Abstract
Background:
The field of clinical research informatics includes creation of clinical data repositories (CDRs) used to conduct quality improvement (QI) activities and comparative effectiveness research (CER). Ideally, CDR data are accurately and directly abstracted from disparate electronic health records (EHRs), across diverse health-systems.
Objective:
Investigators from Washington State’s Surgical Care Outcomes and Assessment Program (SCOAP) Comparative Effectiveness Research Translation Network (CERTAIN) are creating such a CDR. This manuscript describes the automation and validation methods used to create this digital infrastructure.
Methods:
SCOAP is a QI benchmarking initiative. Data are manually abstracted from EHRs and entered into a data management system. CERTAIN investigators are now deploying Caradigm’s Amalga™ tool to facilitate automated abstraction of data from multiple, disparate EHRs. Concordance is calculated to compare data automatically to manually abstracted. Performance measures are calculated between Amalga and each parent EHR. Validation takes place in repeated loops, with improvements made over time. When automated abstraction reaches the current benchmark for abstraction accuracy - 95% - itwill ‘go-live’ at each site.
Progress to Date:
A technical analysis was completed at 14 sites. Five sites are contributing; the remaining sites prioritized meeting Meaningful Use criteria. Participating sites are contributing 15–18 unique data feeds, totaling 13 surgical registry use cases. Common feeds are registration, laboratory, transcription/dictation, radiology, and medications. Approximately 50% of 1,320 designated data elements are being automatically abstracted—25% from structured data; 25% from text mining.
Conclusion:
In semi-automating data abstraction and conducting a rigorous validation, CERTAIN investigators will semi-automate data collection to conduct QI and CER, while advancing the Learning Healthcare System.
Keywords: CERTAIN, Informatics, Quality Improvement, Comparative Effectiveness, Health Information Technology
Introduction
At the Institute of Medicine’s (IOM) 2006 Roundtable on Value & Science-Driven Health Care, the nation’s leading healthcare experts defined the learning healthcare system as one “in which science, informatics, incentives, and culture are aligned to provide continuous improvement and innovation, with best practices embedded in the delivery process and new knowledge captured as a by-product of healthcare delivery”.1 Since then, interest on the part of research scientists and funding agencies in using EHR, and in leveraging clinical informatics and quality improvement (QI) data to conduct comparative effectiveness research (CER) has dramatically increased.2–4 Several early adopters of these clinical data platforms have emerged.5 A newer generation of platforms is under development that is intended to link disparate EHR from even more diverse institutions.6
Many challenges must be addressed when considering the secondary use of data collected for clinical purposes.7 Issues of extraction, quality, and governance abound.8–10 Determining an acceptable level of data quality for use in QI and CER remains unresolved.8,11–12 These challenges are greatly increased when aggregating clinical data across organizations.13 Bridging cultural divides among organizations requires thoughtful and ongoing strategies.14 More fundamentally, whether data collected for clinical care can even be used effectively for secondary purposes remains an open question.
Investigators from Washington State’s Surgical Care and Outcomes Assessment Program Comparative Effectiveness Research Translation Network—SCOAP CERTAIN—are addressing these challenges with a multifaceted approach to automated data extraction from diverse EHR, at multiple health systems, in order to conduct QI and CER. The SCOAP CERTAIN investigators have developed generalizable approaches suitable for use by large and small health systems regarding facility recruitment, data sharing governance, data delivery, and data validation and quality assurance.
Background and Significance
Early investigators assessed clinical data quality within their own institutions, usually by measuring data concordance.16 Some compared across different EHR systems, using varying concepts of gold standards.17–18 Others assessed data accuracy as correctness and completeness.19 These assessments focused on the data. To prepare data for use in QI or CER requires a broader concept of data quality, one that encompasses not only the data but also the setting in which the data are used. These concepts include contextual data quality (ie, relevance, timeliness, completeness), representational quality (ie, concise and consistent representation) and accessibility (ie, ease of access, security).20 Indeed, a pragmatic framework for data quality assessment in EHR has recently emerged.13 This set of broader data quality elements constitutes what has been described as “fitness for use,” specifically for QI and CER.13
Despite these advances, caution is warranted. Recent research warns against using EHR-derived data to measure clinical quality (QI). Parsons assessed the validity of eleven EHR-derived quality measures and found that the EHR undercounted these, thereby underestimating quality.21 Wei evaluated data fragmentation, a phenomenon that occurs when patients receive care from more than one health system, and noted that applying algorithms to data from only one (versus both) EHR contributed to misclassification.22 These reports suggest that additional efforts are needed to prepare and extract EHR data for clinical, QI and CER use.
To realize the efficiencies these large datasets promise, it is critical to assess data validity. The SCOAP CERTAIN Automation and Validation Project is contributing to this growing body of knowledge by making a unique contribution from the perspective of building on an existing QI registry to conduct CER. It is unique in that it uses data captured as a direct byproduct of clinical care rather than augmenting usual clinical data collection with custom forms for a specific QI or CER project. The objective of this manuscript is to describe the approach taken to automate data flow, and the analytic plan for validation of automatically abstracted data, to create an enhanced registry to conduct QI and CER. We report progress to date, describe challenges encountered, and pose potential solutions and next steps.
