Abstract
Introduction:
A key attribute of a learning health care system is the ability to collect and analyze routinely collected clinical data in order to quickly generate new clinical evidence, and to monitor the quality of the care provided. To achieve this vision, clinical data must be easy to extract and stored in computer readable formats. We conducted this study across multiple organizations to assess the availability of such data specifically for comparative effectiveness research (CER) and quality improvement (QI) on surgical procedures.
Setting:
This study was conducted in the context of the data needed for the already established Surgical Care and Outcomes Assessment Program (SCOAP), a clinician-led, performance benchmarking, and QI registry for surgical and interventional procedures in Washington State.
Methods:
We selected six hospitals, managed by two Health Information Technology (HIT) groups, and assessed the ease of automated extraction of the data required to complete the SCOAP data collection forms. Each data element was classified as easy, moderate, or complex to extract.
Results:
Overall, a significant proportion of the data required to automatically complete the SCOAP forms was not stored in structured computer-readable formats, with more than 75 percent of all data elements being classified as moderately complex or complex to extract. The distribution differed significantly between the health care systems studied.
Conclusions:
Although highly desirable, a learning health care system does not automatically emerge from the implementation of electronic health records (EHRs). Innovative methods to improve the structured capture of clinical data are needed to facilitate the use of routinely collected clinical data for patient phenotyping.
Keywords: Learning Health System, Comparative Effectiveness, Data Use and Qualit
Introduction and Background
The principles of evidence-based health care posit that clinical and public health decisions should be made using, among other elements, the best available clinical evidence.1,2 A natural consequence of this is the need to continuously generate new and better evidence for sound decision-making. The same principles argue that—in terms of risk of bias—there is a hierarchy of evidence in which randomized, controlled clinical trials (RCT) and systematic reviews (SR) of such trials lie at the top. Although RCTs and SRs offer the smallest risk of bias, several issues limit their availability, such as cost and time. Next in the hierarchy are quasi-randomized-, cohort-, and case-control studies. These studies rely on the observation of naturally occurring phenomena—either prospective or retrospectively—to draw conclusions on the effectiveness of different clinical interventions. The advantage of these approaches are that they enable answering many more clinical questions at much lower cost. However, these study designs are subject to confounding, given the large number of unmeasured variables and the potential for unbalanced distributions of such variables between treatment and control groups, thereby increasing the risk of biased study results.
The increasing implementation of electronic health records (EHRs)3 is producing an increasing availability of routinely collected clinical data in electronic form that can be used for secondary purposes. The Institute of Medicine (IOM) has proposed the creation of a learning health care system in which rapid learning can happen when analyzing electronic clinical data (ECD) from millions of patients.4 Examples of such secondary uses of clinical data are the conduction of comparative effectiveness research (CER) studies5 and health care Quality Assurance and Quality Improvement projects (QA/QI). CER is not a new type of research but is a systematization of the need to obtain head-to-head comparisons on the effectiveness of plausible, alternative therapeutic approaches.6 QA involves measuring compliance against specific standards and QI involves continuous activities to improve processes to meet such standards. The amount of data being generated through patient care could be a valuable source of information to conduct these kinds of studies while reducing, although not eliminating, some of the limitations of nonexperimental studies. Despite this promise, there are several barriers to conducting CER and QA/QI studies using routinely collected clinical data.
