Abstract
Purpose:
Multi-institutional collaborations are necessary in order to create large and robust data sets that are needed to answer important comparative effectiveness research (CER) questions. Before scientific work can begin, a complex maze of administrative and regulatory requirements must be efficiently navigated to avoid project delays.
Innovation:
Staff from research, regulatory, and administrative teams involved in three HMO Research Network (HMORN) multi-institutional collaborations developed and employed novel approaches: to secure and maintain Institutional Review Board (IRB) approvals; to enable data sharing, and to expedite subawards for two data-only minimal risk studies. These novel approaches accelerated required processes and approvals while maintaining regulatory, human subjects, and institutional protections.
Credibility:
Outcomes from the processes described here are compared with processes outlined in the research and regulatory literature and with processes that have been used in previous multisite research collaborations.
Conclusion and Discussion:
Research, regulatory, and administrative staff are essential contributors to the success of multi-institutional collaborations. Their flexibility, creativity, and effective communication skills can lead to the development of efficient approaches to achieving the necessary oversight for these complex projects. Elements of these specific strategies can be adapted and used by other research networks. Other efforts in these areas should be evaluated and shared. The processes that help develop a “learning research system” play an important and complementary role in sustaining multi-institutional research collaborations.
Keywords: Data agreement, Institutional Review Board, Subcontract, Subaward, Administrative Efficiency
Introduction
Multi-institutional research collaborations offer opportunities to engage diverse scientific expertise to answer important public health questions. The current focus on large-scale comparative effectiveness research (CER) highlights the need for this collaboration. The strength of multi-institutional research efforts is the ability to assemble, from multiple sources, a large, robust electronic data set that is critical to achieve the goals of the research.1 Before scientific work can begin, however, a complex maze of administrative and regulatory requirements must be navigated. Among these requirements are securing Institutional Review Board (IRB) approvals and executing data sharing agreements and subcontracts or subawards.
Improvements in administrative and regulatory procedures have not kept pace with nuances that CER has introduced through the use of large, electronic data sources.2–5 Variance in interpretation and application of human subjects protections and Health Insurance Portability and Accountability Act (HIPAA) regulations can result in site-specific requirements that may compromise consistency and foster delays.6 Anecdotally, the processes for securing subcontracts and data sharing agreements are generally found to be inefficient and inconsistent across sites. The successes achieved in administrative and regulatory areas are seldom reported in the literature, making it difficult to share, adapt, and replicate effective strategies. Further, these steps are often not acknowledged or considered when developing research plans and timelines.
This paper describes how three multi-institutional collaborations within the HMO Research Network (HMORN) managed IRB approvals and data sharing for two data-only, minimal risk studies and subcontracting for a large cancer-research network. The approaches emphasize the importance of developing well-organized, measurable, and replicable processes in order to identify areas where improvements can be made, and they highlight the essential role of research and administrative staff in developing those processes.
Background
As funders’ interest in CER increases, multi-institutional research networks are necessary to assemble the “big data” required for this research. This interest, coupled with the compression of the time-lines for research awards by shortening the traditional five years of an investigator-initiated (R01 NIH Research Project Grant) award, makes administrative and regulatory efficiencies more critical than ever. To expedite the start of a research study, and to maintain progress throughout the life of the study, it is important to document best practices—a crucial step in the quality-improvement process.
Members of research teams know that completing required steps for project initiation can be time-consuming and burdensome,7 often leading to project delays and higher costs.5,8–11 Several examples illustrate the lengthy process of securing IRB approval for a multi-institutional research study:12
a yearlong delay to respond to protocol modifications even though an IRB-approved, standardized protocol was used;9
variability in the type of review, type of consent form, time for IRB approval, changes requested, and the quality of human subjects protection afforded among local IRBs for the same study;5 and
17 percent of total research budget consumed by IRB activities even though these actions had “no discernible impact on human subjects’ protection.”11
Delays have also been attributed to the time it takes research staff to respond to requests for clarification and additional information. HIPAA dictates how institutions can share data for research purposes, but state and local regulations add another layer of consideration that can further complicate multi-institutional research studies, particularly when there are multiple data providers, data recipients, and data sets.
The HMORN comprises 18 research institutes that are embedded in health care delivery organizations.13–15 Since 1994, the HMORN has purposefully fostered multi-institutional research collaborations and trusting relationships by continuously developing and refining tools and processes that are mutually agreed upon by member sites. These mutually developed and accepted processes enable studies to meet regulatory requirements that maintain institutional- and human subjects protections while streamlining the research process.
This history of working together within a network increases funders’ expectations for efficiencies in regulatory as well as scientific domains. In this paper, we demonstrate how processes were developed and applied in the areas of human subjects protections, data sharing, and enhanced business practices (contracting). The strategies described here can be adapted to serve other multi-institutional research endeavors.
Approach and Application of Strategies
Several mechanisms were used to accelerate the completion of grant administration and regulatory requirements for 3 large multi-institutional networks: (1) Scalable Partnering Network (SPAN) for CER: Across Lifespan, Conditions, and Settings;16 (2) Surveillance, Prevention, and Management of Diabetes Mellitus (SUPREME-DM);17 and (3) the HMO Cancer Research Network (CRN).13 These networks include 7–16 institutions that are primarily members of the HMORN. The tools and processes that significantly increased administrative and regulatory efficiencies are described.