Methods
Overview of SCOAP CERTAIN
SCOAP (http://www.scoap.org) is a clinician-led, statewide performance, benchmarking and QI platform for surgical and interventional procedures.23 SCOAP data are of high fidelity, based in “real world” practice environments, captured from clinical rather than billing records, and are collected prospectively, with careful attention to risk-adjustment made during analyses. SCOAP includes patients from diverse practice environments, and almost all priority populations.24 SCOAP interventions have resulted in a reduction in surgical complications and significant savings to the system.25 Because SCOAP hospitals submit registry data and additional patient-reported outcomes (PRO) information, it is also ideal for conducting pragmatic trials of CER.
Since its inception, SCOAP has been limited by lack of: (1) automated data gathering, (2) linkage between data streams, and (3) connections to post-discharge and PRO data. To overcome these limitations, SCOAP investigators launched the CERTAIN initiative (http://www.becertain.org). One of the fundamental goals of CERTAIN is to enhance the existing SCOAP platform by automating data abstraction from disparate EHR across diverse care delivery sites to conduct QI and CER.
Project Overview – Automation and Validation
Currently, SCOAP data are collected by manual chart review and data abstraction of 150 care processes from 55 hospitals and represent 25 procedures. Abstracted cases are recorded in a web-based tool provided by ARMUS (Burlingame, California), the data entry and management system used to manage all SCOAP QI data.26 Upon data entry, each data element is instantly checked for errors, and feedback provided to the abstractor for entries that are missing or out of range. Four to six weeks are required to train each abstractor to correctly abstract data for each type of surgical registry (eg, abdominal, vascular). Ongoing training and standardized data collection processes ensure continuing validity and accuracy of data collection. Abstractors are required to maintain a level of 95% agreement and undergo remedial training if they do not. The data dictionary is updated when clarifications of, and revisions to, abstracting decisions are made; a surgeon QI leader approves changes. Once data are accurately entered, they are forwarded to ARMUS for creation of benchmarking reports, which are then returned to clinicians at participating hospitals.
To automate data abstraction, investigators are installing Caradigm’s Amalga Unified Intelligence System (Bellevue, Washington).27 Amalga retrieves data from all types of EHR using clinical event messages such as HL7 feeds. It creates and stores message queues that permit data manipulation independently from clinical systems. Data can be checked against the original clinical message at any time. The Amalga system uses a federated data model and creates a common platform that can be used to conduct QI activities and CER studies.28 Throughout the project investigators are operating the Amalga abstraction system in parallel with the current manual abstraction system. This creates the opportunity to conduct a two-fold validation process, validating data flowing into Amalga: (1) against manually abstracted SCOAP data, as entered into the ARMUS database (the referent), and (2) against the results of the semi-automated abstraction based on electronic data obtained from the EHR (the gold standard). We hypothesize that employing Amalga will result is an enhanced, efficient “big data” registry that can be used to conduct high fidelity, reliable QI and CER across the learning healthcare system.
Site Engagement
During the first year of the project, CERTAIN leadership identified fourteen candidate SCOAP sites and approached site-specific clinical and informatics leadership about the project. CERTAIN investigators introduced the concept of automation with Amalga, conducted site-specific technical review meetings, and executed data use- and business associate agreements. Site selection was based on the readiness of site leadership to participate, the ability of each site to provide the necessary technical support to configure the required systems, a willingness to allow on-site work by the Amalga team, and a commitment to the three-year project time frame. Sites are incentivized in two ways. First, whether hospitals employ their own SCOAP abstractor or pay for contracted abstracting services, automating collection of these data will reduce costs. Second, sites are offered use of the Amalga system as a hospital-wide clinical data repository (CDR), with the cost of configuring the technical systems covered with support from the Agency for Healthcare Research and Quality (AHRQ). At project completion, each site will have the option of continuing Amalga services directly with Caradigm.
Concurrent with site engagement activities, CERTAIN investigators developed a plan to initiate data flow and to validate these data in a rigorous fashion. Steps include: (1) selecting data for automated abstraction, (2) initiating automated data flow, (3) creating a validation analytic plan to prepare for ”go-live,” and (4) developing policies that ensure adherence to required privacy, security, and human subjects protection.
Selecting Data for Automated Abstraction
The automation project focuses on SCOAP patients undergoing abdominal, oncologic, spine, or non-cardiac vascular-interventional surgical or endovascular procedures. Each of these four SCOAP data collection registries is composed of 700 data elements; some are core to all and some are unique.29 Expert investigators reviewed a spreadsheet of aggregated SCOAP data elements cross-referenced to candidate site EHR sources. Each element was labeled: (1) structured electronic data at all sites (eg, age, admit date, laboratory values); (2) structured electronic data at some sites (eg, lowest intra-operative body temperature); (3) machine-readable text (eg, discharge diagnosis); or (4) not assessed/not feasible. Machine-readable text documents are a rich source of clinical data that are being extracted using natural language processing (NLP) algorithms. It may not be feasible to automate abstraction of certain types of data, for example, data from handwritten notes or for technical specifications that do not allow an interface; data that exist in multiple locations within the EHR and therefore may be inconsistent and require human judgment to reconcile; or data that require complex temporal assessments.
