Abstract
Many ancient parchments are defaced by red or purple maculae associated with localized destruction of collagen fibres. Although the main characteristics of this damage were present in most of the manuscripts analysed by many authors, no common microbial or fungal denominator has been found so far, and little or no correspondence between the microbial or fungal species isolated from materials could be addressed. In this study, culture-independent molecular methods and scanning electron microscopy (SEM) were used to identify fungal and bacterial communities on parchments affected by the purple stains. Protocols for c extraction and nucleic-acid-based strategies were selected for assays examining the community structure of fungi and bacteria on biodeteriorated parchment. Both SEM and molecular analysis detected the presence of bacterial and fungal cells in the damaged areas. Halophilic, halotolerant proteolytic bacterial species were selected by the saline environment provided by the parchment samples. As common microbial denominators, members of the Actinobacteria, mainly Saccharopolyspora spp. and species of Aspergillus, were detected in all investigated cases. It is proposed that a relationship exists between the phenomenon of purple spots on ancient parchments and that of the ‘red heat’ phenomenon, known to be present in some products manufactured with marine salt.
Introduction
Parchment has been used as a writing material for many centuries. Some historical parchments represent the cultural heritage of mankind. Parchment is produced from animal skin and consists of a semi-solid matrix of collagen (Reed,1972,1975). During parchment manufacture, animal skin undergoes a series of procedures which results in a product composed of the dermal skin layer only (Ryder, 1969). In contrast to papermaking, parchment manufacture did not involve industrial-scale production; and so each specimen of the material is practically unique. Parchment manufacture was carried out in different ways, depending on the geographical area and historical period of production. After the fourth century, hair-removal was achieved by immersing the animal skins in a calcium hydroxide solution (Poole and Reed, 1962). Powders and pastes of calcium compounds were also used to help in the removal of grease (Bicchieri et al., 2011), so as to prevent applied inks from running. To give the parchment a smooth, white surface, lime, flour, egg whites and milk could be added to the surface. Salting (mainly with marine salt) could also be used in the first phases of parchment manufacture. In particular, salts (mainly sodium or potassium chlorides, ammonium chloride or sulphate, in addition to lime) were used at the beginning of the manufacturing process to inhibit microbial activity and prevent putrefaction of the hides (Reed, 1972) prior to their transformation into parchment. Sea sodium and potassium chlorides were used mainly in the coastal regions or, at least, wherever sea salt was cheap and available in large quantities. The salting was applied immediately after the skinning of the animal, to preserve the hides before the calcination step.
Parchment is quite a resistant material, but its deterioration can be due to a combination of exposure to light, elevated temperatures, humidity, atmospheric pollutants and microorganisms that cause both chemical and structural modifications (Florian, 2007). Studies conducted by Gallo and Strzelczyk (1971) and by Pinzari and colleagues (2012) showed that the microbial alterations detected more frequently on ancient parchments have the following characteristics: red or purple maculae, with a nucleated peripheral halo, isolate or coalescent (Fig. 1). In addition, it was noted that the development of microbial agents is always more intense on the flesh side, which sometimes appears completely destroyed, while the hair side, in many cases, remains practically intact. In the areas most affected by microorganisms, parchment becomes rough, assumes a diffuse staining and a porous appearance, and sometimes becomes perforated (Karbowska-Berent and Strzelczyk, 2000). Gallo and Strzelczyk (1971) observed that some inks inhibit the growth of microorganisms, and that microbial growth is generally more significant on the margins and on the first and last pages of the codex. However, the reproduction in vitro of the purple spots, by means of new parchment inoculation with different microbial strains, led to different results, with none of the permitted experiments obtaining such darkly pigmented purple stains (Gallo and Strzelczyk, 1971; Karbowska-Berent and Strzelczyk, 2000).
Figure 1.
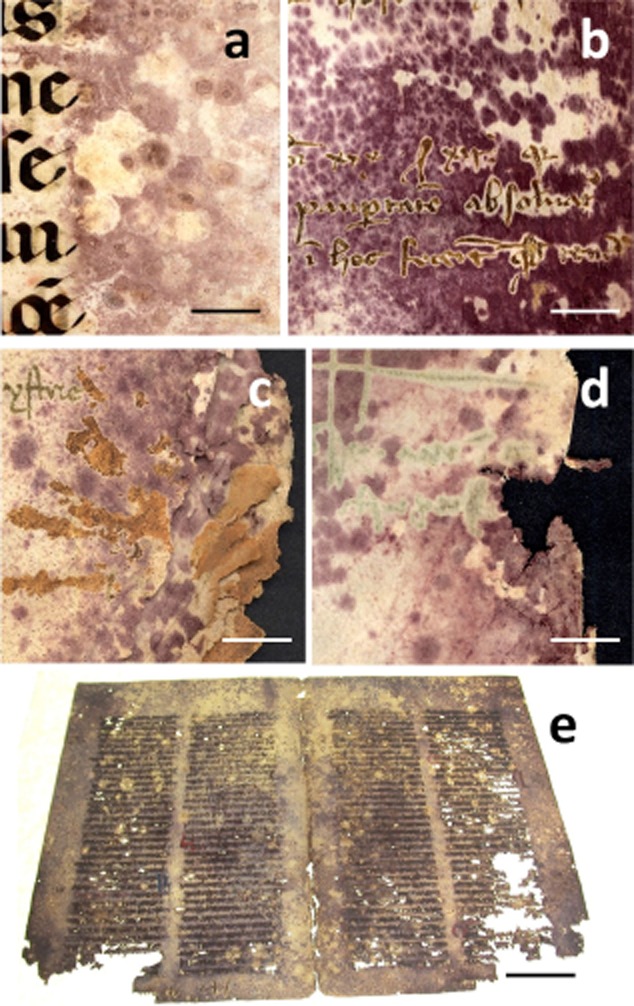
The measles-like purple stains on ancient parchment. In the pictures, five of the case studies analysed are showed:A. MS-492 (bar = 1 cm).B. Ve-21 (bar = 1 cm).C. Bo-Arch (bar = 1 cm).D. Ve-3 (bar = 50 mm).E. 148D (bar = 5 cm).
The attractive features of these alterations led scholars to carry out a series of surveys in Italian and foreign libraries, to see whether the manuscripts on parchment exhibited the same damage (Gallo and Strzelczyk, 1971; Tardieux et al., 1971; Woods et al., 1973; Kowalik, 1980; Voronina et al., 1980; Petushkova et al., 1984; Szabó and Szabó, 1984; Rebrikova and Solovyova, 1987; Polacheck et al., 1989,2005; Beöthy-Kozocsa et al., 1990; Orlita, 1993; Petushkova and Koestler, 1996; Rebrikova and Dmitrieva, 1996; Karbowska-Berent and Strzelczyk, 2000; Polacheck, 1989; Kraková et al., 2012). According to Karbowska-Berent and Strzelczyk (2000), Streptomycetes comprise the bacterial group that plays a major role in the deterioration of many kinds of ancient documents and books made of parchment. These filamentous bacteria are known to produce many types of enzymes, especially collagenases (proteases) that are capable of destroying collagen by their hydrolytic activity. They are alkaliphiles and are, therefore, inclined to develop on parchments (whose surfaces are prepared by rubbing with chalk). All the studies mentioned above reveal that, although the main characters of the damage represented by a dense pattern of purple spots, were present in most of the manuscripts analysed by as many authors, so far a common microbial or fungal denominator has not been found, and little or no correspondence between the microbial or fungal species isolated from materials could be addressed. In our opinion, this may be attributed to the fact that microbiology research carried out in this field up until now has been based mainly on classical cultivation methods. Although these investigations were helpful in demonstrating the importance of microorganisms in deterioration processes, the results obtained covered only those few organisms that could be cultivated. In addition, extensive cultivation strategies require more sample material than could be obtained from such valuable objects.
To overcome these problems, in the last decade, molecular biology has been used and widely applied to the study of cultural assets (Piñar et al., 2001a; Schabereiter-Gurtner et al., 2001; Laiz et al., 2003; González and Saiz-Jimenez, 2005; Michaelsen et al., 2006; Ettenauer et al., 2012). Molecular techniques allow the identification of single microbial species in sample material without the cultivation of the organisms. Most of the experiments carried out in this field are based on ribosomal sequences, or ITS (internal transcribed spacer) regions, which are nested in the rDNA repeat and are used as phylogenetic markers. In addition, the amount of data which can be used to compare the DNA sequences of unknown microorganisms to obtain a phylogenetic identification existing in the databases is growing every day. Another technique used for the analysis of objects of cultural heritage, and which may be used as a valuable approach to examine biodeteriorated parchment, is scanning electron microscopy (SEM) (Pinzari et al.,2010,2012). SEM analyses have revealed two common types of microbial deterioration of parchment: formation of cracks between the bundles of fibrils and disintegration of fibrils as a result of vital activity of microorganisms.
In this study, culture-independent molecular methods in combination with microscopical and microanalytical techniques were used to identify fungal and bacterial communities on parchments affected by the typical purple stains (Fig. 1). Efforts were made to find a microbial denominator and to see whether a correspondence between the bacterial or fungal species detected on the parchments and the observed deterioration phenomenon could be established. As already applied for environmental microbial communities and paper biodeterioration, nucleic-acid-based strategies targeting rRNA-encoding regions were selected for assays examining the community structure of fungi and bacteria on biodeteriorated parchment. The protocols for DNA extraction from parchment were evaluated.