Institutional Review Board (IRB): SPAN and SUPREME-DM as Case Studies
Kaiser Permanente Colorado (KPCO) was the lead site for SPAN and SUPREME-DM, two 11-site, 36-month studies that were funded by the Agency for Healthcare Research and Quality (AHRQ) under the American Recovery and Reinvestment Act of 2009 (ARRA). The objectives of SPAN were to develop a distributed research network that was interoperable across health care systems and to test the network-based research infrastructure by conducting CER studies of obesity and attention deficit hyperactivity disorder. The goal of SUPREME-DM was to develop two longitudinal clinical registries of patients from integrated health care delivery systems with and without diabetes mellitus.
These projects required large data resources amassed from multiple institutions. The project teams knew from the outset that these awards would be ineligible for no-cost extensions, so both studies had to complete work in 36 months. In anticipation of the issues inherent in the research approval process for multi-institutional studies and to achieve project goals within these time constraints, teams at KPCO utilized the HMORN IRB Review of Multi-Site Research process and coupled it with a new innovation to streamline IRB approvals.
HMORN IRB Review of Multi-Site Research Process
In 2008, leadership from the Human Subjects Protections IRB departments of the HMORN responded to the need for more distributed and diversified methods of conducting research across multiple institutions by introducing a research approval process for use among its member organizations for minimal risk, data-only studies.18 This process allows the lead principal investigator (PI) to submit to their local IRB (“lead IRB”) using their local site’s IRB forms and processes. Participating sites’ IRBs then use the application from the lead site to facilitate research review at their institution. Each institution decides if they would like to review and oversee the protocol for their site or cede oversight authority to the lead IRB.18 Only one set of application materials is required for this facilitated review process plus an additional short form, the HMORN Multi-Site Research Application Cover Sheet (Appendix 1) that provides investigator and IRB contact information for each site.
Both SPAN and SUPREME-DM used the HMORN interinstitutional IRB research review process, and all sites were asked to cede IRB oversight to the lead IRB (the KPCO IRB)—a “lead and cede” approach. For SPAN, both HMORN (9) and non-HMORN sites (2) ceded IRB review and oversight to the lead IRB. For the data-only portion of the SUPREME-DM study (11 HMORN sites), all but 2 sites ceded. The interinstitutional process did not supersede the right of local IRBs to make independent determinations. No information was shared or gathered about why these 2 sites elected to retain local oversight. Despite using the HMORN streamlined approach, it took 9 months to obtain initial IRB approval and ceding oversight for the SPAN study and approximately 5.5 months for the SUPREME-DM. While lengthy, this represented an improvement over the average time from submission to project initiation of 12 months that was shown in a review of 121 VA research studies.19,20 Two other studies cite average time to approval at 286 days (median; range, 52–798 days)6 and 81.9 days (mean; range, 13–252 days).5
Data Repository Model
Since these studies involved both infrastructure development as well as the conduct of CER studies, the initial focus was on building a data foundation. During this phase, investigators and analysts progressively identified hypotheses that could be addressed with the data resource. Specific research questions were not fully developed at the time of initial IRB submission. The research team recognized that study activities presented minimal risk by using retrospective data that contained randomly generated study identifiers that prevented reidentification. Therefore, submitting each hypothesis or substudy as a separate IRB protocol would require substantial investigator, project manager, and IRB time and resources across all 11 sites for each study. Furthermore, submitting each as separate IRB protocols would not enhance the patient protections that were already in place (randomly generated study IDs; key kept at local sites and never shared). The research and Human Subjects Protections teams collaborated to develop a comprehensive review process for the collection, storage, and future research use of data stored in these data repositories.
The KPCO Human Subjects Protections team proposed that research accessing data repositories should be reviewed in a similar fashion to how they would evaluate research that involved biorepositories —with one important distinction. Biorepositories store biological samples for prospective research with patients’ consent.21 In this case, the important distinction was that prospective research conducted with data repositories presented minimal risk to study subjects because the data were retrospective and patients’ identities were protected through the assignment of a randomly generated study identifier. This linking file was never shared, thus preventing reidentification of individual patients.
At the time these data repositories were being developed, researchers had a general idea of the types of questions these data repositories would study as well as the methods that would be used. The SPAN repository would be used to study obesity and attention deficit hyperactivity disorder CER; while the SUPREME-DM repository would be used for diabetes surveillance and CER. With this general base of information, the investigators, project managers, and Human Subjects Protections teams collaborated to develop specific application procedures for future research utilizing these repositories. These procedures enabled the study teams to highlight similarities and differences in the proposed “substudies” and efficiently and effectively emphasized areas that could increase the risk of the research. Table 1 lists the substudy elements that were the same as in the main study and the elements that were reviewed for each substudy. As shown, the data source and the risks and benefits to participants for the overall project did not change even though several elements of the substudies did. Concisely emphasizing the differences for each substudy greatly streamlined the IRB review.
Table 1.
IRB Review of Substudy Elements
| Substudy elements same as main study | Substudy elements reviewed for each substudy |
|---|---|
|
|
The cornerstone of these application procedures for future research using data from the repositories was an abbreviated protocol template (Appendix 2). To assist in the administrative oversight of these studies, naming and numbering conventions were developed by the teams (Appendix 3) to track modification decisions, substudy lead PI, participating sites, duration of sub-study, and subcontract changes for both the main study (research repository) and substudies. As part of the review process for these new procedures, site IRBs reaffirmed their ceding arrangement with the lead IRB.
