Abstract
Class Ia phosphoinositide (PI) 3-kinase, an essential mediator of the metabolic actions of insulin, is composed of a catalytic (p110α) and regulatory (p85α) subunit. Here we demonstrate that p85α interacts with X-box binding protein-1 (XBP-1), a transcriptional mediator of the unfolded protein response (UPR), in an ER stress-dependent manner. Cell lines with knockout or knockdown of p85α exhibit dramatic alterations in the UPR including reduced ER stress-dependent accumulation of nuclear XBP-1, decreased induction of UPR target genes and increased rates of apoptosis. This is associated with a decrease activation of IRE1α and ATF6α. Mice with deletion of p85α in liver (L-Pik3r1−/−) display a similar attenuated UPR following tunicamycin administration leading to an increased inflammatory response. Thus, p85α forms a novel link between the PI 3-kinase pathway, which is central to insulin action, and the regulation of the cellular response to ER stress, which can lead to insulin resistance.
Introduction
There is a worldwide epidemic of obesity, type 2 diabetes and metabolic syndrome, all conditions linked to insulin resistance 1,2. While multiple mechanisms contribute to the development of insulin resistance in these disorders, one major mechanism is the activation of cellular stress and inflammatory signaling pathways 3–5. Understanding exactly how these stress pathways are linked to and impact insulin signaling dynamics is of critical importance.
Class Ia phosphoinositide 3-kinases (PI3Ks) are a family of lipid kinases that regulate multiple cellular processes, including cell metabolism and growth, by virtue of their ability to generate the second messenger, phosphatidylinositol-3,4,5-tris phosphate (PIP3) 6. This enzyme exists as an obligate heterodimer, composed of a regulatory (p85α, p85β, or p55γ) and catalytic (p110α, β, or δ) subunit, both of which occur in several isoforms. The central role of this enzyme in mediating the metabolic actions of insulin is apparent from studies where expression of dominant-negative constructs or pharmacological inhibition of PI 3-kinase completely abolish insulin stimulation of glucose transport, lipogenesis and glycogen synthesis 7–9. Alterations in insulin stimulation of PI 3-kinase activity are observed in animal models of obesity, as well as in humans with type 2 diabetes 10,11,12.
The endoplasmic reticulum (ER) forms an interconnected, membranous network that is the major site of synthesis and folding of secreted and integral membrane proteins. Protein folding in the ER lumen is facilitated by a number of molecular chaperones, including BiP and Grp94, and a variety of folding enzymes such as protein disulfide isomerase (PDI) 13,14. Physiological states that increase protein synthesis, or stimuli that disrupt the processes by which proteins obtain their native conformation, create an imbalance between the protein-folding demand and capacity of the ER. This results in the accumulation of unfolded or improperly folded proteins in the ER lumen and a state of ER stress. The cellular response to ER stress, referred to as the unfolded protein response (UPR), results in activation of three linked signal transduction pathways emanating from three principle, ER stress sensors: inositol requiring 1 α (IRE1α), PKR-like kinase (PERK) and activating transcription factor 6α (ATF6α) 15,16. The combined actions of these signaling cascades serve to reduce ER stress through attenuation of translation to reduce protein load and through activation of transcriptional programs that ultimately serve to increase ER protein folding and maturation.
Pathophysiological states including obesity, hyperlipidemia, nutrient deprivation, hypoxia, infection and inflammation have been shown to disrupt ER homeostasis 17–20. For example, mice with genetic or diet-induced obesity exhibit a significant elevation in ER stress with elevated phosphorylation of PERK and IRE1α and enhanced splicing of XBP-1 21. Several markers of ER stress are also elevated in adipose tissue from obese humans 20,22. Conversely, the increase in insulin sensitivity associated with weight loss is associated with a significant reduction in markers of UPR activation23. Furthermore, mice administered chemical chaperones that facilitate protein folding or transgenic mice over-expressing the ER chaperone, ORP150, exhibit improvements in obesity-associated insulin resistance and glucose metabolism 24,25. Mechanistically, activation of the UPR in the obese state contributes to the decrease in insulin sensitivity through IRE1α-dependent activation of JNK, which leads to phosphorylation of insulin receptor substrate-1 (IRS-1) on inhibitory serine residues 26–28.
Additional evidence establishing a mechanistic link between IRE1α and UPR-associated insulin resistance comes from studies of mice haploinsufficient for XBP-1. Normally, in response to ER stress, IRE1α executes site-specific cleavage of XBP-1 mRNA to produce a transcript (XBP-1s) that encodes a potent transcriptional activator of UPR target genes 29,30. When subjected to a high fat diet, XBP-1 heterozygous mice gain more weight and become more insulin resistant than control mice 21. These mice also display an increase in ER stress in adipose tissue with enhanced PERK and IRE1α phosphorylation and activation of JNK. Likewise, XBP-1-deficient fibroblasts exhibit enhanced PERK phosphorylation, hyperactivation of JNK and increased serine phosphorylation of IRS-1 21. Collectively, these data demonstrate how the ER stress response and alterations in XBP-1 can modulate insulin sensitivity.
In the present study, we demonstrate another novel link between insulin signaling and the UPR. We show that the p85α regulatory subunit of PI 3-kinase interacts with XBP-1 in an ER stress-dependent manner and that this interaction is essential in the ER stress response. As a result, cells deficient in p85α or livers with selective inactivation of the p85α gene exhibit a dramatic reduction in ER stress and accumulation of nuclear XBP-1s protein and its downstream target proteins. This link between the regulatory subunit of PI 3-kinase and the cellular response to ER stress provide a novel therapeutic target for the treatment of diseases in which the UPR is activated, such as obesity and type-2 diabetes.
Results
The p85α regulatory subunit of PI 3-kinase associates with an XBP-1-containing protein complex
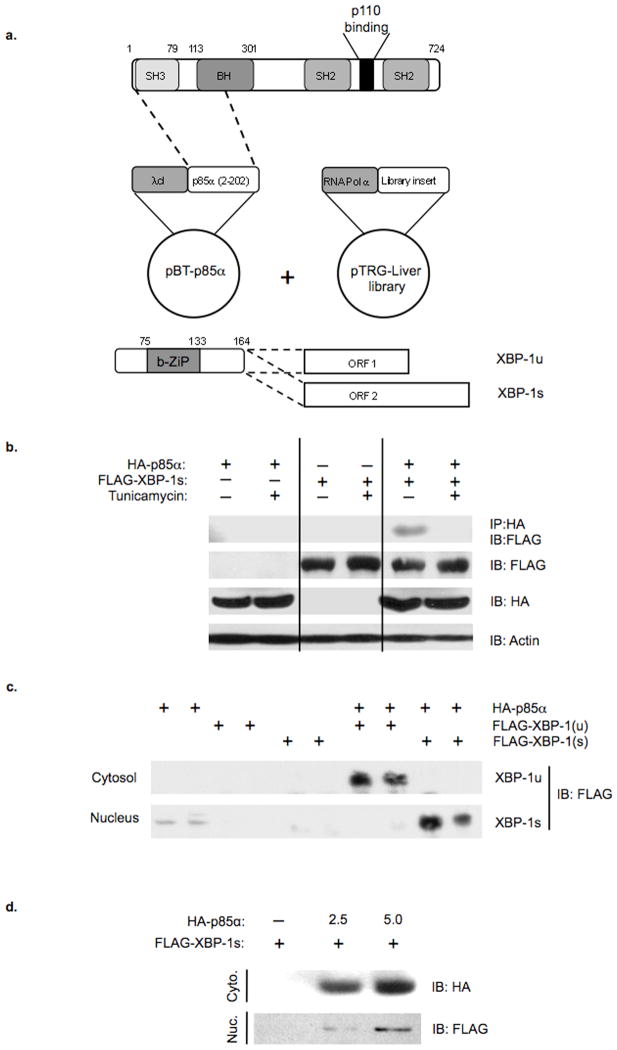
In previous studies, we have shown that deletion of the p85α subunit of PI 3-kinase results in decreased activation of JNK and increased insulin sensitivity, and that this depends on the N-terminal half of p85α and is independent of its role in PI 3-kinase activation 31–33. To identify proteins that interact with the NH2-terminal region of p85α, an interaction screen was performed using a bacterial two-hybrid system and a human liver cDNA library (Fig. 1a). Sequencing of isolated cDNA clones revealed that one novel p85α interacting partner was the transcription factor XBP-1. The XBP-1 interacting fragment included the NH2-terminal portion of the transcript, a region shared between the unspliced (XBP-1u) and spliced (XBP-1s) transcripts of XBP-1 (Fig. 1a, bottom).
