Abstract
Introduction:
A visible example of a successfully disseminated research project in the healthcare space is Informatics for Integrating Biology and the Bedside, or i2b2. The project serves to provide the software that can allow a researcher to do direct, self-serve queries against the electronic healthcare data form a hospital. The goals of these queries are to find cohorts of patients that fit specific profiles, while providing for patient privacy and discretion. Sustaining this resource and keeping its direction has always been a challenge, but ever more so as the ten year National Centers for Biomedical Computing (NCBCs) sunset their funding.
Findings:
Building on the i2b2 structures has helped the dissemination plans for grants leveraging it because it is a disseminated national resource. While this has not directly increased the support of i2b2 internally, it has increased the ability of institutions to leverage the resource and generally leads to increased institutional support.
Discussion:
The successful development, use, and dissemination i2b2 has been significant in clinical research and informatics. Its evolution has been from a local research data infrastructure to one disseminated more broadly than any other product of the National Centers for Biomedical Computing, and an infrastructure spawning larger investments than were originally used to create it. Throughout this, there were two main lessons about the benefits of dissemination: that people have great creativity in utilizing a resource in different ways and that broader system use can make the system more robust. One option for long-term sustainability of the central authority would be to translate the function to an industry partner. Another option currently being pursued is to create a foundation that would be a central authority for the project.
Conclusion:
Over the past 10 years, i2b2 has risen to be an important staple in the toolkit of health care researchers. There are now over 110 hospitals that use i2b2 for research. This open-source platform has a community of developers that are continuously enhancing the analytic capacities of the platform and inventing new functionality. By understanding how i2b2 has been sustained, we hope that other research infrastructure projects may better navigate options in making those initiatives sustainable over time.
Keywords: i2b2, research, medical records
Introduction
A visible example of a successfully disseminated research project in the health care space is Informatics for Integrating Biology and the Bedside (i2b2). The project provides the software that can allow a researcher to do direct, self-serve queries against the electronic health care data from a hospital. The goals of these queries are to find cohorts of patients that fit specific profiles, while providing for patient privacy and discretion. The Institutional Review Board (IRB) is then in control of the detailed data that may be given to the researcher for a scientific analysis through the i2b2 platform.1 Ultimately, tools by the entire community of i2b2 researchers become available to the hospital investigators to view and analyze the patient data in their cohort. Sustaining this resource and keeping its direction has always been a challenge, but it is even more so as the 10-year National Centers for Biomedical Computing (NCBCs) sunset their funding.
Background
In 1999, the Research Patient Data Registry (RPDR) was created at Partners HealthCare System (Partners), based on evaluations of queries against the existing (electronic medical record) EMR database2 and other query-generating tools.3,4 The RPDR is a research data warehouse with medical record information from multiple hospital and outpatient systems at Partners. It includes both data collected into the database and a tool for querying data from the repository. This tool allows research investigators to create cohorts of patients that meet specific criteria, in order to assess the availability of patients and patient data for studies. Once a cohort is defined and queried, patient identifiers and complete EMRs can be obtained according to IRB approval.
After initial pilot studies, the RPDR was released to full production at Partners in early 2002. Since that time the RPDR experienced steady growth in use, and is currently the primary method for clinical researchers at Partners to identify cohorts and access data from electronic health records (EHRs) for research.5
There have been two main effects of the successful implementation and use of the RPDR. First, RPDR use and assessments of benefit have led to sustained institutional support of the RPDR infrastructure.6 Specifically, the RPDR has been linked to funded grants at Partners that are critically dependent on the RPDR, such that the institution continues to fund the RPDR operational costs. Second, the concepts of the RPDR have led to external funding to create i2b2.7
The i2b2 was funded in 2004 as one of four initial National Centers for Biomedical Computing (NCBC).8 One purpose of the i2b2 project was to create software that could be used in research institutions across the nation to query data extracted from EMRs, either for identification of research cohorts or to discover potential clinical knowledge from observational studies using the data. In this way, i2b2 facilitates the use of a research data infrastructure, based on the design and lessons learned from RPDR. The i2b2 has been successfully implemented at over 90 research institutions or academic medical centers throughout the United States, and over 20 additional organizations internationally. It is arguably the most widely used clinical research data infrastructure based on EHRs in the world.9,10
In this paper, we discuss the sustainability experience of i2b2 as a disseminated research data infrastructure, and its effect on the sustainability of research using the RPDR. We describe how i2b2 has expanded in capabilities with its extension of the RPDR and dissemination, how it is effectively used at receiving institutions, how lessons are learned from its use, and projections on how it could ideally be sustained in the future. By understanding how i2b2 has been sustained, we hope that other research infrastructure projects may better navigate options in making those initiatives sustainable over time.
