Abstract
Advances in electron cryo-tomography have provided a new opportunity to visualize the internal 3D structures of a bacterium. An electron microscope equipped with Zernike phase contrast optics produces images with dramatically increased contrast compared to images obtained by conventional electron microscopy. Here we describe a protocol to apply Zernike phase plate technology for acquiring electron tomographic tilt series of cyanophage-infected cyanobacterial cells embedded in ice, without staining or chemical fixation. We detail the procedures for aligning and assessing phase plates for data collection, and methods to obtain 3D structures of cyanophage assembly intermediates in the host, by subtomogram alignment, classification and averaging. Acquiring three to four tomographic tilt series takes approximately 12 h on a JEM2200FS electron microscope. We expect this time requirement to decrease substantially as the technique matures. Time required for annotation and subtomogram averaging varies widely depending on the project goals and data volume.
Keywords: Zernike phase optics, electron cryo-tomography, phase plate evaluation, phase plate alignment, tilt series collection, post-tomographic classification and averaging, 3D structures
Introduction
Electron cryo-tomography (cryoET) has emerged as a promising structural tool to study ultrastructure in individual cells preserved in their native and hydrated conditions without chemical fixing, staining or plastic embedding1,2. A formidable challenge in identifying macromolecular complexes in the tomograms is low contrast, which is partly due to the thickness of the cell (usually in the range of 300 to 1000 nm) and its congested environment. Frozen, hydrated biological samples used for cryoET studies are generally approximated as weak phase objects. Although this approximation is not strictly correct for thicker whole-cells, it is sufficiently accurate that standard transmission electron microscopy (TEM) contrast transfer theory can be applied3. The Contrast Transfer Function (CTF) impacts the image in frequency (Fourier) space and is oscillatory, with very little contrast preserved at low spatial frequencies. This means that the structural information generally useful for identifying the characteristic low-resolution features of the targeted objects may be compromised. Strongly defocusing the microscope (5–15 µm underfocus) can partially compensate for this low frequency damping, by shifting the first oscillatory maximum to lower frequency. However, applying this approach dramatically reduces the amount of higher frequency structural information. In order to restore high-resolution structures, the CTF oscillations have to be computationally corrected for each image, using multiple images with different defocuses (i.e. different CTF oscillation patterns) to fill in the zeros of Fourier components from images having different CTFs.
The electron microscope used in the present protocol is equipped with electron optics designed to remedy this problem: the Zernike phase plate4–8, a thin carbon film with a small circular hole in the middle. When the electron beam passes through the phase plate, the scattered electrons undergo an approximately π/2 phase shift with respect to the unscattered central electron beam, so that the CTF switches from a sine to a cosine function. That is, contrast remains strong at low resolutions (frequencies) close to the origin of the CTF, regardless of defocus. This effect, in principle, enables researchers to achieve nearly in-focus imaging without compromising low resolution contrast and eliminates the need to make corrections for the CTF4,5,8–11. As a test of the Zernike phase plate technology, the electron cryo-microscopy (cryoEM) structures of a 2D crystalline array of bacteriorhodopsin and of an icosahedral virus particle were obtained at ~10–12 Å without CTF correction12. However, since it is practically difficult to set the defocus exactly in-focus, CTF determination and correction would be necessary for structures extending to subnanometer resolution. Images recorded using a Zernike phase plate yielded a 3 to 5-fold low resolution contrast enhancement compared to conventional imaging. Having observed the dramatic improvement for molecular imaging, it is logical to consider using Zernike phase plate for cellular cryoET, with the prospect of achieving dramatically higher contrast of cellular components, without increasing the electron dose to which the specimen is subjected. As a proof of principle, we selected the structural investigation of newly synthesized bacteriophages within the host bacteria as our initial target16. These phages are normally obscured by the thousands of protein complexes in the crowded cytosol. Using Zernika phase contrast (ZPC) cryoET, we demonstrated the ability to identify these bacteriophage particles and classify them at different stages of assembly, from procapsid without DNA to mature virion with DNA and other infection protein complex.
Prior to using the Zernike phase plate, we performed a control experiment, collecting many tomograms of the same specimen at very high defocus values using conventional cryoET, without a phase plate (WD). Though it was possible to localize some phage particles, subsequent attempts to align and average extracted particles failed to produce consistent results. Using the phase plate dramatically improved the visibility of the individual phage particles and enabled reproducible alignment and classification in post-tomographic data processing. Our structural study of newly synthesized bacteriophages within the host bacteria16 demonstrated for the first time that the Zernike phase plate offers a dramatic improvement for cryoET of cells. These results open up many new opportunities for studying subcellular components performing physiological processes in situ. As shown in our study, we were able to identify novel phage assembly intermediates not previously observed, almost certainly due to their transient occurrence in the cell.
Although the physical principle on which it is based is simple, the implementation of ZPC optics in an electron microscope was unsuccessful until recently, primarily because of technical difficulties in manufacturing robust phase plates with good electron dose tolerance. Several electron microscope companies have tried to implement this methodology in their instruments13–15. The implementation of the Zernike phase plate with JEOL microscopes is a recent development as reported here, in that researchers can insert the phase plate with an airlock system similar to a specimen exchange device without breaking the vacuum of the electron microscope. Currently, the quality of the phase plates is not highly reproducible, so rapid phase plate exchange is an important convenience at this stage of technology development. Importantly, our study on intracellular bacteriophage progeny16 demonstrated the feasibility of ZPC cryoET to reveal new biological information, albeit with persistent effort. With further development of phase plate technology by optimizing the choice of materials and fabrication to minimize charging and to increase durability under the beam, we expect that phase plate imaging will be routinely used for cryoET in structural cell biology.
Here, we describe a protocol for ZPC cryoET configuration, imaging conditions, phase plate assessment and alignment, and data processing. This Protocol was used to study the maturation process of cyanophage Syn5 in its host cells, Synechococcus strain WH810916. The principles of illumination configuration we established can be adapted to other microscope models, and the procedures to evaluate and align phase plates for data collection are applicable to a variety of biological samples from macromolecular complexes to prokaryotic and eukaryotic cells. This Protocol is designed to be modular, so that components can be adapted to projects of different biological samples with different goals. We also expect that these procedures will become more streamlined and simplified as the technology improves. We focus on phase plate alignment and assessment components, which often are the keys to yield data that answer biological questions, as exemplified here.
Protocol overview
This Protocol has five components. The first, which is specific to the biological specimen used as an example, describes the steps and conditions for cyanobacteria cell culture, phage infection and cryo-specimen preparation. Components two to four describe the optical illumination conditions and procedures needed for ZPC cryoET data collection (Figs. 1–3). Specifically, in the second component, details of the procedures for phase plate mapping and quality evaluation are provided (Figs. 3–5, Supplementary Figs. 1, 2). In component three, we describe the microscope alignment, including low dose configuration, which is essential for electron tomographic data collection (Fig. 3). Component four describes the steps required for ZPC cryoET tilt series data collection (Figs. 3, 6a, Supplementary Fig. 3), including sample tracking, focusing with the phase plate inserted, and evaluation of phase plate deterioration with electron exposure during tilt series collection. The last component provides directions on image processing and subtomogram alignment (Fig. 7, Supplementary Fig. 4). We provide details on the steps and parameters that are specific to phase plate data.
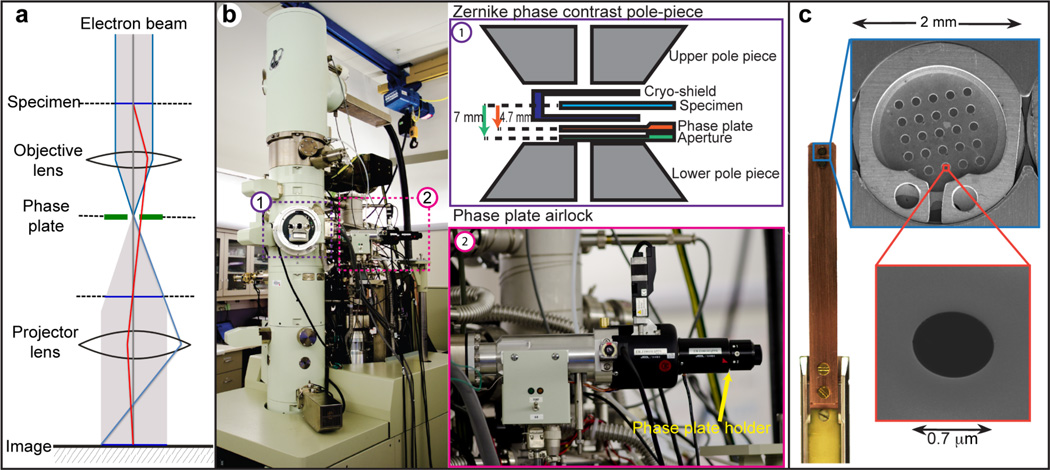
Figure 1. ZPC electron microscope setup and photographs of the phase plate airlock, blade, disc and phase plate hole.