Twelve (six SPAN plus six SUPREME-DM) modifications to the data repositories and 17 new substudies were submitted as modifications to the original approved data-repository protocols over a period of 15 months. The substudies qualified for and underwent expedited review by the lead IRB and all were approved without contingencies. The mean time to obtain approval was 8.8 business days from date of IRB submission to the date the IRB approval letter was received (range, 4–17 days). By way of comparison, a multicenter genetic epidemiology study documented the mean time to obtain approval for an expedited review at 32.3 days (range, 9–72 days).5
Time to complete the substudy modification template was not tracked but, from conversations conducted with substudy lead PIs, it is estimated that this form took from 1 to 2 hours to complete. When compared to a 2002 study that found the range of preparation time for a full research application varied from 2 hours to as many as 40 hours, our estimated timeframe represents a substantial improvement.5 In addition to the obvious benefit of reducing the amount of staff time needed to secure IRB approval, the potential risks to the human subjects were evaluated for each substudy before approval was granted. The study teams developed processes to effectively maintain participant confidentiality throughout the research process. Furthermore, the HMORN-facilitated IRB review process precluded the need for establishing new and redundant ceding arrangements, and processing each substudy as a new application.
Data Sharing: SUPREME-DM as a Case Study
In the SUPREME-DM and SPAN studies, investigators determined that dates of clinical service were needed for scientific analyses. The combination of dates and information from medical records constituted protected health information (PHI) and, thus, defined the data set as limited. According to HIPAA, a data use agreement (DUA) was required: “A data use agreement entered into by both the covered entity and the researcher, pursuant to which the covered entity may disclose a limited data set to the researcher for research, public health, or health care operations” [45 CFR 164.514(e)], Department of Health and Human Services (emphasis added). This case study focuses on data sharing in the SUPREME-DM project.
The HMORN created a DUA template in 2009 to be used in tandem with the HMORN Subaward Agreement Template discussed in the next section. The HMORN DUA template is a short document since most of the legal language has been incorporated into the Subaward Template. Elements of the agreement that are study specific are usually limited to the data set description and the permitted uses and disclosures by the data recipient. Use of the HMORN Subaward and DUA Templates is encouraged for HMORN studies since prenegotiated language often expedites the execution of these agreements.
Historically, DUAs are executed between two sites: a data provider and data recipient. Since CER requires data sharing across multiple sites, executing data agreements is repetitive, time-consuming, and fails to add value or increased institutional or patient protections to the research enterprise. If the traditional process had been followed for SUPREME-DM, each data provider would have needed to execute 10 agreements; a staggering number.
Instead of following this approach, the study staff collaborated with the human subjects protections team to propose a modification to the existing DUA template. This was a single reciprocal agreement that addressed the data elements, uses and disclosures (Appendix 4) and the data flow among the sites involved (Appendix 5). In this case, “reciprocal” meant that all sites agreed to the use and disclosure of limited data sets by all other participating sites. The lead site would be considered the “initial data provider” and, thus, would initiate and draft this modified agreement. The modified agreement allowed for consensus among the sites on the specific data elements that all sites would share and included a diagram illustrating the reciprocal nature of the agreement allowing any of the participating institutions to receive a limited data set for analysis.
It took approximately 12 weeks to execute the reciprocal DUA. This included the time it took for investigators to reach consensus that executing a DUA was necessary to meet the analytic goals of the project and the time to draft, to have each site review and respond with comments, to incorporate comments, and to route for final signatures by research compliance administrators, directors, business administrators, or others who had the authority to enter their institutions into these agreements. Once signed, an investigator from any site could lead a substudy, receive a consolidated and limited data set, and conduct analysis. In addition, the data coordinating center was not responsible for the many concurrent requests for consolidated, limited data sets and analyses that would ensue; this burden could now be shared among the participating sites.
Perhaps even more important than increasing the pace of research and gaining flexibility regarding the division of labor among the sites, the reciprocal DUA allowed analyses to be led by investigators and analytic staff with the greatest content expertise while supporting sites with a data sharing process that met regulatory requirements. In a network comprising over 30 investigators this novel data sharing structure allowed meaningful scientific engagement; maintained autonomy for each institution; and strengthened the SUPREME-DM Network. In addition, the funder benefited from tapping into a wide array of scientific and analytic expertise.
HMORN Subaward Agreement Template: HMO Cancer Research Network (CRN) as a Case Study
The CRN began in 1999 as a National Cancer Institute (NCI) cooperative agreement awarded to Group Health Research Institute (GHRI), the research division of Group Health Cooperative. The overall goal of the CRN has been to conduct collaborative research to determine the effectiveness of preventive, curative, and supportive interventions for major cancers among diverse populations and health systems. When the CRN began, subawards were generally described as a bilateral relationship between a prime and a subrecipient institution. The terms and conditions in the agreements were based on each institution’s contracting preferences, and there was no universally accepted language for how individual subrecipients could collaborate with one another to share research resources and intellectual property.
During the first eight years of the CRN, through two funding cycles, separate negotiations were required with each subrecipient, sometimes on an annual basis (Figure 1). These negotiations had to be conducted during the first six months of the award cycle, generating a significant amount of work for administrative and contracting staff and delaying execution of the subawards. This was burdensome for all sites and did not facilitate timely business and scientific processes across the consortium. Additionally, the contract language negotiated each year did not foster cooperation between subrecipients. As the CRN entered the third cycle of its funding in 2007 (CRN3), an internal HMORN workgroup composed of grant and contract managers, IRB experts, and lawyers developed a standard agreement with the goal of streamlining subcontracting across the HMORN (www.hmoresearchnetwork.org). The CRN grant management team made the strategic decision to field-test the HMORN Subaward Agreement Template from 2007 to 2013 with 13 subrecipients.
Figure 1.