Findings
Informatics for Integrating Biology and the Bedside (i2b2) Development and Extensions
Prior to the NCBC initiative, there were no substantive incentives to distribute the RPDR capability outside Partners, other than reporting its functionality and use through academic publications. With NCBC funding, i2b2 was initially and primarily developed as a direct transfer of functionality from the RPDR to an open-source platform. For example, the query interface, with a hierarchical tree of items, the query panels, and Boolean logic combining the panels were all originally developed as Querytool for the RPDR.5 The security architecture, which obfuscates exact counts of data to prevent identification of single individuals within the system, was also first developed with RPDR.11 The i2b2 data model allows use of different ontologies or vocabularies for creating the tree of items, so that an institution could use clinical data standards other than those used by Partners in the RPDR. But other than an institution’s choice of standards, the functionality of i2b2 was directly migrated from RPDR development. The NCBC funding opportunity also allowed some extensions of i2b2 beyond RPDR that could also enhance the RPDR; e.g., i2b2 was made to be extensible in linking clinical data to genomic information.12
Over time, external grant funding beyond NCBC was received, which also focused more on expanding capabilities of i2b2 than RPDR directly. This was in part due to a competitive advantage in leveraging the NCBC distinction, as well as providing a clearer path for dissemination of ideas outside Partners. Funding has led to expansions in domain areas. For example, Medical Imaging Informatics Bench to Bedside (mi2b2) was an externally funded project that allows the retrieval and use of medical images from a picture archiving and communications system (PACS).13 The mi2b2 does not pull data directly into the i2b2 database, but rather makes specific images available for users to view and extract information. When paired with the i2b2 database, images can be retrieved for a specific patient population based on their clinical indicators from i2b2, thus narrowing the cohort for selection and the efficiency of the image review. The mi2b2 was an important extension of i2b2 because it expanded the i2b2 capabilities to a new realm of data, which would also expand the usefulness of i2b2 to more users. New users of a data infrastructure are best added not by increasing just the amount of data, but rather the number of different data types. Additionally, mi2b2 also allowed the phenotypic richness of medical images to what can be studied as secondary use data through i2b2.
Another externally supported project that extended i2b2 capabilities was the Shared Health Research Informatics Network (SHRINE).14,15 SHRINE was a collaborative development project with i2b2 that allowed the linking of i2b2 instances across different institutions to increase the available population for cohort identification. SHRINE aided in the governance of query distribution, allowing for real-time results that are fully compliant with an institution’s privacy needs. It was important to the expansion of i2b2 to other institutions because it created a structured entry point for using i2b2 with the possibilities of sharing data. In reality, institutions used i2b2 and received value with local queries of local data, but the potential for strategic collaboration and data sharing helped justify initial costs. SHRINE has followed a similar path to i2b2 in extensions after development, moving to dissemination of open-source software and expansion of capabilities through additional external funding.
The Substitutable Medical Applications, Reusable Technologies (SMART) Platforms project also contributed important functionality to i2b2. SMART was part of the Office of the National Coordinator’s (ONC) Strategic Health IT Advanced Research Projects (SHARP) Program, funded by the American Recovery and Re-investment Act (ARRA). SMART developed a platform for “app” (application) technology to construct a mosaic of patient-data visualization tools designed for patient care. The platform was tightly connected to i2b2 due to leveraging the infrastructure. As a result, SMART tools were also adopted by i2b2, providing a patient data visualization capability that was needed in i2b2, and also to RPDR.16
Building on the i2b2 structures has helped the dissemination plans for grants leveraging it because it is a disseminated national resource. While this has not directly increased the support of i2b2 internally, it has increased the ability of institutions to leverage the resource and generally leads to increased institutional support.