(a) Diagram of the ZPC microscope illumination system. The phase plate is inserted at the back focal plane of the objective lens. (b) (1) Diagram of the ZPC pole piece showing the positions of specimen, the phase plate and the objective aperture. (2) Photograph of the phase plate airlock system. (c) The phase plate blade can hold two 2-mm phase plate discs, each consisting of 5 × 5 phase plates. Scanning electron microscopy image of a phase plate shows the 0.7-µm hole in the middle.
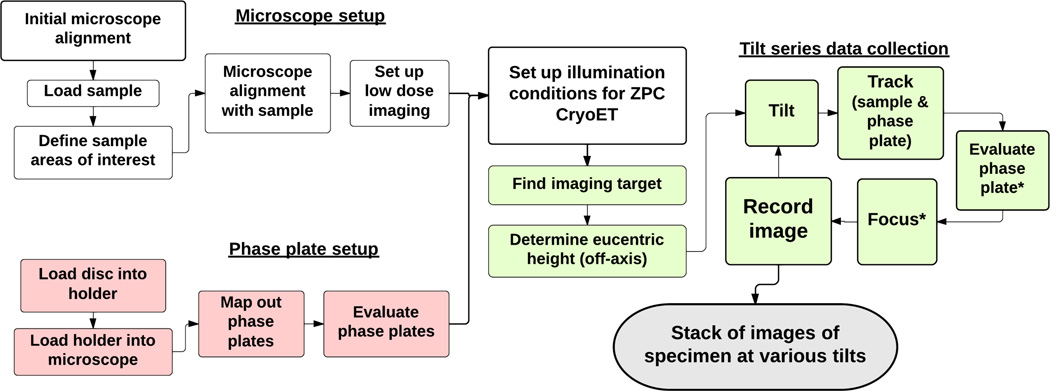
Figure 3. Phase plate setup and microscope alignment for ZPC cryoET imaging.
The flowchart describes the procedural steps involved in components two to four of the protocol. The phase plates are mapped and evaluated by acquiring test images of amorphous carbon film before imaging sessions (component two: steps colored in pink). Component three (white colored steps) covers the basic microscope alignment, sample mapping, and low-dose imaging condition setup. During ZPC cryoET data collection (component four: steps colored in green), stage tilting, sample tracking and phase plate re-centering are done for every tilting step. Focusing and evaluation of the phase plate are performed every three to five tilting intervals, depending on the tilting angle and the quality of images taken (denoted by the star symbols).
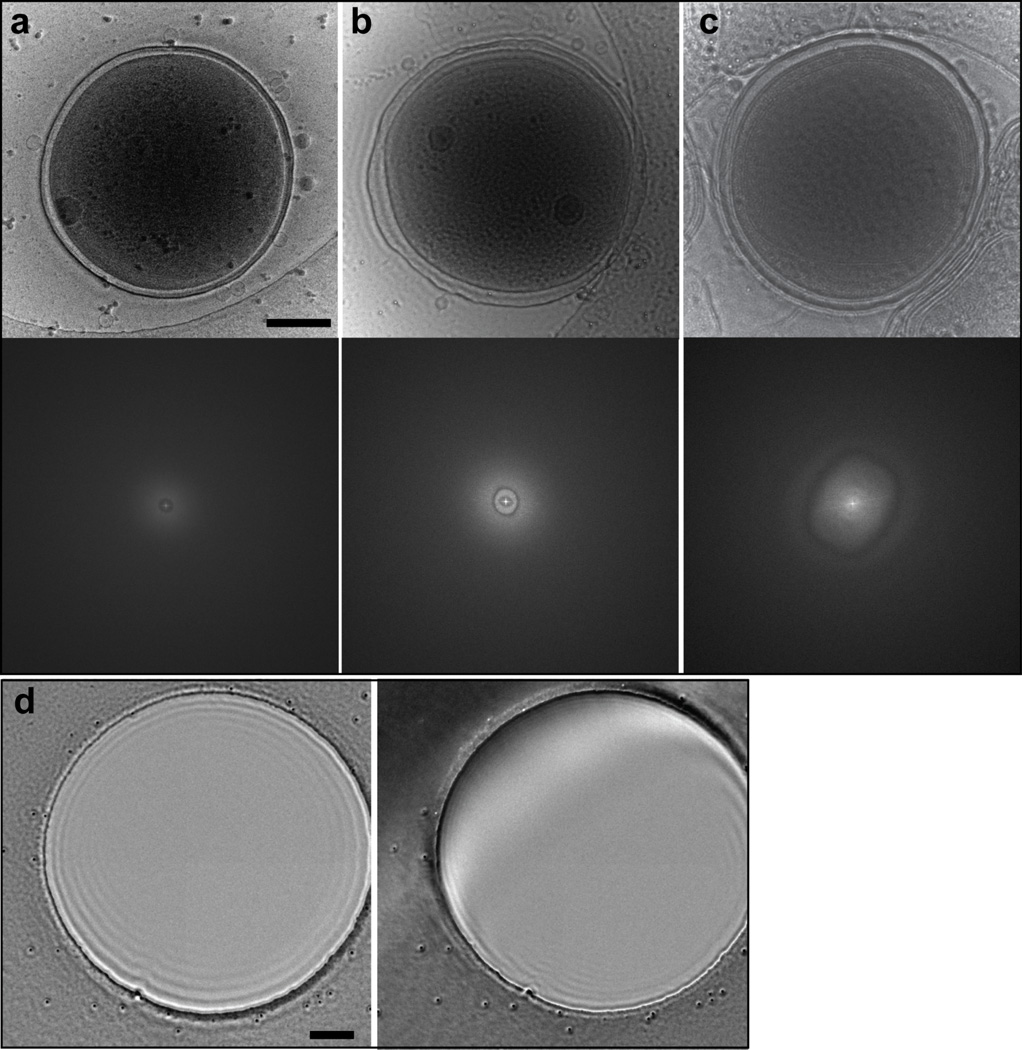
Figure 5. Comparison of ZPC images of WH8109 cells taken with good and charging phase plates, and from centered and off-center phase plates.
(a) A good phase contrast image of a cell shows striking contrast and sharp features. FFT of the image has no CTF rings since the image is taken in-focus. (b) An image of a cell taken with a moderately charging phase plate. Edge of the cell is blurry. FFT shows asymmetric pattern at low spatial frequency. (c) An image of a cell taken with a severely charging phase plate. Some structural features of the cell show inverted contrast. FFT shows aberrant rings. FFT panels in (a) – (c) are shown out to ½ Nyquist frequency. (d) ZPC image of a 1.5-µm Quantifoil grid hole from a phase plate that is properly aligned and centered (left) and from an off-center phase plate (right). Scale bars: 200 nm.
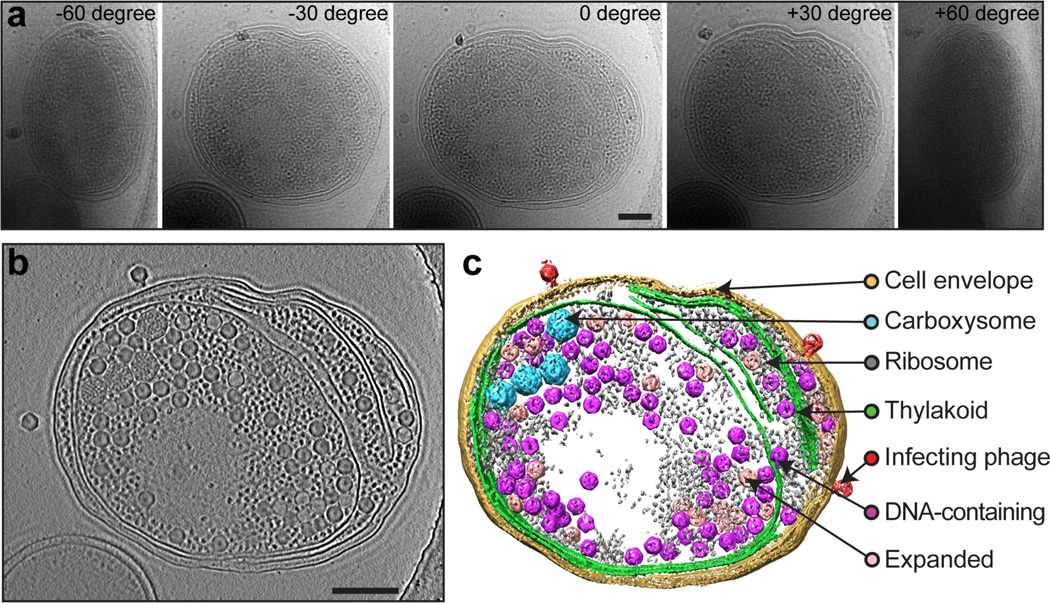
Figure 6. ZPC cryoET of a Syn5-infected WH8109 cell.