Results and discussion
SEM results
Collagen lyses and deep structural damage were documented by SEM imaging on all the samples that presented the purple stains (Fig. 2), suggesting that the organisms that caused the damage were capable of using collagen as carbon source. The comparison between unstained areas and purple/damaged areas showed a typical pattern of degradation, with formation of cracks between the bundles of fibrils, disintegration of the matrix [according to Reed (1972), mainly made of waxes, lipids and some globular proteins] that holds together the fibrils, with the result that the fibres appear loose and their surface no longer smooth and homogeneous (Fig. 2A–G). The physical association and the adhesion of bacterial cells and fungal hyphae to fibres and fibrils is clearly documented in the SEM images obtained from the stained areas, while in unstained areas the presence of microbial and fungal cells was not observed. The distinction between healthy fibres and biodeteriorated ones was possible only in some samples (i.e. Samples Ve-3 and Ve-21) because the studied cases were valuable objects and only small damaged parts from the margins of the pages, which appeared not repositionable during the conservation treatments, could be used for SEM observations.
Figure 2.
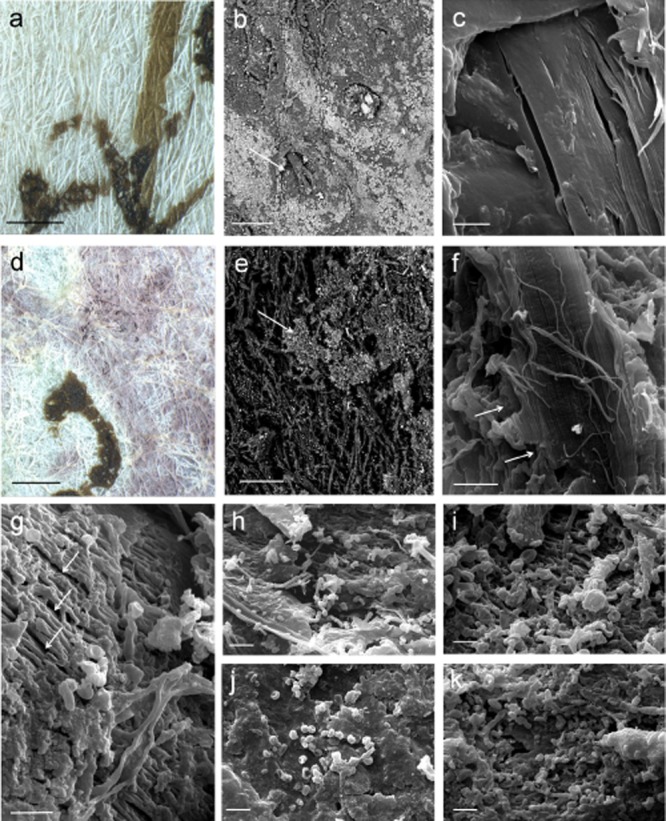
Comparison of healthy and damaged parchment:A. Detail of the surface of unstained Ve-21 parchment obtained with a Leica MZ16 stereomicroscope, the brown sign is the ink of the writing (bar = 1 mm).B. SEM image of a healthy Ve-21 parchment sample. The picture was obtained at variable pressure (50Pa) with a backscattered electron detector on not metallized material. The arrow indicates a hair follicle and a portion of hair. The surface is covered with mineral particles appearing lighter according to their higher atomic number (bar = 100 μm).C. High-vacuum secondary electrons SEM image on gold sputtered samples of a healthy Ve-21 parchment sample. The collagen fibres appear smooth and homogeneous (bar = 2 μm).D. Detail of the surface of purple stained Ve-21 parchment obtained with a Leica MZ16 stereomicroscope, the brown sign is the ink of the writing (bar = 1 mm).E. SEM image of biodeteriorated Ve-21 parchment sample. The picture was obtained at variable pressure (50Pa) with a backscattered electrons detector on not metallized material. The arrow indicates the original surface which was covered with mineral particles appearing lighter according to their higher atomic number (bar = 100 μm).F. High-vacuum secondary electrons SEM image on gold sputtered samples of a biodeteriorated Ve-21 parchment sample. Fibrils of collagen can still be appreciated (arrow), but a general breakdown of the material is present (bar = 2 μm).G. High-vacuum secondary electrons SEM image on gold sputtered samples of a biodeteriorated Ve-3 parchment sample. It can be seen the formation of cracks between the bundles of fibrils, and disintegration of the matrix that holds together the fibrils (arrows) (bar = 2 μm).H–I. Chains of bacterial spores and filaments on sample 148D consistent with Actinomycetales order (a, bar = 3 μm; b, bar = 1 μm).J. Bacterial spores (about 600 nm diameter) on sample Ve-21 (bar = 1 μm).K. Chains of bacterial spores and filaments on sample Ve-21 consistent with Actinomycetales order (bar = 1 μm).
Variable pressure backscattered electrons detector (VP-BSD) SEM imaging allowed the comparison between the patterns showed by salts on parchment surface in damaged and healthy areas, and showed that, in most samples, a sort of dislocation of salts occurred (Fig. 2E). The appearance of the damaged samples indicated that the microbial attack was very often associated with the leaching of the mineral components of the surface of the parchment or, at least, with the consumption of the outer layers of the material.
The observation of gold-coated samples, by means of SEM in high-vacuum mode, showed the presence of both bacterial and fungal cells in the damaged areas (Figs 1–4). A very complex biodeterioration mechanism emerged from the study, with both bacterial and fungal species involved in the deterioration of parchment, or, at least, growing in a succession of events. The bacterial cells can be distinguished from fungal conidia by means of their small dimensions (less than 1 micron in diameter) and their shape (Fig. 2H–K). Both single bacterial cells and chains of bacterial spores could be seen in most of the samples (Fig. 2H–K). Only the sample To800 displayed a prevalent attack by one species of fungus, which caused deep holes in parchment and produced masses of spores (Fig. 3). The samples used in this study had no SEM preparatory treatment because they were already air dried, and so the dehydrated fruiting structures like conidia, conidiophores and spores had already shrunk and become distorted. With this in mind, some fungal species observed in samples (Figs 4 and 3) could be cross-checked with the species list that emerged from the results of molecular cloning and sequencing of the DNA directly extracted from the materials (Fig. 5 and Supporting Information Tables S1 and S2). In the case of sample To800, the fungal structures correspond to a Chaetomium species. The sample To800 consisted of a parchment cover glued to cardboard, together forming the binding of a volume. The presence of C. globosum was initially associated with the presence of cellulose fibres attached to parchment, but, on closer observation, as documented by SEM imaging, an active role of the fungus in parchment spoilage could be assessed. Figure 3C and D shows Chaetomium's fruiting bodies embedded in the surface of the parchment. These structures, which at their production consist of ostiolar perithecia covered in hairs, appear open and partially broken but still recognizable. The fungus mycelia and parchment fibres appear intertwined and the fruiting bodies co-penetrated inside the material.
Figure 4.
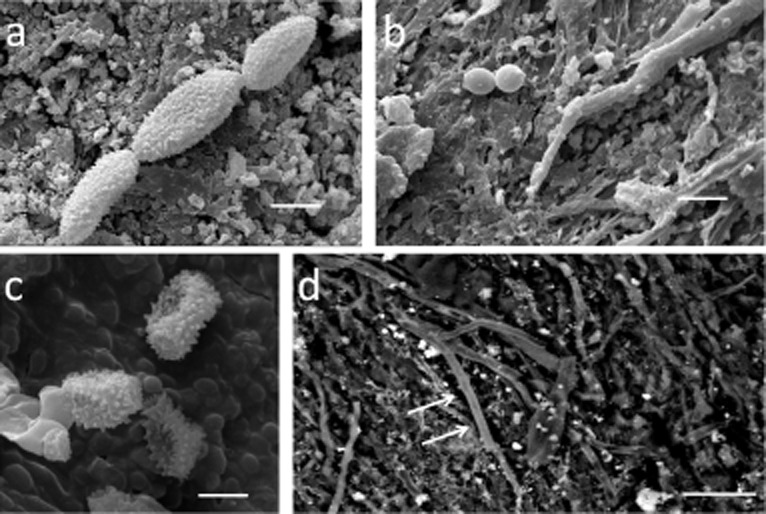
SEM imaging of fungi on parchment samples:A. HV-SEM on gold sputtered samples, chains of verrucolose conidia and consistent with some species of Cladosporium and Toxicocladosporium were observed on MS-492 sample (bar = 5 μm).B. HV-SEM on gold sputtered samples, obovoid conidia with a truncate base, consistent with Phialosimplex, were documented on Ve-3 (bar = 10 μm).C. HV-SEM on gold sputtered samples, echinated conidia and conidiophora resembling a species of Penicillium were documented on samples Ve-3 and Ve-21 (bar = 2 μm).D. VP-BSD-SEM, surface of the parchment sample Bo-Arch with fungal hyphae (arrows) growing adherent to collagen fibres (bar = 20 μm).
Figure 3.