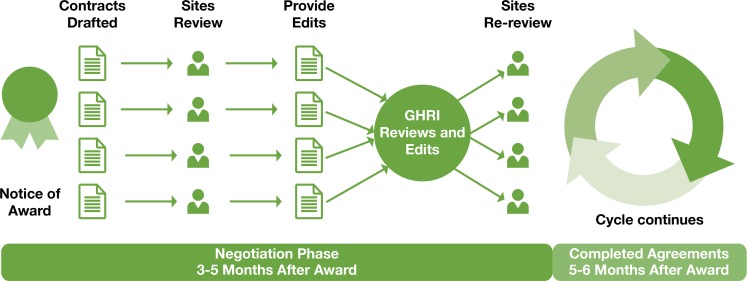
Multiple Contracts Process
The team implemented a new, inclusive process for review and negotiation of a standard template for use across the consortium. Figure 2 shows the improved process flow that starts about 90 days prior to the award of the grant. Providing the proposed contract template for early review by subrecipient sites is a simple step but clearly improves the timeline considerably. Other key elements of the process included: (1) a reasonable timeline that allowed the sites adequate time to review the subaward draft and respond; (2) a request to provide candid feedback on issues felt to be inequitable or unclear; (3) a deadline for input; and 4) an emphasis on the mutual goal of collaboration and timely execution.
Figure 2.
Single Template Process
Each year the GHRI CRN grant management team identified issues and developed approaches to address those issues during the next annual award cycle. The CRN main subaward agreement was executed each year and modified to add newly awarded funds for pilot projects, supplements, and approved carryover. The volume of subawards and modifications for the CRN3 varied from 25 to 69 HMORN and non-HMORN subawards per year. Timely execution of the main agreement was important to establish the terms and conditions of the award, but was also important for the cascading award of subsequent supplements and pilots.
Electronic storage of drafted and completed agreements, communication tools, and official award documents supported the ability to collect data, review metrics, and evaluate performance during this period. Data collection focused on assessing the time it took to initiate and fully execute the main subaward since these agreements contained the material terms and conditions governing the conduct of the project. Initiation time was measured as the interval between award date on the Notice of Award (NOA) or other appropriate award document, and the signature date of GHRI that represented the date the agreement was initiated and sent to the subrecipient for countersignature. Turnaround time at the site was measured by the interval between the signature date of GHRI and signature date of the site. Overall execution time was measured by the interval between award date and the full execution of the subaward represented by the signature date of each subrecipient.
The long interval required to initiate the main subawards in the first year of the pilot (Figure 3) illustrates the difficulty of implementing a new contracting tool and strategy across the broad CRN3 consortium. Key sticking points in the template revolved around the added concept of including all subrecipients as potential collaborators and reaching consensus on the terms governing confidentiality and the sharing and use of intellectual property. Extensive negotiations were required before consensus was reached on template language, but interaction with scientific, technical, and legal professionals shaped the template to address the complex confidentiality, data, and publication needs of the consortium. Review of site input each year revealed fewer requested edits over time, illustrating how HMORN Subaward Agreement Template language became increasingly acceptable to CRN3 HMORN sites.
Figure 3.
Mean Days to Initiate and Fully Execute Subaward
As Figure 3 also shows, the time to initiate and complete the main subawards decreased substantially in the 5 years of CRN3 even though delays occurred in years two, three and five. The 60-day turnaround goal was met once during year four. A close review of the annual process reveals that only one of these delays was related to template use—a second revision of the HMORN Agreement Template in year three. The delays in years two and five were related to budget cuts.
The prenegotiation of template language allowed the agreements to be signed quickly and sent to the sites for countersignature. The average turnaround time between the signature of GHRI and signature of the subrecipients remained stable throughout the 5-year period and took 14–21 days, illustrating that the final sign-off on the agreements did not require additional negotiation with individual sites.
Though use of the HMORN Subaward Agreement Template did not eliminate delays, the process engaged network members to partner in creating a template that addressed key issues and promoted collaboration across the consortium. It also created the mutual benefit of a predictable timeline for subaward execution. In year 3, approximately midway through the CRN3 funding cycle, the HMORN Subaward Agreement Template underwent a full review at all HMORN sites, which led to official acceptance through a Memorandum of Understanding (MOU) that committed members to use the template as the starting point for collaborations involving grants to HMORN institutions. Multidisciplinary collaborators within the HMORN network participated in this review process. Their feedback helped create an agreement that delineated how sites could collaborate with one another while addressing individual institutional concerns and maintaining key protections for health plan patients and their data. Involvement in the review process deepened institutional knowledge of crosscutting issues in multi-institutional collaboration and strengthened support for template use.
Discussion
HMORN members have conducted multi-institutional research for two decades and continue to strengthen their alliance. Through a shared vision and a history of collaboration, trust has been established. This has led to the development of processes and tools that enhance the HMORN’s research mission and support the evolving nature of large multi-institutional studies in which data sharing among many sites is required.
The time that HMORN institutions have spent working together to create and sustain a common culture should not be underestimated22 and makes the Network unique. The successful innovations described here, however, were not simply a result of the Network’s maturity. Rather, the driving force behind these innovations emerged from circumstances external to the Network such as the current focus on CER that requires large data sets to meet research goals. This necessitates interinstitutional collaboration for any network that embarks on research in this area. Additionally, funders expect that research will be conducted more quickly and with fewer resources. Therefore, the process improvements described here evolved because both the scientific and funding environment demanded greater efficiency within and across institutions. These circumstances make these tools and process applicable to other networks and multi-institutional collaborations.
Institutions collaborating for the first time will need to build on the aims and objectives of their research project to establish trust through transparent and clear communication. Adapting tools and templates that have been used successfully by other networks, such as the ones from HMORN, can advance this process. All HMORN tools, templates, standard operations procedures, FAQs, and other documentation are available to the public (www.hmoresearchnetwork.org).