Figure 1.
Identification and characterization of XBP-1 as a p85α interacting protein. a) Schematic diagram of the plasmids used in the bacterial two-hybrid screen. a) (bottom) Schematic of the XBP-1 cDNA and the alternative splice variants that generate open reading frames (ORF) 1 and 2 that encode XBP-1u and XBP-1s, respectively. b) Co-immunoprecipitation experiments were performed on cells transfected with HA-p85α alone or in combination with FLAG-XBP-1u or FLAG-XBP-1s. Anti-HA immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-FLAG antibodies. Anti-actin immunoblots confirmed equal protein was used for each condition. c) HEK 293T cells were transiently transfected with the indicated expression plasmids. Forty-eight hours following transfection, cytosolic and nuclear lysates were prepared and immunoblotting subsequently performed using anti-FLAG antibodies. d) The nucleocytoplasmic shuttling of FLAG-XBP-1s was assessed in HEK293T cells transfected with or without FLAG-XBP-1s and increasing concentrations of HA-p85α expression plasmid. Nuclear and cytoplasmic fractions were resolved by SDS-PAGE and immunoblotted with anti-FLAG or anti-HA antibodies.
To confirm that the association between p85α and XBP-1 observed in vitro would occur in vivo, we performed co-immunoprecipitation experiments using extracts of cells transfected with HA-p85α and FLAG-XBP-1s expression plasmids alone or in combination (Fig. 1b). Although XBP-1 was not observed in HA immunoprecipitates from cells transfected with HA-p85α or FLAG-XBP-1s alone, FLAG-XBP-1s was clearly observed in HA-p85α immunoprecipitates when both proteins were expressed. Interestingly, there was a significant reduction in association between XBP-1s and p85α following treatment with tunicamycin, suggesting that this interaction is regulated by ER stress. Similar results were obtained when rat hepatoma cells were infected with adenoviruses expressing either GFP or p85α with an NH2-terminal tandem affinity purification (TAP) tag containing the streptavidin and calmodulin binding domains (Supplemental Fig. S1) 34,35. These data establish that there is an interaction between p85α and XBP-1 both in vitro and in vivo and demonstrate that the association is modulated by the cellular response to ER stress.
p85α promotes the stabilization and nuclear accumulation of XBP-1
Degradation of XBP-1 involves both ubiquitin-dependent and independent mechanisms, resulting in short half-lives of both XBP-1u and XBP-1s proteins 36. To determine whether p85α altered the stability of XBP-1, cells were transiently transfected with XBP-1 alone or in combination with p85α (Fig. 1c). Co-expression of XBP-1 with p85α led to a substantial increase in the levels of XBP-1u and XBP-1s in the cytoplasm and nucleus, respectively.
In contrast to XBP-1u, XBP-1s is primarily localized to the nucleus. Expression of an N-terminal deletion of XBP-1s leads to a redistribution to both the cytoplasm and nucleus, suggesting that this region of XBP-1s governs its nuclear distribution 36,37. Having established that p85α and XBP-1 interact and that the interaction occurs between the N-terminal portion of p85α and the shared N-terminal fragment of XBP-1 capable of directing nuclear localization, we examined whether p85α was able to alter the cellular distribution of XBP-1. To this end we transiently transfected cells with FLAG-XBP-1s in the absence or presence of increasing amounts of HA-p85α and performed immunoblotting on cytoplasmic and nuclear fractions with anti-FLAG antibodies (Fig. 1d). Interestingly, the dose-dependent increase in HA-p85α resulted in a parallel increase in the nuclear translocation of XBP-1s. Thus, increasing levels of p85α leads to the stabilization of XBP-1 and enhances the nucleocytoplasmic shuttling of XBP-1.
p85α-deficient Fibroblasts Exhibit an attenuated response to ER stress
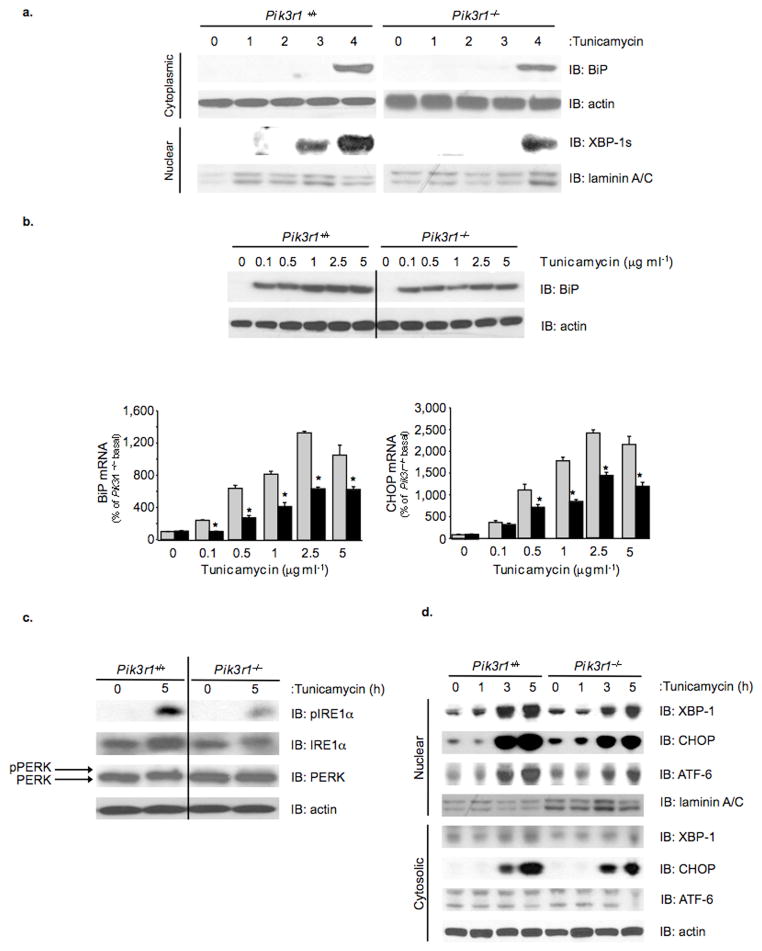
Based upon the role of XBP-1 as a central mediator of the UPR, we sought to determine whether the interaction between p85α and XBP-1 played a role in control of the cellular response to ER stress. To this end, immortalized control (Pik3r1+/+) and p85α-deficient (Pik3r1−/−) brown preadipocyte cell lines were treated with vehicle or tunicamycin, and BiP and XBP-1s protein expression in cytoplasmic or nuclear compartments was assessed (Fig. 2a) 31. As expected, treatment of control preadipocytes with tunicamycin for 4 hr resulted in the appearance of BiP, and immunoblots performed on nuclear lysates revealed a time-dependent accumulation of XBP-1s (Fig. 2a). By contrast, when Pik3r1−/− cells were treated with tunicamycin, BiP induction was reduced by 50% as compared to controls, and there was a marked reduction in nuclear accumulation of XBP-1s with no detectable nuclear XBP-1s at 3 hours and a 40% reduction in nuclear XBP-1s after 4 hours, which did not increase further at five hours (Supplemental Fig. S2). Thus, the absence of p85α alters the cellular response to ER stress with a decrease in nuclear accumulation of XBP-1s and reduced expression of BiP.
Figure 2.
