis factor-α-converting enzyme (TACE) and urokinase-type plasminogen activator (uPA).7 To further demonstrate that detection was amino acid sequence dependent, we compared MMP9 digestion of the sensing peptide (GPLGMWSRKC) and nonsensing peptide (GPLGmWSRKC), where l-Met was replaced with d-Met. As seen from the results in Figure S-1 (Supporting Information), gels incorporating nonsensing peptides did not have an appreciable response when challenged with recombinant MMP9, suggesting specificity of detection. In another specificity study, we mitogenically activated U937 monocytes and primary hepatocytes and then challenged protease sensing peptides with culture media collected from activated cells. As can be seen from Figure S-2 (Supporting Information), only media collected from monocytes resulted in a fluorescence increase. This result was expected, since primary hepatocytes should not be producing proteases, whereas monocyte-like U937 cells are known to release proteases. Taken together with our previous study,7 these experiments support the notion of sensing peptides being specific for MMP9 molecules.
Integration with Reconfigurable Microfluidics Devices
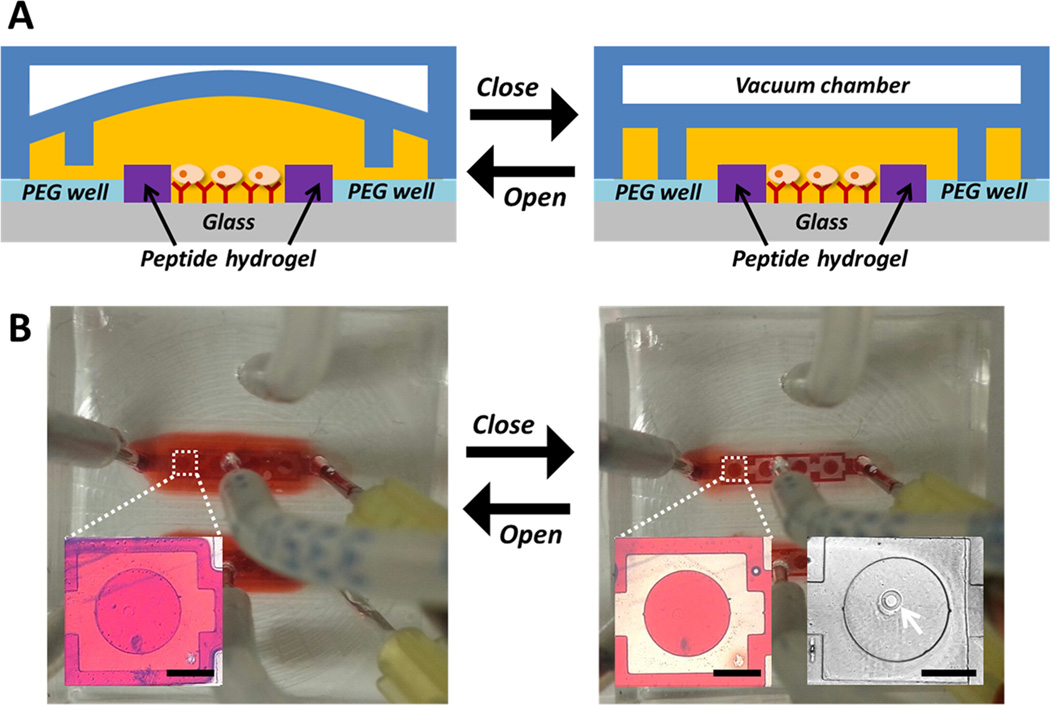
Reconfigurable microfluidic devices with a reversibly collapsible membrane were utilized to reduce the volume around sensing microwells, thereby enhancing the sensitivity during cell secretion experiments. Upon actuation of the device the membrane with microchambers descended on the sensing hydrogel microwells. Parts A and B of Figure 3 show the working principle of a reconfigurable device integrated with a protease sensing hydrogel structure. When vacuum is applied to the upper layer of the microfluidic device, the reversibly collapsible membrane is raised and red dye is distributed throughout (open state). When the vacuum is released (closed state), the roof collapses, making a small chamber with a volume of ~40 nL that limits the diffusion of analytes, amplifying the signal.
Figure 3.