Even within the HMORN, it often requires time for these innovations to diffuse. In the case of SPAN, the lead and cede model for IRB approval was ultimately adopted by the two sites that were not HMORN members. Longtime members describing the process and sharing experiences from their institutions led to the successful adoption of this process by the nonmember sites. As noted in Hagen, et al.,23 developing policy guidelines for data sharing and management of intellectual property is one of the key elements of a successful network. Though these resources clearly provide advantages for the HMORN, use of the tools and processes remains voluntary.
Some institutions have policies that prevent them from ceding IRB oversight, which may lengthen the review process. The same is true if a site, such as a non-HMORN site, has institutional requirements for using its own IRB application forms. The HMORN IRB Review of Multi-Site Research process works well for data-only studies because the process was developed for those types of studies. Since the SPAN and SUPREME-DM studies underwent IRB approval, the process has been adapted for epidemiologic and health services research and network-wide consent form templates are being considered.24
Although reciprocal DUAs may allow data sharing across multiple sites in one document, they may also have limitations that restrict their use in specific circumstances. Changes to a reciprocal DUA require that all site signatories review and resign the document. When drafting the reciprocal DUA for SUPREME-DM, data contributing sites required a provision that restricted the sharing of limited data sets with sites that were data contributing partners. This meant that investigators from sites not contributing data could receive only deidentified data; thus limiting the types of analyses these investigators could perform.
Further, this DUA promoted sites’ autonomy by requiring investigators to opt in or opt out of each substudy. In SUPREME-DM, investigators excluded their sites from substudies for reasons such as the following: competing demands for their time or the time needed from programming or analytic staff, similar work being done by a fellow investigator at their institution using these data, or because they felt the research question had already been answered. Site autonomy is a foundational value of the HMORN and the experience of SUPREME-DM demonstrates how supporting sites’ autonomy plays out in a collaborative research effort.
The HMORN Subaward Agreement Template and the associated DUA are important tools, but a team of volunteers is required to periodically update, negotiate, and agree on final versions that are sent to HMORN sites for comment, review, and eventual adoption. Questions have been raised about the value and effectiveness of these tools compared to the effort it requires to update and maintain them. Though the investment is significant, the data in this CRN3 case study shows that use of the HMORN Agreement Template and a prenegotiation strategy successfully streamlined the research contracting process. More importantly, the template has evolved into a document with well-accepted, reciprocal guidelines for data sharing and management of intellectual property between sites in a multi-institutional consortium, facilitating cross-site collaboration. For example, in the fourth competitive award for the CRN, the prime award recipient was moved from Group Health to Kaiser Permanente Northern California, necessitating new institutional arrangements. The substantial administrative work on template development and harmonization of procedures among participating HMORN sites under the previous awards resulted in relatively rapid implementation of IRB approvals and completion of subawards. Additionally, the HMORN Agreement Template has been externally validated through its acceptance at institutions outside the HMORN. The ability to attract new partners facilitates opportunities for CER and other multi-institutional collaborations that can facilitate the journey of HMORN institutions to becoming learning health care systems.25,26
Conclusion and Next Steps for the Community
These case studies show how collaborations between investigators and administrative teams can lead to the development of new processes and tools to accelerate multi-institutional research, while broadening the ability of scientists at participating sites to contribute as content and methods experts. This not only reinforces the infrastructure for sustainability efforts in a multi-institutional collaboration but can also increase value to the funder by expanding the scientific capacity of scientific networks. Standardizing processes and procedures fosters relationships, trust, and the capability for continual improvement while maintaining necessary protections to meet regulatory requirements. These case studies also show how research and administrative staff, not just scientific investigators, can innovate and create value for their local institutions and their research networks.
The experience of the HMORN demonstrates how a network of researchers can partner with research administrators to create, continually evaluate, and improve its tools and systems to meet the needs of its member institutions and others. Useful tools and processes, however, cannot take the place of time working together on multiple studies to build trust and transparency in multi-institutional collaborations. Because much of the HMORN’s research depends on successful multisite collaborations, it is in the Network’s best interest to further develop tools and processes that result in administrative and regulatory efficiencies. To advance continual learning and improvement in regulatory and administrative processes, it is necessary to share experiences and information gained in adapting and using HMORN tools. Encouraging one another to publish successes and challenges will help study teams ensure administrative requirements, and approvals are secured in a timely manner.
Tracking administrative efficiency was not a research objective of these studies. As such, outcome metrics were extracted from administrative systems developed for organizational purposes. If study teams specify at the outset how they will measure the time necessary to obtain IRB approval, execute a subcontract or DUA, or submit a study modification, they will be able to proactively deploy metrics to identify barriers and opportunities for improvement.
Although much of what has been presented here focuses on multi-institutional collaborations, internal efforts of research project teams to reach out to their local institutions’ administrative and regulatory colleagues cannot be overestimated. Just as it takes time and effort to build trust with other sites, building trust at the local level empowers staff to explore innovative ways to streamline administrative processes and facilitate consistency and efficiency in their current system that can be used to inform future enhancements and create predictable timelines.
Multi-institutional research requires significant administrative and scientific infrastructure. It takes time for scientists and grant staff to build consensus and foster collaboration around administrative, regulatory, and scientific processes. Research networks should recognize the need for, and invest adequate time to develop, these systems and processes. Furthermore, developing standard metrics to quantify progress and to identify areas for improvement increases our accountability to funders and helps research networks react more quickly to changing regulations at the federal, state, and local levels.
In summary, ongoing dialogue between scientists and administrators, careful measurement of administrative processes, and dissemination of learnings from these “experiments” are strategic steps toward achieving important goals of learning health care systems.