Dissemination and Expansion of i2b2
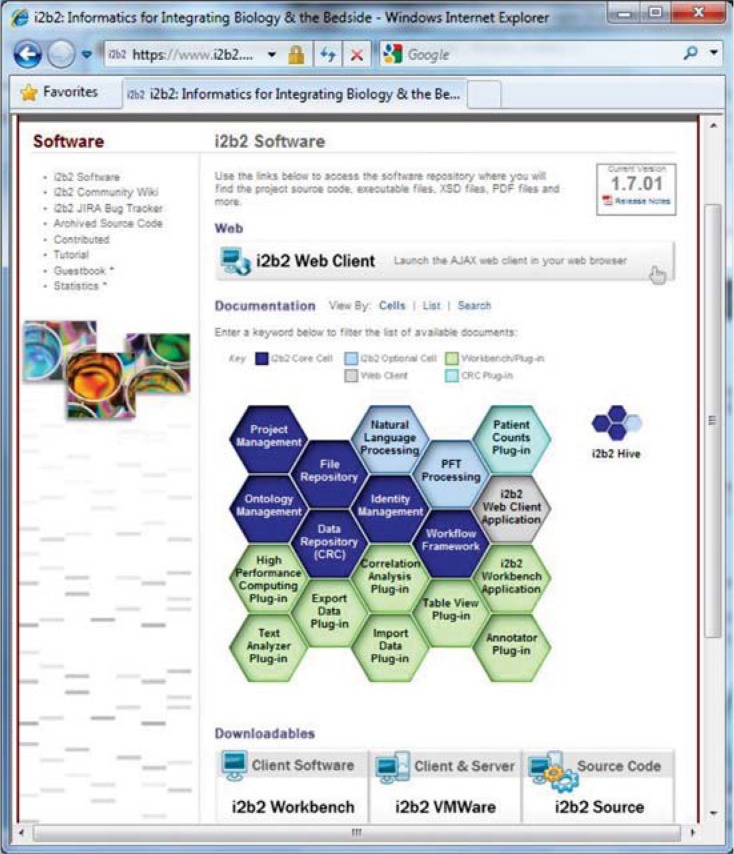
As mentioned above, i2b2 has been disseminated to over 110 institutions worldwide. The software includes the main client application—Java’s “workbench,” in addition to an easily distributable web-based client application. As functionality of i2b2 has expanded, the software has evolved to include other modules. The core software and modules are organized into “cells” of the i2b2 “hive” (see Figure 1).
Figure 1.
Core Software and Modules of i2b2
Notes: The distribution page of i2b2 (at www.i2b2.org/software) is shown with the i2b2 hive, which is arranged as a graphical depiction of how the modules (cells) of the hive can be brought together by different developers to create an entire framework. Clicking on the cells will bring up descriptions and installation instructions.
Critical cells are organized as core components (e.g., data repository, identity management), with backend or “workbench” plug-ins (e.g., text analyzer, export data plug-in) that give expanded functionality to the hive (e.g., natural language processing, pulmonary function test processing). While some of these optional cells have been developed by the core i2b2 team, other cells have been designed and developed by outside organizations that have implemented i2b2 and needed extensions to functionality in different areas. These are available on the i2b2 community web site at https://community.i2b2.org
Another way that functionality has expanded through i2b2 dissemination is through collaborative developments across the i2b2 user community. Formal examples of these are the i2b2 Challenges, where institutions are requested to address a specific issue for expanding i2b2 functionality, by designing and developing solutions.17–19 For example, the Medication Extraction challenge led to improvements in how i2b2 can use natural language processing to extract medication information from discharge summaries. The various i2b2 challenges have led to advancements in both i2b2 and related scientific fields—over 100 publications in research journals or conferences were enabled by i2b2 challenges (REF www.i2b2.org/NLP/DataSets/Publications.php).20
Expansions in functionality that come from other institutions improve the sustainability of the system by spreading development costs outside the host organization. Other institutions have helped spread other costs. At the University of Utah, researchers using i2b2 were able to perform evaluations of how the software could be used for self-service queries,21 and then tracked improvements in utility over time.22 Other researchers have disseminated how they have used the system, thus increasing resources for training.23–26 The use of technology to create collaborative information resources about i2b2 can also decrease education and training needed for implementation.