A qualitative study conducted by the AcademyHealth Electronic Data Methods (EDM) Forum revealed several barriers to conducting CER studies using routinely collected EHR data. Among these, the authors described substantial efforts to establish data sharing practices, the limitations of informatics tools, the need for methods to assess data quality, and the need to generate patient and consumer engagement.7
One issue that significantly affects ECD quality—its availability in particular—is the abundance of free-text used to capture key patient attributes. Since the primary purpose of EHRs is to support individual patient care—and this is a task predominantly performed by clinicians—free-text is frequently used to capture patient information using clinical notes, operative reports, and discharge summaries, among others. However, as we mentioned above, this hampers the reuse of such data. Information systems that allow structured clinical documentation have been implemented to address this issue, but this is not innocuous as they can significantly interfere with clinical workflow.8 As Rosenbloom et al.9 report, the “continuous tension between structure and expressiveness” of clinical notes remains to be solved. To this date, organizations with significant experience in secondary use of clinical data still rely on manual extraction of information from clinical notes.10
Natural Language Processing (NLP) is the currently accepted method to overcome this barrier. However, despite the increasing accuracy of NLP engines, their classification precision is still not ideal. Harkema et al. conducted a study in 2011 in which they designed an NLP engine to process colonoscopy reports and extract procedural quality measures.11 The authors concluded that the accuracy of their system was sufficient for less than 50 percent of all the measures studied. In addition to insufficient accuracy, the resources and time required to develop a single NLP classifier and its limited transferability to alternative settings12 have limited its widespread adoption.13
Without precise methods to extract information from free-text, there remains a significant barrier in the reuse of ECD. The extent to which the prevalence of free-text affects or impedes the conduction of CER and QA/QI studies using EHR data is unknown. Moreover, since the implementation of an EHR can be significantly customized to a specific organization’s needs, the amount of data stored in unstructured formats might vary significantly among organizations. The aims of this study are the following: (1) to describe the extent to which data needed for a specific pre-existing QA/QI and CER project are stored in noncomputable or hard to compute formats, (2) to identify differences among institutions regarding storage of clinical data for CER and QA/QI, and (3) to suggest possible ways to overcome the lack of availability of key data for CER and QA/QI.
Methods
Setting
This study was conducted in the context of the Surgical Care and Outcomes Assessment Program (SCOAP).14 SCOAP is a clinician-led, performance benchmarking, and QI registry for surgical and interventional procedures. Created by a community of clinicians in Washington State,15 SCOAP operates under the Foundation for Health Care Quality (FHCQ), a nonprofit 501(c)(3) corporation, as a Washington State approved Coordinated Quality Improvement Program. Hospitals hire and train staff to review medical records and abstract clinical data using SCOAP data collection forms and data dictionaries. The clinical data are manually entered into the SCOAP registry using a web-based form. Participant hospitals receive quarterly QI performance reports showing their data alongside benchmarks and peer performance. The SCOAP continuous data collection and feedback loop is proven to improve the quality and safety of surgical and interventional care while decreasing costs.16,17 There are five SCOAP registries: gastrointestinal and general surgical procedures, oncologic surgical procedures, pediatric surgical procedures, spine surgical procedures, and vascular surgical and interventional health care procedures. The number of clinical questions in each form to document cases included in each registry ranges from approximately 580 to 880.
One of the barriers that restrict the scale of SCOAP is the cost of manual data collection. To address this problem with registries in general, the Agency for Healthcare Research and Quality (AHRQ) funded the Comparative Effectiveness Research and Translation Network (CERTAIN)18,19,20 in a three-year project to analyze SCOAP data for CER and enhance the ability to advance capture of SCOAP data, while improving its clinical quality. A key component of this was the SCOAP automation project. To accomplish this, CERTAIN investigators implemented a central data repository in which data were automatically retrieved from the EHRs at their original source hospitals and stored for later analysis. To achieve this, investigators recruited hospitals already participating in SCOAP, in which EHRs were available for automated data abstraction. This was seen as an opportunity to assess the availability of structured clinical data for CER and QA/QI and its variability across institutions.
Site Selection
The project leadership contacted 19 hospitals, 15 of which provided project support at the Chief Medical Officer (CMO) or Chief Information Officer (CIO) level. We selected the following participating sites for data analysis.
Two of the hospitals are part of a large, integrated academic health system in King County, Washington. Hospital 1 is a 450-bed university tertiary referral hospital; and Hospital 2 is a 385-bed county hospital and level I trauma center. Both institutions are supported by a single health IT organization (Health IT Group A). Both hospitals use individual instances of an EHR but are centrally managed by the IT department.
Four of the hospitals operate in Spokane County, Washington. Hospital 3 is a 307-bed private for-profit general and medical hospital, Hospital 4 is a 272-bed private not-for-profit general hospital; Hospital 5 is a 628-bed private not-for-profit multispecialty hospital and level II pediatric trauma center; and Hospital 6 is a 123-bed private hospital and level III trauma center. All four institutions are supported by a single health IT organization (Health IT Group B). All four hospitals use a single instance of an EHR that is centrally managed by the IT department.