(a) A tilt series of a Syn5-infected WH8109 cell. Five of the image frames at –60°, –30°, 0°, 30° and 60° are shown. (b) A 50-nm slab taken from center of the tomogram reconstructed from the tilt series shown in (a). (c) Annotated view of the tomogram. The infecting phage, phage progeny and cellular components are labeled on the right. Scale bars: 250 nm.
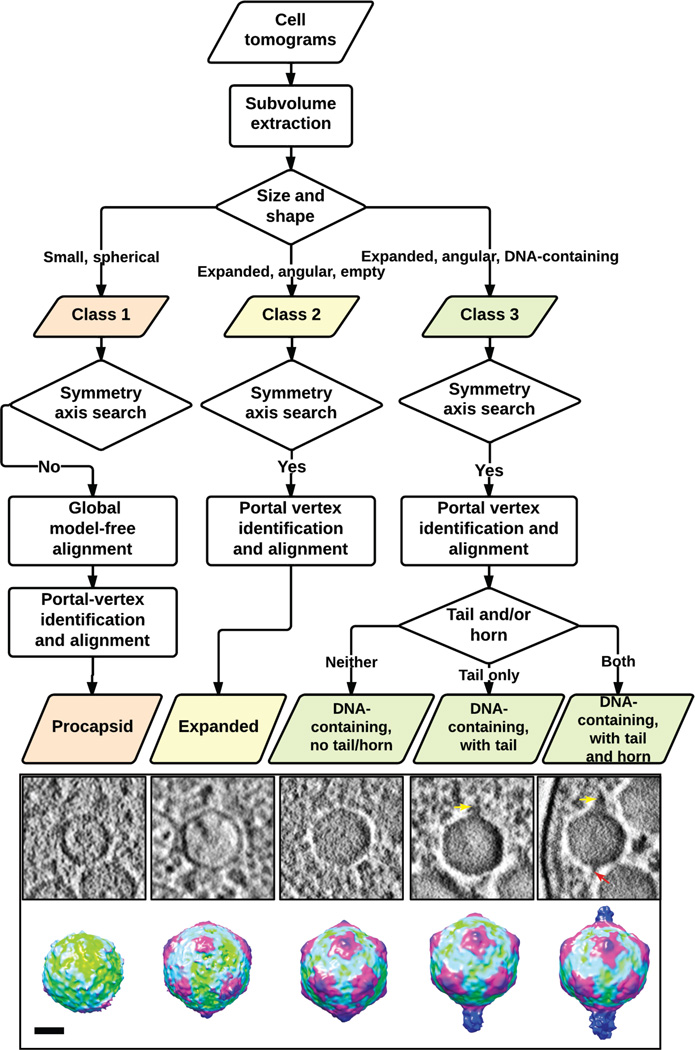
Figure 7. Flowchart of subtomogram classification and alignment.
Visual inspection, symmetry axis determination and a model-free all-vs-all alignment procedure are applied to the extracted intracellular phage progeny subtomograms to classify them into three classes and to align particles in each class to obtain averaged maps. The central slices and the averaged maps are adapted from Nature, 502, 707–710. Scale bar denotes 25 nm for the averaged maps.
Materials
REAGENTS
MilliQ water
Zinc sulfate heptahydrate (ZnSO4·7H2O; Sigma, CAS no. 7446-20-0)
Manganese (II) chloride tetrahydrate (MnCl2·4H2O; Sigma, CAS no. 13446-34-9)
Cobalt (II) chloride hexahydrate (CoCl2·6H2O; Sigma, CAS no. 7791-13-1)
Sodium molybdate dihydrate (Na2MoO4·2H2O; Sigma, CAS no. 10102-40-6)
Citric acid (Sigma, CAS no. 77–92-9)
Ferric citrate (C6H5FeO7; Sigma, CAS no. 3522-50-7)
Sodium carbonate (Na2CO3; Sigma, CAS no. 497-19-8)
Ethylenediaminetetraacetic acid disodium salt dihydrate (Na2EDTA·2H2O; Sigma, CAS no. 6381-92-6)
Sodium chloride (NaCl; Sigma, CAS no. 7647-14-5)
Magnesium chloride hexahydrate (MgCl2·6H2O; Sigma, CAS no. 7791-18-6)
Potassium chloride (KCl; Sigma, CAS no. 7447-40-7)
Sodium nitrate (NaNO3; Sigma, CAS no. 7631-99-4)
Magnesium sulfate (MgSO4; Sigma, CAS no. 7487-88-9)
Calcium chloride dihydrate (CaCl2·2H2O; Sigma, CAS no. 10035-04–8)
Potassium phosphate dibasic trihydrate (K2HPO4·3H2O; Sigma, CAS no. 16788-57-1)
Sodium bicarbonate (NaHCO3; Sigma, CAS no. 144-55-8)
Tris (Sigma, GE17-1321-01)
Hydrochloric acid (HCl; Sigma, CAS no. 7647-01-0)
Liquid ethane: Air Liquide, Catalog no. 4230-7
Cyanobacteria Synechococcus strain WH810917,18: kindly provided by Dr. Jonathan A. King, Massachusetts Institute of Technology
Cyanophage Syn519: kindly titered and provided by Dr. Jonathan A. King, Massachusetts Institute of Technology. Store the phage sample at 4 °C until the infection experiment. At 4 °C, the sample is stable for about 2 weeks.
BSA tracer (10 nm gold fiducial marker): Electron Microscopy Sciences (EMS) catalog number: 25486. Gold particles are coated with BSA protein.
REAGENT SETUP
Trace metal solution 1000× stock
Prepare in MilliQ water a solution containing 0.77 mM ZnSO4·7H2O, 7.0 mM MnCl2·4H2O, 0.14 mM CoCl2·6H2O, 30 mM Na2MoO4·2H2O, 30 mM citric acid, 5 mM ferric citrate.
Supplementary salt solutions 1000× stocks
Prepare in MilliQ water two stock solutions containing100 mM Na2CO3 and 15 mM Na2EDTA·2H2O, respectively.
Artificial sea water (ASW)
Dissolve nine salts (428 mM NaCl, 9.8 mM MgCl2·6H2O, 6.7 mM KCl, 17.8 mM NaNO3, 14.2 mM MgSO4, 3.4 mM CaCl2·2H2O, 0.22 mM K2HPO4·3H2O, 5.9 mM NaHCO3, 9.1 mM Tris) in MilliQ water, and adjust the pH to 8.0 with 6M HCl. After autoclaving, cool down the medium and store it at room temperature (20–25 °C). At room temperature, this medium is stable for ~1 month. Before starting the cell culture, add the trace metal solution (1:1000 volume ratio) and the supplementary salt solutions (1:1000 volume ratio) to the medium to supplement the trace minerals19,20. Once the trace and supplementary salt solutions are added, the medium should be used immediately.
EQUIPMENT
General
Infection light chamber (Orbital/Reciprocal Shaking Bath Model ORS-200, Boekel/Grant)
Hotplate (Talboys 700 series advanced hotplate stirrer, VWR)
Controlled environmental cell culture chamber: home-made chamber with cool fluorescent lights installed at the top and on one of the side panels. Temperature is controlled by a hotplate inside the chamber, and maintained at 28 °C.
Steris Amsco Renaissance Remanufactured 3031 steam sterilizer (Steris)
Milli-Q Academic: Millipore Corporation.
Milli-Q filter: Millipore Millipak 40 0.22 µm.
Petroff-Hausser counting chamber (EMS, Catalog no. 60512-20)
Transmission electron microscope model JEM2200FS (JEOL)
Phase plate airlock system (JEOL)
Phase plates (Terabase Inc., Japan)
Single tilt liquid nitrogen cryo-specimen holder and holder transfer system model 626 (Gatan Inc.)
US4000 Charge Coupled Device (CCD) camera model 895 (Gatan Inc.)
Vacuum glow discharger (Ernest F. Fullam Inc.)
Desktop centrifuge (Eppendorf centrifuge 5417R)
FEI Vitrobot Mark III (FEI)
Filter paper (Whatman Number 1, Qualitative circles, 55 mm diameter, Catalog no. 1001-055)
Quantifoil holey grids (R 2.0/1.0, Cu, 200 mesh, Quantifoil, Germany)
Carbon evaporator (Emitech K950)
Carbon rods (EMS, Catalog no. 70220-02)
Mica sheets (EMS, Catalog no. 71855-05)
Software
PPCont: phase plate controller provided by JEOL.
TEMCom: TEM controller provided by JEOL.
Digital Micrograph: image acquisition and processing software from Gatan Inc., (http://www.gatan.com/scripting/downloads.php).
IMOD21: tomographic alignment and reconstruction package developed by the Boulder Laboratory for 3D Electron Microscopy of Cells (http://bio3d.colorado.edu/imod/). This package also contains programs for tomogram display and structure modeling.