Expansion of functionality by other institutions through i2b2 extensions have generally happened by three methods. One process is that individuals from other institutions would look through the i2b2 code and make suggestions (e.g., bug fixes), which would then be fixed by i2b2 developers. A second process was i2b2 sponsored projects, where funds were contributed for external institutions to help develop components of i2b2. Examples are development of the i2b2 web client and the integration of the National Center for Biomedical Ontology (NCBO) web services.27 The third method was i2b2 related projects, where extensions were developed independently of the i2b2 team, but later contributed to the projects. While i2b2 has funded 2 sponsored projects, there have been over 30 related projects contributed, making it the most productive and sustainable method of i2b2 extension development.28 Other than providing the platform for development and the communication medium for users and developers from different institutions (the i2b2 community wiki), the central i2b2 leadership did not have to provide any additional support for these related projects to be developed and shared. Overall, these plug-ins grew organically as there were needs at local institutions. The sharing of the plug-ins allowed the effort form one institution to be reused at another, but this sharing activity was almost entirely supported through the goodwill of the researchers and developers involved.
Implementation support has also been an area for collaboration with industry on the i2b2 project. The original Recombinant Data was the first commercial company to provide support services for the i2b2 environment, helping many Clinical and Translational Science Award (CTSA) recipients to facilitate its deployment and use.29 Other companies have also worked to improve its technical implementation.30
Discussion
Lessons Learned from i2b2
The successful development, use, and dissemination of the RPDR and i2b2 have been significant in clinical research and informatics. Its evolution has been from a local research data infrastructure to one disseminated more broadly than any other NCBC product, and an infrastructure spawning larger investments than were originally used to create it. Throughout this, there were two main lessons about the benefits of dissemination. The lessons learned from the experience and observations of critical factors should also be disseminated as they are understood.
Lesson 1: People Have Great Creativity in Doing Different Things with a Resource
While some use of i2b2 was expected to mimic the utility at Partners that led to its consideration as a project to promote outside the institution, the varied uses of the infrastructure have been way beyond that expected or imagined by the original development teams. Researchers at other institutions have successfully applied i2b2 to new and challenging domains, such as cancer research31 and meaningful use,32,33 even though it was not initially envisioned for those domains. Additions in functionality provided by collaborators have also pushed the use of i2b2 in different ways and domains. Integrated analyses such as survival plots give important additional capabilities to the platform.34 Other projects mentioned above have extended the capabilities of the system when different needs prompted extensions and new developments.34–48
Lesson 2: Broader System Use Can Make It More Robust
Users of i2b2 make the system more robust by hitting upon its limitations. When different institutions deployed i2b2, circumstances that did not match those at Partners could often cause problems, which then had to be addressed to further generalize its use. Sometimes these issues were also issues with the RPDR data structure, but had just not been recognized yet since the use case had not yet presented itself. Because the technical skill level of the teams deploying i2b2 almost necessarily needed to be fairly high, often the teams were also able to identify potential solutions to problems that were discovered. Typically complaints about functionality would come with suggestions of how it could be fixed.
Projections on Sustainability
While i2b2 as a project has been successfully developed, disseminated, and expanded, and continues to be supported both at deploying institutions and centrally from the NCBC at Partners, it is not yet self-sustaining. Partners, as the host organization, continues to be the host authority for the project and drives many of the activities that lead to its success. This includes promoting challenges, hosting conferences, expanding functionality, and prioritizing development. Most development was done by a small team of six individuals (an informatics lead, a managing developer, three programmers, and an analyst). A concern is that if there were no centralized authority for the project—if i2b2 were no longer supported by Partners or supported effectively—the project as a single open-source software development could cease to exist. Instead, several offshoots, some perhaps proprietary, would likely be all that remained. Many of the benefits of the dissemination structure, including the stability and interoperability of the hive, would be lost, as well as the community.
One option for long-term sustainability of the central authority would be to translate the function to an industry partner. The problem is that there might be misalignment with the missions of i2b2, perhaps not in the beginning, but developing as financial pressures mount. The pressure to perform financially will lead to gravitation toward the most lucrative alternative. Often, this will be away from research, as traditionally the field does not have rapid returns in investment, or the returns in investment can be difficult to measure. For example, Recombinant Data and i2b2 were successful in forming a cooperative environment, but the two entities had no official relationship, with the i2b2 leadership retaining governance authority over the project and its development roadmap. A partnership that shared governance authority might have been less successful in allowing i2b2 full flexibility of development or in its ability to embrace and support many types of related projects, which was critical to the full expansion of its functionality.