Data Analysis
Analysis of data availability was conducted using the SCOAP data collection form for gastrointestinal and general surgical procedures (General SCOAP). This 15-page form is routinely used to manually collect data generated during general surgical procedures such as appendectomies, colectomies, and bariatric surgeries; it usually takes approximately 30 minutes to complete. The General SCOAP abstraction form includes multiple data elements that range from simple demographic information such as age, gender and insurance type, to more complex data such as whether there was an unplanned intensive care unit stay in the postoperative period. A subset of the form contains data elements common to all types of procedures (core) and includes information such as the date of the procedure. The remainder of the data elements are specific to each type of procedure, such as the presence or absence of a colostomy in the case of a procedure involving the colon.
Using the core of the General SCOAP data abstraction form, investigators constructed a spreadsheet in which every data element was stored in a single row. Next, investigators met with local information technology (IT) and clinical personnel to determine the system in which each data element could be found, the format in which it was stored, and the file format or messaging standard in which the content of a field could be transmitted to the central data warehouse. Depending on the format in which the data were stored, investigators determined whether each data element was easy, moderate, or difficult to extract. The investigators used a team consensus process to assign classification values to each element, consulting frequently with local IT and clinical personnel to verify assumptions and clarify questions. The classification schema used is described in Table 1.
Table 1.
Scheme Used to Classify Data Elements Necessary to Complete the General SCOAP Data Abstraction Form According to Ease of Automatic Extraction
| Classification | Description | Example |
|---|---|---|
| Easy | Data element stored as a structured and accessible database field. | Age stored as an integer, smoking status stored as “yes/no,” diagnosis stored as an ICD-9 code. |
| Moderate | Data element that requires the use of more than one structured database field to calculate. | “Antibiotics administered within 60 minutes of surgical incision,” for which the antibiotics administration date/time and surgical incision date/time are needed as well as logic to compare the temporality of the events. |
| Complex | Data element stored as free-text, or that needs human interpretation to abstract. | “Comorbidities” captured within the admission notes and stored as free-text, or an “unplanned ICU stay” for which human interpretation is needed to decide whether or not an ICU stay was planned. |
After classifying every data element, investigators (a clinical data abstractor, a clinical data repository administrator, and a biomedical informatician) qualitatively assessed the sources of complexity for the data elements classified as “moderate” and “complex.” For example, a data element classified as moderate would then be assigned a source of such complexity, such as “data in multiple, non-integrated clinical systems.” Finally, to provide suggestions about ways to improve the availability of structured data for CER and QA/QI, investigators annotated these moderate and complex data elements according to the time the information was available with respect to the hospitalization that prompted the data collection, and whether it was stored as free-text in a single clinical system and, thus, suitable for NLP extraction. For example, “pre-existing hypertension” is information available before the surgical episode that would be suitable for NLP extraction if stored as free-text. On the other hand, “highest intraoperative blood glucose” is information not available before the surgery and, since usually stored as structured data, it would not be necessary to extract using NLP.
Investigators used simple descriptive statistics such as means and proportions to summarize findings, and the Chi2 test to assess differences in proportions of data elements classifications between sites.
Results
The SCOAP automation project began recruiting hospitals in Washington State in November 2010, and by 2014 it has recruited five individual sites, four of which are currently submitting patient data to the project’s data repository. The present analysis was conducted on Health IT Group A and Health IT Group B data.
The core of the General SCOAP spreadsheet consisted of 185 unique clinical data elements. Those elements covered multiple dimensions of a surgical episode such as:
Patient demographics (i.e., age, gender, insurance type);
Risk factors (i.e., smoking status, home mobility device use, home oxygen use);
Comorbidities (i.e., hypertension, diabetes, asthma);
Operative events (i.e., incision time, skin preparation, lowest intraoperative body temperature);
Perioperative interventions (i.e., epidural injection administered within 24 hours of incision time, statins administered after surgery, nasogastric tube placement); and
Postoperative events (i.e., stroke, myocardial infarction, urinary tract infection).