(A) Drawing of the reconfigurable microfluidic device, and (B) photographs and microscope images showing the working principle using red dye. The white arrow in B indicates peptide sensing hydrogel with an Ab-modified region. Scale bar: 500 µm.
Monitoring MMP9 Secretion of Lymphoma Cells
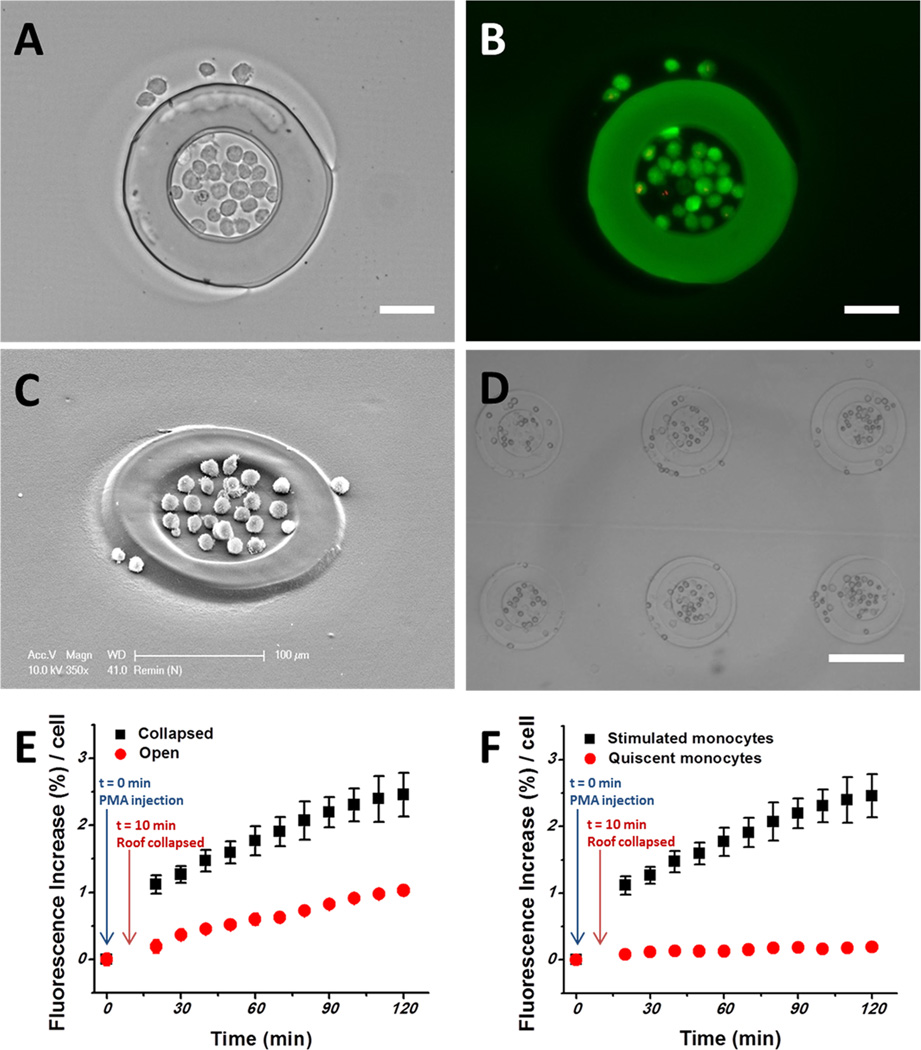
Sensing of cell-secreted MMP9 with FRET-peptide containing hydrogel microwells was investigated. The microwells were modified with antibodies to capture U-937 cells, model lymphoma cells, which have been known to secrete MMP9 under mitogenic activation.2,5 A small group of U-937 cells (24 ± 4) was captured onto the Ab-modified regions surrounded by 100 µm diameter hydrogel microwells (Figure 4A–D). The viability of captured U-937 cells was determined to be 95% ± 1.2 by using a live/dead viability/cytotoxicity assay, as shown in Figure 4B.
Figure 4.
Monitoring MMP9 secretory activities. (A) U-937 lymphoma cells captured inside the sensing hydrogel microwell. This hydrogel microwell (inner diameter 100 µm) could capture 24 ± 4 cells (n = 4). Scale bar: 50 µm. (B) Live/dead staining (green, live; red, dead). Scale bar: 50 µm. (C, D) Scanning electron microscope and optic images of U-937 cells captured mostly inside the peptide-containing hydrogel microwell. (E) Change in fluorescence signal due to cell secretion in the collapsed (black) vs open (red) microfluidic device. (F) Detection from activated (black) vs quiescent cells in the collapse state of the reconfigurable microfluidic device. Change in fluorescence for mitogenically activated cells (black) and quiescent cells (red).