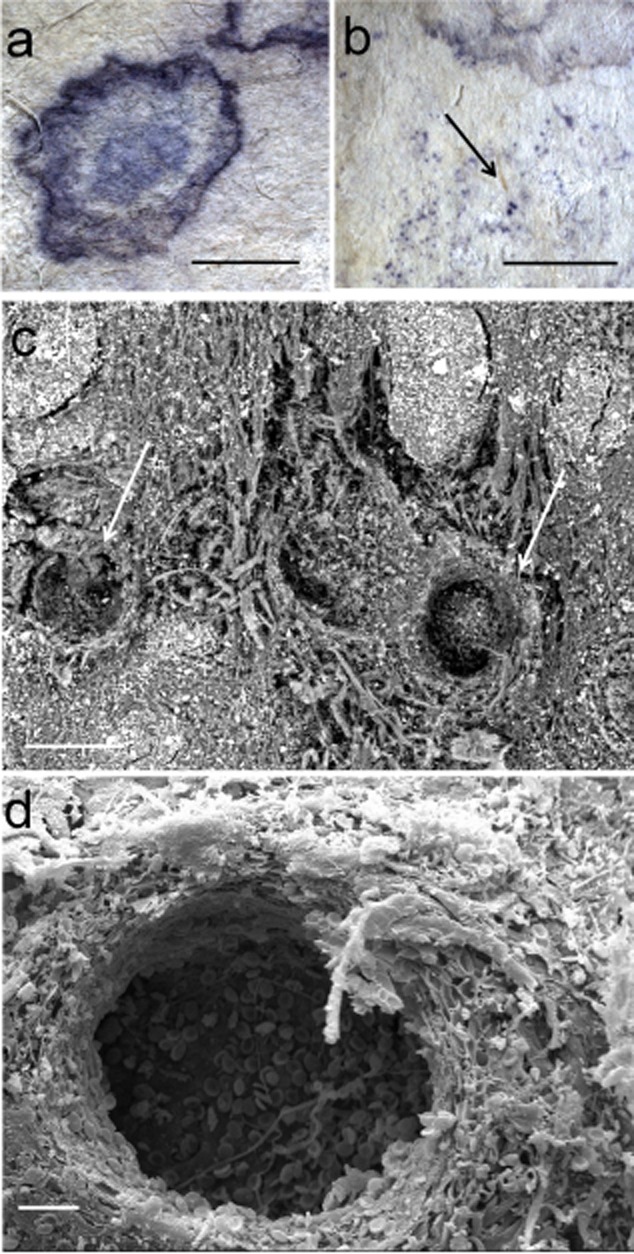
Purple stains from sample To800:A–B. Details of the stains obtained with a Leica MZ16 stereomicroscope (bar = 2 mm); The arrow in panel B indicates spots corresponding to Chaetomium sp. peritecia, immersed in the substrate (bar = 5 mm).C. SEM picture obtained at variable pressure (50Pa) with a backscattered electrons detector on not metallized material. Chaetomium fruiting structures immersed in parchment (bar = 100 μm).D. HV SE SEM image on a gold sputtered sample of the fractured fungal peritecia immersed in parchment (bar = 20 μm). The hole is produced by the fungus and consists of the inner walls of the fruiting body. On the on the bottom of the crater there are some ascospores.
Figure 5.
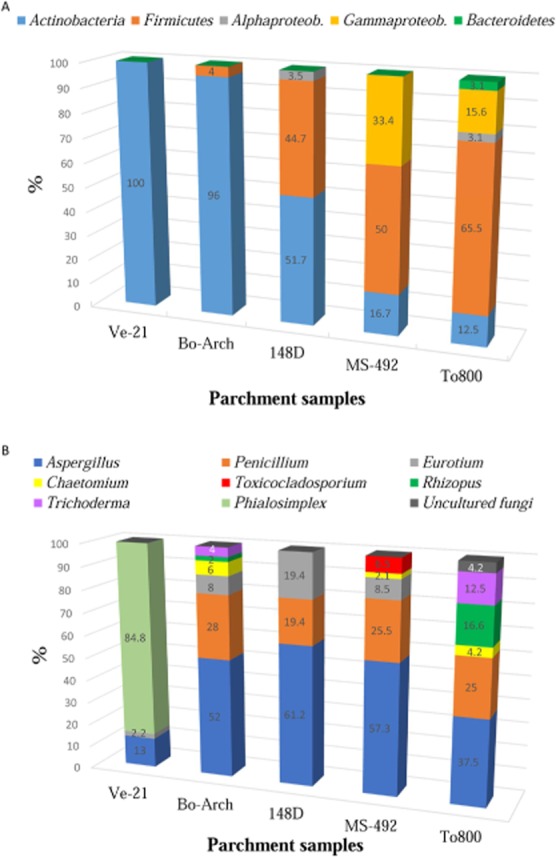
Distribution of major phylogenetic groups on the different parchment samples (in percentages):A. Bacteria.B. Fungi.
Chains of verrucolose conidia and ramoconidia consistent with some species of Cladosporium and Toxicocladosporium genera were observed (Fig. 4A and Supporting Information Table S2). Conidia in chains, smooth, pyriform, obovoid with a truncate base, consistent with Phialosimplex, were documented on Ve-3 (Fig. 4B and Supporting Information Table S2). Echinated conidia and conidiophora resembling a species of Penicillium were also documented on samples Ve-3 and Ve-21 (Fig. 4C).
Moreover, samples 148D and Ve-21 showed similar damage at the level of collagen fibres, and both the samples were covered with a mass of bacterial spores mainly in chains, mixed with very thin (less than 1 μm in diameter) pseudohyphae, resembling the typical structure of some Actinomycetes (Fig. 2I). The description of Saccharopolyspora salina (see Supporting Information Table S1) is consistent with some bacterial structures documented by high-voltage (HV)-SEM imaging. The genus Saccharopolyspora belongs to the family Pseudonocardiaceae (order Actinomycetales) and it is known for producing antibiotics such as vancomycin, erythromycin and rifamycins. Members of the genus Saccharopolyspora are characterized by aerobic, extensively branched hyphae that fragment into rod-shaped elements and aerial hyphae that segment into bead-like chains of spores. These organisms grow well between 6–18% NaCl concentrations, and the absence of NaCl in the medium inhibits their growth.
Energy dispersive X-ray spectroscopy EDS results
Table 1 reports the average values, by percentage weight, of chemical elements present on all the surface samples analysed. Although EDS is a not quantitative analytical method because of the non-uniform density of the analysed material (parchment), a comparison performed on a statistically substantial number of measurements, conducted by careful standardization of the instrumental parameters, may still hold value for the description of the samples. A comparison between damaged areas and integer parchment surfaces could be performed only in the case of the specimen Ve-3, where unstained areas (V-3i, intact) could be distinguished from the damaged ones (V-3b, biodeteriorated) in the material that could be collected.
Table 1.
Microanalysis (EDS) results
| C | O | Na | Mg | Al | Si | P | S | Cl | K | Ca | Fe | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 148D | 56.91 | a | 31.05 | a | 0.04 | a | 0.67 | a | 0.29 | a | 1.21 | a | 0.61 | b | 0.83 | a | 0.62 | c | 0.21 | a | 7.34 | b | 0.23 | a | ||||||||||
| Bo-Arch | 60.79 | a | 29.72 | a | 0.63 | c | 0.22 | a | 0.72 | a | 1.23 | a | 0.27 | a | 1.83 | a | b | 0.61 | b | c | 1.04 | a | b | 2.61 | a | 0.31 | a | |||||||
| MS-492 | 61.75 | a | 30.27 | a | 0.31 | b | 0.33 | a | 0.62 | a | 0.96 | a | 0.07 | a | 0.65 | a | 0.44 | b | c | 0.61 | a | 3.62 | a | 0.38 | a | |||||||||
| To800 | 63.83 | a | 30.93 | a | 0.03 | a | 0.25 | a | 0.53 | a | 0.57 | a | 0.18 | a | 1.04 | a | 0.01 | a | 0.22 | a | 2.35 | a | 0.04 | a | ||||||||||
| Ve-21 | 58.66 | a | 30.71 | a | 0.09 | a | 0.38 | a | 0.14 | a | 0.69 | a | 0.17 | a | 2.30 | a | b | 0.62 | c | 1.94 | b | 4.27 | a | 0.02 | a | |||||||||
| Ve3_i | 61.23 | a | 31.08 | a | 0.46 | b | c | 0.11 | a | 0.48 | a | 0.45 | a | 0.11 | a | 0.85 | a | 0.30 | a | b | c | 0.53 | a | 3.77 | a | 0.63 | a | |||||||
| Ve3_b | 60.88 | a | 29.97 | a | 0.33 | b | 0.02 | a | 0.00 | b | 0.01 | b | 0.13 | a | 3.23 | b | 0.28 | a | b | 0.37 | a | 4.78 | a | b | 0.00 | a |
Comparison of elemental composition of parchment samples. Data are reported as weight %. One-way ANOVA/Tukey's t-test comparisons are reported: Statistically significant differences (P < 0.001) in elemental composition is marked with different letters (a, b, c). Sample ‘Ve-3’ was measured in unstained areas (V-3i, intact) and in damaged ones (V-3b, biodeteriorated).
Nitrogen could not be measured, owing to the well-known important matrix effect in the determination of nitrogen by SEM-EDS, which makes it impossible to construct a calibration curve that correlates the signal directly with the nitrogen content for any type of sample (Goldstein et al., 2003).
Almost all the samples exhibited the presence of the following elements: Na, Mg, Al, Si, P, S, Cl, K, Ca and Fe. The most abundant element was Ca; significantly higher in the sample 148D (around 7 w %), and between 2 w% and 4 w % in the other samples. The presence of Ca is attributable to the treatments based on lime and Ca oxides and hydroxides (CaO, Ca(OH)2) for de-hairing, and chalk (CaCO3) used to give parchment a whiter surface (Bowden and Brimblecombe, 2000,2002). Sulphur (S) was also an important presence, and it was higher in the samples Bo-Arch, Ve-3 and Ve-21. Na and Cl were present as traces in 148D, To800 and Ve-21, while being present in statistically significant higher concentrations in the other samples. Traces of Fe were found in all the samples, except in the biodeteriorated Ve-3 sample, although the observed differences were not statistically significant.
The microanalysis showed a meaningful difference between the chemical content of damaged and healthy areas of sample Ve-3. In fact, a higher content of Si and Al in the integer surface areas of parchment was measured. A lower concentration of salts in the biodegraded areas could be discerned in the micrographs obtained from the backscattered electron detector. The physical appearance of the damaged samples indicated that the microbial attack was associated with the leaching of the mineral components of the surface of the parchment (Fig. 2E). The presence of Si and Al can be attributed to the manufacturing of parchment, which involved abrading the skins on the flesh and hair side surfaces with pumice stone (composed of SiO2 and Al2O3), and whitening both sides of the parchment by applying thin pastes of chalk or mixtures of lime, flour, egg whites and milk.