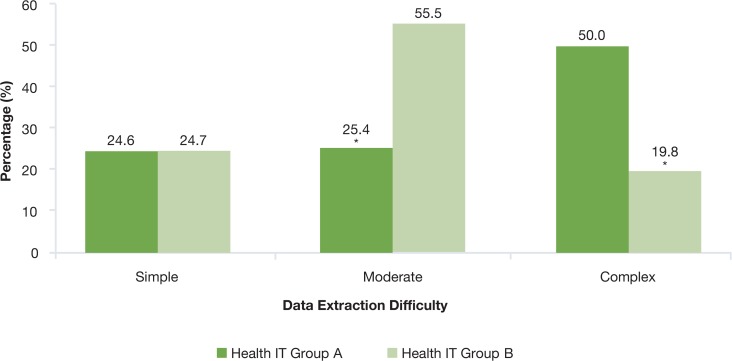
On average, considering both Health IT Groups, 25 percent of all data elements were considered easy to extract, 43 percent moderate, and 32 percent complex. When comparing Health IT Group A with Health IT Group B, the proportion of easy-to-extract data elements was similar, with 25 percent each. However, there were marked differences between both organizations in the proportion of moderately complex to extract (25% versus 56%, p < 0.0001), as well as the proportion of complex to extract (50% versus 20%, p < 0.0001) (Figure 1).
Figure 1.
Complexity of Extraction of Electronic Clinical Data for Comparative Effectiveness Research and Quality Improvement across Two Hospital Groups
* p <0.01 for the comparison
A large proportion of the easily extractable data elements were demographic variables: 91 percent at Health IT Group A and 67 percent at Health IT Group B. Those that constituted the moderately complex and complex categories also differed across both organizations. Most cases of moderately complex elements for the Health IT Group A were constituted by information obtained in the perioperative period (42.4%), such as “beta blockers restarted within 24 hours after surgery” or “epidural injection administered within 24 hours before surgery.” In the case of Health IT Group B, the most frequently reported data elements in the moderate category was information obtained in the postoperative period (31%), such as the presence of a “Clostridium difficile infection after surgery” or an episode of “deep venous thrombosis” after surgery.
In the case of data elements classified as complex to extract, in the Health IT Group A’s database the most prevalent were operative information (45%) followed by postoperative events (28%). For Health IT Group B, the most prevalent were postoperative events (70%) followed by perioperative events. A detailed comparison between categories across sites is depicted in Table 2.
Table 2.
Type of Data and Complexity of Extraction across Both Health IT Groups
| Moderate (%) | Complex (%) | |||
|---|---|---|---|---|
| Health IT A | Health IT B | Health IT A | Health IT B | |
| Demographics | 18.2 | 30.0 | 0.0 | 0.0 |
| Risk Factors | 24.2 | 11.0 | 3.1 | 0.0 |
| Operative | 6.1 | 0.0 | 44.6 | 0.0 |
| Intraoperative | 9.1 | 0.0 | 6.2 | 11.1 |
| Perioperative | 42.4 | 19.0 | 9.2 | 19.4 |
| Comorbidities | 0.0 | 9.0 | 9.2 | 0.0 |
| Postoperative | 0.0 | 31.0 | 27.7 | 69.4 |
Sources of Complexity
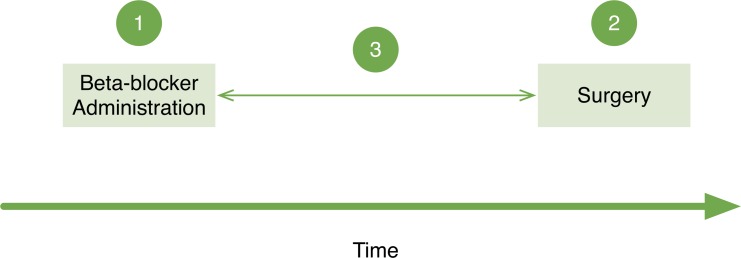
Moderate complexity was primarily attributed to the need to perform calculations using two or more additional data elements, such as assessing relative temporal relations. For example, to determine whether an eligible patient had received a beta-blocker within 24 hours after the surgical procedure, it was necessary to obtain the time interval during which the patient underwent the operation, the time the beta-blocker was reordered postoperatively, and then calculate whether the time difference between those two was shorter than or equal to 24 hours (Figure 2). These relative temporal relations required building fairly complex database queries so they were frequently assessed manually instead.