MMP9 secretion from U-937 cells that were mitogenically activated by PMA2,5 was monitored for 2 h and analyzed quantitatively by percent fluorescence increase per cell. To characterize signal enhancement achieved with a reconfigurable microfluidic device, fluorescence of MMP9-sensing hydrogels was monitored in open and collapsed microfluidic channels. Figure 4E shows that sensing hydrogels in the collapsed state generate 2-fold higher signals than those in the open state, highlighting the advantage of volume control enabled by reconfigurable microfluidics. Figure 4F shows that, in comparison to unstimulated controls, mitogenically activated cells produced a 5.5-fold higher signal. Additional experiments were conducted to vary the number of cells captured inside individual sensing wells. As seen from Figure S-3 (Supporting Information), a detectable signal was observed from 11 cells. We believe that with further refinement of the sensing gels and microfluidic devices single-cell protease detection may be possible.
Modeling To Quantify Cell-Secreted MMP9
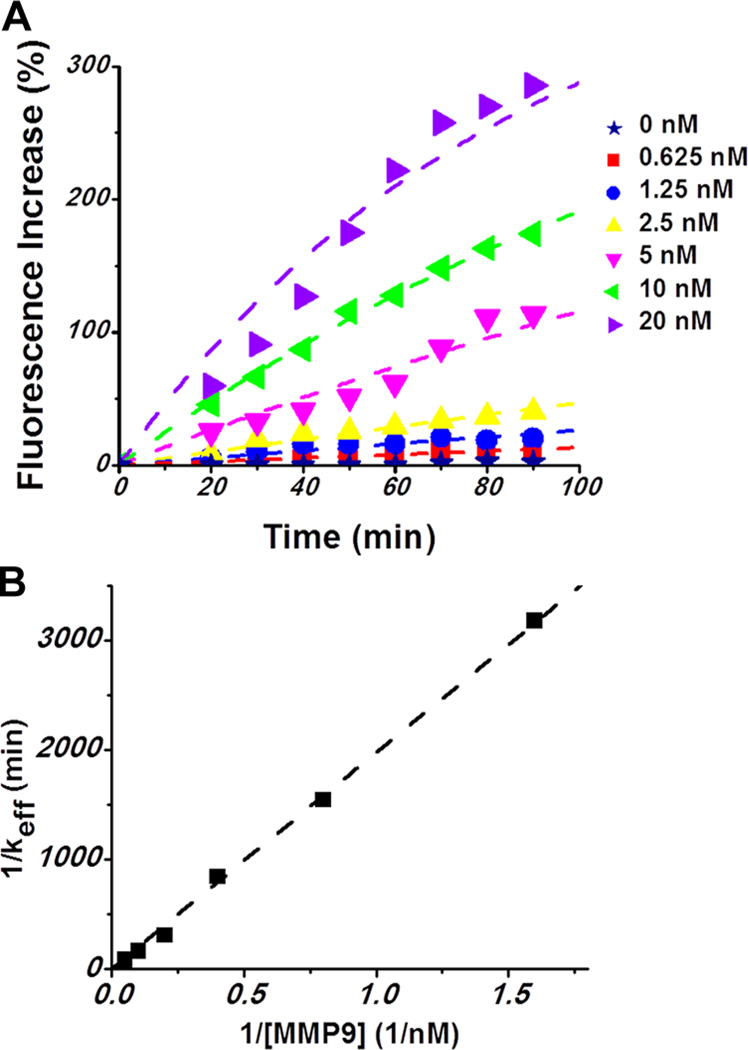
While simple calibration curves were constructed and gross differences in fluorescence due to cellular secretion of MMP9 were reported in Figures 2 and 4, these results stopped short of quantitating cell-secreted MMP9 and determining cell secretion rates. For example, cell-secreted signals in Figure 4E,F are a function of enzyme diffusion from cells and its activity at the site of the sensor. Therefore, to determine cell secretion rates, we needed to first obtain parameters associated with enzymatic activity of MMP9 and then to incorporate these parameters into a diffusion-reaction model. Figure 5A demonstrates responses of sensing hydrogels to varying MMP9 concentrations measured over time. On the basis of these data and Figure 2D, the limit of detection was determined to be 0.6 nM with a linear range extending to 40 nM. Figure S-4 (Supporting Information) shows a typical time-resolved sequence of fluorescence images, from which fluorescence intensity values for Figure 5A were generated. The fluorescence intensity from sensing hydrogels was measured every 10 min for 90 min: the shortest time to produce detectable signals (percent fluorescence increase ≥25%) was 20 min when [MMP9] = 5 nM (average concentration of produced MMP9 by 24 cells for 90 min). We estimated the limit of detection on the basis of signal-to-noise characteristics (S/N = 3)19 at t = 30 min, and the linear range was achieved before fluorescence reached the point of saturation.20 Moreover, the data in Figure 5A were used to determine parameters (constants) describing enzyme kinetics.