Another option currently being pursued by a committee of i2b2 stakeholders is to create a foundation that would be a central authority for the project. The committee would establish governance with a rotating group of directors to steer the foundation. Such an approach requires an initial investment, but would have the advantage that it could be influenced directly by academic users and have its path set inexorably toward academic goals. Although this is likely to be able to bridge gaps in funding, it is unlikely to sustain i2b2 over a long period. The ability to grow and change to serve new national goals and to adapt to a changing national environment will likely always need support from government agencies to prevent the software from changing from national interests to niche- or specialty domains due to demand from individual or industry funders.
Conclusion
Over the past 10 years, i2b2 has risen to be an important staple in the toolkit of health care researchers. There are now over 110 hospitals that use i2b2 for research. This open-source platform has a community of developers that are continuously enhancing the analytic capacities of the platform and inventing new functionality. The i2b2 is being rapidly adapted as a central component of the informatics infrastructure for a supermajority of CTSA institutions, such that the CTSAs and other national initiatives have become somewhat dependent on i2b2, and would suffer if it were not sustained. Because of this dependence, sustaining this resource should be a national priority.
Acknowledgments
This work was funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54LM008748.
References
- 1.Murphy SN, Gainer V, Mendis M, Churchill S, Kohane I. Strategies for maintaining patient privacy in i2b2. J Am Med Inform Assoc JAMIA. 2011 Dec;18(Suppl 1):i103–108. doi: 10.1136/amiajnl-2011-000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy SN, Morgan MM, Barnett GO, Chueh HC. Optimizing healthcare research data warehouse design through past COSTAR query analysis. Proc AMIA Annu Symp AMIA Symp. 1999:892–6. [PMC free article] [PubMed] [Google Scholar]
- 3.Safran C, Porter D, Lightfoot J, Rury CD, Underhill LH, Bleich HL, et al. ClinQuery: a system for online searching of data in a teaching hospital. Ann Intern Med. 1989 Nov 1;111(9):751–6. doi: 10.7326/0003-4819-111-9-751. [DOI] [PubMed] [Google Scholar]
- 4.Banhart F, Klaeren H. A graphical query generator for clinical research databases. Methods Inf Med. 1995 Sep;34(4):328–39. [PubMed] [Google Scholar]
- 5.Murphy SN, Gainer V, Chueh HC. A visual interface designed for novice users to find research patient cohorts in a large biomedical database. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2003:489–93. [PMC free article] [PubMed] [Google Scholar]
- 6.Nalichowski R, Keogh D, Chueh HC, Murphy SN. Calculating the benefits of a Research Patient Data Repository. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2006:1044. [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, et al. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2007:548–52. [PMC free article] [PubMed] [Google Scholar]
- 8.Miller K. NCBCs Take Stock and Look Forward: Fruitful Centers Face Sunset. Biomedical Computing Review. 2012 Fall;:19–27. [Google Scholar]
- 9.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc JAMIA. 2010 Apr;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy SN, Dubey A, Embi PJ, Harris PA, Richter BG, Turisco F, et al. Current state of information technologies for the clinical research enterprise across academic medical centers. Clin Transl Sci. 2012 Jun;5(3):281–4. doi: 10.1111/j.1752-8062.2011.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy SN, Chueh HC. A security architecture for query tools used to access large biomedical databases. Proc AMIA Annu Symp AMIA Symp. 2002:552–6. [PMC free article] [PubMed] [Google Scholar]