Acknowledgments
The authors wish to acknowledge the contributions of James E. McNerney, JD RAC CIP (Kaiser Permanente); Ella E. Thompson, BS (Group Health Research Institute); Rick Perrault (Group Health Research Institute); Susan Bennett (Group Health Research Institute); Edward H. Wagner, MD, MPH (Group Health Research Institute); and Barbara McCray (Kaiser Permanente Colorado). We would also like to thank the members of the HMORN contracting template workgroup and HMORN IRB workgroup.
The Academy Health Electronic Data Methods (EDM) Forum, a project supported by the Agency for Healthcare Research and Quality (AHRQ) through the American Recovery & Reinvestment Act of 2009, Grant U13 HS19564-01. Additional support provided by Grants R01HS019912 and R01HS019859 from the Agency for Healthcare Research and Quality. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality. Initial data collection from the Cancer Research Network across Health Care Systems project (CRN3) was supported by Grant U19 CA079689 from the National Cancer Institute.
Appendix 1. Multi-Site Research Application Cover Sheet
Appendix 2. Sub-Study IRB Template

(Example of)
SUPREME-DM Sub-Study Modification Template
Modification Number: (Would correspond to number on SUPREME-DM tracking sheet)
Sub-Study Title:
Lead Investigator:
Participating Sites:
Participating co-Investigators:
Lead Analytic Site:
Brief Background and Rationale:
Study Aims or Question(s)/Research Hypotheses:
Research Design and Methods
- How does this sub-study meet 45CFR§46 part 111 criteria for IRB approval of research
- (1) Risks to subjects are minimized: The minimal risk present is the potential loss of confidentiality. This minimal risk is further mitigated by assigning randomly generated study identifiers before de-identified or limited data are shared. The linking file is kept resident at each site and is never shared. Data managers at each of the participating sites have successfully completed HIPAA and human subjects research trainings as required by their home institutions. In addition, project staff will comply with their institutions’ policies for sharing data elements, destroying identifiers, and destroying data sets. This method has been approved by this IRB in the past and has been proven effective in securely transmitting de-identified and LDS. Data will then be analyzed according to the sub-study protocol outlined in the approved modification.
- (2) Risks to subjects are reasonable in relation to anticipated benefits, if any, to subjects, and the importance of the knowledge that may reasonably be expected to result. This is a retrospective, data-only study that presents only minimal risk (loss of confidentiality). As described above, this minimal risk is further mitigated by employing proven procedures for maintaining patients’ anonymity and keeping linking files at local sites, stored behind firewalls. This minimal risk is offset by the value of study findings that will be widely distributed in order to improve patient care and outcomes.
- (3) Selection of subjects is equitable. Because the data in the SUPREME-DM DataLink are retrospective, each site will create a dataset of all health plan members that includes anyone with any health plan eligibility 1/1/1998-12/31/2013. It will contain all the necessary flags, dates, and values for identifying individuals with (possible) diabetes by any definition. In addition, we will create flags, dates and values for categories of increased risk for diabetes (impaired fasting glucose, impaired glucose tolerance, at-risk A1C). These data will be used for research and surveillance purposes only. Selection for each research analysis will be equitable, in that all exclusions will be based on scientifically justifiable criteria. Inclusion of data from certain vulnerable groups such as children, pregnant women, individuals with mental disabilities or socioeconomic disadvantages is likely to address specific questions about the effect of diabetes on those individuals. These vulnerable populations will be specifically noted in the relevant applications, and all steps outlined to preserve the confidentiality of their data will be followed.
- (4) Informed consent will be sought from each prospective subject or the subject’s legally authorized representative, in accordance with, and to the extent required by §46.116. A waiver of informed consent and a waiver of HIPAA Authorization were granted when the DataLink was first approved in February 2010 since the DataLink research: involves millions of patients (research could not be practically carried out without the waiver); research questions posed are answered using retrospective data (no patient contact); and the research activities present minimal risk. The minimal risk present is the potential loss of confidentiality and this minimal risk is mitigated by employing proven processes as explained in (1) above.
- (5) Informed consent will be appropriately documented, in accordance with, and to the extent required by §46.117. Not applicable since a waiver of informed consent and a waiver of HIPAA Authorization were granted for the SUPREME-DM DataLink.
- (6) When appropriate, the research plan makes adequate provision for monitoring the data collected to ensure the safety of subjects. Not applicable because this is a retrospective, data only study (no patient contact).
- (7) When appropriate, there are adequate provisions to protect the privacy of subjects and to maintain the confidentiality of data. The privacy of study subjects is protected by implementing proven methods to obfuscate individuals’ identifers (sharing only de-identified or limited data sets that have assigned randomly generated study IDs; keeping linking files resident at local sites behind firewalls; data managers at each of the participating sites have successfully completed HIPAA and human subjects research trainings as required by their home institutions; and project staff comply with their institutions’ policies for sharing data elements, destroying identifiers, and destroying data sets.).
- (b) When some or all of the subjects are likely to be vulnerable to coercion or undue influence, such as children, prisoners, pregnant women, mentally disabled persons, or economically or educationally disadvantaged persons, additional safeguards have been included in the study to protect the rights and welfare of these subjects. This is not applicable since this is a retrospective, data only endeavor that does not include any patient contact.