Evaluation of ER stress and UPR signaling dynamics in p85α-deficient fibroblasts. a) Immunoblots were performed on cytoplasmic and nuclear protein fractions to assess the induction of BiP or XBP-1s protein, respectively, over a time-course of tunicamycin (2 μg ml−1) treatment. b) (top) Control and Pik3r1−/− fibroblasts were treated with the indicated concentrations of tunicamycin for five hours and anti-BiP immunoblots were performed on whole cell lysates. Anti-actin immunoblots were performed to ensure equal loading. b) (bottom) Quantitative RT-PCR was performed on cDNA derived from control and Pik3r1−/− fibroblasts treated with the indicated concentrations of tunicamycin for five hours. c) Control and Pik3r1−/− fibroblasts were treated with vehicle or tunicamycin (2 μg ml−1) for five hours, and immunoblots performed on whole lysates using IRE1α, pIRE1α and PERK-specific antibodies. Anti-actin immunoblots were performed to confirm equal protein loading. d) Control and Pik3r1−/− fibroblasts were treated with tunicamycin (2 μg ml−1) over the indicated time-course. Immunoblot analysis on nuclear lysates was performed using XPB-1, ATF6 and CHOP antibodies. Anti-laminin A/C immunoblots were performed to confirm equal loading. Data are presented as the means ± SEM, and asterisks indicate statistical significance determined by student’s t test (*p < 0.05, **p < 0.001, n.s.: non-significant)
To exclude the possibility that the defect observed in Pik3r1−/− cells was due to reduced sensitivity to UPR activation, a tunicamycin dose-response was performed. As expected, Pik3r1+/+ cells demonstrated a dose-dependent increase in BiP expression following treatment with 0.1–5 μg ml−1 of tunicamycin (Fig. 2b; top). In Pik3r1−/− cells treated with tunicamycin, BiP protein expression showed a dose-dependent increase, however, BiP levels were 50–60% lower at all tunicamycin concentrations. Likewise there was a dose-dependent 10- to 20-fold increase in BiP and CHOP mRNA levels following tunicamycin treatment in control cells, whereas p85α-deficient cells displayed a 50–60% reduction in BiP and CHOP mRNA at all concentrations (Fig. 2b, bottom). A similar reduction in the UPR targets BiP and CHOP was observed at the mRNA and protein level when thapsigargin was used to activate the UPR (Supplemental Fig. S2, S3, data not shown). These effects were independent of PI 3-kinase enzyme activity: pharmacological inhibition of PI 3-kinase using LY294002 had no effect on the induction of BiP expression following tunicamycin or thapsigargin treatment (Supplemental Fig. S4).
Multiple UPR Pathways are altered in p85α-deficient Fibroblasts
From the above it is clear that p85α-deficiency severely impairs the cellular response to ER stress with reduced expression of BiP and nuclear accumulation of XBP-1s in response to UPR activation. p85α deficiency was also associated with alterations in IRE1α and ATF6α-dependent signaling pathways. Thus, while control cells responded to tunicamycin treatment with a significant up-regulation of IRE1α protein and an enhancement of IRE1α phosphorylation, Pik3r1−/− cells exhibited an ~20–40% reduction of IRE1α expression in the basal state and after tunicamycin-treatment, and an associated reduction in IRE1α phosphorylation (Fig. 2c). Interestingly, although an alteration in IRE1α activation was readily observed, there were no differences in PERK levels or activation between control and Pik3r1−/− cells as assessed by alterations in PERK mobility or phosphorylation (Fig. 2c, data not shown). Activation of ATF6α, as assessed by immunoblot analysis of the processed form of ATF6α in nuclear lysates, was also significantly attenuated in Pik3r1−/− cells at the 3 and 5 hour time-points, and paralleled the blunted response in accumulation of nuclear XBP-1s and induction of CHOP (Fig. 2d). Thus, p85α deficiency attenuates the cellular response to ER stress through multiple mechanisms involving alterations in the induction of XBP-1s and activation of IRE1α and ATF6-dependent pathways.
Altered XBP-1 splicing and UPR target gene expression in p85α-deficient fibroblasts
To examine whether alterations in IRE1α pathway led to a reduction in XBP-1 splicing capacity in Pik3r1−/− fibroblasts, we performed an XBP-1 splicing assay. As expected, untreated control fibroblasts produced primarily the XBP-1u transcript with only a small amount of XBP-1 migrating at the expected size of the spliced transcript (Supplemental Fig. 5a). Treatment of these cells with tunicamycin resulted in a significant decrease in XBP-1u and concomitant increase in XBP-1s transcript levels. In contrast, Pik3r1−/− cells exhibited a reduction in XBP-1u transcript in the basal state and a relatively modest increase in spliced XBP-1 transcript following treatment with tunicamycin. qPCR analysis demonstrated a 2.5-fold increase in total XBP-1 transcript following tunicamycin treatment in control cells (Supplemental Fig. 5b, left panel, p < 0.05). In p85α-deficient cells, total XBP-1 transcript was reduced by 50% in the basal state, and there was a significantly attenuated, tunicamycin-dependent increase when compared to controls. Likewise in control cells, tunicamycin produced a dramatic, ~30-fold increase (100 ± 2.45 vs. 3067 ± 778) in XBP-1s, and this response was markedly blunted in p85α-deficient cells (91 ± 29 vs.1181 ± 193) (Supplemental Fig. 5b, right panel). Thus, p85α-deficiency decreases IRE1α-dependent, XBP-1 splicing in response to ER stress.
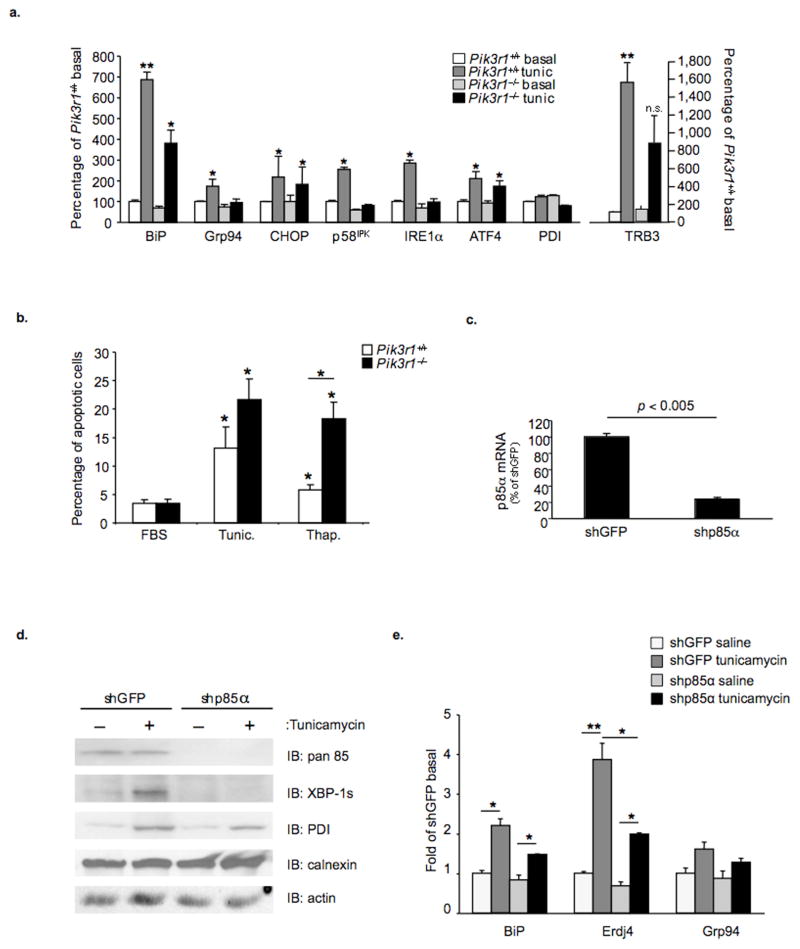
We next sought to establish the extent to which the defects in IRE1α protein and phosphorylation, and decreased transcription and splicing of XBP-1 mRNA observed in Pik3r1−/− cells altered induction of UPR target genes. Analysis of control fibroblasts treated with tunicamycin for five hours revealed 50–60% increases in multiple UPR target genes, including BiP, Grp94, CHOP, p58IPK, IRE1α, ATF4 and TRB3 (Fig. 3a). In contrast, the transcriptional profile in Pik3r1−/− cells could be classified into two distinct categories. The first contained genes such as BiP, CHOP and ATF4 that were up-regulated by tunicamycin, but to a much lower level than achieved in control cells. The second included Grp94, p58IPK and IRE1α and TRB3 that were significantly elevated in control cells following induction of ER stress, but displayed a non-significant increase in Pik3r1−/− cells following treatment with tunicamycin.