UCSF Chimera22: an extensible molecular modeling and visualization system developed in the University of California at San Francisco (http://www.cgl.ucsf.edu/chimera/).
Avizo: 3D analysis and visualization software for scientific and industrial data developed by Visualization Science Group, FEI (http://www.vsg3d.com/avizo/overview).
EMAN123 and EMAN224: 3D single particle reconstruction and image processing packages developed at NCMI (http://blake.bcm.edu/emanwiki/EMAN1, http://blake.bcm.edu/emanwiki/EMAN2).
The GNU Scientific Library (GSL): a numerical library for C and C++ programming (http://www.gnu.org/software/gsl/).
EQUIPMENT SETUP
Microscope phase plate airlock system
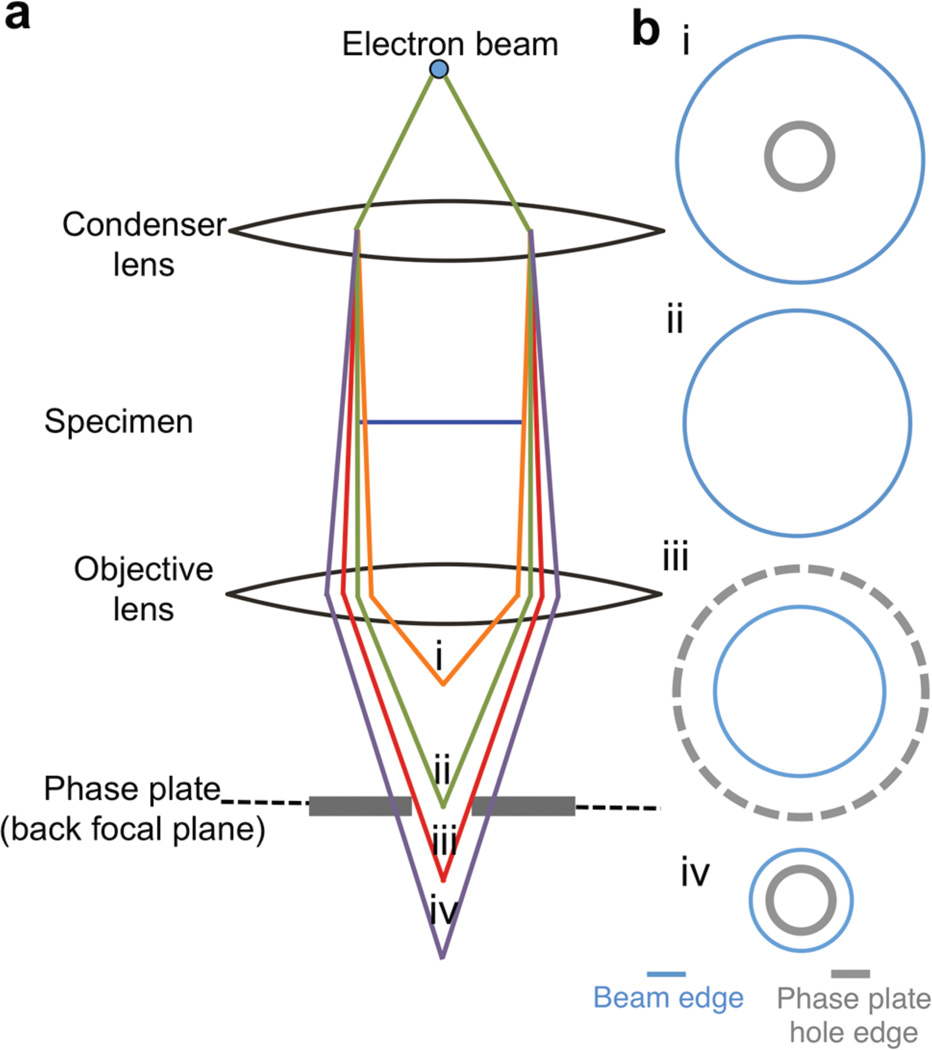
One of the key requirements for setting up a ZPC system on the JEM2200FS microscope is positioning the Zernike phase plate at the back focal plane of the objective lens5,8,25 (Figs. 1, 2). Our JEM2200FS, equipped with the phase plate system, has a specialized pole piece (ZPC-type) with a longer focal length (5.2 mm) and wider space (Fig. 1b) than a standard cryo-EM pole piece (focal length = 2.8 mm). In the back focal plane of the objective lens, frequency is related both to the radial distance from the unscattered beam and to the focal length. The cut-on frequency (c) can be calculated by the formula: c = r/(f λ), where f is the focal length, λ is the electron wavelength, and r is the radius of the phase plate hole. Thus, a longer focal length permits the use of larger, easier to manufacture, phase plate holes to achieve the same cut-on frequency7,25. If the cut-on frequency becomes too large, the contrast enhancement is reduced, and additional artifacts appear in the image. However, smaller phase plate holes are substantially more difficult to align, and misalignment can cause additional artifacts. Our electron microscope is equipped with an airlock system to permit rapid exchange of the phase plate discs (Fig. 1c), as opposed to the previous configuration4,12, which required breaking the microscope column vacuum every time the disc was changed. A typical phase plate disc contains 25 phase plates; each phase plate consists of a ~250-Å thick carbon film with a circular 0.7-µm central hole (Fig. 1c). The holder also heats the phase plate to minimize contamination in the column. The heating element can maintain a steady temperature of up to 500 °C. In order to protect the cryo-specimen from the heat and contamination generated by the heating element, the cold trap in the column provides additional thermal shielding between the cryo-specimen and the phase plate holder (Fig. 1b). The x-y movement of the phase plate stage can be controlled by software or gamepad for coarse adjustment and a piezomotor for fine steps. The condenser lens system of the microscope is optimized to provide parallel illumination, such that the back focal plane of the objective lens becomes coincident with the position of the phase plate. This setup is termed “on-plane conditions” (Fig. 2)5,8,25. Due to effects of the cut-on frequency, this requirement is much more stringent than the alignment requirement for the objective aperture, which can be fairly far from the back focal plane with no adverse effects (Fig. 1b). The on-plane requirement limits the possible range of beam intensities available at the specimen while maintaining the parallel illumination conditions required for high resolution.
Figure 2. Zernike phase plate on-plane condition.
To achieve on-plane condition, the illuminating beam has to be set to cross over at the back focal plane of the objective lens, which is coincident with the location of the phase plate; this objective is achieved by adjusting the convergence angle of the illumination on the specimen. The schematic diagram (a) shows the ray diagram at on-plane condition (ii) and when the beam crossover is above (i) or below the back focal plane (iii, iv). In (b) the spread of the illumination beam and the appearance of the phase plate hole at each condition are shown.
Preparation of gas dispersion bottles
Use a 1 M HCl solution to soak the gas dispersion bottles overnight, and rinse them with MilliQ water six times before autoclaving.
Critical: WH8109 cells are sensitive to heavy metal contamination, so gas dispersion bottles need to be treated beforehand as described to minimize the presence of trace heavy metals.
Preparation of amorphous thin carbon grids
Evaporate a thin layer of carbon film on a freshly cleaved mica sheet, and float the carbon film on water. Dip the Quantifoil holey grids in water under the floating carbon and lift the grids up so that the carbon film is deposited on top of the grid. Dry the grids on filter paper with the carbon film facing up.
PROCEDURE
Sample preparation TIMING 5–6 days
-
1.
Marine Synechococcus represents a major group of photosynthetic cyanobacteria found throughout temperate and tropical oceans17,18. They are gram-negative coccoid cells between 0.6 µm and 1.6 µm in size. For the infection experiment, culture the cells in ASW in gas dispersion bottles (treated to minimize trace heavy metal contamination according to the Equipment setup) under continuous cool white fluorescent light with a light intensity of 1200 lux19. The cells grow optimally when cultured with aeration at 28 °C, and usually reach the exponential phase after 5 days in a fresh culture medium under the above conditions. Begin a new culture from cells in the exponential phase by transferring 10 ml of cells to a bottle with 190 ml of fresh ASW and culture them as just detailed.
-
2.
When the cells reach the exponential phase (usually 4–5 days from a new culture, at conditions described in Step 1), count the cells in a Petroff-Hauser chamber. At exponential phase, cell concentration is in the range of ~1–4×108/ml. Infect the cell with titered Syn5 cyanophage at a multiplicity of infection (MOI) of 5 (five phages per bacterium) in 50 ml conical tubes. Incubate the infected cells in a controlled water bath at 28 °C under continuous, cool white fluorescent light at an intensity of 1200 lux and a shaking speed of 110 rpm. With the progression of infection, the color of the culture will change from light pink to light orange, a property that can be used to visually monitor the infection. Under these conditions, the majority of the cells will reach intermediate to late stages of infection after 65–75 min.
-
3.