Figure 2.
Three Steps Involved in a Relative Temporal Query to Determine Whether a Dose of Beta-Blockers Was Administered Within 24 Hours of a Surgery
At least three steps are involved in a relative temporal query to determine whether a dose of beta-blockers was administered within 24 hours of a surgical procedure. This is a relative temporal relationship since there is no absolute relationship, such as a specific date or time, between the events and the timeline.
In the case of data elements classified as complex to extract, an overwhelming majority was information stored as free-text within the electronic medical record. As examples, the determination of whether a patient had undergone a surgical wound revision was made by reviewing the information contained in daily clinical plans or in operative reports of such procedures. In addition to free-text, a case worth noting was when information was simply not available within the organization’s electronic medical record, such as the occurrence of a post discharge death.
Alternative Sources for Extraction
For Health IT Group A data classified as moderate or complex to extract, 36 percent was information produced before the index hospitalization, and 50 percent of those data elements could be obtained directly from the patient. Strategies to capture that information using preoperative patient surveys might increase the proportion of structured data for CER and QA/QI. On the other hand, 64.1 percent of the data elements consisted of information produced during the index hospitalization, of which all except one were information stored as free-text and thus suitable for extraction using NLP.
Discussion
Our findings suggest that a significant proportion of the data available to phenotype patients for quality improvement and CER studies are not available in an easily extractable, structured format. In this study, only one-fourth of the data required to populate the SCOAP automated central data repository were available in an easily extractable format. This scenario, if we assume it is representative of the realities across multiple health care systems, severely limits the ability to build a learning health care system in which routinely collected clinical data can be used to generate new evidence about what works in health care—without implementing innovative methods to improve either the extraction or the structured capture of clinical data.
This study’s main finding was the difference encountered when analyzing data from the different study sites. At Health IT Group A, the majority of data elements required to populate the SCOAP automated central data repository were considered too complex to extract automatically. Most of the data were stored in the form of free-text or as information that needed human interpretation to be adequately abstracted. At Health IT Group B, a minority of all the data elements fell into that same category and this organization’s data were mostly classified as moderately complex to extract.
Although we didn’t formally explore the causes behind these differences, during work meetings it became apparent that Health IT Group B was able to successfully mandate a greater proportion of structured documentation from their providers. These documentation practices might be the result of tacit or explicit decisions made at the time of implementing such health information systems. This possible source of the observed differences highlights the importance of having secondary uses of clinical data in mind when implementing health information systems.21
One obvious question that emerges from this study is: how can we increase the proportion of data elements that are captured in a structured and computable format? There is probably not a single right answer for this, but there are multiple strategies to address this. First, as we mentioned above, organizations need to have secondary uses in mind when designing and implementing their information systems. Second, there is a need for balance between the expressiveness of free-text documentation and the benefit of documenting these in structured formats; this principle should guide decisions on how to capture individual data elements. For example, in Health IT Group A, “smoking status” was captured using free-text, and the option to automate its extraction was to build a NLP engine; whereas in Health IT Group B it was captured as a structured field. In this example, where the information has little or no ambiguity and probably does not require expressive language to document, it is amenable to capture it in a structured format. In contrast, “ICU readmission post discharge” was categorized as complex to extract in both institutions; unless there is a standard definition for this concept, it is unlikely that it could be captured using a yes or no answer since the definition might change for any given study. For instance, two consecutive ICU stays may be interpreted differently. An ICU patient that goes to the operating room and back to the ICU may not count as a re-admission, whereas a surgical ward patient that reenters the ICU due to a complication may count as one. This interpretability of the concept “ICU readmission post discharge” renders it unlikely to be captured primarily as a structured field and more likely to require interpretation according to the type of secondary use.