Initiating Data Flow
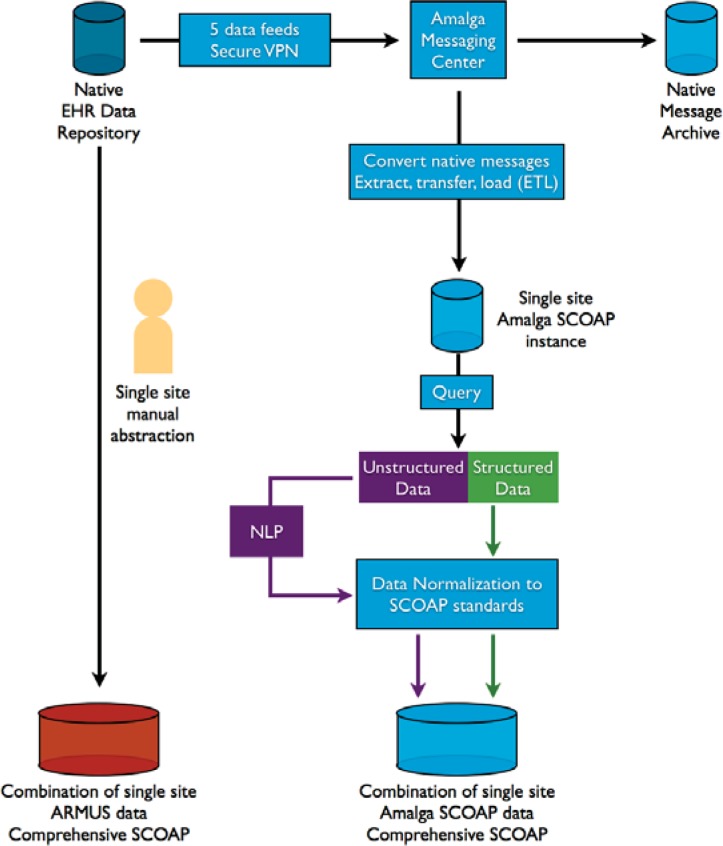
Figure 1 illustrates project data flow. There is a native EHR repository at each site. A virtual private network (VPN) is then established, through which only data from the selected feeds will flow to the Amalga messaging center. All data are archived in native message format, preserving a shadow copy of the original data from each EHR. For sites delivering data on all inpatients using HL7 messages, the original native messages are extracted and converted into the requisite SCOAP data elements, then transformed and loaded into the common data platform. A query is run to identify SCOAP patients and to define each structured data element according to the SCOAP data dictionary. These data are normalized to SCOAP standards. This entails transforming units of measurement into one common unit for each data element, looking for outliers, and cleaning data. The resulting aggregated data serve as the centralized SCOAP-Amalga common data platform. Site-specific QI representatives are authorized to access site-specific Amalga data tables through their password protected, VPN connection.
Figure 1.
Data Flow Diagram for the CERTAIN Automation and Validation Project
CERTAIN=Comparative Effectiveness Research Translation Network; EHR=electronic health record; NLP=natural language processing; SCOAP=Surgical Care Outcomes & Assessment Program; VPN=virtual private network. The illustration is consistent with all existing data use agreements, business associate agreements, and memoranda of understanding between CERTAIN investigators and staff, and participating sites.
Errors in data flow can occur at several points in the data flow process: (1) extraction from the VPN feeds, (2) message handling, (3) when native messages are extracted, transformed and loaded, (4) when queries run through NLP algorithms misclassify data elements, and (5) when data are normalized. Investigators troubleshoot at each juncture until technical and clinical personnel are assured data are flowing correctly. All problems and solutions are documented for later aggregation for additional validation and for publication. Data in the centralized SCOAP-Amalga CDR are then ready to be validated against the manually abstracted data housed in the corresponding ARMUS database for the same time period.
Validation Analytic Plan and Performance Measures
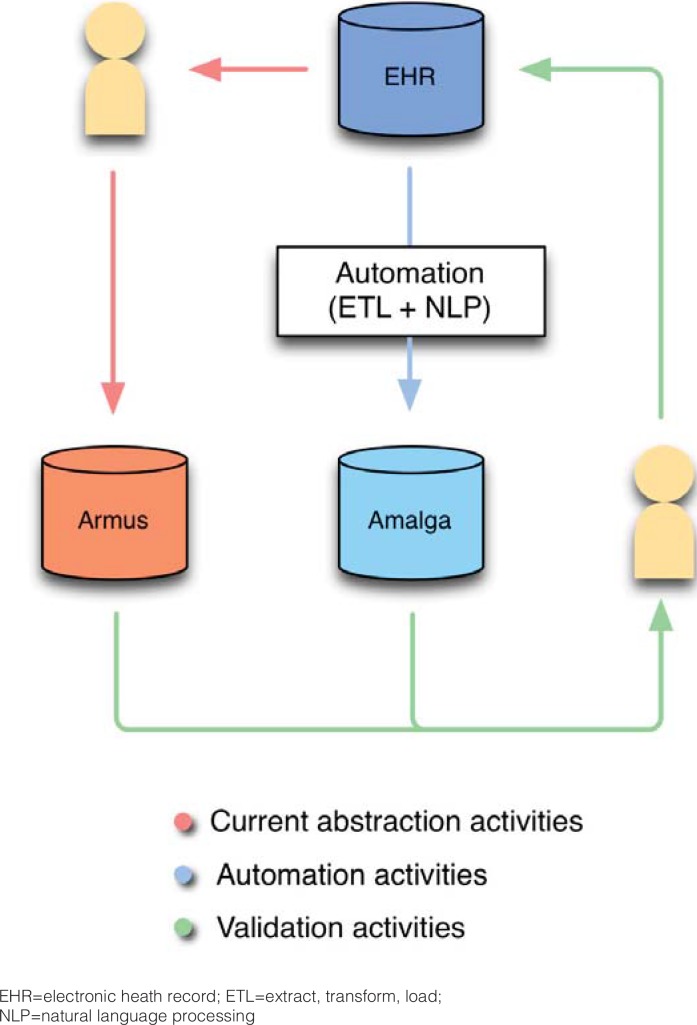
The final step illustrated in Figure 1 involves the creation of comparator databases. The referent database (ARMUS) contains data that are manually abstracted. The comparator is the data that has automatically flowed into the Amalga CDR for the same time frame. The gold standard is data from the EHR. To carry out the validation plan, investigators combine the data from these two databases into one dataset. By comparing these, investigators identify errors that could have occurred at any previous steps in the process.