The elements Mg and K were also present in all the samples, also as a result of manufacturing processes, in particular from chlorides that were used as preservatives in the drying conservation process. Mg and K, together with Fe, can also originate from the hide itself, since they occur naturally at low concentrations in leather and parchment (Larsen, 2002).
Overall, the samples had a high content of salts, which lent the material a specific nature, combining a prevalent principal proteic component (collagen), together with a series of mineral compounds to form a rather selective microhabitat for specialized microbial species.
DNA extraction, amplification and denaturing gradient gel electrophoresis (DGGE) fingerprinting
A total of seven samples, collected from different parchment documents (Fig. 1), were analysed by culture-independent techniques. It is worth remarking on the challenge of DNA extraction from parchments, due to the complex nature of the material and the contaminants, being salt used for their preparation, which interfere with the final purity of the DNA. As mentioned above, the presence of several elements was shown by EDS analysis in all the samples (Table 1), although in variable amounts. Na, Mg, Al, Si, P, S, Cl, K, Ca and Fe can be present in different chemical combinations to form salts and ionic forms that could have a negative effect on the protocols used in DNA extraction and, in any case, introduce a variability factor in the procedures.
The main problem observed when extracting DNA from parchments was the strong coloration of the supernatants throughout the whole DNA extraction, which could be eliminated only after a second purification step using the QIAamp Viral RNA mini kit, as explained in the section of Experimental Procedures. Before this purification step, the ratios A260/A230, assessed using a NanoDrop® ND-1000 Spectrophotometer, were determined to be < 2.0, indicating a contamination either with organic compounds or with salts present in the parchment samples. After this purification step, this ratio was determined to be > 2.0, the supernatants were colourless and all samples yielded pure DNA extracts with concentrations of 53.79–69.50 ng DNA μl−1. The DNA extracts were amplified by PCR, with primers targeting the 16S rDNA of bacteria and archaea as well as the ITS regions of fungi. PCR analysis using eubacterial-specific and fungal-specific primers yielded positive results for all samples, whereas archaeal-specific primers yielded no amplification of the extracted DNAs, confirming the absence of these microorganisms on the analysed parchment samples (data not shown).
The bacterial 16S rDNA and the fungal ITS amplified fragments were further analysed using DGGE fingerprints. This technique allowed an estimation of the most abundant organisms inhabiting the parchment samples and showed the putative differences and similarities in microbial composition among the different parchments analysed. The DGGE profiles obtained are shown in Supporting Information Fig. S1A (bacteria) and Supporting Information Fig. S1B (fungi). DGGE fingerprints derived from bacterial and fungal sequences revealed almost identical patterns for samples 148D and MXIV (see Supporting Information Fig. S1A and B, lanes 1 and 2 respectively). Therefore, only one of these samples was selected for further phylogenetic analyses: the sample 148D (Fig. 1E). From the two volumes of the collection ‘Grazie’, only the volume Ve-21 (Fig. 1B) was selected for further phylogenetic analysis.
Phylogenetic identification of the microbial communities colonizing parchment
To accomplish phylogenetic identification of the bacterial and fungal communities inhabiting the parchments, clone libraries containing the 16S rDNA or the ITS fragments were generated from five selected samples: 148D, MS-492, To800, Bo-Arch and Ve-21. Clones were screened by DGGE and those displaying different fingerprints were grouped. Finally, one representative of each group was selected for sequencing. The obtained sequences were compared with those of known microorganisms in the NCBI database. Supporting Information Tables S1 and S2 show the phylogenetic affiliations of the selected bacterial and fungal clones sequenced respectively and their percentages in the clone libraries. The comparative sequence analyses revealed similarity values ranging from 82–100% and 94–100% with sequences from the NCBI database for bacteria and fungi respectively.
Bacterial communities
Sequence analyses revealed that members of the phylum Actinobacteria were present in all samples (Fig. 5A), and showed to be dominant on samples Ve-21 (100% of the screened clones), Bo-Arch (96%) and 148D (51.7%). Nevertheless, on samples MS-492 and To800, this phylum accounted only for 16.7% and 12.5% of the screened clones respectively (see Supporting Information Table S1). Species of the genus Saccharopolyspora (Tang et al., 2009; Jurado et al., 2010) were present in all samples except sample To800, and dominated on sample 148D (all actinobacterial sequences) and on samples Ve-21 and Bo-Arch. However, in the case of sample Ve-21, some of these sequences (2.2%) also affiliated, with a higher similarity, with an uncultured Pseudonocardiaceae bacterium clone detected on skin from deteriorated mummies (Piñar et al., 2013) and with an uncultured bacterium clone (6.7%) from mould-colonized, water damaged building material (Schäfer et al., 2010) (see Supporting Information Table S1). The same was observed on sample Bo-Arch, where 68% of the sequences were most related to uncultured Pseudonocardiaceae bacterium clones and with a lower similarity (93–96% similarity) to Saccharopolyspora sp., 16% of the sequences showed to be most affiliated with Saccharopolyspora sp., and the rest of the actinobacterial sequences (12%) affiliated with Pseudonocardia sp. (Lee et al., 2000). On sample MS-942, the sequences related to Saccharopolyspora sp. accounted only for 4.2% of the screened clones and the rest of the actinobacterial sequences (12.5%) were related to Arthobacter sp. Sample To800 showed to be different, and sequences related to the phylum Actinobacteria were related to uncultured clones detected on skin microbiome, associated with disease in children with atopic dermatitis (Kong et al., 2012) and with cultured strains of Propionibacterium acnes, a commensal of human skin (Brüggemann et al., 2004) and related to acne (Fitz-Gibbon et al., 2013) (see Supporting Information Table S1).
Members of the phylum Firmicutes were present in all samples except sample Ve-21 (Fig. 5A). They dominated on samples To800 and MS-492 (65.5% and 50% of the screened clones respectively) and were also abundant on sample 148D (44.7% of clones). However, on sample Bo-Arch, members of this phylum accounted only for 4% of the screened clones. Species of the genus Bacillus were present in all these samples and accounted for 46.8%, 44.7%, 43.8% and 4% of the screened clones respectively for samples To800, 148D, MS-492 and Bo-Arch (see Supporting Information Table S1). These sequences affiliated with uncultured Bacillus sp. clones detected in stone monuments in Spain (Ettenauer et al., 2011) and with cultured Bacillus sp. isolated from saline environments (Berrada et al., 2012). In addition, sequences related to uncultured clones retrieved from skin microbiome (Grice et al., 2009), as well as to the cultured Geomicrobium halophilum (Echigo et al., 2010), were detected on sample To800 (18.7%). Furthermore, salt-tolerant strains of Halobacillus spp. (Piñar et al., 2001b,2009) were detected on sample MS-492 (6.2%).
The Proteobacteria (Alpha- or Gamma-classes) were found on samples 148D, MS-492 and To800 (Fig. 5A). Alphaproteobacteria were detected on samples 148D and To800 (3.5% and 3.1% of the screened clones respectively). These sequences were related to uncultured clones previously detected on skin microbiome, which were associated with disease in children with atopic dermatitis (Kong et al., 2012) and with cultured Methylobacterium spp. Members of the Gammaproteobacteria were detected on samples MS-492 and To800 (33.4% and 15.6% of the screened clones respectively). In the case of sample MS-492, these sequences were related to Stenotrophomonas maltophilia (12.5%), to halophilic Halomonas sp. (16.7%) and to Pseudomonas putida (4.2%). The sequences retrieved from sample To800 affiliated with Acinetobacter spp.
Finally, the Bacteroidetes phylum was detected only in sample To800 (Fig. 5A), accounting for 3.1% of the screened clones, and being the sequences most related to uncultured clones detected on skin microbiome, associated with disease in children with atopic dermatitis (Kong et al., 2012).
Fungal communities
Molecular analyses revealed that, among fungi, sequences related to Aspergillus and Penicillium spp. dominated on almost all investigated samples (Fig. 5B and Supporting Information Table S2), especially on samples 148D (61.2% and 19.4% of the screened clones respectively for Aspergillus and Penicillium spp.), MS-492 (57.3% and 25.5% respectively), Bo-Arch (52% and 28% respectively) and To800 (37.5% and 25% respectively). Nevertheless, on sample Ve-21, species of the genus Aspergillus accounted only for 13% of the screened clones, and species of the genus Penicillium were not detected. In addition, Eurotium spp. were detected on samples 148D (19.4%), MS-492 (8.5%), Bo-Arch (8%) and Ve-21 (2.2%), but not on sample To800 (see Supporting Information Table S2). Chaetomium globosum strains were detected on samples Bo-Arch (6%), To800 (4.2%) and MS-492 (2.1%).
The genus Aspergillus includes saprophytic species, but also organisms capable of living as opportunistic pathogens, it includes mesophilic species as well as extremophiles. Among the sequences that identified Aspergillus units at the species level, the A. versicolor sequences, occurring on different samples, and the A. niger sequences are noteworthy. Both these species have been associated with the production of extracellular alkaline proteases and could, therefore, have had a role in parchment spoliation (Kalpana Devi et al., 2008; Choudhary, 2012). A. versicolor was found to be associated with parchment and leather degradation by many authors (Gallo and Strzelczyk, 1971; Kowalik, 1980; Voronina et al., 1980; Kraková et al., 2012), and often found to be co-occurring with bacteria species. The retrieval of Penicillium chrysogenum can be readily associated with airborne contamination of the samples, since the density of the spores of this fungus in every sampled region of the planet, including Antarctica and the upper atmosphere, makes it among the most numerous eukaryotic species on Earth (Samson et al., 2002). Besides its very wide biogeography and occurrence, P. chrysogenum could have an active role in parchment deterioration since it is a fungus that can produce alkaline proteases and exhibit strong proteolytic attitudes (Haq et al., 2006). Moreover, the P. chrysogenum sequences detected in this study showed to be most affiliated with mycotoxin-producing strains on dry-cured ham in Spain (non-published), with penicillin-producing species (Houbraken et al., 2012) and with pathogenic moulds (Rakeman et al., 2005).