Centrifuge cells at 8,500 g for 5 min at 28 °C to remove free phages and excess volume of the medium. Gently resuspend the cell pellet in fresh ASW. We usually use 1/100 of the initial infected cell culture volume so that the cells are concentrated 100 fold.
-
4.
Place 2.0/1.0-µm Quantifoil holey grids on a clean glass slide, with the carbon layer facing up. Place the slide in a glass petri dish, and glow-discharge the grids for 10 s in the vacuum glow discharger to make the grid surface hydrophilic.
-
5.
Load one grid in the Vitrobot with the carbon layer of the grid facing the sample-loading window.
-
6.
Just before applying the sample to the grid, mix 10-nm gold fiducial markers with the Syn5 phage–infected cells at a 1:4 volume ratio to facilitate alignment of tilt series during data processing. Apply an aliquot of 3.5 µl of sample just prepared to the front face of the grid. Keep the relative humidity of the chamber above 90% to avoid excessive sample evaporation. Blot the grid from both sides for 3 s to remove excess liquid before plunge-freezing in a liquid ethane reservoir26.
Critical step: Blotting time is determined by the nature of the sample and properties of the grids. It may require testing several grid types and blotting settings to obtain optimal ice thickness.
Troubleshooting
Phase plate setup and evaluation TIMING ~4 h
-
7.
Load the phase plate into the microscope. A phase plate disc contains 5 × 5 phase plates. Please note that some phase plate discs (e.g. made by Terabase) are pre-mounted on the blade, which is attached to the end of the phase plate holder (Fig. 1c). Other phase plate discs (e.g. made by JEOL) must be loaded by the microscope operator on the tip of the blade and clipped in place with a C-ring. Set the phase plate heater temperature to 250 °C to minimize contamination.
-
8.
To minimize mechanical drift of the phase plates during imaging, wait for at least 40 minutes after the phase plate tip reaches the target temperature before commencing data acquisition.
-
9.
Determine the positions of the 5 × 5 phase plates on the disc using the PPCont software. At low magnification (200×), use the PPCount program to move the phase plate blade, and find and record the coordinates of two marker points on the phase plate disc: the origin point (the central phase plate on the disc) and the adjacent phase plate. Since the size of the phase plate and the spacing between two neighboring phase plates are constant, the software generates a table listing the location of the 5 by 5 grid of phase plate holes based on the locations of those two marker points.
-
10.
Before data collection, evaluate the initial performance of each phase plate on the disc by acquiring a test image of amorphous carbon film at a moderate dose (~5 e−/Å2) and 25,000× magnification and checking the computed Fast Fourier Transform (FFT) of the image acquired by a CCD camera for any signs of charging. Currently, charging of the phase plate is the biggest obstacle for getting high quality image data of the specimen. Each phase plate has a different usable lifespan due to various possible causes8,27,28. Some plates show signs of charging in the very first exposure to the electron beam, whereas others deteriorate only gradually after exposure5,8 (Fig. 5, Supplementary Fig. 1). The manner and extent of changes in the FFT of charging phase plates vary depending on the initial quality of the phase plate and the cumulative electron exposure to the phase plate. Phase plate charging often manifests as an aberrant pattern or unusual Fourier oscillations at low spatial frequency that do not match the zeros of the CTF for any defocus value27 (Fig. 5b–c). If a phase plate hole produces images with good contrast, sharp edges, and proper CTF ring patterns at under-defocus, it is marked as suitable for subsequent use in imaging experiments (Supplementary Fig. 2). Phase plates that show signs of charging or have breaks or contamination in the carbon film are marked as unsuitable and are not used.
Critical step: Expose the phase plates to as little electron dose as possible during evaluation to avoid radiation damage, and extend their useful lifetime.
Microscope configuration TIMING ~2 h
-
11.
Set up the microscope at the magnification and illumination conditions for imaging of the sample. To image Syn5 phage–infected cyanobacteria, use spot size 1, a 70-µm condenser aperture, and a 60-µm objective aperture. At this setting, the specimen dose rate is about 3 e−/Å2/s. These conditions should be suitable for other cellular tomography imaging experiments, but would not be appropriate, for example, for high-dose single particle experiments. Adjust illumination conditions for different specimens based on the size and electron dose tolerance of the specific sample, and the targeted dose rate.
-
12.
Align the condenser lens aperture. Correct the condenser astigmatism using condenser lens stigmator so that the beam is circular.
-
13.
Align the spot displacement by iterating between spot size 1 and 5 using the gun shift and beam shift, respectively, so that the beam position remains unchanged between the spot sizes.
-
14.
Align gun tilt using the anode wobbler so that the beam expands and contracts concentrically.
-
15.
Align pivot points using tilt and shift compensators to minimize beam tilt when applying beam shift, and vice versa.
-
16.
Transfer the plunge-frozen grids to the Gatan cryo-specimen holder via the cryo-transfer station, and insert the holder into the microscope column. This process is no different from that used for conventional cryo-TEM imaging.
-
17.
Pre-scan the entire grid in Low Mag mode (200× magnification) to select grid squares that have suitable ice thickness and indication (black dots at this magnification) that there are cyanbocateria present. Record the coordinates of candidate imaging areas in the microscope controller software, TEMCom. Designate one specimen area for microscope alignment by recording the coordinates of a region of uniform ice adjacent to the imaging areas. Identify and record the coordinates of an open area of broken carbon film to perform CCD camera gain normalization and phase plate position calibration.
-
18.
Set the optimal objective lens excitation by pressing the Standard Focus button. Go to the alignment area (identified in Step 17) and determine the eucentric height of the holder by locating a feature at 8,000× magnification, tilting the stage to 25° and adjusting the Z-height to re-center the feature. Repeat this process for –25°. The stage is eucentric when the feature remains centered as the stage is tilted.
-
19.
Turn on the objective lens wobbler to force the objective lens current to fluctuate. Adjust the condenser tilt so that movement of the image is minimal during fluctuation.
-
20.
Acquire the gain reference correction using the Digital Micrograph software. Under parallel illumination conditions, the intensity of any pixel on the CCD camera should be uniform. To perform the CCD camera gain reference correction, move to the open area on the sample grid (identified in Step 17) and run Digital Micrograph gain reference correction. Magnification and illumination conditions for gain reference correction should be the same as those for the actual image recording. This assumes a Gatan CCD camera. This protocol should not be sensitive to different detector choices, but this step will be detector-specific.
-
21.
In the specimen alignment area, insert and adjust the objective aperture in diffraction mode. Correct the objective lens astigmatism on an area with carbon support at 150,000 to 200,000× magnification using the live view mode and live FFT in Digital Micrograph so that the CTF rings are circular.
-
22.
The JEM2200FS microscope is equipped with an in-column omega energy filter, which can further enhance the image contrast, particularly for thick specimens. For the ZPC cryoET of Syn5 phage–infected WH8109 cells, a slit width of 20 eV works well. We expect this setting will also be appropriate for other cellular tomography applications. First, set the image shift coils and projector lens deflectors to default values. Then degauss the filter. In spectroscopy mode, insert the energy slit and center the slit position so that there is no shadow either at the center or at the outer edges of the beam.
-
23.
Use the MDS panel on the JEM2200FS to set up low dose tomography. Use Photo mode (25,000× magnification) to record low dose images, Focus mode (25,000× magnification) for focusing, and Search mode (defocused diffraction, so-called shadow image) for specimen tracking (Supplementary Fig. 3). Focus and Search modes are also used for centering the phase plate hole, and are described in the following steps.
-
24.
Set up Photo mode at a dose rate of 3 e−/Å2/s with the 70-µm condenser aperture, spot size 1.
-
25.
Set up off-axis Focus mode 2–3 µm away from the area set in Photo mode along the tilt axis of the holder. Adjust the beam diameter in Focus mode to less than 1 µm so that it will not overlap with the sample area in Photo mode.
-
26.
Set up Search mode in diffraction mode with camera length 60 cm, spot size 5 (first condenser lens current setting), and defocus the diffraction spot. Use the Intermediate Lens stigmators to minimize the image distortion of the defocused diffraction spots for Search mode if necessary.
-
27.
Align Search mode to Photo mode. In Photo mode, find a feature on the grid as a reference and move this feature to the center of view. In Search mode, adjust the projector lens deflectors to center the same feature.
-
28.
Move the grid to the open area without any specimen (as identified in Step 17), and insert the phase plate disc into the column. Calibrate the phase plate position in Search mode, option A, or Focus mode, option B. During the tilt series, use either Focus mode or Search mode to make any adjustments to the centering of the phase plate. Focus mode will enable the user to center the phase plate during focusing, whereas Search mode will enable phase plate centering with minimal electron beam exposure to the phase plate.
Critical step: After inserting the phase plate holder into the microscope, and heating up the tip to minimize the contamination, wait at least 40 min for the phase plate to be stable before beginning data collection. Phase plates with unstable temperature may have large drift during tilting and usually require re-centering.