In the combined dataset, each data element for each patient (patient-data pair) forms a row, with a specific patient-level identifier, a hospital-level identifier, and a unique date-time stamp. All patient-level data are coded and de-identified; the link is maintained separately. Patient-data element pairs, and their classification (eg, patient #3, smoking status=yes) are recorded in two adjacent columns, one containing data from the ARMUS database, the other from Amalga, blinding the original source of each. A third column exists alongside these two as a place to record the gold standard of the same patient-data element pair, obtained from the EHR. (Figure 2, Table 1)
Figure 2.
Comparison of the ARMUS and Amalga Databases for the SCOAP CERTAIN Automation and Validation Project
EHR=electronic heath record; ETL=extract, transform, load;
NLP=natural language processing
Table 1.
Example of data elements in combined ARMUS and Amalga dataset for specified time frame
| Hospital-level ID | Patient-level ID | Data Element | Date element recorded | Source #1 | Source #2 | EHR |
|---|---|---|---|---|---|---|
| 1 | 1 | HbA1c | 1/1/13 | 7.4 | 7.3 | 7.4 |
| 1 | 1 | SCr | 1/1/13 | 0.7 | 0.8 | 0.8 |
| 2 | 3 | Age | 2/3/13 | 26 | 26 | 26 |
| … | ||||||
| 3 | 375 | Smoker | 3/31/13 | Yes | No | Yes |
HbA1c=hemoglobin A1c; SCr=serum creatinine; ID=Identifier.
Source #1 or #2=data from ARMUS or Amalga, each blinded.
Note: Identifiers are at the hospital and patient level but are coded and do not reflect actual hospital or patient identifiers.
Investigators have established acceptable margins of discordance for each variable, and use the Kappa statistic to estimate concordance and discordance within each pair of binary or categorical data elements, and the intra-class correlation coefficient within each pair of continuous data elements. Statistical algorithms are used to examine all patient-data element pairs, identifying which are discordant between the two, blinded sources. Investigators will return to each EHR to determine the correct recording of these data elements, as well as a random sample of all concordant pairs. This is an iterative process that takes place under the umbrella of QI. Employing statistical algorithms to estimate these performance metrics avoids bias that would exist were human reviewers to conduct the comparison.
Comparing against the EHR enables investigators to determine the validity of each pair by comparing automated abstracting in Amalga to the gold standard. (The same exercise can compare ARMUS data with the EHR.) Returning to the EHR enables investigators to employ methods from the diagnostic test literature to calculate performance using measures commonly used in the information retrieval literature: sensitivity, specificity, positive predictive value (PPV), positive likelihood ratio (LR+), negative likelihood ratio (LR-), and the F-measure—the weighted harmonic mean of PPV and sensitivity. These are estimated for each data element, aggregated over patient and time. (Table 2)
Table 2.
Example of performance measures of validity, for each time frame
| Data element | Sensitivity (Recall) (%) | Specificity (%) | LR+ (%) | LR− (%) | F-measure (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source #1 to EHR | Source #2 to EHR | Source #1 to EHR | Source #2 to EHR | Source #1 to EHR | Source #2 to EHR | Source #1 to EHR | Source #1 to EHR | Source #1 to EHR | Source #1 to EHR | |
| HbA1c | 67 | 60 | 91 | 90 | 7 | 6 | 0.4 | 0.4 | 9 | 7 |
| SCr | 73 | 53 | 92 | 92 | 49 | 7 | 0.3 | 0.2 | 10 | 8 |
| Age | 67 | 80 | 91 | 91 | 7 | 9 | 0.4 | 0.5 | 10 | 12 |
| Gender | 91 | 90 | 67 | 60 | 3 | 2 | 0.1 | 0.2 | 48 | 47 |
EHR=electronic health record; HbA1c=hemoglobin A1c; LR+=likelihood ratio positive; OR−=likelihood ratio negative; SCr=serum creatinine; Source #1 or #2=data from ARMUS or Amalga, each blinded
To accomplish these analyses investigators vary the unit of analysis. We begin with each patient-data element pair and proceed to use packets of data elements, each packet consisting of elements logically grouped together (eg, sociodemographic data, laboratory data). We aggregate over patients and relevant time periods. We first stratify analyses by hospital and then combine results across health systems, while controlling for health-system specific indicators. Default summaries weight each element equally. Investigators also calculate summaries wherein each data element is weighted according to the importance of the element under investigation. Missing data and extreme outliers not corrected in the normalization phase are segregated a priori and managed separately.