Figure 3.
Evaluation of XBP-1 target-gene transcription, UPR-dependent gene expression and apoptosis. a) Quantitative PCR was performed to determine UPR target-gene expression in Pik3r1+/+ or Pik3r1 −/− cells treated with vehicle or tunicamycin for five hours. b) Cells were grown to confluence and maintained in 10% FBS supplement with or without tunicamycin (2 μg ml−1) or thapsigargin (100 nM) for 24 hours. Floating and attached cells were collected, pooled and incubated with both annexin V PE and propidium iodide (PI) for 15 minutes at room temperature. FACS analysis was then performed. The percentage of apoptotic cells (PI negative, annexin-PE positive cells) was determined. Data are mean ± S.E. from 3 independent experiments. c) Quantitative PCR was performed on control and shp85α cell lines using p85α-specific primers. d) Immunoblot analysis was performed using whole cell lysates prepared from shGFP and shp85α cell lines treated with vehicle or tunicamycin (2 μg ml−1) for five hours. e) Quantitative PCR was performed to quantify BiP, Erdj4 and Grp94 mRNA expression in shGFP and shp85α cell lines before or following treatment with tunicamycin (2 μg ml−1) for five hours. Data are presented as the means ± SEM, and asterisks indicate statistical significance determined by student’s t-test (*p < 0.05; **p < 0.001; n.s. = non-significant)
Lastly, to evaluate the outcome of blunted UPR activation in p85α-deficient cells, apoptosis was measured in control and Pik3r1−/− pre-adipocytes following treatment with tunicamycin and thapsigargin (Fig. 3b and Supplemental Fig. 6). As expected, treatment of Pik3r1+/+ and p85α-deficient cells for 24 hours with tunicamycin or thapsigargin increased the rates of apoptosis when compared to cells grown in normal medium. Interestingly, a higher proportion of p85α-deficient cells became apoptotic following treatment with tunicamycin (13.1 ± 3.7% vs. 21.6 ± 3.5%) and thapsigargin (5.8 ± .9% vs. 18.2 ± 3.0%). Thus, by all criteria, p85α-deficient cells exhibit a failure to normally activate the UPR and resolve ER stress. This leads to deleterious outcomes including a propensity to enter apoptotic programs.
Reduced expression of p85α in the human hepatoma cell line alters the ER stress response
Similar effects of p85α deficiency were observed in human Huh7 hepatoma cells following lentiviral-mediated shRNA knockdown of p85α mRNA. Using this approach, we achieved a 78% decrease in p85α mRNA levels when compared to shGFP-expressing control cells (Fig. 3c), and this was confirmed by immunoblot analysis using anti-p85α antibodies (Fig. 3d, top). Control cells exposed to tunicamycin also displayed a marked increase in XBP-1s protein, whereas no XBP-1 was detected in cells after p85α knockdown. In addition, there was an attenuated increase in PDI in p85α-deficient cells. Similarly, treatment of shGFP cells with tunicamycin led to a robust transition from the XBP-1u to XBP-1s transcript, while splicing of the XBP-1 transcript following tunicamycin was diminished in shp85α cells as reflected by lower levels of XBP-1s transcript and an associated increase in XBP-1u mRNA when compared with controls (Supplemental Fig. 5c). As a result, the tunicamycin-dependent induction of the UPR target genes, ERdj4 and BiP, was attenuated in shp85α cells when compared to controls (Fig. 3e). Thus, p85α-deficiency alters the cellular response to ER stress independent of cell type or stimulus indicating an important role of the interaction between p85α and XBP-1 in ER stress induction and the UPR.
A blunted UPR is observed in mice with a liver-specific deletion of p85α
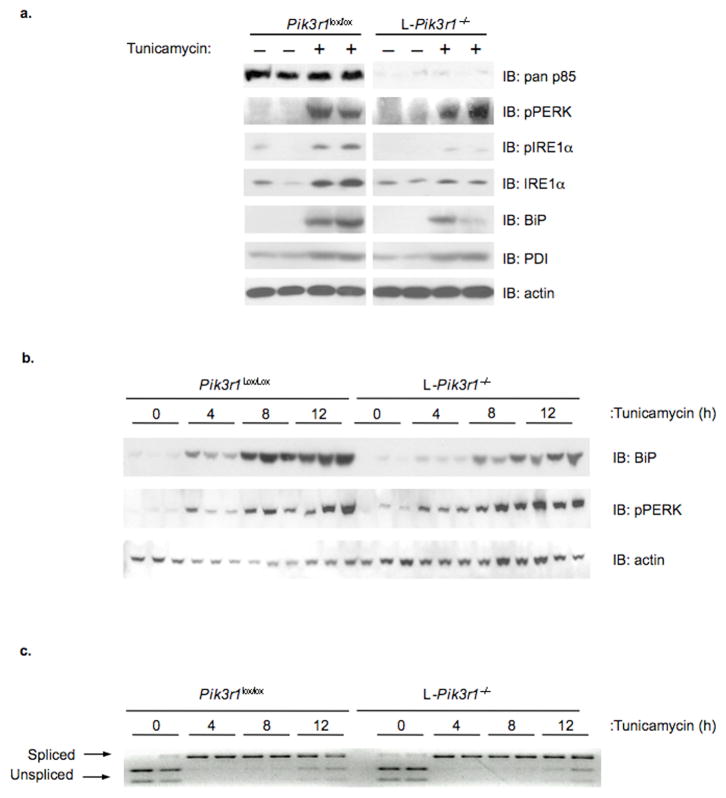
To determine whether the effects observed in vitro would be recapitulated in vivo, we utilized mice with a liver-specific deletion of the p85α gene (L-Pik3r1−/−). At two months of age, male mice were administered vehicle or tunicamycin (2.5 μg/g B.W.) and after 72 hours, mice were euthanized, and livers collected for analysis. Immunoblots confirmed the absence of p85α in lysates from vehicle and tunicamycin treated L-Pik3r1−/− animals (Fig. 4a). As anticipated, PERK and IRE1α phosphorylation was low in vehicle treated p85α lox/lox animals, but increased dramatically following tunicamycin administration (Fig. 4a, left). In contrast, livers from L-Pik3r1−/− mice had undetectable basal and drastically reduced, tunicamycin-dependent increases in IRE1α phosphorylation with no significant alteration in activation of the PERK pathway (Fig. 4a, right). There was also a dramatic, tunicamycin-dependent increase in total IRE1α and BiP protein levels in control animals, and these responses were severely blunted in L-Pik3r1−/− mice. A similar blunting of the UPR and reduced activation of the ATF6α pathway was seen 18-hours following administration of tunicamycin (Supplemental Fig. S7). Likewise, as early as 4 hours after tunicamycin administration, the increase in BiP protein was significantly blunted in p85α-deficient livers when compared to controls, despite normal activation of the PERK pathway (Fig. 4b). Thus, activation of the IRE1α and ATF6α pathways and the ER stress response pathways are impaired in the livers of p85α-deficient mice. In vivo, this occurred independent of alterations in XBP-1 splicing (Fig. 4c).
Figure 4.
Evaluation of the UPR in livers from control and L-Pik3r1−/− mice. At eight weeks of age, male Pik3r1lox/lox or L-Pik3r1−/− mice were administered vehicle or tunicamycin (2.5 μg/g B.W.) by intraperitoneal injection. a) Following 72 (n = 4 per group) or b) a short time-course of tunicamycin administration, livers were collected and tissue lysates prepared. Following separation by SDS-PAGE and transfer to PVDF, immunoblotting was performed using the indicated antibodies. Actin immunoblots were performed to confirm equal protein loading. c) XBP-1 splicing was evaluated by performing PCR on cDNA derived from livers of control and L-Pik3r1−/− mice treated with vehicle or tunicamycin for 4, 8 or 12 hours. Primers were designed to flank the 26 nt intron excised from the XBP-1s transcript. Resultant PCR products were digested with Pst I restriction endonuclease and resolved by agarose gel electrophoresis.