Figure 5.
Fluorescence detection of recombinant MMP9 and computational modeling of peptide cleavage. (A) Monitoring fluorescence increase of FRET-peptide containing sensing hydrogels in various concentrations of MMP9 solutions (0–20 nM) over time. Symbols indicate experimental data, and dotted lines indicate stmulated data. (B) Lineweaver–Burk plot of 1/keff versus 1/[MMP9] for determination of kcat and Km.
Assuming Michaelis–Menten kinetics, the enzyme reaction may be described as
When FRET peptides (S) in the hydrogel are exposed to free MMP9 molecules (E) in solution, they form the intermediate complex (C) and subsequently dissociate into MMP9 and cleaved peptides (P). Peptides are cleaved into DABCYL-containing fragments and FITC-containing fragments, which are immobilized onto hydrogel surfaces. In this work, cleaved peptides (P) indicate FITC-containing peptide fragments retained in the hydrogel after enzyme digestion. This heterogeneous proteolytic reaction7,21 is described by the equation
| (1) |
where [S]0 is the initial concentration of cleavable peptides on sensing hydrogel surface and Km is the Michaelis–Menten constant, which is defined as (kcat + k−1)/k1. The initial concentration of peptide, [S]0, was determined to be 61 pmol/hydrogel (n = 48) using fluorescence-based indirect quantitation methods. In this approach, the fluorescence of peptides that remained in solution after immobilization experiments was quantified. The amount of gel-bound peptide was determined by subtracting the amount of unbound peptide from the peptide present in solution initially. The solution for the first-order homogeneous differential eq 1 was obtained as
| (2) |
where
| (3) |
On the assumption that [P]/[S]0 is linearly related to the fluorescence signal, keff was estimated by fitting eq 2 to the experimental data from Figure 5A using MATLAB. Consequently, the Lineweaver–Burk plot in Figure 5B was generated by taking the reciprocal of eq 3 and used to determine catalytic constant (kcat) and Michaelis–Menten constant (Km). From the intercept and the slope of Figure 5B, kcat was determined to be 0.1118 min−1 and Km was 220.1 nM. The ratio kcat/Km, the specificity constant, for the FRET-peptide GPLGMWSRKC was 0.8 × 104 M−1 s−1, which is comparable to the value of 2.4 × 104 M−1 s−1 determined previously with electrochemistry for a similar species (GPLGMWSRC).7
On the basis of the experimental data in Figure 4F, the rate of MMP9 secretion from lymphoma cells was simulated using COMSOL Multiphysics. To determine the change in [E] over time, the reaction–diffusion model was set up. While only diffusion of MMP9 molecules secreted from lymphoma cells occurs in solution, both diffusion and proteolytic reaction take place inside the sensing hydrogel matrix, as shown in Figure 6A. Diffusion of MMP9 in solution is described as
| (4) |
where DE,sol is the diffusion coefficient of MMP9 in solution, taken to be 8.18 × 10−7 cm2/s.22 Note that MMP9 secretion occurs only at the cell capture region with the rate of Rsec (pg/h/cell).7 On the other hand, the reaction–diffusion equations for MMP9 and cleaved peptides (P) in the sensing hydrogel matrix can be described as23
| (5) |
| (6) |
where DE,gel and DP,gel are the diffusion coefficients of MMP9s and cleaved peptides inside the hydrogel matrix. The diffusivity of MMP9 in a hydrogel matrix, DE,gel, was taken to be 4.36 × 10−8 cm2/s on the basis of literature reports.24 Because peptide fragments accounted for in our model were covalently linked to the hydrogel, DP,gel was assumed to be negligible. The concentration of the intermediate complex, [C], is assumed to be at equilibrium, resulting in ∂[C]/∂t being equal to zero. kcat[C] in eq 6 can be expressed as (kcat[E]([S]0 − [P]))/Km on the assumption that Km is much larger than [S]0. Equations 3–6 were solved numerically by COMSOL multiphysics to obtain the concentration profiles for protease release from stimulated and quiescent cells.