These procedures are now being completed. As the system iteratively improves over time, with corrections being made along the way, intervals between comparisons will lengthen. Calculating concordance, as iterative improvements are made in Amalga, enables investigators to determine how Amalga is “learning” over time. Concordance in 95% of pairs will constitute success and enable “go-live.” This benchmark is consistent with the current IRR performance of the manual abstraction system.
Data Privacy, Security and Human Subjects Protection
The CERTAIN Automation and Validation Project is a prototype of a project wherein the lines between QI and research become blurred. To ensure adequate protection of data, investigators met with leadership from three institutional committees that oversee these two activities: the University of Washington (UW) Medicine Quality and Safety Executive Committee, the Central Data Repository Oversight Committee, and the Human Subjects Committee. Together, investigators and leaders thoughtfully created a framework that addresses the challenges of privacy and security inherent in the project. The framework places a virtual firewall between QI and research activities, and assigns each project investigator and staff to only one side. Those serving on the QI side are authorized, a priori, to view patient-level data for QI purposes. Those serving on the research side see only aggregated data. The framework has been codified in a memorandum of understanding (MOU). The project now serves as an institutional prototype for future projects that require use of large datasets that span QI and research activities.
The framework is consistent with the primary purpose of the project, which is to semi-automate extraction of EHR data to improve the timeliness and quality of data abstraction to conduct hospital related QI activities, for example, determining the number of patients per month whose perioperative blood glucose exceeds 200 mg/dL (11.0 mmol/L), and for whom insulin is ordered. SCOAP is an approved Coordinated Quality Improvement Program (CQIP) subject to protections against legal disclosure and discoverability of data provided by Washington State law (RCW 43.70.510).30 The use of protected health information is acceptable under the Health Insurance Portability and Accountability Act laws and regulations as part of clinical QI.31 All patient-level analyses are performed under this protective umbrella. (Table 1) Any QI data used outside of a protected CQIP is open for public use, whether in a legal case or in other external publications. As an AHRQ-funded project, data used for research may have additional protections. Even so, adopting a conservative approach, the MOU requires that investigators and staff serving on the research side see data solely in anonymized, aggregated form. (Table 2) As an anonymized dataset, the UW Institutional Review Board (IRB) does not consider the automation and validation project human subjects research. We anticipate that patient-level data will be used for specific CER projects conducted in the future; these will receive IRB approval as a limited dataset.
Progress to Date
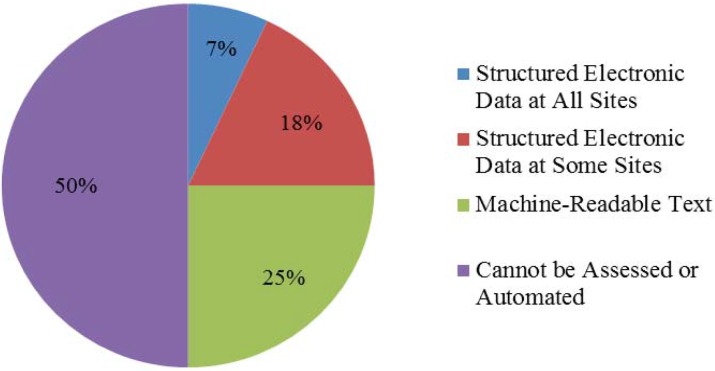
The technical analysis plan was distributed to 14 sites, and completed at six. Five sites executed participation agreements, each contributing data from two to three surgical registries, totaling 13 end-user test cases. Fifteen to 18 unique data feeds are contributing data. Common electronic feeds are registration, laboratory, transcription/dictation, radiology, and medications. The alpha phase of the common data platform is complete; the beta phase was released in May 2013. We are abstracting approximately 50% of the 1,320 data elements from the SCOAP data collection forms that are required for SCOAP CERTAIN projects. Figure 3 shows that structured electronic data at all sites represent 92 elements (7%); structured electronic data at some sites, 238 elements (18%). Machine-readable text represents 330 elements (25%), for which 68 are currently being extracted using NLP extraction that employs both rules-based and statistical approaches. The remaining 660 (50%) data elements have not been assessed or cannot be automated at this time (e.g., non-machine-readable text in nursing notes).
Figure 3.
Characteristics of SCOAP Data Collection Elements
Amalga has now been programmed from SCOAP open-source registry data dictionaries for four SCOAP surgical registries: general, oncologic, vascular and interventional procedures, and orthopedic spine. To further the goal of improved timeliness and quality of clinical data extraction for QI, we have programmed functions within Amalga to generate automatic case lists of eligible registry patients and queries to extract QI data elements for the SCOAP registry. NLP algorithms have been created to pull additional data from text documents that typically are not accessible electronically. At the end of the study, NLP algorithms will become available through open-source libraries.
Using the process illustrated in Figure 1 and described above, investigators are now calculating the validity of data flowing from Amalga, and will soon be comparing those data with data entered into the ARMUS system. In future publications we will report our performance measures and illustrate the performance of Amalga over time.
Once validation is complete, Amalga will be deployed directly to the five project sites during year one following the end of the AHRQ grant. License fees for the additional year are underwritten directly by Caradigm at no additional cost to project sites. During this period, project sites will be trained and expected to use Amalga to semi-automate SCOAP QI data collection. Investigators will evaluate site use and productivity of Amalga through periodic assessment of direct workflow observation after introduction of semi-automation, and through administration of the Post-Study System Usability Questionnaire.32 Longer term program sustainability beyond this additional year is dependent upon successful application within project sites. Other products from this project will become available immediately to the SCOAP and CERTAIN community upon completion of data validation. These products include open source text-mining software programming language, and detailed reports of SCOAP data collection forms that will aid hospitals desiring to improve their QI data extraction processes, or investigators considering CER projects.