Eurotium species are sexual states of Aspergillus species, in particular those belonging to the Aspergillus glaucus group. The genus Eurotium contains several species that can grow well at low water activities. They are common in foods preserved with high concentrations of NaCl or sugar. Eurotium species are also important mycotoxin-producers (Samson and Von der Lustgraaf, 1978). The sample 148D showed the presence of E. halophilicum, which is a strongly xerophilic fungus with a marked tolerance to water stress. The observed water activity (Aw) for its germination and growth is 0.67, one of the lowest in the species from the Eurotium genus (Christensen et al., 1959). The occurrence of this fungus is associated by some authors with air-dust. (Samson and Von der Lustgraaf, 1978). However, some authors (Michaelsen et al., 2010; Montanari et al., 2012) reported its occurrence in library materials. E. chevalieri, according to Butinar and colleagues (2005), is an opportunistic airborne contaminant, while E. amstelodami probably represents part of the indigenous fungal community in salterns all over the world.
Chaetomium species are dematiaceous (dark-walled) ascomycetes in the Chaetomiaceae family. Members of this genus produce fruiting bodies both inside the materials and also on the surface, consisting of ostiolar perithecia, covered in hairs. These fungi have well-known cellulolytic abilities. In particular, Chaetomium globosum is frequently associated with paper spoilage. In this study, C. globosum strains were detected on three parchment samples (MS-492, To800 and Bo-Arch). As well as being an airborne contaminant, Chaetomium sp. is also encountered as a causative agent of infections in humans and animals, and as a parasite on a large variety of phytopathogenic fungi. Chaetomium species produces and releases different hydrolytic enzymes and antibiotics which can inhibit fungal growth and can also exhibit bacteriolytic attitudes (Imada et al., 1973). Chitinases from C. globosum have also been widely characterized, and cDNA sequences of a subtilisin-like serine protease gene were recently constructed from C. globosum. The proteolytic attitudes of this fungus were also reported by Florian (2007), who associated this species with some forms of silk biodeterioration. Brewer and colleagues (1968) reported the isolation of a purple pigment, which was termed cochliodinol, from isolates of Chaetomium cochliodes and C. globosum. Furthermore, the antifungal properties of cochliodinol have also been documented (Meiler and Taylor, 1970).
In addition to the above-mentioned genera, on sample MS-492, the remaining 6.5% of the screened clones showed to be affiliated with members of the order Capnodiales (Fig. 5B and Supporting Information Table S2), namely with a Toxicocladosporium irritans strain, which was previously isolated from a mouldy paint (Crous et al., 2007). This fungus was also found as one of the predominant species in skin infections in patients with atopic dermatitis (Zhang et al., 2011).
On sample To800, besides the sequences related to species of the genera Aspergillus, Penicillium and Chaetomium, 4.2% of sequences showed to be related to an uncultured fungus clone from urban air; 16.6% of sequences to Rhizopus oryzae strains, well known for the production of organic acids, such as fumaric, lactic and malic acids, as well as ethanol (Abe et al., 2007), and as a human pathogen being the major agent of mucormycosis (Nagao et al., 2005), and 12.5% of sequences affiliated with Trichoderma spp., as Trichoderma pseudokoningii strains with proteolytic activities (Chen et al., 2009). Sequences related to Rhizopus oryzae and Trichoderma spp. were also detected on sample Bo-Arch, representing 2% and 4% of the screened clones respectively (Fig. 5B and Supporting Information Table S2).
On sample Ve-21, besides the sequences related to species of the genera Aspergillus and Eurotium (Fig. 5B and Supporting Information Table S2), the rest of the sequences affiliated with uncultured and cultured Phialosimplex spp., being dominant in this sample (84.8% of screened clones). Species of this genus have been associated with human and animal infections (Gené et al., 2003; Sigler et al., 2010), and, more recently, associated with the biodeterioration of collagen-rich materials collected from human remains (Piñar et al., 2013).
In summary, the microbial communities inhabiting the investigated parchment samples were determined by the environment offered by the material itself. The infection and colonization of parchment by fungal and bacterial cells, spores and propagules take place mainly through air-dispersion, although direct inoculation of both fungi and bacteria by human handling or by insects and mites as vectors can occur. The parchment has a high hygroscopicity, i.e. it has a high affinity for water, which means that water is scarcely available for microorganisms (Florian, 2007). This property is mainly because of collagen, its main constituent, which is able to bind water by means of hydrogen bonds, because of the presence of many polar groups. Furthermore, the process of degradation of parchment occurs when the environmental humidity exceeds 65% and when the water content in the material exceeds 15% (Florian, 2007). In addition, the presence of hygroscopic salts, such as the minerals used in parchment manufacturing, give the material a peculiar quality in which the presence of compounds, such as chlorides and sulphates, makes the microenvironment even more selective against halophilic and osmophilic species. To further complicate the scenario, parchment pH can be strongly alkaline following the sizing procedures during manufacturing. The parchment microenvironment, therefore, can be selective also for haloalkalophiles, which are extremophilic microorganisms adapted to both saline and alkaline conditions. Haloalkalophilic microorganisms are usually characterized by peculiar adaptive mechanisms which avoid the denaturing effect of salts and balance their interior pH. Enzymes from archaeal and eubacterial haloalkalophilic organisms have unique properties. They have activity and stability at broad pH ranges and high concentrations of salts, at which levels many other proteins are denaturated. It has been reported that halophilic bacteria have a role in damage occurring in brine-cured hides with their high proteoliytic activity (Bailey and Birbir, 1993). Many Bacillus species are known to produce alkaline and saline stable proteases, amylases, cellulases, lipases, pectinases and xylanases (Martins et al., 2001; Berrada et al., 2012).
Therefore, the occurrence of halotolerant and halophilic species of the phyla Actinobacteria, as Saccharopolyspora spp., of the Firmicutes, as halophilic species of Bacillus, previously detected and/or isolated from saline environments and salt-tolerant strains of Halobacillus spp., and of the Gammaproteobacteria (as Halomonas) is widely explained by the chemical and physical characteristics of the microenvironment offered by the surface of the parchment. All of them, and also the detected species of the genera Arthrobacter and Methylobacter, which are often red-pigmented, may be responsible for the measles-like parchment discoloration as a result of the very common pigmentation of these species. Their cell membranes contain carotenoid pigments, such as β-carotene, α-bacterioruberin, and derivatives that appear primarily to protect the cells against photooxidative damage (Oren, 2009). These pigments may be related to the observed purple spots. In addition, it is worth remarking on the presence of sequences related to microorganisms from the human skin microbiome, even associated with skin diseases, such as some uncultured clones belonging to different phyla detected in this study and described as being associated with diseases in children with atopic dermatitis (Kong et al., 2012), as well as uncultured clones related to the deterioration of the skin of mummies (Piñar et al., 2013), and, in addition, sequences related to strains of Propionibacterium acnes, a commensal of human skin and associated with acne (Fitz-Gibbon et al., 2013). Furthermore, our results also showed a specific colonization of parchment materials by specialized microorganisms. Our sequences affiliated strongly with sequences of microorganisms well known for their proteolytic activities, as with Bacillus spp., showing extracellular hydrolytic enzyme activities, including proteolytic enzymes (Berrada et al., 2012) and with Stenotrophomonas maltophilia, a well-known keratinolytic and proteolytic bacterium (Cao et al., 2009). These microorganisms may be directly related to the deterioration of the parchments.
The identified Saccharopolyspora seems to be a common denominator in all the samples with purple stains having the features already described. The only sample in which sequences related to species of Saccharopolyspora were absent was the sample To800, but on closer inspection, the coloured spots that mark the surface of the material of this sample (Fig. 3A) are very different from those documented in the other case studies. In the first place, the colour in To800 is more bluish, and also the contours are less rounded, and the spot appearance not nucleated. In addition, the sample To800, as previously mentioned, is profoundly altered by the presence of C. globosum, a fungus capable of producing pigments from pink to blue as a result of the production of coloured metabolites.
One hypothesis that we can advance about the measles-like parchment discoloration is that there is, in fact, a common denominator, namely Saccharopolyspora species and allied species related to the presence of salt, NaCl in particular, in the material. A very similar phenomenon is the so called ‘red heat’, a typical red colouration found as a pitting of the surface on the flesh side of salted hides after storage during leather manufacturing. Some authors established that it is a damage caused by aerobic salt-tolerant (halophilic) bacteria (Lochhead, 1934), which come from the salt, being typical inhabitants of sea salt deposits, and which grow very slowly, producing rhodopsin-like pigment (Grote and O'Malley, 2011). This pigment is a light-sensitive protein which provides chemical energy for the bacterial cell by using sunlight to pump protons out of the cell. The origin of salt may influence the development of red heat, with marine salt containing high levels of halophilic bacteria compared to rock salt (Trüper and Galinski, 1986). Materials other than hides can also exhibit the red-pitted putrescence phenomena, having in common some form of contact with sea salt, such as the salted summer herring studied by Luijpen and colleagues (1953).