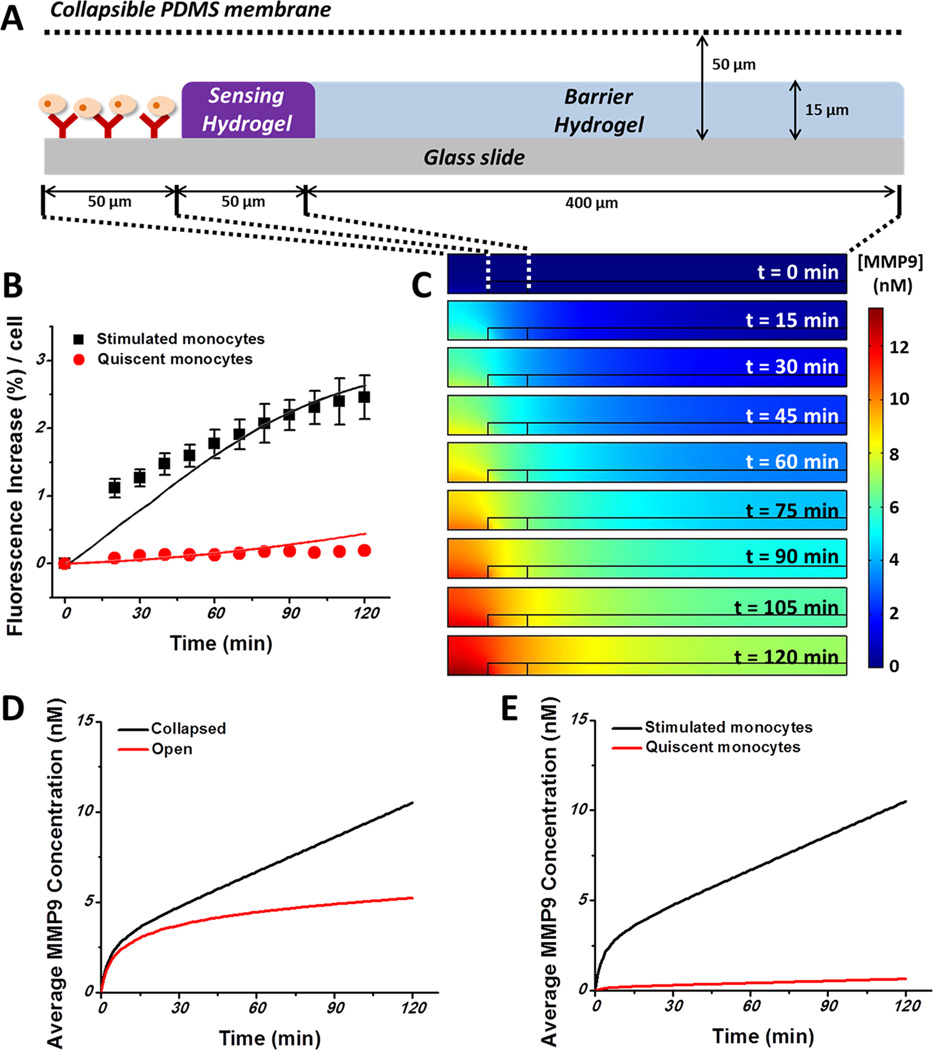
Figure 6.