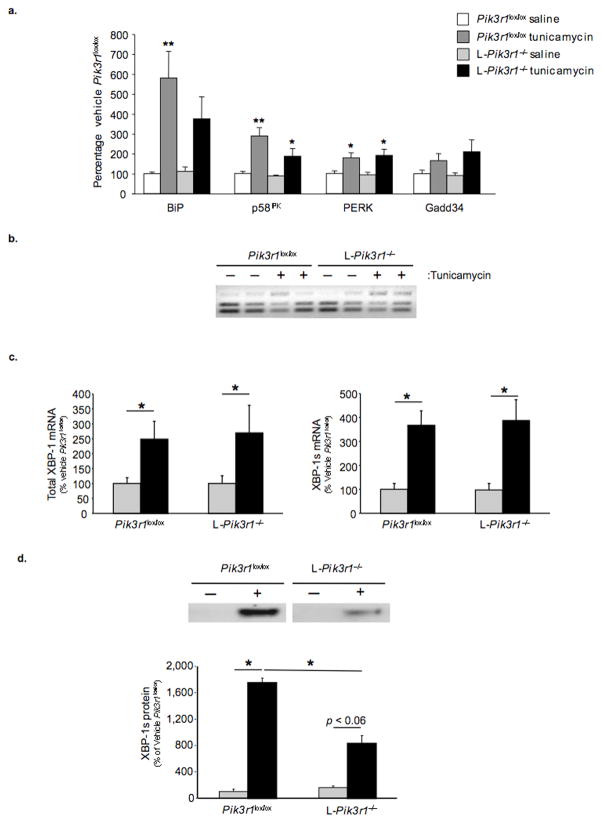
The attenuated response to ER stress observed in livers from L-Pik3r1−/− mice was accompanied by altered the expression of UPR-dependent target genes. In liver of control mice, BiP mRNA increased five-fold (100 ± 8.2 vs. 579 ± 131) in response to tunicamycin treatment, whereas in L-Pik3r1−/− mice, the increase in BiP was reduced by ~50% and failed to reach statistical significance (Fig. 5a). Likewise, in control animals, tunicamycin elicited a 3-fold increase in p58IPK mRNA (100 ± 10.1 vs. 288 ± 39.6, p < 0.001), and this response was blunted in the livers of L-Pik3r1−/− mice (88.2 ± 5.2 vs.187 ± 39, p < 0.05).
Figure 5.
Analysis of gene expression, XBP-1 splicing and XBP-1 stability in livers from Pik3r1lox/lox and L-Pik3r1−/− mice following 72 hours of vehicle or tunicamycin treatment. (n=5 per group) a) UPR-dependent target gene expression was determined by performing quantitative PCR on cDNA derived from Pik3r1lox/lox and L-Pik3r1−/− livers using gene-specific primers. b) A PCR-based assay was used to evaluate XBP-1 splicing. c) XBP-1 splicing was assessed in the livers from saline and tunicamycin-treated (2.5 μg/g BW) mice by quantitative PCR as previously described. d) Nuclear lysates from Pik3r1lox/lox and L-Pik3r1−/− mice following 18 hours of tunicamycin (2.5 μg/g BW) treatment were resolved by SDS-PAGE and immunoblotted with XBP-1-specific antibodies (top panel). Quantification of nuclear XBP-1s protein was achieved by densitometry using the Image J software. Data are presented as the means ± SEM, and asterisks indicate statistical significance determined by student’s t-test (*p < 0.05; **p < 0.001)
Some differences were observed between in vivo and in vitro studies. Thus, in vivo, PERK and Gadd34 mRNA levels were elevated to a similar extent in livers from L- Pik3r1lox/lox and L-Pik3r1−/− mice following tunicamycin treatment. In addition, tunicamycin administration in vivo caused a comparable increase in XBP-1 splicing in both controls and L-Pik3r1−/− mice (Fig. 5b, c). On the other hand, as in vitro, the robust 17-fold increase (100 ± 39.4 vs. 1749 ± 74.4) in the nuclear XBP-1s protein observed in controls following tunicamycin was blunted by > 50% (160 ± 25.9 vs. 830 ± 117) in livers of L-Pik3r1−/− mice (Fig. 5d). Thus, deletion of p85α does not affect all UPR-dependent pathways equally, and the extent of pathway involvement can be affected by other aspects of cellular context.
L-Pik3r1−/− mice exhibit markers of inflammation associated with a failure to resolve ER stress
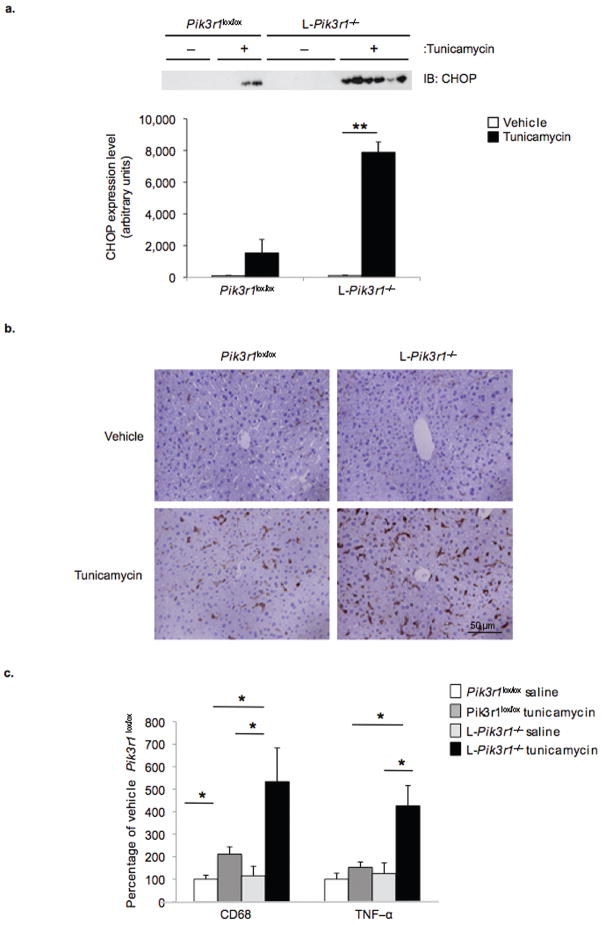
In order to evaluate the outcomes associated with impairment in the UPR of L-Pik3r1−/− mice apoptosis was assessed in liver following tunicamycin administration. Surprisingly, despite the increase in CHOP induction in L-Pik3r1−/− relative to controls following 18 (11.7 ± 7.3 vs. 54 ± 17.7) or 36 (1.5 ± .4 vs. 16.8 ± 6.8) hours of treatment, no increases in apoptosis could be detected by TUNEL or by immunoblot analysis of cleaved caspase-3 (Fig. 6a, data not shown). Histological analysis, however, revealed striking changes in livers from tunicamycin treated L-Pik3r1−/− mice including edema and dilation of the sinusoids and bile ducts when compared to vehicle-treated controls (Supplemental Fig. S8). In contrast, the livers of Pik3r1lox/lox mice treated with vehicle or tunicamycin appeared normal.
Figure 6.
Analysis of liver from control and L-Pik3r1−/− mice following tunicamycin treatment. a) CHOP protein expression was assessed by performing SDS-PAGE on liver lysates followed by immunoblotting with anti-CHOP antibodies. Protein quantification was achieved by densitometry using NIH Image J software. b) Livers from control and L-Pik3r1−/− mice were fixed in 4% paraformaldehyde, paraffin embedded and sections stainied with Hematoxylin & Eosin. Representative images are shown. (n = 5 per group) c) The expression of CD68 and TNFα was determined by performing quantitative PCR on cDNA derived from livers from control and L-Pik3r1−/− mice 36 hours following administration of vehicle or tunicamycin (2 μg/g body weight). Data are presented as the means ± SEM, and asterisks indicate statistical significance determined by student’s t-test (*p < 0.05)
One pathological outcome of unresolved stress in the liver is an inflammatory response. Immunohistochemistry of liver sections using antibodies to the macrophage marker F4/80 revealed only sporadic staining in vehicle treated control and L-Pik3r1−/− livers (Fig. 6b). After tunicamycin, there was a modest increase in F4/80 positive cells in the livers of Pik3r1lox/lox mice, whereas there was a substantial increase in the number of F4/80 positive cells in L-Pik3r1−/− livers when compared to controls. Gene expression analysis revealed a parallel changes in the macrophage marker, CD68, and the inflammatory cytokine TNFα (Fig. 6c). Consistent with increased macrophage recruitment, livers from L-Pik3r1−/− mice also exhibited greater activation of the NFκB pathway compared to controls as revealed by a reduction in IκB expression (36.6% vs. 18.6%) (Supplemental Fig. S8). These data suggest that p85α deficiency alters the fundamental ability of the liver to resolve an ER stress, leading to pathological states of inflammation.