Conclusions
The strategy used in this study, combining microscopical and molecular analyses, proved to be a reliable tool to monitor the microbiota inhabiting different parchment samples and to establish a relationship between the observed types of deterioration suffered by the parchments investigated.
Results evidenced the following conclusions: As common microbial or fungal denominators, members of the Actinobacteria and species of Aspergillus were detected in all investigated samples. The salty environment provided by the parchment samples selected halophilic and halotolerant microorganisms. These halophilic microorganisms may be responsible for the purple stain discoloration observed on the parchments. In particular, Saccharopolyspora species was shown to be a constant presence on most of the samples examined (the only sample in which the Saccharopolyspora species was not detected actually showed a different pattern of discolouration), and an unequivocal role of a Chaetomium species in altering the surface of the parchment. Among the fungi, A. versicolor appeared strongly associated with deteriorated parchment samples, suggesting a sort of co-occurrence or even an ecological relationship between its role and that of halophilic bacteria.
A role of some salts in the creation of a particular environment in parchment, that can promote the occurrence of the purple spots, seems plausible on the basis of the data. In this study, for the first time, a relationship between the phenomenon of purple spots on ancient parchments and that of the ‘red heat’ phenomenon, identified in leather and other salted products, is proposed. Further studies will be conducted on the in vitro reproduction of the measles-like parchment discoloration, on possible bacteria–fungus interactions in damage production, and on the understanding of the mechanisms of colonization of ancient parchments by halophilic bacteria.
Experimental procedures
Case studies
Seven different parchment manuscripts affected by purple spots were selected for this study. Samples were obtained following techniques designed to minimize invasive action on objects made from parchment and from historical documents (Larsen, 2002; Pinzari et al., 2010). Before any sampling operation was carried out, the points most suitable for this purpose were evaluated. Some fragments of parchment (2–3 mm2) were collected, mainly from the margins of the most degraded pages, using watchmaker tweezers.
The manuscripts selected for investigation were the following (Fig. 1):
Sample MS-492 (Fig. 1A) – This case study was conducted on a Pontifical manuscript conserved in the Museum of the Dioceses in Salerno (Italy). The manuscript is illuminated and very precious. It is dated between the end of the 13th century and the beginning of 14th century. The importance of the volume is multidisciplinary because it reports several aspects of the liturgics performed in the church of Salerno before the Council of Trento. Several pages of the manuscript are discoloured with purple spots, sometimes accompanied by a weakening of the substrate that caused the detachment of the miniatures (Carrarini and Casetti Brach, 2006).
Sample To800 (Fig. 3) – This case study was conducted on the cover, made of parchment, of a volume belonging to the Archives of the Library of Turin (Italy). The cover was discarded during the restoration of the Official Register of Students for the Faculty of Surgery of the years 1833–1836. The interest in this case is due to the type of damage which is represented by large purple stains covering both the parchment and the inner pages made of paper. The Library of Turin experienced a terrible fire at the beginning of the 20th century, and most of the volumes, which were soaked with water during the fire-extinction procedures, presented a similar damage.
Samples Ve-3 and Ve-21 (Fig. 1D) – The collection ‘Grazie’ is a wide archival group of volumes dating back to the period 1299–1767, and representing a register of great historical value. The volumes report all the public activities of the administration of the ‘Comune Veneciarum’ (Venice, Italy) from the 13th to the 18th century. The whole collection exhibited serious biological damage that defaced several pages of the volumes to the point of rendering the writing no longer legible. The damage is represented by a very dense pattern of purple spots that could not be attributed to a particular species of fungus or bacteria by means of culture-dependent techniques.
Bo-Arch (Fig. 1C) – The volume belongs to a wide collection from the 13th century conserved in the Archive of Bologna (Italy). The volume is representative of the whole collection which is widely affected by purple spots of biological origin.
Sample 148D (Fig. 1E) – The ‘Novum Testamentum’ volume of a 13th century Bible supported on parchment and conserved at the Vatican Library (Italy). The volume is heavily affected by biological damage consisting of dark purple stains, holes and an unusual powdery consistency of pages.
Sample M-XIV – Inc. IV. 117, ‘Plinius Caecilius Secundus, Epistolae, ed. Pomponius Laetus’, 1490, conserved at the Vatican Library (Italy). A few pages at the front of the volume were affected by biological damage consisting of dark purple stains.
DNA extraction and purification
DNA extraction was performed directly from all seven parchment samples by using the FastDNA Spin Kit for Soil from MP Biomedicals (Illkrich, France). The kit combines a mechanical lysis, using bead beating, and chemical lysis of the cells. The protocol of the manufacturer was slightly modified. About 10–20 mg (2–3 mm2) of parchment were placed in the Lysing Matrix E Tubes with the appropriate buffers, and then processed twice in the Fast Prep FP120 Ribolyzer (Thermo Savant, Holbrook, NY, USA) for 30 s at a speed of 5.5 (m s−1), with 5 min intervals on ice. The Lysing Matrix E Tubes were centrifuged at 14,000× g for 15 min, and the supernatants were transferred to clean 2 ml tubes. The PPS reagent and the binding matrix solution were applied to the supernatant; the suspension was transferred to the provided spin filter and centrifuged at 14,000× g for 1 min, following the instructions of the manufacturer. DNA was washed twice with 500 ml of the SEWS-M solution and eluted from the binding matrix with 100 ml of DNase/Pyrogen Free Water.
After the DNA extraction, the concentration and quality of the DNA extracts were assessed using a NanoDrop® ND-1000 Spectrophotometer (peqLab Biotechnologie GmbH, Linz, Austria). The analyses were performed according to the manufacturer's protocol and the extracted DNA was analysed in duplicate. Because of the low ratios of A260/A230 obtained, the DNA crude extracts were further purified prior to PCR amplification with the QIAamp Viral RNA mini kit (Qiagen, Hilden, Germany) with modifications as follows: the washing step was performed twice with 750 μl buffer AW1/AW, rolling the column to allow more contact with the cartridge and leaving the tubes to stand for 2 min with the buffer, prior to centrifugation. The final elution step was repeated twice with 100 μl of 80°C preheated ddH2O (Sigma Aldrich, St Louis, MO, USA) letting the tubes stand for 2 min before centrifugation. The purified DNA was used directly for PCR amplification.
PCR amplification of extracted DNA
For all PCR reactions 2x PCR Master Mix from Promega (Vienna, Austria) [50 units ml−1 of TaqDNA Polymerase supplied in an appropriate reaction buffer (pH 8.5), 400 μM dATP, 400 μM dGTP, 400 μM dCTP, 400 μM dTTP, 3 mM MgCl2] was diluted to 1x, and 12.5 pmol μl−1 of each primer (stock: 50 pmol μl−1; VBC-Biotech, Austria) were added. In a total volume of 25 μl, 400 μg ml−1 of BSA (stock: 20 mg ml−1; Roche, Diagnostics Gmbh, Germany) and 2.5 μl of DNA template were added. PCR was performed in an MJ Research PTC-200 Peltier Thermal Cycle.
For the analysis of fungal sequences, fragments of 450–600 bp in size, corresponding to the ITS1, the ITS2 region and the adjacent 5.8S rRNA gene, were amplified with the primer pair ITS1 and ITS4 (White et al., 1990). For DGGE analysis, a nested PCR was performed with the PCR product of the first round as template DNA using the primers ITS1GC with a 37-base GC clamp attached to the 5′ end (Muyzer et al., 1993) and ITS2. All reactions were carried out as described by Michaelsen and colleagues (2006).
For the amplification of bacterial 16S rRNA sequences, DNA was amplified with the primer pair 341f/985r (Muyzer et al., 1993; Heuer et al., 1999). For DGGE analysis, 200 bp fragments of the 16S rDNA were amplified with a nested PCR using the eubacterial specific primer 341f-GC with a 40-bp GC clamp added to its 5′ end (Muyzer et al., 1993) and the universal consensus primer 518r (Neefs et al., 1990). PCR conditions were as described by Schabereiter-Gurtner and colleagues (2001).
For the amplification of archaeal 16S rRNA sequences, the primer pair ARC344f/ ARC915r (Raskin et al., 1994) that amplify a PCR fragment of 590 bp was used. PCR conditions were as described by Piñar and colleagues (2001a). All PCR products were analysed by electrophoresis in a 2% (w/v) agarose gel.
Denaturing gradient gel electrophoresis
DGGE was performed as previously described (Muyzer et al., 1993) using a D-Code system (Bio-Rad) in × 0.5 TAE [20 mM Tris, 10 mM acetate, 0.5 mM Na2 EDTA; pH 7.8 with 8% (w/v) acrylamide]. Gels were run at a constant temperature of 60°C with a voltage of 200 V for a period of 3.5 h for bacteria, and 5 h for fungal fingerprints. The linear chemical gradient of denaturants used in this study [100% denaturing solution contains 7 M urea and 40% (v/v) formamide] are indicated at the legend of figures.
After completion of electrophoresis, gels were stained in a 1 μg ml−1 ethidium bromide solution [stock: 10 mg ml−1] for 20 min and afterwards visualized by a UVP documentation system (Bio-Rad Transilluminator, Universal Hood; Mitsubishi P93D-printer).
Creation of clone libraries and sequence analysis
To obtain a detailed phylogenetic identification of the microbial community members, clone libraries containing either ITS fungal regions (fungal community) or 16S rRNA fragments (bacterial communities) were carried out. For the construction of clone libraries 2 × 3 μl DNA templates of each sample were amplified in 2 × 50 μl reaction volumes using the following primer pair combinations: for fungal clone libraries, the DNA template was amplified using the primers ITS1/ITS4, as mentioned above. For bacterial clone libraries, the primer pair 341f/985r was used as mentioned above. The PCR products were purified using the QIAquick PCR Purification Kit Protocol (Qiagen, Hilden, Germany) and resuspended in ddH2O water.