Numerical simulation of MMP9 release and diffusion over time based on the diffusion reaction modeling. (A) 2D cylindrical coordinates of a sensing element with the origin at the center of the Ab-modified region. (B) Comparison of simulation results (solid lines) with experimental data (symbols) showing changes in fluorescence increase over time for stimulated/quiescent cells. RMS deviations: 7.19% for activated cells and 0.46% for quiescent cells. (C) Simulation results showing the MMP9 concentration profile over time. MMP9s are secreted by lymphoma cells located at the center and diffused to the sensing hydrogel. (D, E) Changes in average MMP9 concentration inside the sensing hydrogel matrix: (D) collapsed devices vs open devices; (E) stimulated cells vs quiescent cells.
Simulation results were achieved on the basis of the cylindrical coordinates with the axis at the center of the cell capture region (radius 50 µm) surrounded by sensing gel (inner radius 50 µm, outer radius 100 µm). The geometry simulated, 50 µm in the z direction and 0.5 mm in the r direction, was based on the geometry of microchambers in the collapsible membrane. With assumed MMP9 secretion rate Rsec, the peptide concentration, [P], can be obtained by solving eqs 3–6, generating a theoretical fluorescence signal which is linearly related to the total amount of [P]/[S]0 inside the sensing hydrogel matrix. The MMP9 secretion rate Rsec was obtained using iterative methods to match experimental data (symbols in Figure 6B) to the diffusion reaction model with a root mean square (RMS) deviation of ~10%. According to our simulation, activated cells secrete MMP9 at a rate of 0.56 pg/h/cell, while for quiescent cells the rate is 0.035 pg/h/cell. These simulation results are consistent with our previous work, which reported the values of 0.65 and 0.036 pg/h/cell for activated and quiescent cells, respectively.7 The slight difference may be due to the fact that experiments in this work were performed at room temperature, while previous work was carried out at 37 °C. More active cellular secretory activity is expected at physiological temperature. In Figure 6B, we compare the experimental data (symbols) to theoretical fluorescence signals (solid lines) generated on the basis of secretion rate modeling discussed above. As seen from these data, modeling and experiments were in close agreement (RMS deviations of 7.19% and 0.46% for activated and quiescent cells, respectively). On the basis of estimated MMP9 release rate from cells, we calculated concentration profiles of MMP9s released from cells (Figure 6C). As seen from Figure 6C, MMP9 molecules released from cells diffused into the hydrogel, creating a concentration gradient. Such modeling is important, as it allows designing the geometry (spacing) of sensing microwells where cross-talk is eliminated. We confirmed that no cross-talk by diffusion through the gel occurred for adjacent wells.
Cell secretion rates were also used to obtain concentration profiles for both open vs closed reconfigurable microfluidic devicea (Figure 6D) and activated vs quiescent cell releasea in closed microdevices (Figure 6E). It is important to note that modeling results of Figure 6D predict an ~2-fold higher concentration of MMP9 in a collapsed microfluidic device, which is in good agreement with experimental data (Figure 4E). Figure 6E indicates that activated lymphoma cells secrete MMP9s at a 16-fold higher secretion rate than quiescent cells. Overall, combining experimental results with diffusion reaction modeling allows (1) quantification of cell secretion rates and (2) determination of analyte concentration gradients emanating from the cells. The first allows the determination of quantitative descriptions of cell function that may be connected to pathology/diagnosis, and the second may be used for rational design of sensing surfaces.
CONCLUSIONS
This paper describes the development of hydrogel microwells that may be used for sequestering small groups of cells and sensing cell-secreted proteases. The latter was accomplished by designing protease-cleavable peptides to contain donor–acceptor chromophore pairs and then incorporating these sensing elements inside the hydrogel microstructures. When they were calibrated with recombinant MMP9, sensing hydrogels had a limit of detection of 0.6 nM with the linear range extending to 40 nM. To address the possible problem of diffusion overlap between the adjacent wells and to enhance the sensitivity of detecting MMP9 from cells, we developed a reconfigurable microfluidic device. This device contained a microstructured roof that descended onto the sensing surfaces, confining hydrogel microwells within separate 40 nL chambers. This strategy allowed us to enhance the cell-secreted signal by 2.5 times. In these experiments, the FRET pair of Dabcyl/FITC was chosen on the basis of cost and ease of modification by commercial peptide synthesis companies. However, in the future, the sensitivity of the hydrogel may be further enhanced by using fluorophores such as AlexFluor488, which has a better quantum yield than physiological pH or by using other quenchers such as Iowa black and gold nanoparticles (diameter ≤4.5 nm).