-
Centering the phase plate in Search mode
Use the PPCont software to find a pre-selected phase plate from the calibrated map (as described in Step 10), and scan across the phase plate to find the hole. Roughly center the hole in Search mode with X and Y movement of the phase plate holder. Switch to Photo mode and fine-tune the position of the phase plate hole to the center again using a combination of the PPCont and Piezo X and Y.
Switch back to Search mode and mark the location of the phase plate hole for reference.
-
Centering the phase plate hole in Focus mode
Select phase plate offset option in the PPCont software. When the phase plate hole is centered with condenser tilt in off-axis Focus mode, the illumination system will apply the same offset to center the phase plate hole in Photo mode. In other words, users can center the phase plate hole in Focus mode without irradiating the targeted specimen (Supplementary Fig. 3).
-
29.
Achieve on-plane condition5,8,25 by mechanically positioning the z-height of the phase plate holder close to the back focal plane during its installation and by adjusting the Brightness knob of the microscope so that the edge of the phase plate is no longer visible (Fig. 2).
-
30.
Determine conditions to achieve a specimen exposure of 1 e−/Å2 per image. On our microscope this objective was achieved with a 70-µm condenser aperture followed by adjustment of the first condenser lens with spot size 1. Measure the dose rate in this condition and determine the exposure time required for every image frame. Ideally the 1 e−/Å2 can be achieved in an exposure of 1 second or less. The specific adjustments will depend on the condition of the electron gun in the specific microscope and instrument calibration.
Tilt series setup and data collection with phase plate TIMING Variable, on average 2–3 h for one tilt series
-
31.
It is essential to have the image area at eucentric height so that the specimen area remains centered during tilting. To achieve this objective, fine-tune the eucentricity at each imaging area before beginning tilt series data collection. In Search mode, on the area selected for data collection, manually tilt the stage and adjust the Z-height to re-center the area. Adjust fine eucentricity by tilting the stage at least ± 25° and changing the Z-height to re-center specimen area.
-
32.
In Search mode, scan the grid (loaded into the microscope in Step 16) and go to each imaging area selected in Step 17 to find cells at the right stage of infection (when they have phage progeny inside). Do not image areas too far from the tilt axis, due to limited available tilt range, areas too close to a grid-bar, due to the grid-bar obscuring the specimen at high tilt, or areas with visible ice contamination.
-
33.
Tilt the stage to a starting angle of –60° and center both the sample area and the phase plate hole. Wait for 2 min to allow the cryo-specimen holder to stabilize prior to image recording.
-
34.
The imaging area may move away from the center of the screen as the specimen holder is tilted. The magnitude of this movement depends on the accuracy of the eucentricity and the stability of the stage. To combat this shift of the imaging area, use Search mode to track the image area, and move the stage to re-center the imaging area at each angle during the tilt series. Also in Search mode, verify that the phase plate hole is still centered and, if necessary, use the Piezo controller (X and Y) to re-align the position to the reference marker (see Step 28). If a more extensive re-centering is required, move the grid to the alignment area, to avoid exposing the specimen area to electrons, and check the phase plate position in Photo mode. First center the beam shift and then mechanically adjust the position of the phase plate hole. Then, make any necessary adjustments to Bright Tilt in Focus mode so that the phase plate hole is again centered in both Focus and Photo modes. Then return to Search mode and update the phase plate reference position (Supplementary Fig. 3).
Critical step: To avoid radiation damage of the sample, assign an alignment area (for each imaging area) for phase plate evaluation and centering during the tilt series data collection. This area is usually in the same grid square of the imaging area, with flat and appropriate thickness of ice. We often choose a region that is along the tilt axis, and at least 5–6 µm away from the imaging area.
Troubleshooting
-
35.
In Focus mode, adjust the defocus to in-focus in order to achieve the maximal contrast improvement over a wide range of spatial frequencies6. To estimate a focus setting close to the in-focus condition, switch between underfocus and overfocus by ± 5–10 µm with continuous beam illumination. This method requires an experienced user with a good eye for defocus assessment. An alternative method is to use live FFT in Digital Micrograph to assess defocus; however, this method of focusing requires a lot of electron exposure, which leads to premature deterioration of the phase plate. A third alternative is to record a moderate dose image in Focus mode, measure the defocus in Digital Micrograph, and iterate until the desired value is achieved. Focus using one of the procedures described above at the beginning of the tilt series (60° tilt), and then at every three tilt steps at high tilt (60°–30°), and every five tilt steps at low tilt (30°–0°).
-
36.
Take a Photo mode image of the cell and evaluate the contrast, the sharpness of cell features, such as the cell membrane, and the corresponding FFT of the image. If the image of the cell shows good contrast and features (Figs. 4, 5a, Supplementary Fig. 1a), and no CTF ring is seen in the FFT, save the image and tilt the stage by the increment (usually 3°) for the next frame.
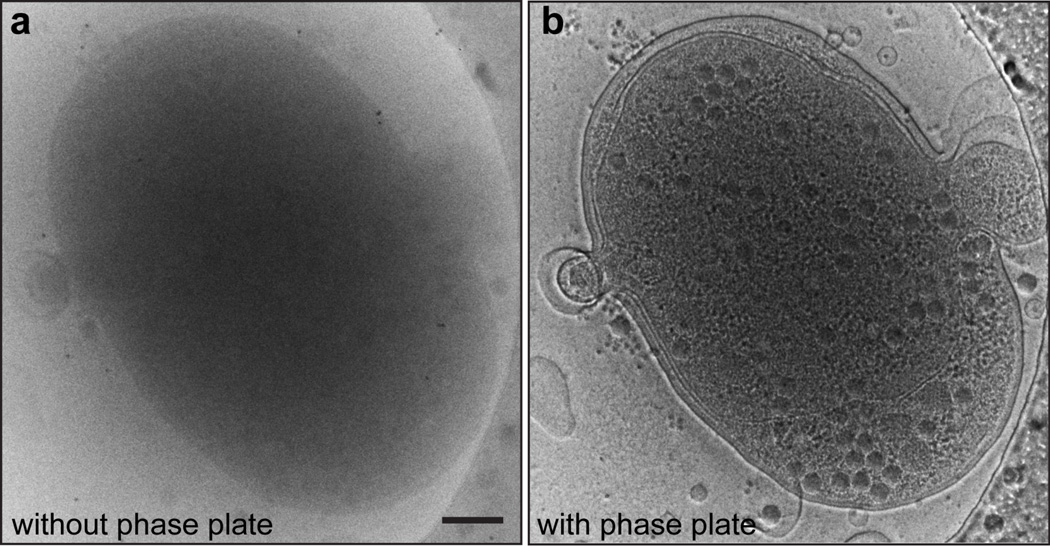
Figure 4. Contrast Improvement by Zernike phase plate using Syn5-infected WH8109 cells as an example specimen.
(a) A conventional cryoEM image of an ice-embedded Syn5 phage infected cell. (b) A ZPC image of the same cell as shown in a under the same imaging conditions. Scale bar: 200 nm.
Troubleshooting
-
37.
Continue the tilt series by tilting to the next angle, tracking the cell location, centering the phase plate hole, focusing adjustment and evaluating the phase plate if needed, and acquiring an image (Steps 34–36). Wait ~2 minutes after each tilt to allow the specimen stage to stabilize. The completed tilt series should normally range from – 60° to +60°.
-
38.
Due to the deteriorating effects of the electron beam on the phase plate, phase plates may no longer produce good images after a certain level of electron exposure12,29 (Fig. 5, Supplementary Fig. 1). As mentioned in Step 35, use the FFT of the Focus mode images to assess the degree of phase plate deterioration (Fig. 5). If FFT of acquired images show an aberrant pattern or oscillations, typically accompanied by blurry real-space images, move to another good phase plate from the list obtained from phase plate evaluation step (Step 10). The lifespan of phase plates is variable; currently only ~10% of phase plates last for an entire tilt series (>100 e−/A2 including dosage used for focusing and tracking). Often, to complete one tilt series, multiple phase plates are required.
Critical step: Focusing, phase plate centering and evaluation are done every three or five tilting intervals, depending on the tilt angles. It is important to monitor the quality of the images closely during tilt series data collection. If any visual signs of phase plate charging emerge or out-of-focus images are produced, stop the tilt series and examine the defocus or phase plate by looking at the real space image and FFT of the image. Change to a new phase plate if the one in use is no longer producing high-quality images.
Data processing TIMING ~6 months
-
39.
Align the frames of the tilt series (Fig. 6a) in IMOD21 using fiducial marker-based alignment. To achieve this goal, first, align all image frames in the tilt series by cross correlation based on global features of the images. Then select 10 to 20 gold fiducials from each frame of the pre-aligned tilt series as fixed markers for fine alignment of the stack (see IMOD tutorials at http://bio3d.colorado.edu/imod/doc/tomoguide.html). Reconstruct the final tomogram by back-projection.
-
40.