Discussion
In this manuscript we describe the CERTAIN Automation and Validation Project. The goal of the project is to improve within hospital data capture of QI data for submission of metrics of in-patient surgical encounters to SCOAP registries. The project does not replace an individual institution’s electronic data warehouse. Rather, the goal is to improve the efficiency of data collection to create a benchmarking database across all participating institutions. This will achieve both improved QI through SCOAP, and CER through CERTAIN, as SCOAP data are a primary source of data for CERTAIN studies. Further, the purpose of the validation project is to validate data coming into the CDR from each EHR. Reconciling patient-reported data with historical, clinician reported data was outside the scope of this project.
The strength of the project is in employing a federated data model that captures HL7 standard messages in real time, enabling investigators to assess whether clinical data collected as a by-product of healthcare delivery can be secondarily used to conduct QI and CER. This detailed description of automation and abstraction methods illustrates the complexities of abstracting high fidelity data to achieve these goals.
What has slowed the number of participating sites is their competing investment of time in implementing their own core EHR systems, in meeting the Meaningful Use requirements promulgated by the Health Information Technology for Economic and Clinical Health (HITECH) Act; and in adopting International Classification of Diseases 10th Revision (ICD-10) coding.33 These organizational priorities are higher than is participating in a regional QI initiative. To address these barriers, CERTAIN investigators negotiated compromises to ease the time required for participation. One such option was delivering previously parsed data as historic datasets that could be batch-loaded into Amalga. Another was adding additional feeds from existing sites, to further test the robustness of Amalga. The most significant limitation of the project is that, at this time, only a portion of data elements can feasibly be extracted electronically— that we may be able to achieve, at best, “semi-automation.”
What we have learned is consistent with the state of the field for creating a clinical data infrastructure to conduct CER.34 Many and significant efforts are underway to achieve this goal, and challenges abound.6,35–36 One recent report of the Electronic Data Methods Forum suggests that a substantial level of effort is required to establish and sustain data sharing partnerships.35 Investigators are addressing methodological challenges in case identification, validation, and accurate representation of data and are reporting that “the devil is in the details.”37–38 Still others are focusing on data protection and security, developing new approaches to address the concerns of investigational review boards.39–41
Further, since inception of the SCOAP CERTAIN project, the field has advanced. Future initiatives to create an electronic infrastructure for QI and CER will be aligned with Stage 3 Meaningful Use criteria.42 New initiatives will focus around major vendor-purchased EHR, around which national purchases are coalescing.43 And the need to pursue regulatory and governance revisions to streamline approvals for use of data for QI and research is now an issue of national prominence.44
In sum, the CERTAIN Automation and Validation Project is a first step in enhancing the existing SCOAP registry to advance QI and to create an electronic clinical data platform for CER. In successfully completing the CERTAIN project, we will have created a technological system that semi-automates data extraction across multiple institutions and disparate EHR by using a common data platform for categorizing surgical and interventional care processes and outcomes. Employing a rigorous and systematic approach to data validation will ensure data quality. Through the CERTAIN Project clinicians and investigators are creating a prototype of the Learning Healthcare System for Washington State.
Acknowledgments
This work was supported by Agency for Healthcare Research and Quality (AHRQ) Grant Number 1 R01 HS 20025-01 and Academy Health/AHRQ Grant Number U13 HS19564-01. The Surgical Care and Outcomes Assessment Program (SCOAP) is a Coordinated Quality Improvement Program of the Foundation for Health Care Quality. CERTAIN is a program of the University of Washington, the academic research and development partner of SCOAP.
All authors listed are a part of the CERTAIN Collaborative. Personnel contributing to this study: Marisha Hativa, MSHS; Kevin Middleton; Megan Zadworny, MHA.
Footnotes
Disciplines Health Services Research
References
- 1.Olsen LA, Aisner D, McGinnis JM, editors. The Learning Healthcare System. Workshop Summary. IOM Roundtable on Evidence-Based Medicine. Available at: http://www.nap.edu/catalog/11903.html. Accessed August 7, 2013. [PubMed]
- 2.D’Avolio LW, Farwell WR, Fior LD. Comparative effectiveness research and medical informatics. Am J Med. 2010 Dec;123(12 Suppl 1):e-32–7. doi: 10.1016/j.amjmed.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Embi PJ, Payne PR. Clinical research informatics: challenges, opportunities and definition for an emerging domain. J Am Med Inform Assoc. 2009 May-Jun;16(3):316–27. doi: 10.1197/jamia.M3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etheredge LM. Creating a high-performance system for comparative effectiveness research Health Aff (Millwood) 2010 Oct;29(10):1761–7. doi: 10.1377/hlthaff.2010.0608. [DOI] [PubMed] [Google Scholar]
- 5.Gluck ME, Ix M, Kelley B. Early glimpses of the learning health care system The potential impact of Health IT. AcademyHealth. Available at: https://www.academyhealth.org/Programs/ProgramsDetail.cfm?itemnumber=5091. Accessed August 7, 2013.