- 12.Miller K, On Your, Mark Get, Set Build. Infrastructuer: The NCBC Launch. Biomedical Computing Review. 2005 Jun;:16–27. [Google Scholar]
- 13.Murphy S. SIIM 2011. Washington, DC: 2011. New tools for integrating clinical images into research studies. [Google Scholar]
- 14.Weber GM, Murphy SN, McMurry AJ, Macfadden D, Nigrin DJ, Churchill S, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc JAMIA. 2009 Oct;16(5):624–30. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMurry AJ, Murphy SN, MacFadden D, Weber G, Simons WW, Orechia J, et al. SHRINE: enabling nationally scalable multi-site disease studies. PloS One. 2013;8(3):e55811. doi: 10.1371/journal.pone.0055811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandl KD, Mandel JC, Murphy SN, Bernstam EV, Ramoni RL, Kreda DA, et al. The SMART Platform: early experience enabling substitutable applications for electronic health records. J Am Med Inform Assoc JAMIA. 2012 Aug;19(4):597–603. doi: 10.1136/amiajnl-2011-000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ware H, Mullett CJ, Jagannathan V. Natural language processing framework to assess clinical conditions. J Am Med Inform Assoc JAMIA. 2009 Aug;16(4):585–9. doi: 10.1197/jamia.M3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doan S, Bastarache L, Klimkowski S, Denny JC, Xu H. Integrating existing natural language processing tools for medication extraction from discharge summaries. J Am Med Inform Assoc JAMIA. 2010 Oct;17(5):528–31. doi: 10.1136/jamia.2010.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamon T, Grabar N. Linguistic approach for identification of medication names and related information in clinical narratives. J Am Med Inform Assoc JAMIA. 2010 Oct;17(5):549–54. doi: 10.1136/jamia.2010.004036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Publications Enabled by i2b2 Challenges, 2006–2012. Partners HealthCare; Boston, Mass.: https://www.i2b2.org/about/index.html. Accessed September 8, 2014. [Google Scholar]
- 21.Deshmukh VG, Meystre SM, Mitchell JA. Evaluating the informatics for integrating biology and the bedside system for clinical research. BMC Med Res Methodol. 2009;9:70. doi: 10.1186/1471-2288-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huser V, Deshmukh VG, Wilcox A, Lowe H. Going Beyond Cohort Discovery: Current Limitations, Advanced Methodologies and Future Trends. AMIA 2012 Joint Summits on Translational Science; San Francisco, CA: 2012. [Google Scholar]
- 23.Klann JG, Porter A, Wattanasin N, Murphy SN. Importing Continuity of Care Documents into i2b2 and SMART. AMIA Summits Transl Sci Proc AMIA Summit Transl Sci. 2014 [Google Scholar]
- 24.Dong X, Chukhman M, Sadhu E, Johnson R, Sharma H, Hynes D. Leveraging Big Data Technology within i2b2 Platform. AMIA Summits Transl Sci Proc AMIA Summit Transl Sci. 2014 [Google Scholar]
- 25.Wattanasin N, Mendis M, Porter A, Ubaha S, Bickel J, Mandl KD, et al. Components and Workflow for Patient Identification using i2b2 for Clinical Trials (i2b2-CT) AMIA Summits Transl Sci Proc AMIA Summit Transl Sci. 2014 [Google Scholar]
- 26.Bradford R, Farrag A, Mostafa J. Implementing i2b2 as a Research Portal to the Carolina Data Warehouse through OpenFurther. AMIA Summits Transl Sci Proc AMIA Summit Transl Sci. 2014 [Google Scholar]
- 27.Collaboration: Informatics for Integrating Biology and the Bedside (i2b2) [Internet] Available from: http://www.bioontology.org/Biology%20and%20the%20Bedside%20%28i2b2%29.
- 28.i2b2 Wiki [Internet] Available from: https://community.i2b2.org/.
- 29.Abend A, Housman D, Johnson B. Integrating Clinical Data into the i2b2 Repository. Summit Transl Bioinforma. 2009;2009:1–5. [PMC free article] [PubMed] [Google Scholar]
- 30.i2b2 Collaborations: Opportunities for Industry [Internet] Available from: https://www.i2b2.org/work/industry.html.