Perform section-by-section visualization of the cell tomograms (Fig. 6b) in 3dmod, a specialized visualization and modeling program in the IMOD package.
-
41.
Perform 3D isosurface visualization of the cellular tomograms and subvolumes in UCSF Chimera22 (Fig. 6c).
-
42.
Perform segmentation of the cellular components, the infecting phage, and the phage progeny from the cell tomograms in Avizo.
-
43.
To facilitate identification of features, apply a Gaussian low pass filter (using EMAN224 program e2proc3d.py with the -process=filter.lowpass.gauss:cutoff_freq=0.03 option) to cell tomograms. Record coordinates of center of individual progeny phages, found visually inside infected WH8109 cells (Fig. 6) in the IMOD model mode. Then extract phage progeny subvolumes from the raw unfiltered cell tomograms (using IMOD command boxstartend).
-
44.
Perform an initial classification of the intracellular phage progeny by visually inspecting the sizes and features of the particles (see Fig. 7). The procapsid has a diameter of 59 nm and a scaffolding core in the middle of the capsid. The expanded and the DNA-containing particles are the same size as mature phage, with a diameter of 66 nm. The expanded particles have low density inside the capsid shell. For the higher-density DNA-containing phage progeny, we further divide the class into three subtypes based on the presence or absence of the tail and/or the horn at the opposite vertex. This sub-classification is carried out after all particles in this class are aligned to the icosahedral symmetry axis16 (see the following Step).
-
45.
Align symmetric particles to their symmetry axes. For objects with symmetry, symmetry-based alignment offers an unbiased alternative to the typical reference-based alignment procedures in subtomogram averaging. To facilitate achieving this aim, we developed a symmetry based alignment program, now available in EMAN224, called e2symsearch3d.py. This algorithm aligns the particle to its symmetry axes if the symmetry is present. The procedure used internally in this program is outlined in Supplementary Fig. 4. Briefly, n randomized initial orientations are applied to the particle. For each of these orientations, apply an icosahedral symmetry to the particle. The program will then compute the normalized cross correlation score between the symmetrized and unsymmetrized particle, accounting for the missing wedge30–32. A simplex minimizer software is then applied to optimize the alignment. If the particle does possess icosahedral symmetry, and the particle orientation is right at the icosahedral symmetry axes, the symmetrized particle’s icosahedral features will be enhanced, and the cross correlation will produce a high score. If the particle does not have icosahedral symmetry, or if the particle’s current orientation is far from the icosahedral symmetry axes, symmetrization will generate a featureless ball, and the algorithm will generate poor cross correlation scores. Repeat the above process for the n randomized initial orientations, and define the global best orientation as the highest score out of the n local best orientations.
Critical step: Determine the number of initial random orientations n based on the targeted symmetry, the contrast of the images, and the features of the subtomograms. We used n=10 for the phage progeny subtomograms to search for the icosahedral symmetry axis. Lower contrast or lower symmetry will require larger n values, which will need to be determined by trial and error. Using a larger n will take more computing time, but yields fewer false negatives (i.e. missing the true symmetry axis).
-
46.
For DNA-containing particles, once the particles are aligned to the symmetry axis, use a model-free all-vs-all alignment program, tomohunter (a tomographic subtomogram alignment module in EMAN123), to align all the particles and create averaged maps of phage assembly intermediates30,31,33. Since all the particles in this class have been aligned to the icosahedral symmetry axis, constrain the search and alignment to varying the 12 vertices only, aiming at identifying and registering the unique portal vertex (the vertex with the portal ring structure through which DNA enters and exits the capsid) of all the particles to the positive z-axis32,34.
-
47.
Carry out a subclassification of the DNA-containing particles by visually inspecting the individual particles. First, apply a high-pass filter to all the particles to remove the cut-on artifact as manifested by fringes around the objects (more details in Step 50) due to the presence of a central hole in the phase plate. Then apply a low pass filter to remove the high frequency noise. All of these steps are performed using EMAN2 e2proc3d.py program. Examine all particles using the UCSF Chimera software package22. Look for the tail along the positive Z-axis and the horn along the negative Z-axis. The high contrast of the ZPC cryoET subvolumes enables researchers to further partition the particles into three subtypes: with neither tail nor horn, with tail only, and with tail and horn on opposite vertices (Fig. 7).
-
48.
For this Syn5 phage, the symmetry axis search algorithm finds about 70% of the expanded particles to have icosahedral symmetry in their capsid shell (Step 45). For particles with identified icosahedral symmetry axis, use the all-vs-all alignment scheme, ‘constrained’ to search the 12 vertices, to identify and bring the portal vertex in each particle along the positive Z-axis.
-
49.
The procapsid particles have a smooth and spherical shell without apparent angularity35. The symmetry search algorithm fails to identify icosahedral symmetry axes in particles in this class. Use the model-free all-vs-all alignment procedure in tomohunter to search the entire rotational space to obtain an initial model for the procapsid class. Iteratively refine the orientations of the particles in the dataset to the model until no further improvement is observed.
-
50.
Correct the cut-on frequency effects on averaged maps of DNA-containing particles. The cut-on frequency effects of phase contrast images are caused by the central hole in the Zernike phase plate where the unscattered electrons pass through. In the images, this effect is manifested as fringes next to densities with high intensity29,36 (see Fig. 4b). The cut-on frequency (and thus the spacing of the fringes) is determined by the size of the phase plate hole, whose edge forms an abrupt step (Fig. 1c). In our experiments, most of the phase plates are in the range of 0.7 to 1 µm in size, yielding a cut-on frequency of about 1/300 Å−1. Cut-on frequency correction is done to minimize the fringes in the images and to boost the signal below the cut-on frequency to a magnitude comparable to that of the frequency beyond the cut-on frequency region. To correct the cut-on artifacts, first compute a 1D structure factor from known conventional single particle reconstruction of purified mature Syn5 by rotationally averaging power spectrum of the particle structure. Then use this 1D structure factor as a reference to rescale the power spectrum of the averaged DNA-containing phage progeny maps. By doing this, the cut-on frequency spectrum is approximately scaled to match the structure factor (using EMAN1 proc3d command with the option of the pixel size and the name of the structure factor file). Note that no correction is attempted for subvolume averages of procapsid or expanded capsid because no appropriate structure factor is available.
TIMING
Steps 1–6, sample preparation: Synechococcus WH8109 cells are ready for infection 4– 5 days after starting a fresh culture. The infection experiments require 65–70 min to bring over 50% of the cells to intermediate/late stage of infection. Preparation of the grids takes half a day.
Steps 7–10, phase plate setup and evaluation: With the phase plate airlock system in our JEM2200FS microscope, phase plate discs can be exchanged without breaking the microscope vacuum. Loading of the phase plate holder into the microscope takes 20 min. After the temperature of the phase plate holder tip reaches 250 °C, stabilization at this temperature on phase plates takes ~1 h. The phase plate controller software generates the phase plate position map in 5 min once the two position markers are designated. Evaluation of 25 phase plates on a disc usually requires 2 h. Steps 11–30, microscope configuration: Basic microscope alignment takes 30 min for an experienced microscopist. Loading the sample into the microscope and stabilizing the holder in column is a 40-min process. Detailed configuration of condenser and objective lenses, acquiring a gain reference for the camera, aligning the in-column energy filter, and setting up low dose mode takes roughly 40 min.
Steps 31–38, tilt series setup and data collection with phase plate: Time required for this component of the Protocol is variable depending on the quality of the phase plates. A tilt series collected from a single phase plate needs less time than the one that requires multiple phase plates. On average, acquisition of a tilt series requires 2 to 3 hours. Steps 39–50, data processing: Tilt series alignment and tomographic reconstruction are usually quick and straightforward because of the high contrast of the image frames in the tilt series. Subtomogram extraction and visual inspection of 500 phage progeny subvolumes required approximately 1 month after all the tomograms were reconstructed. Testing and setting up the appropriate parameters and applying them for phage progeny classification, alignment, and averaging took about 3 months. Visualization and annotation of 20 tomograms required another 2 months of work.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 6 | Ice too thin | Humidity in the sample blotting chamber is too low |
Check the water level in the humidifier reservoir to make sure there is enough water. Close the sample loading window so that the chamber is completely closed, and wait until the humidity reaches about 90% before loading the sample |
| Ice too thick or uneven |
Blotting paper too wet from high humidity in the chamber. This usually happens in extended sample freezing sessions |
Use a new pair of blotting papers | |
| 34 | Big image target movement during tilting |
Bad eucentricity | Move to alignment area and adjust the eucentricity |
| Sample drift | Make sure the holder has good Vacuum. Stabilize the stage for 1–2 min after every tilt |
||
| Phase plate drift | The temperature of the heated phase plate blade is not stabilized |
Wait longer for the phase plate to stabilize |
|
| 36 | Shadow at the center or the edge of images |
Energy filter misalignment | Degauss the filter and then center it again |
| Image has improved contrast but the features are blurry |
Phase plate charging. Sometimes a charging phase plate can produce a normal FFT, but the images taken from that phase plate have lost high- resolution information |
Replace phase plate with a new one | |
| Phase plate edge shown on image |
Phase plate is far from back- focal plane |
Turn the brightness to a wider beam | |
| Un-even fringe around features in images (Fig. 5d) |
Phase plate is not properly centered |
Center the phase plate using the Piezo control |
ANTICIPATED RESULTS
This Protocol focuses on microscope setup and imaging conditions for ZPC cryoET. Application of this Protocol will generate phase contrast tilt series and 3D tomograms of biological sample of your choice. If the implementation is done correctly, the images will be of high contrast. Annotation of the tomogram (Fig. 6c) and classification, alignment and averaging of the subsequent subvolumes (Fig. 7) will be straightforward. The cut-on frequency correction step should be performed for appropriate visualization and interpretation of the data.