- 6.Sittig DF, Hazlehurst BL, Brown J, et al. A survey of informatics platforms that enable distributed comparative effectiveness research using multi-institutional heterogenous clinical data. Med Care. 2012 Jul;:S49–S59. doi: 10.1097/MLR.0b013e318259c02b. 50 (Electronic Data Methods (EDM) Forum Special Supplement): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holve E, Segal C, Hamilton Lopez M, Rein A, Johnson BH. The Electronic Data Methods (EDM) Forum for Comparative Effectiveness Research (CER) Medical Care. 2012 Jul;:S7–S10. doi: 10.1097/MLR.0b013e318257a66b. 50 (Electronic Data Methods (EDM) Forum Special Supplement): [DOI] [PubMed] [Google Scholar]
- 8.Rein A. Finding value in volume: An exploration of data access and quality challenges. A Health IT for Actionable Knowledge Report, Academy Health. Available at: https://www.academyhealth.org/Programs/ProgramsDetail.cfm?itemnumber=5091. Accessed August 7, 2013.
- 9.McGraw D, Leiter A. Legal and policy challenges to secondary uses of information from electronic health records. A Health IT for Actionable Knowledge report, Academy Health. Available at: https://www.academyhealth.org/Programs/Programs-Detail.cfm?itemnumber=5091. Accessed August 7, 2013. [Google Scholar]
- 10.Gliklich RE, Dreyer NA, editors. Registries for Evaluating Patient Outcomes: A User’s Guide. Beyond the patient: protection of registry data from litigation and other confidentiality concerns for providers, manufacturers, and health plans. Rockville MD: Agency for Healthcare Research and Quality; Mar, 2012. Draft white paper for the 3rd edition (Prepared by Outcome DEcIDE Center [Outcome Sciences, Inc. dba Outcome] under Contract No. HHSA290200500351 T01.) AHRQ Publication 07-EHC001-1. [Google Scholar]
- 11.Gliklich RE, Dreyer NA, editors. Registries for evaluating patient outcomes: A User’s Guide. Analytic challenges in studies that use administrative databases or medical registries. 3rd edition. Rockville MD: Agency for Healthcare Research and Quality; Mar, 2012. Draft white paper for the. (Prepared by Outcome DEcIDE Center [Outcome Sciences, Inc. dba Outcome] under Contract No. HHSA290200500351 T01.) AHRQ Publication 07-EHC001-1. [Google Scholar]
- 12.Weiner MG, Embi PJ. Toward reuse of clinical data for research and quality improvement: the end of the beginning? Ann Intern Med. 2009 Sep;151(5):360–1. doi: 10.7326/0003-4819-151-5-200909010-00141. [DOI] [PubMed] [Google Scholar]
- 13.Kahn MG, Raebel MA, Glanz JM, Riedlinger K, Steiner JF. A pragmatic framework for single-site and multisite data quality assessment in electronic health record-based clinical research. Med Care. 2012 Jul;:S21–S29. doi: 10.1097/MLR.0b013e318257dd67. 50 (Electronic Data Methods (EDM) Forum Special Supplement): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abernethy AP, et al. Rapid-learning system for cancer care. J Clin Onc. 2010 Sep 20;28(27):4268–74. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinbrook R. The NIH Stimulus – The Recovery Act and biomedical research. New Engl J Med. 2009 Apr 9;360(15):1479–81. doi: 10.1056/NEJMp0901819. [DOI] [PubMed] [Google Scholar]
- 16.Stein HD, Nadkarni P, Erdos J, et al. Exploring the degree of concordance of coded and textual data in answering clinical queries from a clinical data repository. J Am Med Inform Assoc. 2000 Jan-Feb;7(1):42–52. doi: 10.1136/jamia.2000.0070042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aronsky D, Haug J. Assessing the quality of clinical data in a computer-based medical record for calculating the pneumonia severity index. J Am Med Inform Assoc. 2000;7:55–65. doi: 10.1136/jamia.2000.0070055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahn MG, Eliason BB, Bathurst J. Quantifying clinical data quality using relative gold standards. AMIA Annu Symp Proc. 2010 Nov 13;:356–60. [PMC free article] [PubMed] [Google Scholar]
- 19.Hogan WR, Wagner MM. Accuracy of data in computer-based patient records. J Am Med Inform Assoc. 1997 Sep-Oct;4(5):342–55. doi: 10.1136/jamia.1997.0040342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang RY, Strong DM. Beyond accuracy: What data quality means to data consumers. Journal of Management Information Systems. 1996;12:5–34. [Google Scholar]
- 21.Parsons A, McCullough C, Wong J, Shih S. Validity of electronic health record-derived quality measurement for performance monitoring. J Am Med Inform Assoc. 2012 Jul-Aug;19(4):604–9. doi: 10.1136/amiajnl-2011-000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei WQ, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J Am Med Inform Assoc. 2012 Mar-Apr;19(2):219–24. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flum DR, Fisher N, Thompson J, et al. Washington State’s approach to surgical variability in surgical processes/outcomes: Surgical Clinical Outcomes Assessment Program (SCOAP) Surgery. 2005 Nov;138(5):821–8. doi: 10.1016/j.surg.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 24.Agency for Healthcare Research and Quality. Effective Healthcare Program. Patient Population Priorities. Available at: http://www.effectivehealthcare.ahrq.gov/index.cfm/submit-a-suggestion-for-research/how-are-research-topics-chosen/. Accessed: August 7, 2013.