Discussion
The ER stress response has been strongly implicated in the pathophysiology of diabetes, affecting both insulin sensitivity in liver and fat, and the survival of pancreatic β-cells 38–41. Adipose tissues from obese, insulin-resistant mice and humans exhibit a persistent low level of inflammation and activation of ER stress pathways, including induction of the UPR 42,4,21. Identifying points of regulatory convergence that can serve as therapeutic targets to improve insulin sensitivity and relieve ER stress associated with obesity are therefore of great importance.
Previous studies have demonstrated that PI 3-kinase is central to the actions of insulin, as well as other hormones and growth factors 7,8,43,44. While induction of ER stress has been shown to reduce insulin-stimulated AKT activation and decrease IRS tyrosine phosphorylation 21, to date, there has been no evidence establishing a direct link between the PI 3-kinase pathway and ER stress. In this study, we demonstrate a novel and unexpected role for the p85α regulatory subunit of PI 3-kinase as a critical modulator of the cellular response to ER stress. This occurs via a mechanism involving p85α-dependent regulation of XBP-1s protein expression, XBP-1s nuclear translocation and ATF6α activation. Since the level of p85α can change in some insulin resistant states, including obesity, pregnancy 45 and states of growth hormone excess 46,47, as well as in some cancers 48,49, a link between p85α and the cellular response to ER stress has important implications for our understanding of a broad range of UPR-associated diseases, including diabetes, cancer and a variety of other disorders.
Our data demonstrate that p85α and XBP-1 physically associate and that this interaction is both modulated by and modulates the cellular response to ER stress. The identification of XBP-1 as a p85α interacting partner using the bacterial two-hybrid system indicates that the association is direct. Co-precipitation experiments reveal that p85α and XBP-1s interact in a protein complex that dissociates following treatment with tunicamycin, indicating that this interaction is dynamically regulated by ER stress. While the precise interaction domains remain to be determined, they clearly involve the NH2-terminal portion of p85α and a domain shared by both XBP-1 isoforms. The NH2-terminal region of p85α has also been shown to bind to several other protein partners, including c-Cbl, the small GTPase Rac1 and Cdc42, indicating a number of important roles for the p85α regulatory subunit of PI 3-kinase in addition to its role as a partner in the p85/p110 heterodimer 50,51.
The interaction between p85 and XBP-1s acts to alter the UPR through several mechanisms. Enforced expression of p85α leads to an increase in XBP-1 protein stability and enhanced nuclear trafficking of XBP-1s. Conversely, deficiency or reduction in p85α expression in cultured cells results in a significantly attenuated UPR. This includes a reduction in IRE1α and ATF6 pathway activation and a concomitant reduction in UPR targets at the mRNA and protein level. This is accompanied by a dramatic reduction in nuclear accumulation of XBP-1s following treatment with tunicamycin. Collectively, these changes are associated with a relative failure of p85α-deficient cells to resolve ER stress as indicated by a significant increase in apoptosis following induction of ER stress.
Alterations in the UPR are also observed in mice with a liver-specific deletion of p85α, including reduced induction of the UPR targets BiP, Grp94 and CHOP at both the mRNA and protein level. L-Pik3r1−/− mice also exhibit a reduction in IRE1α expression and activation, and a dramatic reduction in accumulation of XBP-1s in the nucleus following induction of ER stress. In contrast to isolated cells, no alteration in XBP-1 splicing was observed in the livers of L-Pik3r1−/− mice following acute stimulation with tunicamycin. These data are in accord with the study of Park et. al. which indicates that enforced expression of either p85α or p85β leads to an increase in XBP-1s nuclear localization, whereas knock-down of both regulatory subunits leads to a reduction in the nuclear accumulation of XBP-1s, providing further evidence for a critical role of PI3-kinase regulatory subunits in modulating the UPR 52. Thus reducing p85α can modify the UPR by several mechanisms including a reduction in XBP-1 protein due to altered protein stabilization, decreased nuclear translocation, a reduction in total and activated IRE1α, and a decrease in ATF6α pathway activation. Recent analysis of ATF6α-deficient fibroblasts has revealed that ATF6α action is critical for the UPR-dependent transcription of ER chaperones, and that heterodimerization of ATF6α and XBP-1 mediate transcriptional induction of ERAD genes 53. Thus, further studies exploring the interplay between p85α, ATF6α activation and XBP-1 will be of importance in identifying other points of regulatory convergence.
The magnitude of the decrease in XBP-1s observed in response to decreases in p85α are comparable to or greater than that observed in XBP-1 heterozygote knockout mice, suggesting that the UPR perturbations due to reduction in p85α expression may have a similar underlying mechanism, i.e., a decline in XBP-1 expression. XBP-1 heterozygous knockout mice have been shown to display signs of elevated ER stress and have reduced insulin signaling and an associated decrease in whole body insulin sensitivity when placed on a high fat diet 21. However, XBP-1 haploinsufficient mice fed a normal diet exhibit normal insulin, C-peptide and glucose levels, suggesting that an additional lesion is required to unmask a phenotype 21. By comparison, mice with heterozygous deletion of p85α, homozygous inactivation of p85β or targeted deletion of p85α in liver exhibit enhanced, rather than diminished, insulin sensitivity 54–56. These data suggest that XBP-1 and p85α haploinsufficiency regulate insulin sensitivity through multifactorial and at least partially distinct pathways or that obesity may be required to unmask XBP-1-dependent insulin resistance.
Over the past decade, it has become clear that inflammation is a common feature of obesity and Type-2 diabetes 42,57,58. In obese mice, there is a ~50% reduction in p85α expression in liver 59. These mice also display signs of heightened ER stress, including enhanced PERK phosphorylation, increased BiP expression and JNK1 activation 21. While there are many factors contributing to the insulin resistance in obesity, these data suggest a model where decreases in p85α lead to a reduced cellular response to ER stress via a reduction in XBP-1 stability and/or nuclear translocation and a concomitant decrease in ATF6α pathway activation.
One might predict that the failure of L-Pik3r1−/− mice to adequately resolve ER stress could, over long periods of time, lead to an enhanced inflammatory response. In this regard, we have found that on prolonged follow-up, L-Pik3r1−/− mice develop progressive inflammatory changes in the liver that culminates in the development of hepatoma (Taniguchi, et al, data unpublished). UPR dysregulation may underlie the ability of tumors to escape hypoxia-induced apoptosis and also play an important role in other diseases, including Huntington’s, Parkinson’s, amyotrophic lateral sclerosis, and Alzheimer’s disease. Further studies will be needed to determine if the UPR in these disorders is regulated in a p85-dependent fashion, and if altering the levels of p85 expression may be a therapeutic approach to reducing the effects of the UPR in disease pathogenesis.
Methods
Chemicals and materials
Tunicamycin, protease inhibitor cocktail (AEBSF, pepstatin A, E-64, bestatin, leupeptin, aprotinin) and streptavidin agarose was purchased from Sigma Aldrich. Thapsigargin was purchased from Santa Cruz Biotechnology.
Antibodies
Rabbit anti-BiP, anti-phospho-PERK (pPERK), anti-calnexin, anti-IRE1α, anti-PDI, and anti-Grp94 were from Cell Signaling Technologies. Rabbit anti-Calreticulin and anti-PERK were from Abcam. Rabbit polyclonal anti-phospho-IRE1α (S724) was from Novus Biologicals. Rabbit polyclonal anti-XBP-1 CT was from Biolegend. Monoclonal anti-ATF6 was from Imgenex. Monoclonal Anti-FLAG M2 antibody was from Sigma Aldrich. HRP-conjugated anti-actin was from Santa Cruz Biotechnology, Inc.