Purified PCR products were ligated into the pGEM-T easy Vector system (Promega) following the instructions of the manufacturer. The ligation products were transformed into One shot TOP10 cells (Invitrogen). These cells allow the identification of recombinants (white colonies) on an indicator Luria–Bertani medium containing ampicilline (100 μg ml−1), streptomycine (25 μg ml−1) and X-Gal (5-bromo-4-chloro-3-indolyl-β-1-galactopyranoside; 0.1 mM) (Sambrook et al., 1989).
Fifty clones per each clone library were screened in a DGGE gel as described by Schabereiter-Gurtner and colleagues (2001). Selected clones were externally sequenced by Sanger sequencing with a fleet of 16 ABI 3730xl (GATC Biotech, Germany). Comparative sequence analysis was performed by comparing pair-wise insert sequences with those available in the public online database NCBI using the BLAST search programme (Altschul et al., 1997). The resulting sequences of the bacterial and fungal clones have been deposited at the GenBank genetic sequence database at the National Center for Biotechnical Information.
SEM-EDS technique
Parchment samples of 1–3 mm in diameter were analysed using a variable pressure SEM instrument (EVO50, Carl-Zeiss Electron Microscopy Group) equipped with a detector for electron backscattered diffraction (BSD). A chemical characterization of the inorganic constituents of the samples was performed by means of electronic dispersion spectroscopy (EDS), which allows for an X-ray area scanning of what is brought into focus in SEM images, and creates a compositional map of the parchment surface. Only after having observed samples with SEM in variable pressure mode, at 20 keV, with the BSD were some of the samples covered with gold with a Baltec Sputter Coater for a further analysis in high-vacuum mode (Goldstein et al., 2003). The sputtering was performed under an Argon gas flow, at 50 mm working distance with 0.05 mbar of pressure and a current of 40 mA for 60 s to obtain a film of gold of about 15 nm. Reference elemental intensities acquired from pure compounds (standards) are commonly utilized for calibrating SEM-EDS systems. In the case study shown in this paper, conventional ZAF correction integrated into Oxford INCA 250 microanalysis package was applied to the spectrum dataset (Oxford Instruments). More than 60 points in each sample were analysed by means of the EDS probe, in order to gain a significant number of values to be used in a statistical comparison. The microanalysis was performed on all samples except the sample M-XIV which, due to its very small size, was devoted entirely to the molecular analysis.
Data analysis
Microanalysis data obtained for the different parchment samples were analysed by means of one-way ANOVA, and the statistical significance of differences between samples were tested (Tukey's test with 95% confidence interval). The statistical package included in XLStat (Addinsoft, XLStat 2009 Paris, France) was used.
Acknowledgments
This study and G. Piñar were financed by the Austrian Science Fund (FWF) project ‘Elise-Richter V194-B20’. The authors are grateful to the staff of the ICRCPAL conservation laboratory for providing samples and some of the pictures of the studied manuscripts.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. DGGE fingerprints derived from seven different parchment samples showing:
A. Bacterial communities.
B. Fungal communities.
The linear chemical gradient of denaturants used was 35–60% for bacteria and 20–50% for fungi. Lane 1: 148D; lane 2: MXIV; lane 3: MS-492; lane 5: To800; lane 6: Bo-Arch; lane 7: Ve-3; lane 8: Ve-21; lanes + : positive controls for bacteria and fungi respectively.
Table S1. Bacteria on parchment: phylogenetic affinities of partial 16S rRNA coding sequences.
Table S2. Fungi on parchment: phylogenetic affinities of the fungal ITS coding sequences.
References
- Abe A, Oda Y, Asano K. Sone T. Rhizopus delemar is the proper name for Rhizopus oryzae fumaric-malic acid producers. Mycologia. 2007;99:714–722. doi: 10.3852/mycologia.99.5.714. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W. Lipman JD. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DG. Birbir M. A study of the extremely halophilic microorganisms found on commercially brine-cured cattle hides. J Am Leather Chem Assoc. 1993;88:285–293. [Google Scholar]
- Beöthy-Kozocsa I, Sipos-Richter T. Szlabey G. Parchment codex restoration using parchment and cellulose fibre pulp. Restaurator. 1990;11:95–109. and. Denmark. [Google Scholar]
- Berrada I, Willems A, De Vos P, El fahime EM, Swings J, Bendaou N, et al. Diversity of culturable moderately halophilic and halotolerant bacteria in a marsh and two salterns a protected ecosystem of Lower Loukkos (Morocco) Afr J Microbiol Res. 2012;6:2419–2434. [Google Scholar]
- Bicchieri M, Monti M, Piantanida G, Pinzari F. Sodo A. Non-destructive spectroscopic characterization of parchment documents. Vib Spectrosc. 2011;55:267–272. [Google Scholar]
- Bowden DJ. Brimblecombe P. Sulphur distribution in parchment and leather exposed to sulphur dioxide. J Soc Leath Technol Chem. 2000;84:177–186. [Google Scholar]
- Bowden DJ. Brimblecombe P. Sulphur inclusions within parchment and leather exposed to sulphur dioxide. In: Larsen R, editor; Microanalysis of Parchment. London, UK: Archetype Publications; 2002. pp. 45–51. [Google Scholar]
- Brewer D, Jerram WA. Taylor A. The production of cochliodinol and a related metabolite by Chaetomium species. Can J Microbiol. 1968;14:861–866. doi: 10.1139/m68-145. [DOI] [PubMed] [Google Scholar]
- Brüggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, et al. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science. 2004;305:671–673. doi: 10.1126/science.1100330. [DOI] [PubMed] [Google Scholar]
- Butinar L, Zalar P, Frisvad JC. Gunde-Cimerman N. The genus Eurotium – members of indigenous fungal community in hypersaline waters of salterns. FEMS Microbiol Ecol. 2005;51:155–166. doi: 10.1016/j.femsec.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Cao ZJ, Zhang Q, Wei DK, Chen L, Wang J, Zhang XQ. Zhou MH. Characterization of a novel Stenotrophomonas isolate with high keratinase activity and purification of the enzyme. J Ind Microbiol Biotechnol. 2009;36:181–188. doi: 10.1007/s10295-008-0469-8. [DOI] [PubMed] [Google Scholar]
- Carrarini R, editor; Casetti Brach C, editor. Libri & Carte Restauri e analisi diagnostiche. Roma, Italy: Istituto Centrale di Patologia del Libro, Gangemi editore; 2006. and (eds) (. Quaderni 1. [Google Scholar]
- Chen LL, Liu LJ, Shi M, Song XY, Zheng CY, Chen XL. Zhang YZ. Characterization and gene cloning of a novel serine protease with nematicidal activity from Trichoderma pseudokoningii SMF2. FEMS Microbiol Lett. 2009;299:135–142. doi: 10.1111/j.1574-6968.2009.01746.x. [DOI] [PubMed] [Google Scholar]
- Choudhary V. Production, isolation and characterization of alkaline protease from Aspergillus versicolor PF/F/107. J Acad Indus Res. 2012;1:272–277. [Google Scholar]
- Christensen CM, Papavizas GC. Benjamin CR. A new halophilic species of Eurotium. Mycologia. 1959;51:636–640. [Google Scholar]
- Crous PW, Braun U, Schubert K. Groenewald JZ. Delimiting Cladosporium from morphologically similar genera. Stud Mycol. 2007;58:33–56. doi: 10.3114/sim.2007.58.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echigo A, Minegishi H, Mizuki T, Kamekura M. Usami R. Geomicrobium halophilum gen. nov., sp. nov., a moderately halophilic and alkaliphilic bacterium isolated from soil. Int J Syst Evol Microbiol. 2010;60:990–995. doi: 10.1099/ijs.0.013268-0. [DOI] [PubMed] [Google Scholar]
- Ettenauer J, Piñar G, Sterflinger K, Gonzalez-Muñoz MT. Jroundi F. Molecular monitoring of the microbial dynamics occurring on historical limestone buildings during and after the in situ application of different bio-consolidation treatments. Sci Total Environ. 2011;409:5337–5352. doi: 10.1016/j.scitotenv.2011.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettenauer J, Piñar G, Lopandic K, Spangl B, Ellersdorfer G, Voitl C. Sterflinger K. Microbes on building materials – Evaluation of DNA extraction protocols as common basis for molecular analysis. Sci Total Environ. 2012;439:44–53. doi: 10.1016/j.scitotenv.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133:2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian MLE. Protein Facts. Fibrous Proteins in Cultural and Natural History Artifacts. London, UK: Archetype Publications; 2007. [Google Scholar]
- Gallo F. Strzelczyk A. Indagine preliminare sulle alterazioni microbiche della pergamena. Bollettino dell'Istituto di Patologia del Libro. 1971;30(Fasc. 1/2):71–87. [Google Scholar]
- Gené J, Blanco JL, Cano J, Garcia ME. Guarro J. New filamentous fungus Sagenomella chlamydospora responsible for a disseminated infection in a dog. J Clin Microbiol. 2003;41:1722–1725. doi: 10.1128/JCM.41.4.1722-1725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J, Newbury D, Joy D, Lyman C, Echlin P, Lifshin E, et al. Scanning Electron Microscopy and X-ray Microanalysis. 3rd edn. New York, USA: Springer Science, Business Media; 2003. [Google Scholar]