- 25.SCOAP Collaborative Writing Group for the SCOAP Collaborative. Kwon S, Florence M, Gragas P, et al. Creating a learning healthcare system in surgery: Washington State’s Surgical Care and Outcomes Assessment Program (SCOAP) at 5 years. Surgery. 2012 Feb;151(2):146–52. doi: 10.1016/j.surg.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ARMUS™ Corporation. Burlingame CA. Available at: http://www.ARMUS.com/. Accessed August 7, 2013.
- 27.Amalga Unified Intelligence System (UIS)™. Caradigm™ Corporation. Bellevue, WA: Available at: http://www.caradigm.com/Pages/en-us/default.aspx. Accessed August 7, 2013. [Google Scholar]
- 28.Louie B, Mork P, Martin-Sanchez F, et al. Data integration and genomic medicine. J Biomed Inform. 2007 Feb;40(1):5–16. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 29.SCOAP data collection forms. Available at: http://www.scoap.org/documents. Accessed August 7, 2013.
- 30.Washington State Department of Health. Coordinated Quality Improvement Program. Available at: http://www.doh.wa.gov/PublicHealthandHealthcareProviders/HealthcareProfessionsandFacilities/CoordinatedQualityImprovement.aspx. Accessed August 7, 2013.
- 31.Health Information Privacy Summary of the HIPAA Privacy Rule. US Department of Health and Human Services. Available at: http://www.hhs.gov/ocr/privacy/hipaa/understanding/summary/index.html. Accessed August 7, 2013.
- 32.Lewis JR. Psychometric evaluation of the PSSUQ using data from five years of usability studies. International Journal of Human-Computer Interaction. 2002;14:463–88. Available from URL: http://drjim.0catch.com/PsychometricEvaluation-OfThePSSUQ.pdf. Accessed: August 7, 2013. [Google Scholar]
- 33.Being a meaningful user of electronic health records The Office of the National Coordinator of Health Information Technology, US Department of Health and Human Services. Available at: http://healthit.hhs.gov/portal/server.pt?CommunityID=2998&spaceID=42&parentname=&control=SetCommunity&parentid=&in_hi_userid=12059&Page-ID=0&space=CommunityPage. Accessed August 7, 2013.
- 34.Holve E, Segal C, Lopez MH. Opportunities and challenges for comparative effectiveness research(CER) with electronic clinical data. A perspective from the EDM Forum. Med Care. 2012 Jul;50(Electronic Data Methods (EDM) Forum Special Supplement):S11–S18. doi: 10.1097/MLR.0b013e318258530f. [DOI] [PubMed] [Google Scholar]
- 35.Lopez MH, Holve E, Sarkar IN, Segal C. Building the informatics infrastructure for comparative effectiveness research. A review of the literature. Med Care. 2012 Jul;50(Electronic Data Methods Forum Special Supplement):S38–S48. doi: 10.1097/MLR.0b013e318259becd. [DOI] [PubMed] [Google Scholar]
- 36.Randhawa GS, Slutsky JR. Building sustainable multi-functional prospective electronic clinical data systems. Med Care. 2012 Jul;50(Electronic Data Methods (EDM) Forum Special Supplement):S3–S6. doi: 10.1097/MLR.0b013e3182588ed1. [DOI] [PubMed] [Google Scholar]
- 37.Desai JR, Wu P, Nichols GA, Lieu TA, O’Connor PJ. Diabetes and asthma case definition, validation, and representativeness when using electronic health data to construct registries for comparative effectiveness and epidemiologic research. Med Care. 2012 Jul;50(Electronic Data Methods Forum Special Supplement):S30–S35. doi: 10.1097/MLR.0b013e318259c011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glasgow RE. Commentary: Electronic health records for comparative effectiveness research. Med Care. 2012 Jul;50(Electronic Data Methods (EDM) Forum Special Supplement):S19–S20. doi: 10.1097/MLR.0b013e3182588ee4. [DOI] [PubMed] [Google Scholar]
- 39.Luft HS. Commentary: Protecting human subjects and their data in multi-site research. Med Care. 2012 Jul;50(Electronic Data Methods (EDM) Forum Special Supplement):S74–S76. doi: 10.1097/MLR.0b013e318257ddd8. [DOI] [PubMed] [Google Scholar]
- 40.Marsolo K. Approaches to facilitate institutional review board approval of multicenter research studies. Med Care. 2012 Jul;50(Electronic Data Methods (EDM) Forum Special Supplement):S77–S81. doi: 10.1097/MLR.0b013e31825a76eb. [DOI] [PubMed] [Google Scholar]
- 41.Kushida CA, Nichols DA, Jadrnicek R, Miller R, Walsh JK, Griffin K. Strategies for de-identification and anonymization of electronic health record data for use in multicenter research studies. Med Care. 2012 Jul;50(Electronic Data Methods (EDM) Forum Special Supplement):S82–S101. doi: 10.1097/MLR.0b013e3182585355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Office of the National Coordinator for Health Information Technology. Health IT Policy Committee: Request for comment summary. Available at: http://www.healthit.gov/sites/default/files/stage_3_rfc_responses_summary_v3.pdf. Accessed August 7, 2013.
- 43.Simborg DW, Detmer DE, Berner ES. The wave has finally broken: now what? J Am Med Inform Assoc. 2013;20:e21–25. doi: 10.1136/amiajnl-2012-001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.US Department of Health and Human Services. Quality Improvement Activities – FAQs. Available at: http://answers.hhs.gov/ohrp/categories/1569. Accessed August 7, 2013.