Appendix 3. IRB Tracking Sheet for Data Repository Model

SPAN Modifications and Sub-Studies Tracking Sheet
| Study or sub-study # | Lead PI | Lead Analytic Site | Participating Sites & Investigators | Study Name | Sub-Study Type | IRB Approval Date | Project Status |
|---|---|---|---|---|---|---|---|
| 001 | |||||||
| 001ContRev2 | |||||||
| 001ContRev3 | |||||||
| 001ContRev4 | |||||||
| Study Modifications | |||||||
| 001Mod1 | |||||||
| 001Mod2 | |||||||
| 001Mod3 | |||||||
| 002Mod1 | |||||||
| 002Mod2 | |||||||
| 005Mod1 | |||||||
| 005Mod2 | |||||||
| Sub-Studies | |||||||
| 002 | |||||||
| 003 | |||||||
| 004 | |||||||
| 005 | |||||||
| 006 | |||||||
| 007 | |||||||
| 008 | |||||||
Appendix 4. Permitted Uses and Permitted Disclosures from a Reciprocal Data Use Agreement (DUA)
The study, [insert study title], [insert grant number], [insert IRB number], has [insert number of participating sites] as described in Appendix A [insert and appendix that lists names of participating institutions].
Permitted Uses:
Use One (by each participating site as described in Appendix A): All participating sites will use PHI to develop data sets up to and including a Limited Data Set (LDS) as described in Appendix B. Each site will assign a unique Study ID to identifiable patient information. The linking file for re-identification of the LDS will be securely stored at each participating site and will not be disclosed with the LDS. Each participating site may send their coded, LDS to any other participating site as named in Appendix A.
Use Two (by each participating as described in Appendix A): Any site listed in Appendix A above may lead an analytic effort (Lead Analytic Site) The Lead Analytic Site will use the LDS received from the other participating sites in order to create a Consolidated Data Set (CDS) and perform analysis on the CDS (see data flow diagram in Appendix D).
[if the project has a data coordinating center] Use Three (by Data Coordinating Center [DCC]): The Lead Analytic Site, which may be any participating site as described in Appendix A, may choose to have the DCC create and distribute the SAS code, receive the sites’ LDS, create the CDS and/or conduct quality assurance work. In this case, the DCC would create and distribute the SAS code, would receive and use the LDS received from the participating sites to create a CDS, and/or would perform data quality checks as required by the Lead Analytic Site.
Permitted Disclosures:
Disclosure One: Each participating site as described in Appendix A will disclose a coded LDS. This coded LDS may be disclosed to any other participating site that acts as a Lead Analytic Site as described in Appendix A. This includes disclosing the LDS to the DCC for the purpose of creating a CDS and/or quality assurance work (see Permitted Use Three described above).
Disclosure Two: If the DCC creates a CDS and/or conducts quality assurance work, it will disclose the CDS to the Lead Analytic Site which may be any participating site as described in Appendix A.
Disclosure Three: The Lead Analytic Site which may be any participating site as described in Appendix A, may disclose a deidentified CDS to:
[If there are participating institutions which are not permitted to receive an LDS but can receive a deidentified data set; list them here.]
Appendix 5. Data Flow Diagram from a Reciprocal Data Use Agreement (DUA)
Insert name of project, IRB number, Grant number
Lead Analytic (can be any one of the [insert number of sites] institutions participating sites as described in Appendix A) sends SAS code to sites choosing to participate. Code is generated/distributed by Lead Analytic Site or by DCC.
Sites choosing to participate run the SAS code and create the LDS.
Participating sites send LDS to the Lead Analytic Site (or to the DCC).
LDS from each site are pooled into a CDS (by Lead Analytic Site or by DCC); data quality checks performed, and analyzed.
- Lead Analytic Site (can be any one of the 11 participating sites as described in Appendix A) may send a deidentified, CDS to [insert institutions allowed to receive only deidentified data sets] as described in Appendix C, Disclosure 3.
- DCC = data coordinating center
- LDS = limited data set
- CDS = consolidated data set
 = Optional (functions executed by DCC: code generated/distributed; data quality checks performed; CDS; and/or Lead Analytic site sends deidentified CDS sent to [insert institutions allowed to receive only deidentified data sets] as described in Appendix C, Disclosure 3.)
= Optional (functions executed by DCC: code generated/distributed; data quality checks performed; CDS; and/or Lead Analytic site sends deidentified CDS sent to [insert institutions allowed to receive only deidentified data sets] as described in Appendix C, Disclosure 3.)