Finally, innovative methods of data capture that do not disrupt clinical workflows and leveraging of other sources of information, such as patient reported data and outcomes, are necessary to increase the availability of structured and computable data for secondary uses of ECD. In all cases, the cost of changing established documentation practices or capturing data de novo is probably higher than deciding to capture data in a structured format from the beginning. A natural consequence is the need to move from meaningful use of health information technologies to meaningful implementation. Although there is a large body of research describing the reasons behind inadequate ECD quality,22 to our knowledge this is the first comparison of the availability of structured ECD across multiple sites.
The main limitation of the study is the method selected to assess the complexity of extracting specific elements of the SCOAP form. To our knowledge there is no standardized method to assess such complexity so we created this classification schema specifically for this study. To reduce variability, the same group of investigators applied the scheme to both Health IT Groups. The proposed method, when applied by the same investigator as was the case in this study, should allow the comparison between sites but may not be transferrable to other organizations when applied by a different set of researchers. As a consequence, we highlight that the generalizable findings are the differences we encountered between organizations rather than the exact prevalence of complex-to-extract data elements.
Future Work
Considering these findings, our group will continue working on a generalizable tool to formally assess the complexity of extracting clinical data for CER and QA/QI. Such a tool could be used to test “readiness” across organizations that wish to collaborate on such secondary uses of clinical data. Finally, in order to achieve the vision of a learning health care system, the health informatics community and health care delivery organizations must develop tools that allow data capture in ways that preserve rich descriptions of patient phenotypes, as well as their easy computation for secondary use. These tools should probably include combinations of natural language processing, capture of patient reported data, and novel human-computer interfaces.
Acknowledgments
This project was supported by grant number 1 R01 HS 20025-01 from the Agency for Healthcare Research and Quality. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality. The Surgical Care and Outcomes Assessment Program (SCOAP) is a Coordinated Quality Improvement Program of the Foundation for Health Care Quality. CERTAIN is a program of the University of Washington, the academic research and development partner of SCOAP.
Personnel contributing to this study: Centers for Comparative and Health Systems Effectiveness (CHASE Alliance), University of Washington, Seattle, WA: Daniel Capurro, MD, PhD; Allison Devlin, MS; E. Beth Devine, PharmD, MBA, PhD; Marisha Hativa, MSHS; Prescott Klassen, MS; Kevin Middleton; Michael Tepper, PhD; Peter Tarczy-Hornoch, MD; Erik Van Eaton, MD; N. David Yanez III, PhD; Meliha Yetisgen-Yildiz, PhD, MSc; Megan Zadworny, MHA.
Footnotes
Disciplines
Health Information Technology | Health Services Research
References
- 1.Guyatt G, Rennie D. Users’ Guides to the Medical Literature: A Manual for Evidence-based Clinical Practice. McGraw-Hill Professional Publishing; 2008. [Google Scholar]
- 2.Sackett DL, Rosenberg W, Gray J, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn’t. BMJ. British Medical Journal Publishing Group. 1996;312(7023):71. doi: 10.1136/bmj.312.7023.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jha A, DesRoches C, Campbell E, Donelan K, Rao S, Ferris T, et al. Use of electronic health records in US hospitals. New England Journal of Medicine. 2009;360(16):1628. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- 4.National Research Council . The Learning Healthcare System: Workshop Summary (IOM Roundtable on Evidence-Based Medicine) Washington, DC: The National Academies Press; 2007. [PubMed] [Google Scholar]
- 5.Holve E, Segal C, Lopez MH, Rein A, Johnson BH. The Electronic Data Methods (EDM) forum for comparative effectiveness research (CER) Medical Care. 2012 Jul;50(Suppl):S7–10. doi: 10.1097/MLR.0b013e318257a66b. [DOI] [PubMed] [Google Scholar]