Supplementary Material
Acknowledgements
This research has been supported by NIH grants (P41GM103832 and R01GM080139) and the Robert Welch Foundation (Q1242). We thank Desislava Raytcheva, Cameron Haase-Pettingell, Jonathan A. King for the supplies of Syn5 phage and much assistance to prepare the samples for the experiments; Kuniaki Nagayama for suppling the phase plates; John Flanagan for computational software; Ryan H. Rochat for photographs and part of the illustration in Figure 1; Xiangan Liu for programs for cut-on frequency correction; Masahiro Kawasaki for many helpful discussions.
Footnotes
STAND-FIRST: Equipping an electron microscope with Zernike phase contrast optics dramatically increases the contrast of the images. Dai et al. describe how to successfully apply this technology for the acquisition and analysis of electron tomograms.
Author contributions statements
W.D. carried out the Synechococcus WH8109 cell culture and Syn5 infection experiments. W.D. and C.F. prepared all the grids for ZPC cryoET imaging. C.F. and H.A.K. established the ZPC illumination configuration and the airlock system for phase plate disc exchange. W.D. collected all ZPC cryoET tilt series. S.J.L and M.F.S. developed subtomogram processing programs. W.D. performed the data processing and subtomogram alignment. W.D., M.F.S. and W.C. performed the data analysis and interpretation. All authors contributed to the preparation of this paper.
The authors declare that they have no competing financial interests.
Contributor Information
Wei Dai, Email: wei.dai@bcm.edu.
Caroline Fu, Email: cjfu@bcm.edu.
Htet A. Khant, Email: hkhant@bcm.edu.
Steven J. Ludtke, Email: sludtke@bcm.edu.
Michael F. Schmid, Email: mschmid@bcm.edu.
References
- 1.Barcena M, Koster AJ. Electron tomography in life science. Semin. Cell Dev. Biol. 2009;20:920–930. doi: 10.1016/j.semcdb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerrero-Ferreira RC, Wright ER. Cryo-electron tomography of bacterial viruses. Virology. 2013;435:179–186. doi: 10.1016/j.virol.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thon F. Electron microscopy in material sciences. Academic Press; 1971. pp. 570–625. [Google Scholar]
- 4.Danev R, Nagayama K. Single particle analysis based on Zernike phase contrast transmission electron microscopy. J. Struct. Biol. 2008;161:211–218. doi: 10.1016/j.jsb.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Danev R, Glaeser RM, Nagayama K. Practical factors affecting the performance of a thin-film phase plate for transmission electron microscopy. Ultramicroscopy. 2009;109:312–325. doi: 10.1016/j.ultramic.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danev R, Kanamaru S, Marko M, Nagayama K. Zernike phase contrast cryo-electron tomography. J. Struct. Biol. 2010;171:174–181. doi: 10.1016/j.jsb.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Danev R, Nagayama K. Optimizing the phase shift and the cut-on periodicity of phase plates for TEM. Ultramicroscopy. 2011;111:1305–1315. doi: 10.1016/j.ultramic.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Marko M, Leith A, Hsieh C, Danev R. Retrofit implementation of Zernike phase plate imaging for cryo-TEM. J. Struct. Biol. 2011;174:400–412. doi: 10.1016/j.jsb.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi M, Danev R, Nishiyama K, Sugawara K, Nagayama K. Zernike phase contrast electron microscopy of ice-embedded influenza A virus. J. Struct. Biol. 2008;162:271–276. doi: 10.1016/j.jsb.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Guerrero-Ferreira RC, Wright ER. Zernike phase contrast cryo-electron tomography of whole bacterial cells. J. Struct. Biol. 2014;185:129–133. doi: 10.1016/j.jsb.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda Y, Fukazawa Y, Danev R, Shigemoto R, Nagayama K. Tuning of the Zernike phase-plate for visualization of detailed ultrastructure in complex biological specimens. J. Struct. Biol. 2009;168:476–484. doi: 10.1016/j.jsb.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Murata K, et al. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure. 2010;18:903–912. doi: 10.1016/j.str.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton B, et al. In-focus electron microscopy of frozen-hydrated biological samples with a Boersch phase plate. Ultramicroscopy. 2011;111:1696–1705. doi: 10.1016/j.ultramic.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Gamm B, Schultheiss K, Gerthsen D, Schroder RR. Effect of a physical phase plate on contrast transfer in an aberration-corrected transmission electron microscope. Ultramicroscopy. 2008;108:878–884. doi: 10.1016/j.ultramic.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Majorovits E, et al. Optimizing phase contrast in transmission electron microscopy with an electrostatic (Boersch) phase plate. Ultramicroscopy. 2007;107:213–226. doi: 10.1016/j.ultramic.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Dai W, et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature. 2013;502:707–710. doi: 10.1038/nature12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller NJ, et al. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the red sea. Appl. Environ. Microbiol. 2003;69:2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocap G, Distel DL, Waterbury JB, Chisholm SW. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pope WH, et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine synechococcus. J. Mol. Biol. 2007;368:966–981. doi: 10.1016/j.jmb.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raytcheva DA, Haase-Pettingell C, Piret JM, King JA. Intracellular assembly of cyanophage Syn5 proceeds through a scaffold-containing procapsid. J. Virol. 2011;85:2406–2415. doi: 10.1128/JVI.01601-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kremer JR, Mastronarde DN, McIntosh JR. Computer visulasualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 22.Pettersen EF, et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 24.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Danev R, Nagayama K. Phase plates for transmission electron microscopy. Methods Enzymol. 2010;481:343–369. doi: 10.1016/S0076-6879(10)81014-6. [DOI] [PubMed] [Google Scholar]
- 26.Taylor KA, Glaeser RM. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol. 2008;163:214–223. doi: 10.1016/j.jsb.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marko M, Meng X, Hsieh C, Roussie J, Striemer C. Methods for testing Zernike phase plates and a report on silicon-based phase plates with reduced charging and improved ageing characteristics. J. Struct. Biol. 2013 doi: 10.1016/j.jsb.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagayama K. Another 60 years in electron microscopy: development of phase-plate electron microscopy and biological applications. J. Electron Microsc. 2011;60:S43–S62. doi: 10.1093/jmicro/dfr037. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda Y, Nagayama K. Zernike phase contrast cryo-electron tomography of whole mounted frozen cells. J. Struct. Biol. 2012;177:484–489. doi: 10.1016/j.jsb.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Schmid MF. Single-particle electron cryotomography (cryoET) Adv. Protein Chem. Struct. Biol. 2011;82:37–65. doi: 10.1016/B978-0-12-386507-6.00002-6. [DOI] [PubMed] [Google Scholar]
- 31.Schmid MF, Booth CR. Methods for aligning and for averaging 3D volumes with missing data. J. Struct. Biol. 2008;161:243–248. doi: 10.1016/j.jsb.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid MF, et al. A tail-like assembly at the portal vertex in intact herpes simplex type-1 virions. PLoS Pathog. 2012;8:e1002961. doi: 10.1371/journal.ppat.1002961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 2007;81:2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rochat RH, et al. Seeing the portal in herpes simplex virus type 1 B capsids. J. Virol. 2011;85:1871–1874. doi: 10.1128/JVI.01663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen DH, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. USA. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosogi N, Shigematsu H, Terashima H, Homma M, Nagayama K. Zernike phase contrast cryo-electron tomography of sodium-driven flagellar hook-basal bodies from Vibrio alginolyticus. J. Struct. Biol. 2011;173:67–76. doi: 10.1016/j.jsb.2010.08.004. [DOI] [PubMed] [Google Scholar]
PRIMARY ARTICLES
- 1.Dai W, et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature. 2013;502:707–710. doi: 10.1038/nature12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murata K, et al. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure. 2010;18:903–912. doi: 10.1016/j.str.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rochat RH, et al. Seeing the portal in herpes simplex virus type 1 B capsids. J. Virol. 2011;85:1871–1874. doi: 10.1128/JVI.01663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.