Footnotes
Disciplines
Health and Medical Administration | Health Services Research
References
- 1.Maro JC, Platt R, Holmes JH, et al. Design of a national distributed health data network. Ann Intern Med. 2009;151:341–344. doi: 10.7326/0003-4819-151-5-200909010-00139. PM:19638403. [DOI] [PubMed] [Google Scholar]
- 2.Kahn MG, Raebel MA, Glanz JM, Riedlinger K, Steiner JF. A pragmatic framework for single-site and multisite data quality assessment in electronic health record-based clinical research. Med Care. 2012;50(Suppl):S21–S29. doi: 10.1097/MLR.0b013e318257dd67. PM:22692254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oakes M. Effect Identification in Comparative Effectiveness Research. eGEMs (Generating Evidence & Methods to improve patient outcomes) 2013;1 doi: 10.13063/2327-9214.1004. Article 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toh S, Gagne JJ, Rassen JA, Fireman BH, Kulldorff M, Brown JS. Confounding adjustment in comparative effectiveness research conducted within distributed research networks. Med Care. 2013;51:S4–10. doi: 10.1097/MLR.0b013e31829b1bb1. PM:23752258. [DOI] [PubMed] [Google Scholar]
- 5.McWilliams R, Hoover-Fong J, Hamosh A, Beck S, Beaty T, Cutting G. Problematic variation in local institutional review of a multicenter genetic epidemiology study. JAMA. 2003;290:360–366. doi: 10.1001/jama.290.3.360. PM:12865377. [DOI] [PubMed] [Google Scholar]
- 6.Greene SM, Geiger AM. A review finds that multicenter studies face substantial challenges but strategies exist to achieve Institutional Review Board approval. J Clin Epidemiol. 2006;59:784–790. doi: 10.1016/j.jclinepi.2005.11.018. PM:16828670. [DOI] [PubMed] [Google Scholar]
- 7.Emanuel EJ, Wood A, Fleischman A, et al. Oversight of human participants research: identifying problems to evaluate reform proposals. Ann Intern Med. 2004;141:282–291. doi: 10.7326/0003-4819-141-4-200408170-00008. PM:15313744. [DOI] [PubMed] [Google Scholar]
- 8.Greene SM, Geiger AM, Harris EL, et al. Impact of IRB requirements on a multicenter survey of prophylactic mastectomy outcomes. Ann Epidemiol. 2006;16:275–278. doi: 10.1016/j.annepidem.2005.02.016. PM:16005245. [DOI] [PubMed] [Google Scholar]
- 9.Schneider EC, Malin JL, Kahn KL, Emanuel EJ, Epstein AM. Developing a system to assess the quality of cancer care: ASCO’s national initiative on cancer care quality. J Clin Oncol. 2004;22:2985–2991. doi: 10.1200/JCO.2004.09.087. PM:15284249. [DOI] [PubMed] [Google Scholar]
- 10.Silverman H, Hull SC, Sugarman J. Variability among institutional review boards’ decisions within the context of a multi-center trial. Crit Care Med. 2001;29:235–241. doi: 10.1097/00003246-200102000-00002. PM:11246299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphreys K, Trafton J, Wagner TH. The cost of institutional review board procedures in multicenter observational research. Ann Intern Med. 2003;139:77. doi: 10.7326/0003-4819-139-1-200307010-00021. PM:12834327. [DOI] [PubMed] [Google Scholar]
- 12.Marsolo K. Approaches to facilitate institutional review board approval of multicenter research studies. Med Care. 2012;50(Suppl):S77–S81. doi: 10.1097/MLR.0b013e31825a76eb. PM:22692264. [DOI] [PubMed] [Google Scholar]
- 13.Wagner EH, Greene SM, Hart G, et al. Building a research consortium of large health systems: the Cancer Research Network. J Natl Cancer Inst Monogr. 2005:3–11. doi: 10.1093/jncimonographs/lgi032. PM:16287880. [DOI] [PubMed] [Google Scholar]
- 14.Durham ML. Partnerships for research among managed care organizations. Health Aff (Millwood) 1998;17:111–122. doi: 10.1377/hlthaff.17.1.111. PM:9455021. [DOI] [PubMed] [Google Scholar]
- 15.Greene SM, Hart G, Wagner EH. Measuring and improving performance in multicenter research consortia. J Natl Cancer Inst Monogr. 2005:26–32. doi: 10.1093/jncimonographs/lgi034. PM:16287882. [DOI] [PubMed] [Google Scholar]
- 16.Toh S, Platt R, Steiner JF, Brown JS. Comparative-effectiveness research in distributed health data networks. Clin Pharmacol Ther. 2011;90:883–887. doi: 10.1038/clpt.2011.236. PM:22030567. [DOI] [PubMed] [Google Scholar]
- 17.Nichols GA, Desai J, Elston LJ, et al. Construction of a multisite DataLink using electronic health records for the identification, surveillance, prevention, and management of diabetes mellitus: the SUPREME-DM project. Prev Chronic Dis. 2012;9:E110. doi: 10.5888/pcd9.110311. PM:22677160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene SM, Braff J, Nelson A, Reid RJ. The process is the product: a new model for multisite IRB review of data-only studies. IRB. 2010;32:1–6. PM:20590050. [PubMed] [Google Scholar]
- 19.Green LA, Lowery JC, Kowalski CP, Wyszewianski L. Impact of institutional review board practice variation on observational health services research. Health Serv Res. 2006;41:214–230. doi: 10.1111/j.1475-6773.2005.00458.x. PM:16430608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kupersmith J, Atkins D. Seven ways for helath services research to lead health system change. 2013. [Cited 2013 April 3]. Available from: http://healthaffairs.org/blog/2013/05/30/seven-ways-for-health-services-research-to-lead-health-system-change/.
- 21.Ginsburg GS, Burke TW, Febbo P. Centralized biorepositories for genetic and genomic research. JAMA. 2008;299:1359–1361. doi: 10.1001/jama.299.11.1359. PM:18349099. [DOI] [PubMed] [Google Scholar]
- 22.Steiner JF, Paolino AR, Thompson EE, Larson E. Sustaining Research Networks: the Twenty-Year Experience of the HMO Research Network. eGEMS (Generating Evidence & Methods to IMprove Patient Outcomes) 2014;2 Article 1 http://repository.academyhealth.org/egems/vol2/iss2/1. [PMC free article] [PubMed] [Google Scholar]
- 23.Hagen NA, Stiles CR, Biondo PD, et al. Establishing a multi-centre clinical research network: lessons learned. Curr Oncol. 2011;18:e243–e249. doi: 10.3747/co.v18i5.814. PM:21980256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene SM, Braff J, Nelson AF, Reid RJ. HMO group uses alternate IRB model to review low-risk studies. IRB Advisor. 2011 Mar;11:30–31. [Google Scholar]
- 25.Greene SM, Reid RJ, Larson EB. Implementing the learning health system: from concept to action. Ann Intern Med. 2012;157:207–210. doi: 10.7326/0003-4819-157-3-201208070-00012. PM:22868839. [DOI] [PubMed] [Google Scholar]
- 26.Olsen LA, Aisner D, McGinnis JM. The Learning Healthcare System: Workshop Summary. Washinton, D.C: National Academies Press (US); 2007. [PubMed] [Google Scholar]