- 6.Sox HC, Greenfield S. Comparative effectiveness research: a report from the Institute of Medicine. Annals of Internal Medicine. 2009;151(3):203–205. doi: 10.7326/0003-4819-151-3-200908040-00125. [DOI] [PubMed] [Google Scholar]
- 7.Holve E, Segal C, Hamilton Lopez M. Opportunities and Challenges for Comparative Effectiveness Research (CER) With Electronic Clinical Data: A Perspective From the EDM Forum. Medical Care. 2012 Jul;50(Suppl):S11–8. doi: 10.1097/MLR.0b013e318258530f. [DOI] [PubMed] [Google Scholar]
- 8.Embi PJ, Yackel TR, Logan JR, Bowen JL, Cooney TG, Gorman PN. Impacts of computerized physician documentation in a teaching hospital: perceptions of faculty and resident physicians. Journal of the American Medical Informatics Association : JAMIA. 2004 Jun;11(4):300–9. doi: 10.1197/jamia.M1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: a perspective on the tension between structure and flexible documentation. Journal of the American Medical Informatics Association. 2011 Mar 8;18(2):181–6. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCahill LE, Single RM, Aiello Bowles EJ, Feigelson HS, James TA, Barney T, et al. Variability in Reexcision Following Breast Conservation Surgery. JAMA. 2012 Jan 31;307(5):467–75. doi: 10.1001/jama.2012.43. [DOI] [PubMed] [Google Scholar]
- 11.Harkema H, Chapman WW, Saul M, Dellon ES, Schoen RE, MEHROTRA A. Developing a natural language processing application for measuring the quality of colonoscopy procedures. Journal of the American Medical Informatics Association. BMJ Publishing Group Ltd. 2011;18(Suppl 1):i150–6. doi: 10.1136/amiajnl-2011-000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hripcsak G, Austin JHM, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002 Jul;224(1):157–63. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 13.Stanfill MH, Williams M, Fenton SH, Jenders RA, Hersh WR. A systematic literature review of automated clinical coding and classification systems. Journal of the American Medical Informatics Association. 2010;17(6):646–651. doi: 10.1136/jamia.2009.001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.SCOAP: Surgical Clinical Outcomes Assessment Program [Internet]. SCOAP; [cited 2013 Mar 15]. Available from: http://www.scoap.org/ [Google Scholar]
- 15.Flum DR, Fisher N, Thompson J, Marcus-Smith M, Florence M, Pellegrini CA. Washington State’s approach to variability in surgical processes/Outcomes: Surgical Clinical Outcomes Assessment Program (SCOAP) Surgery. 2005;138(5):821–828. doi: 10.1016/j.surg.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Kwon S, Florence M, Grigas P, Horton M, Horvath K, Johnson M, et al. Creating a learning healthcare system in surgery: Washington State’s Surgical Care and Outcomes Assessment Program (SCOAP) at 5 years. Surgery. 2011 doi: 10.1016/j.surg.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flum DR, Pellegrini CA. The Business of Quality in Surgery. Ann Surg. 2012;255(1):6–7. doi: 10.1097/SLA.0b013e31824135e4. [DOI] [PubMed] [Google Scholar]
- 18.Devine E, Alfonso-Cristancho R, Devlin A, Edwards T, Farrokhi E, Kessler L, et al. A Model for Incorporating Patient and Stakeholder Voices in a Learning Healthcare Network: Washington State’s Comparative Effectiveness Research Translation Network (CERTAIN) J Clin Epidemiol 2013. 2013 Apr;66:S122–S129. doi: 10.1016/j.jclinepi.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flum D, Alfonso-Cristancho R, Devine E, Devlin A, Farrokhi E, Tarczy-Hornoch P, et al. Implementation of a “Real World” Learning Healthcare System: Washington State’s Comparative Effectiveness Research Translation Network (CERTAIN) Surgery In Press. doi: 10.1016/j.surg.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Comparative Effectiveness Research and Translation Network. Available from: http://www.becertain.org/
- 21.Blumenthal D, Tavenner M. The“ Meaningful Use” Regulation for Electronic Health Records. New England Journal of Medicine. 2010 Aug 5;363(6):501–4. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 22.Hersh WR, Weiner MG, Embi PJ, Logan JR, Payne PR, Bernstam EV, Lehmann HP, Hripcsak G, Hartzoh TH, Cimino JJ, Saltz JH. Caveats for the use of operational electronic health record data in comparative effectiveness research. Medical Care. 2013;51:S30–S37. doi: 10.1097/MLR.0b013e31829b1dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]