Mammalian tissue culture, transfection and sample preparation
Mouse brown pre-adipocytes, Huh7 hepatoma cells and Human Embryonic Kidney 293T (HEK 293T) cells were cultured (37°C and 5% CO2) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with streptomycin/penicillin and 10% FBS. FAO cell were cultured under identical conditions in Coon’s modified Ham’s F12 media supplemented with streptomycin/penicillin and 10% FBS. Transient transfections were performed on HEK293T cells at 80% confluence using Transit-Express as outlined by the manufacturer (Mirus Bio.). Following treatment, cells were washed two times in ice cold PBS (pH 7.4), collected by centrifugation at 2500 rpm for 5 minutes and resuspended in lysis buffer (50 mM Tris-HCl pH7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.1% SDS, 0.1% Triton X-100) supplemented with protease inhibitors. Cell extracts were spun at 14,000 rpm for 10 minutes at 4° to remove insoluble material.
Immunoprecipitation and streptavidin precipitation
Immunoprecipitations were performed on whole cell lysates prepared as previously described. Cleared lysates were diluted 10-fold in immunoprecipitation buffer (10 mM HEPES pH 7.9, 100 mM NaCl, 5% glycerol, 0.5% NP-40, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 10 mM β-glycerophosphate, 10 mM NaF) supplemented with protease inhibitors. Following two hours of incubation with HA-conjugated agarose, immunoprecipitates were washed three times with immunoprecipitation buffer and resuspended in Laemeli sample buffer prior to resolution by SDS-PAGE, transfer to PVDF and immunoblotting performed using the indicated antibodies. Streptavidin precipitations were performed using the same protocol with the exception that streptavidin conjugated agarose was used to precipitate TAP-p85α.
Analysis of gene expression by quantitative RT-PCR
Total RNA was purified using the RNeasy mini kit (Qiagen) with inclusion of an on-column DNase digestion. Two micrograms of total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). A portion of diluted cDNA was amplified with specific primers using a SYBR green PCR master mix (Applied Biosystems) on an ABI Prism 7500HT instrument. For each gene, mRNA expression was calculated relative to TBP as an internal control. Assay fidelity was assessed for each primer pair by performing melting-curve analysis. The nucleotide sequence of primers used for quantitative PCR are available upon request.
Recombinant adenovirus
The control GFP adenovirus was purchased from CellBioLabs. To create the TAP-p85α recombinant adenovirus, the full-length cDNA of human p85α was ligated into pNTAP, and adenovirus produced according to the standard AdEasy (Stratgene) protocol.
Analysis of apoptosis
Cells were incubated with growth medium alone or containing 2 μM tunicamycin or 100 μg/ml−1 thapsigargin for 24 hours. At the end of the incubation period, medium containing the floating cells was collected. Cells were rinsed with PBS, which was then added to the medium containing the floating cells. Attached cells were resuspended using trypsin and pooled with the floating cells. Cells were collected by centrifugation, washed once in PBS, and then resuspended in 400 μl of binding buffer (0.1 M HEPES, pH 7.4, 1.4 M NaCl, 25 mM CaCl2). Cells were then incubated with 5 μl annexin V coupled to PE (BD Biosciences, San Jose, CA) and 2.5 μg/ml−1 propidium iodide for 15 minutes at room temperature and submitted to FACS analysis.
Animals
All animals were housed on a 12-hour light-dark cycle and fed a standard rodent chow. All protocols for animal use, tunicamycin administration and euthanasia were approved by the Animal Care Use Committee of the Joslin Diabetes Center and Harvard Medical School in accordance with National Institutes of Health guidelines. All mice used in the study were on a 129Sv-C57BL/6 mixed genetic background. Tunicamycin was administered by intraperitoneal injection at a final dose of 2.5 μg/g BW in saline/150 mM glucose solution as previously described 60. Control animals were administered saline/150mM glucose by intraperitoneal injection.
Liver immunoblot analysis
Liver protein was prepared in a tissue homogenization buffer (25 mM Tris-HCl, (pH 7.4), 10 mM Na3VO4, 100 mM NaF, 50 mM Na4P2O7, 10 mM EGTA, 10 mM EDTA, 1% NP-40, 0.1% SDS) supplemented with protease inhibitors. Insoluble protein was cleared by centrifugation at 55,000 rpm, and protein concentrations of cleared lysates were determined by the method of Bradford. All protein expression data were quantified by densitometry using NIH Image J software.
Statistics
Data are presented as ± S.E.M. Two-tailed Student’s t-test were performed for statistical analysis between two groups.
Supplementary Material
Acknowledgments
We would like to that D. Ron (Rockefeller) for providing the FLAG-XBP-1u and FLAG-XBP-1s expression plasmids. This work was supported by NIH grant DK55545 and the NIH training grant DK07260-30, as well as Core laboratory support from the Joslin DERC grant DK36836. We would also like to thank Dr. Umut Ozcan for useful advice and discussion, and Sarah Green and Shannon Flaherty for assistance in preparation of this manuscript.
Abbreviations Used
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor 6
- BiP
glucose-regulated protein, 78kD
- CHOP
C/EBP homologous protein
- CD68
scavenger receptor class D, member 1
- ER
endoplasmic reticulum
- ERdj4
endoplasmic reticulum DnaJ homolog 4
- GADD34
growth arrest and DNA-damage-inducible 34
- GRP 95
glucose-regulated protein, 95kD
- IRE1α
inositol requiring 1α
- p58IPK
Protein kinase inhibitor of 58 kDa
- PDI
protein disulfide isomerase
- PERK
double-stranded RNA-dependent protein kinase (PKR)-like ER kinase
- PI 3-kinase
phosphatidylinositol-3 kinase
- TNF
tumor necrosis factor
- TRB3
Tribbles homolog 3
- UPR
unfolded protein response
- XBP-1
x-box binding protein 1
Footnotes
Author Contributions
J.W. created the hypothesis, designed and performed the experiments, analyzed the data and wrote the manuscript. J.B. and M.M. designed and performed the experiments and analyzed the data. K.U. created the hypothesis and generated reagents. C.R.K. created the hypothesis, helped analyze the data and wrote the manuscript.