In addition to designing sensing gels, we developed a diffusion reaction model for determining cell secretion rates and concentrations of MMP9. The secretion rates were found to be 0.56 and 0.035 pg/h/cell for activated and quiescent cells, respectively. Given that a wide range of biorecognition molecules (enzymes, antibodies, and aptamers) have been incorporated into hydrogels, the strategy of monitoring cellular secretion with sensing hydrogel microwells described here may be easily extended to other analytes. Subsequent refinements of this technology may include single-cell detection and function-based cell sorting.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Tingrui Pan (Department of Biomedical Engineering, UC Davis) for the use of the mask aligner. We also thank Prof. Laura Marcu (Department of Biomedical Engineering, UC Davis) for the use of the fluorescence microscope. This work was supported by an NSF EFRI grant. Additional funding came from “Research Investments in Science and Engineering from UC Davis”. T.K. was supported by NIH Fellowship T32-NIBIB 5T32EB003827
Footnotes
ASSOCIATED CONTENT
S Supporting Information
Text and figures giving details of the specificity of peptide beacons for detection of MMP9, effect of captured lymphoma cell numbers on detection of MMP9 secretion, fluorescence-based detection of peptide cleavage, and analysis of cross-talk between adjacent sensing wells. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Chambers AF, Matrisian LM. J. Natl. Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 2.Welgus HG, Campbell EJ, Cury JD, Eisen AZ, Senior RM, Wilhelm SM, Goldberg GI. J. Clin. Invest. 1990;86:1496–1502. doi: 10.1172/JCI114867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egeblad M, Werb Z. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 4.Ranuncolo SM, Armanasco E, Cresta C, Bal De Kier Joffe E, Puricelli L. Int. J. Cancer. 2003;106:745–751. doi: 10.1002/ijc.11288. [DOI] [PubMed] [Google Scholar]
- 5.Newby AC. Arterioscler., Thromb., Vasc. Biol. 2008;28:2108–2114. doi: 10.1161/ATVBAHA.108.173898. [DOI] [PubMed] [Google Scholar]
- 6.Di Nezza LA, Misajon A, Zhang J, Jobling T, Quinn MA, Ostor AG, Nie G, Lopata A, Salamonsen LA. Cancer. 2002;94:1466–1475. doi: 10.1002/cncr.10355. [DOI] [PubMed] [Google Scholar]
- 7.Shin DS, Liu Y, Gao YD, Kwa T, Matharu Z, Revzin A. Anal. Chem. 2013;85:220–227. doi: 10.1021/ac302547p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Yan AT, Yan RT, Spinale FG, Afzal R, Gunasinghe HR, Stroud RE, McKelvie RS, Liu PP. Eur. J. Heart Failure. 2008;10:125–128. doi: 10.1016/j.ejheart.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Banfi C, Cavalca V, Veglia F, Brioschi M, Barcella S, Mussoni L, Boccotti L, Tremoli E, Biglioli P, Agostoni P. Eur. Heart J. 2005;26:481–488. doi: 10.1093/eurheartj/ehi073. [DOI] [PubMed] [Google Scholar]
- 9.(a) Shoji A, Kabeya M, Sugawara M. Anal. Biochem. 2011;419:53–60. doi: 10.1016/j.ab.2011.07.031. [DOI] [PubMed] [Google Scholar]; (b) Wu S-H, Lee K-L, Chiou A, Cheng X, Wei P-K. Small. 2013;9:3532–3540. doi: 10.1002/smll.201203125. [DOI] [PubMed] [Google Scholar]
- 10.(a) Fudala R, Ranjan AP, Mukerjee A, Vishwanatha JK, Gryczynski Z, Borejdo J, Sarkar P, Gryczynski I. Curr. Pharm. Biotechnol. 2011;12:834–838. doi: 10.2174/138920111795470967. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fudala R, Rich R, Mukerjee A, Ranjan AP, Vishwanatha JK, Kurdowska AK, Gryczynski Z, Borejdo J, Gryczynski I. Curr. Pharm. Biotechnol. 2012 doi: 10.2174/138920101413140605111109. [DOI] [PubMed] [Google Scholar]; (c) Xia ZY, Xing Y, So MK, Koh AL, Sinclair R, Rao JH. Anal. Chem. 2008;80:8649–8655. doi: 10.1021/ac801562f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Shi LF, De Paoli V, Rosenzweig N, Rosenzweig Z. J. Am. Chem. Soc. 2006;128:10378–10379. doi: 10.1021/ja063509o. [DOI] [PubMed] [Google Scholar]