- 31.London JW, Balestrucci L, Chatterjee D, Zhan T. Design-phase prediction of potential cancer clinical trial accrual success using a research data mart. J Am Med Inform Assoc JAMIA. 2013 Dec;20(e2):e260–266. doi: 10.1136/amiajnl-2013-001846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klann JG, McCoy AB, Wright A, Wattanasin N, Sittig DF, Murphy SN. Health care transformation through collaboration on open-source informatics projects: integrating a medical applications platform, research data repository, and patient summarization. Interact J Med Res. 2013;2(1):e11. doi: 10.2196/ijmr.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klann JG, Murphy SN. Computing health quality measures using Informatics for Integrating Biology and the Bedside. J Med Internet Res. 2013;15(4):e75. doi: 10.2196/jmir.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segagni D, Ferrazzi F, Larizza C, Tibollo V, Napolitano C, Priori SG, et al. R engine cell: integrating R into the i2b2 software infrastructure. J Am Med Inform Assoc JAMIA. 2011 May 1;18(3):314–7. doi: 10.1136/jamia.2010.007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson N, Abend A, Mandel A, Geraghty E, Gabriel D, Wynden R, et al. Implementation of a deidentified federated data network for population-based cohort discovery. J Am Med Inform Assoc JAMIA. 2012 Jun;19(e1):e60–67. doi: 10.1136/amiajnl-2011-000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherry C, Zhu X, Martin J, de Bruijn B. A la Recherche du Temps Perdu: extracting temporal relations from medical text in the 2012 i2b2 NLP challenge. J Am Med Inform Assoc JAMIA. 2013 Oct;20(5):843–8. doi: 10.1136/amiajnl-2013-001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doan S, Collier N, Xu H, Pham HD, Tu MP. Recognition of medication information from discharge summaries using ensembles of classifiers. BMC Med Inform Decis Mak. 2012;12:36. doi: 10.1186/1472-6947-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jindal P, Roth D. Using domain knowledge and domain-inspired discourse model for coreference resolution for clinical narratives. J Am Med Inform Assoc JAMIA. 2013 Apr;20(2):356–62. doi: 10.1136/amiajnl-2011-000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang N, Afzal Z, Singh B, van Mulligen EM, Kors JA. Using an ensemble system to improve concept extraction from clinical records. J Biomed Inform. 2012 Jun;45(3):423–8. doi: 10.1016/j.jbi.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Livne OE, Schultz ND, Narus SP. Federated querying architecture with clinical & translational health IT application. J Med Syst. 2011 Oct;35(5):1211–24. doi: 10.1007/s10916-011-9720-3. [DOI] [PubMed] [Google Scholar]
- 41.Natter MD, Quan J, Ortiz DM, Bousvaros A, Ilowite NT, Inman CJ, et al. An i2b2-based, generalizable, open source, self-scaling chronic disease registry. J Am Med Inform Assoc JAMIA. 2013 Jan 1;20(1):172–9. doi: 10.1136/amiajnl-2012-001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segagni D, Tibollo V, Dagliati A, Napolitano C, G Priori S, Bellazzi R. CARDIO-i2b2: integrating arrhythmogenic disease data in i2b2. Stud Health Technol Inform. 2012;180:1126–8. [PubMed] [Google Scholar]
- 43.Segagni D, Tibollo V, Dagliati A, Zambelli A, Priori SG, Bella-zzi R. An ICT infrastructure to integrate clinical and molecular data in oncology research. BMC Bioinformatics. 2012;13(Suppl 4):S5. doi: 10.1186/1471-2105-13-S4-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang B, Cao H, Wu Y, Jiang M, Xu H. Recognizing clinical entities in hospital discharge summaries using Structural Support Vector Machines with word representation features. BMC Med Inform Decis Mak. 2013;13(Suppl 1):S1. doi: 10.1186/1472-6947-13-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. J Am Med Inform Assoc JAMIA. 2011 Oct;18(5):580–7. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H, AbdelRahman S, Lu Y, Denny JC, Doan S. Applying semantic-based probabilistic context-free grammar to medical language processing--a preliminary study on parsing medication sentences. J Biomed Inform. 2011 Dec;44(6):1068–75. doi: 10.1016/j.jbi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, Wang Y, Sun J-T, Zhang J, Tsujii J, Chang E. Building large collections of Chinese and English medical terms from semi-structured and encyclopedia websites. PloS One. 2013;8(7):e67526. doi: 10.1371/journal.pone.0067526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang G-Q, Siegler T, Saxman P, Sandberg N, Mueller R, Johnson N, et al. VISAGE: A Query Interface for Clinical Research. AMIA Summits Transl Sci Proc AMIA Summit Transl Sci. 2010;2010:76–80. [PMC free article] [PubMed] [Google Scholar]