References
- 1.Gotto AM, Jr, et al. The metabolic syndrome: a call to action. Coron Artery Dis. 2006;17:77–80. doi: 10.1097/00019501-200602000-00013. [DOI] [PubMed] [Google Scholar]
- 2.Ford ES, Giles WH, Mokdad AH. Increasing prevalence of the metabolic syndrome among u.s. Adults. Diabetes Care. 2004;27:2444–2449. doi: 10.2337/diacare.27.10.2444. [DOI] [PubMed] [Google Scholar]
- 3.Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol. 2004;25:4–7. doi: 10.1016/j.it.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 5.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest. 2003;112:1785–1788. doi: 10.1172/JCI20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanhaesebroeck B, Stein RC, Waterfield MD. The study of phosphoinositide 3-kinase function. Cancer Surv. 1996;27:249–70. 249–270. [PubMed] [Google Scholar]
- 7.Cheatham B, et al. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma PM, et al. Inhibition of phosphatidylinositol 3-kinase activity by adenovirus-mediated gene transfer and its effect on insulin action [In Process Citation] J Biol Chem. 1998;273:18528–18537. doi: 10.1074/jbc.273.29.18528. [DOI] [PubMed] [Google Scholar]
- 9.Alessi DR, Downes CP. The role of PI 3-kinase in insulin action. Biochim Biophys Acta. 1998;1436:151–164. doi: 10.1016/s0005-2760(98)00133-7. [DOI] [PubMed] [Google Scholar]
- 10.Cusi K, et al. Insulin resistance differentially affects the PI 3-kinase- and MAP kinase-mediated signaling in human muscle. J Clin Invest. 2000;105:311–320. doi: 10.1172/JCI7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heydrick SJ, Gautier N, Olichon-Berthe C, Van Obberghen E, Le Marchand-Brustel Y. Early alteration of insulin stimulation of PI 3-kinase in muscle and adipocyte from gold thioglucose obese mice. Am J Physiol. 1995;268:E604–612. doi: 10.1152/ajpendo.1995.268.4.E604. [DOI] [PubMed] [Google Scholar]
- 12.Bandyopadhyay GK, Yu JG, Ofrecio J, Olefsky JM. Increased p85/55/50 expression and decreased phosphotidylinositol 3-kinase activity in insulin-resistant human skeletal muscle. Diabetes. 2005;54:2351–2359. doi: 10.2337/diabetes.54.8.2351. [DOI] [PubMed] [Google Scholar]
- 13.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobson CM. Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Schroder M, Kaufman R. ER stress and the unfolded protein response. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 16.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 17.Elouil H, et al. Acute nutrient regulation of the unfolded protein response and integrated stress response in cultured rat pancreatic islets. Diabetologia. 2007;50:1442–1452. doi: 10.1007/s00125-007-0674-4. [DOI] [PubMed] [Google Scholar]
- 18.Lee AS. Mammalian stress response: induction of the glucose-regulated protein family. Curr Opin Cell Biol. 1992;4:267–273. doi: 10.1016/0955-0674(92)90042-b. [DOI] [PubMed] [Google Scholar]
- 19.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 20.Boden G, et al. Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Diabetes. 2008;57:2438–2444. doi: 10.2337/db08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozcan U, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 22.Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev. 2008;29:42–61. doi: 10.1210/er.2007-0015. [DOI] [PubMed] [Google Scholar]
- 23.Gregor MF, et al. Endoplasmic Reticulum Stress is Reduced in Tissues of Obese Subjects after Weight Loss. Diabetes. 2008 doi: 10.2337/db08-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozawa K, et al. The endoplasmic reticulum chaperone improves insulin resistance in type 2 diabetes. Diabetes. 2005;54:657–663. doi: 10.2337/diabetes.54.3.657. [DOI] [PubMed] [Google Scholar]
- 25.Ozcan U, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanti JF, et al. Alteration in insulin action: role of IRS-1 serine phosphorylation in the retroregulation of insulin signalling. Ann Endocrinol (Paris) 2004;65:43–48. doi: 10.1016/s0003-4266(04)95629-6. [DOI] [PubMed] [Google Scholar]
- 27.Gao Z, et al. Inhibition of insulin sensitivity by free fatty acids requires activation of multiple serine kinases in 3T3-L1 adipocytes. Mol Endocrinol. 2004;18:2024–2034. doi: 10.1210/me.2003-0383. [DOI] [PubMed] [Google Scholar]
- 28.Herschkovitz A, et al. Common inhibitory serine sites phosphorylated by IRS-1 kinases, triggered by insulin and inducers of insulin resistance. J Biol Chem J Biol Chem. 2007;282:18018–18027. doi: 10.1074/jbc.M610949200. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 30.Ron D, Hubbard SR. How IRE1 reacts to ER stress. Cell. 2008;132:24–26. doi: 10.1016/j.cell.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 31.Ueki K, Fruman DA, Yballe CM, Fasshauer M, Klein J. Positive and Negative Roles of p85α and p85β Regulatory Subunits of Phosphoinositide 3-Kinase in …. Journal of Biological Chemistry. 2003 [Google Scholar]
- 32.Terauchi Y, et al. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat Genet. 1999;21:230–235. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi CM, et al. The p85alpha regulatory subunit of phosphoinositide 3-kinase potentiates c-Jun N-terminal kinase-mediated insulin resistance. Mol Cell Biol. 2007;27:2830–2840. doi: 10.1128/MCB.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forler D, et al. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotechnol. 2003;21:89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
- 35.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 36.Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J Biol Chem. 2006;281:5852–5860. doi: 10.1074/jbc.M509061200. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fonseca SG, Lipson KL, Urano F. Endoplasmic reticulum stress signaling in pancreatic beta-cells. Antioxid Redox Signal. 2007;9:2335–2344. doi: 10.1089/ars.2007.1790. [DOI] [PubMed] [Google Scholar]
- 39.Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes. 2002;51 (Suppl 3):S455–461. doi: 10.2337/diabetes.51.2007.s455. [DOI] [PubMed] [Google Scholar]
- 40.Eizirik DL, Cardozo AK, Cnop M. The Role for Endoplasmic Reticulum Stress in Diabetes Mellitus. Endocrine Reviews. 2007 doi: 10.1210/er.2007-0015. [DOI] [PubMed] [Google Scholar]
- 41.Nakatani Y, et al. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem. 2005;280:847–851. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- 42.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shepherd PR, Withers DJ, Siddle K. Phosphoinositide 3-kinase: the key switch mechanism in insulin signalling. Biochem J. 1998;333:471–490. doi: 10.1042/bj3330471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duronio V, Scheid MP, Ettinger S. Downstream signalling events regulated by phosphatidylinositol 3-kinase activity. Cell Signal. 1998;10:233–239. doi: 10.1016/s0898-6568(97)00129-0. [DOI] [PubMed] [Google Scholar]
- 45.Barbour LA, et al. Human placental growth hormone increases expression of the p85 regulatory unit of phosphatidylinositol 3-kinase and triggers severe insulin resistance in skeletal muscle. Endocrinology. 2004;145:1144–1150. doi: 10.1210/en.2003-1297. [DOI] [PubMed] [Google Scholar]
- 46.Khalfallah Y, Sassolas G, Borson-Chazot F, Vega N, Vidal H. Expression of insulin target genes in skeletal muscle and adipose tissue in adult patients with growth hormone deficiency: effect of one year recombinant human growth hormone therapy. J Endocrinol. 2001;171:285–292. doi: 10.1677/joe.0.1710285. [DOI] [PubMed] [Google Scholar]
- 47.Barbour LA, et al. Human placental growth hormone increases expression of the p85 regulatory unit of phosphatidylinositol 3-kinase and triggers severe insulin resistance in skeletal muscle. Endocrinology. 2004;145:1144–1150. doi: 10.1210/en.2003-1297. [DOI] [PubMed] [Google Scholar]
- 48.Luo J, Cantley LC. The negative regulation of phosphoinositide 3-kinase signaling by p85 and it’s implication in cancer. Cell Cycle. 2005;4:1309–1312. doi: 10.4161/cc.4.10.2062. [DOI] [PubMed] [Google Scholar]
- 49.Yeates LC, Gallegos A, Kozikowski AP, Powis G. Down regulation of the expression of the p110, p85 and p55 subunits of phosphatidylinositol 3-kinase during colon cancer cell anchorage-independent growth. Anticancer Res. 1999;19:4171–4176. [PubMed] [Google Scholar]
- 50.Fang D, Liu YC. Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat Immunol. 2001;2:870–875. doi: 10.1038/ni0901-870. [DOI] [PubMed] [Google Scholar]
- 51.García Z, et al. A PI3K activity-independent function of p85 regulatory subunit in control of mammalian cytokinesis. EMBO J. 2006;25:4740–4751. doi: 10.1038/sj.emboj.7601324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park SW, Zhou Y, Lee J, Lu A, Sun C, Chung J, Ueki K, Ozcan U. Regulatory subunits of PI3K, p85alpha and p85 beta, interact with XBP1 and increase its nuclear translocation. Nat Med. 2010 doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto K, et al. Transcriptional Induction of Mammalian ER Quality Control Proteins Is Mediated by Single or Combined Action of ATF6α and XBP1. Developmental Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 54.Mauvais-Jarvis F, et al. Reduced expression of the murine p85alpha subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. J Clin Invest. 2002;109:141–149. doi: 10.1172/JCI13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen D, et al. p50alpha/p55alpha phosphoinositide 3-kinase knockout mice exhibit enhanced insulin sensitivity. Mol Cell Biol. 2004;24:320–329. doi: 10.1128/MCB.24.1.320-329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brachmann SM, Ueki K, Engelman JA, Kahn RC, Cantley LC. Phosphoinositide 3-kinase catalytic subunit deletion and regulatory subunit deletion have opposite effects on insulin sensitivity in mice. Mol Cell Biol. 2005;25:1596–1607. doi: 10.1128/MCB.25.5.1596-1607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hotamisligil G. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 59.Kerouz NJ, Hörsch D, Pons S, Kahn CR. Differential regulation of insulin receptor substrates-1 and -2 (IRS-1 and IRS-2) and phosphatidylinositol 3-kinase isoforms in liver and muscle of the obese diabetic (ob/ob) mouse. J Clin Invest. 1997;100:3164–3172. doi: 10.1172/JCI119872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marciniak SJ, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.