- 11.Pham W, Choi YD, Weissleder R, Tung CH. Bioconjugate Chem. 2004;15:1403–1407. doi: 10.1021/bc049924s. [DOI] [PubMed] [Google Scholar]
- 12.Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Nat. Mater. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 13.(a) Liu Y, Dong Y, Jauw J, Linman MJ, Cheng Q. Anal. Chem. 2010;82:3679–3685. doi: 10.1021/ac1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yan J, Pedrosa VA, Simonian AL, Revzin A. ACS Appl. Mater. Inter. 2010;2:748–755. doi: 10.1021/am9007819. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liu Y, Yan J, Howland MC, Kwa T, Revzin A. Anal. Chem. 2011;83:8286–8292. doi: 10.1021/ac202117g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Yan J, Pedrosa VA, Enomoto J, Simonian AL, Revzin A. Biomicrofluidics. 2011;5:032008. doi: 10.1063/1.3624739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Moorthy J, Burgess R, Yethiraj A, Beebe D. Anal. Chem. 2007;79:5322–5327. doi: 10.1021/ac070226l. [DOI] [PubMed] [Google Scholar]; (b) Jang E, Kim S, Koh WG. Biosens. Bioelectron. 2012;31:529–536. doi: 10.1016/j.bios.2011.11.033. [DOI] [PubMed] [Google Scholar]; (c) Appleyard DC, Chapin SC, Srinivas RL, Doyle PS. Nat. Protoc. 2011;6:1761–1774. doi: 10.1038/nprot.2011.400. [DOI] [PubMed] [Google Scholar]; (d) Srinivas RL, Chapin SC, Doyle PS. Anal. Chem. 2011;83:9138–9145. doi: 10.1021/ac202335u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Hynd MR, Frampton JP, Burnham MR, Martin DL, Dowell-Mesfin NM, Turner JN, Shain W. J. Biomed. Mater. Res. A. 2007;81A:347–354. doi: 10.1002/jbm.a.31002. [DOI] [PubMed] [Google Scholar]
- 15.(a) Revzin A, Maverakis E, Chang HC. Biomicrofluidics. 2012;6:021301. doi: 10.1063/1.4706845. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pedrosa VA, Yan J, Simonian AL, Revzin A. Electroanalysis. 2011;23:1142–1149. [Google Scholar]
- 16.Yan J, Sun YH, Zhu H, Marcu L, Revzin A. Biosens. Bioelectron. 2009;24:2604–2610. doi: 10.1016/j.bios.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) You J, Shin DS, Patel D, Gao Y, Revzin A. Adv. Healthcare Mater. 2013 [Google Scholar]; (b) Shah SS, Kim M, Cahill-Thompson K, Tae G, Revzin A. Soft Matter. 2011;7:3133–3140. [Google Scholar]
- 18.(a) Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]; (b) Mosadegh B, Kuo CH, Tung YC, Torisawa YS, Bersano-Begey T, Tavana H, Takayama S. Nat. Phys. 2010;6:433–437. doi: 10.1038/nphys1637. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rhee M, Burns MA. Lab Chip. 2008;8:1365–1373. doi: 10.1039/b805137b. [DOI] [PubMed] [Google Scholar]
- 19.Long GL, Winefordner JD. Anal. Chem. 1983;55:A712–&. [Google Scholar]
- 20.Waters JC. J. Cell Biol. 2009;185:1135–1148. doi: 10.1083/jcb.200903097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez OA, Chavez M, Lissi E. Anal. Chem. 2004;76:2664–2668. doi: 10.1021/ac049885d. [DOI] [PubMed] [Google Scholar]
- 22.Tyn MT, Gusek TW. Biotechnol. Bioeng. 1990;35:327–338. doi: 10.1002/bit.260350402. [DOI] [PubMed] [Google Scholar]
- 23.Tzafriri AR, Bercovier M, Parnas H. Biophys. J. 2002;83:776–793. doi: 10.1016/S0006-3495(02)75208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amsden B. Macromolecules. 1998;31:8382–8395. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.