- González JM. Saiz-Jimenez C. Application of molecular nucleic acid-based techniques for the study of microbial communities in monuments and artworks. Int Microbiol. 2005;8:189–194. [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote M. O'Malley MA. Enlightening the life sciences: the history of halobacterial and microbial rhodopsin research. FEMS Microbiol Rev. 2011;35:1082–1099. doi: 10.1111/j.1574-6976.2011.00281.x. [DOI] [PubMed] [Google Scholar]
- Heuer H, Hartung K, Wieland G, Kramer I. Smalla K. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl Environ Microbiol. 1999;65:1045–1049. doi: 10.1128/aem.65.3.1045-1049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Seifert KA, Overy DP, Tuthill DM, Valdez JG. Samson RA. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia. 2012;29:78–100. doi: 10.3767/003158512X660571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq IU, Mukhtar H. Umber H. Production of protease by Penicillium chrysogenum through optimization of environmental conditions. J Agri Soc Sci. 2006;2:23–25. [Google Scholar]
- Imada A, Nakahama K, Igarasi S. Isono M. A bacteriolytic enzyme from Chaetomium globosum, a marine-isolate. Arch Mikrobiol. 1973;91:41–54. doi: 10.1007/BF00409537. [DOI] [PubMed] [Google Scholar]
- Jurado V, Porca E, Pastrana MP, Cuezva S, Fernandez-Cortes A. Saiz-Jimenez C. Microbiological study of bulls of indulgence of the 15th-16th centuries. Sci Total Environ. 2010;408:3711–3715. doi: 10.1016/j.scitotenv.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Kalpana Devi M, Rasheedha Banu A, Gnanaprabhal GR, Pradeep BV. Palaniswamy M. Purification, characterization of alkaline protease enzyme from native isolates Aspergillus niger and its compatibility with commercial detergents. Indian J Sci Technol. 2008;1:1–6. [Google Scholar]
- Karbowska-Berent J. Strzelczyk A. The role of Streptomycetes in the biodeterioration of historic parchment. Acta Microbiol Pol. 2000;49:177–178. [Google Scholar]
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik R. Microbiodeterioration of library materials, Part 2. Microbiodecomposition of basic organic library materials, Chapter 4. Restaurator. 1980;4:200–208. Denmark. [Google Scholar]
- Kraková L, Chovanová K, Selim SA, Šimonovičová A, Puškarová A, Maková A. Domenico P. A multiphasic approach for investigation of the microbial diversity and its biodegradative abilities in historical paper and parchment documents. Int Biodeter Biodegr. 2012;70:117–125. [Google Scholar]
- Laiz L, Piñar G, Lubitz W. Saiz-Jimenez C. Monitoring the colonisation of monuments by bacteria: cultivation versus molecular methods. Environ Microbiol. 2003;5:72–74. doi: 10.1046/j.1462-2920.2003.00381.x. [DOI] [PubMed] [Google Scholar]
- Larsen R. Microanalysis of Parchment. London, UK: Archetype Publications LTD; 2002. [Google Scholar]
- Lee SD, Kim ES. Hah YC. Phylogenetic analysis of the genera Pseudonocardia and Actinobispora based on 16S ribosomal DNA sequences. FEMS Microbiol Lett. 2000;182:125–129. doi: 10.1111/j.1574-6968.2000.tb08885.x. [DOI] [PubMed] [Google Scholar]
- Lochhead AG. Bacteriological studies on the red discoloration of salted hides. Can J Res. 1934;10:275–286. [Google Scholar]
- Luijpen AFMG, Mossel DAA. Broek CJH. Bacteriological investigation of a case of red discolouration in Dutch salted summer herring. Antonie Van Leeuwenhoek. 1953;19:78–82. doi: 10.1007/BF02594833. [DOI] [PubMed] [Google Scholar]
- Martins RF, Davids W, Al-Soud WA, Levander F, Radström P. Hatti-Kaul R. Starch-hydrolyzing bacteria from Ethiopian soda lakes. Extremophiles. 2001;5:135–144. doi: 10.1007/s007920100183. [DOI] [PubMed] [Google Scholar]
- Meiler D. Taylor A. The effect of cochliodinol, a metabolite of Chaetomium cochliodes on the respiration of microspores of Fusarium oxysporum. Can J Microbiol. 1970;17:83–86. doi: 10.1139/m71-014. [DOI] [PubMed] [Google Scholar]
- Michaelsen A, Pinzari F, Ripka K, Lubitz W. Piñar G. Application of molecular techniques for identification of fungal communities colonizing paper material. Int Biodeter Biodegr. 2006;58:133–141. [Google Scholar]
- Michaelsen A, Piñar G. Pinzari F. Molecular and microscopical investigation of the microflora inhabiting a deteriorated Italian manuscript dated from the 13th century. Microb Ecol. 2010;60:69–80. doi: 10.1007/s00248-010-9667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanari M, Melloni V, Pinzari F. Innocenti G. Fungal biodeterioration of historical library materials stored in Compactus movable shelves. Int Biodeter Biodegr. 2012;75:83–88. [Google Scholar]
- Muyzer G, De Waal EC. Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao K, Ota T, Tanikawa A, Takae Y, Mori T, Udagawa S. Nishikawa T. Genetic identification and detection of human pathogenic Rhizopus species, a major mucormycosis agent, by multiplex PCR based on internal transcribed spacer region of rRNA gene. J Dermatol Sci. 2005;39:23–31. doi: 10.1016/j.jdermsci.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Neefs JM, Van de Peer Y, Hendriks L. De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990;18:2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Microbial diversity and microbial abundance in salt-saturated brines: why are the waters of hypersaline lakes red? Nat Resour Environ Issues. 2009;15:247–255. Art. 49. [Google Scholar]
- Orlita A. Biodeterioration of leather materials especially book-leather bindings and parchments. In: Mukerji KG, editor; Garg KL, Garg N, editors. Recent Advances in Biodeterioration and Biodegredation. Vol. 1. Calcutta, India: Publisher Naya Prokash; 1993. pp. 259–299. Biodeterioration of Cultural Heritage. [Google Scholar]
- Petushkova JP. Koestler RJ. Biodeterioration studies on parchment and leather attacked by bacteria in the commonwealth of socialist states. In: Munafò P, editor; Federici C, editor. International Conference on Conservation and Restoration of Archival and Library Materials. I. Palermo, Italy: G.B. Palumbo Publisher on behalf of Assessorato Regionale dei Beni Cultuali, Ambientali e P.I; 1996. pp. 195–211. and (eds)., and, Vol. [Google Scholar]
- Petushkova JP, Shemyakina TM. Cherdyntseva TA. Proteolytic activity of bacteria isolated from the surface of parchment manuscripts and handmade leather articles. Mikrobiologya. 1984;53:399–403. [Google Scholar]
- Pinzari F, Montanari M, Michaelsen A. Piñar G. Analytical protocols for the assessment of biological damage in historical documents. Coalition. 2010;19:6–13. [Google Scholar]
- Pinzari F, Cialei V. Piñar G. A case study of ancient parchment biodeterioration using variable pressure and high vacuum scanning electron microscopy. In: Mongiatti A, editor; Meeks N, Cartwright C, Meek A, editors. Historical Technology, Materials and Conservation: SEM and Microanalysis. London, UK: Archetype Publications – International Academic Projects; 2012. pp. 92–98. and (eds)., and. [Google Scholar]
- Piñar G, Gurtner C, Lubitz W. Rölleke S. Identification of Archaea in objects of art by DGGE analysis and shot gun cloning. Methods Enzymol. 2001a;336:356–366. doi: 10.1016/s0076-6879(01)36601-6. [DOI] [PubMed] [Google Scholar]
- Piñar G, Ramos C, Rölleke S, Schabereiter-Gurtner C, Vybiral D, Lubitz W. Denner EBM. Detection of indigenous Halobacillus populations in damaged ancient wall paintings and building materials: molecular monitoring and cultivation. Appl Environ Microbiol. 2001b;67:4891–4895. doi: 10.1128/AEM.67.10.4891-4895.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñar G, Ripka K, Weber J. Sterflinger K. The micro-biota of a sub-surface monument: the medieval chapel of St. Virgil (Vienna, Austria) Int Biodeter Biodegr. 2009;63:851–859. [Google Scholar]
- Piñar G, Piombino-Mascali D, Maixner F, Zink A. Sterflinger K. Microbial survey of the mummies from the Capuchin Catacombs of Palermo, Italy: biodeterioration risk and contamination of the indoor air. FEMS Microbiol Ecol. 2013;86:341–356. doi: 10.1111/1574-6941.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck I, Salkin IF, Schenav D, Ofer L, Magen M. Haines JH. Mycopathologia. Vol. 106. 1989. Damage to an ancient parchment document by Aspergillus; pp. 89–93. [Google Scholar]
- Polacheck I. Fungi and cultural heritage. In: Adriaens A, editor; Gunneweg J, Greenblatt C, editors. Bio- and material cultures at Qumran. In COST Action G8 Working Group Meeting, Israel. Stuttgart, Germany: Fraunhofer IRB Verlag; 2005. pp. 267–270. [Google Scholar]
- Poole JB. Reed R. Technol Cult. Vol. 3. 1962. The preparation of leather and parchment by the Dead Sea scrolls community; pp. 1–26. [Google Scholar]
- Rakeman JL, Bui U, Lafe K, Chen YC, Honeycutt RJ. Cookson BT. Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J Clin Microbiol. 2005;43:3324–3333. doi: 10.1128/JCM.43.7.3324-3333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin L, Stromley JM, Rittmann BE. Stahl DA. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebrikova NL. Dmitrieva MB. Experimental investigation of parchment manuscripts with traces of microbiological damage. In: Munafò P, editor; Federici C, editor. International Conference on Conservation and Restoration of Archival and Library Materials. I. Palermo, Italy: G.B. Palumbo Publisher on behalf of Assessorato Regionale dei Beni Cultuali, Ambientali e P.I; 1996. pp. 275–283. and (eds)., and, Vol. [Google Scholar]
- Rebrikova NL. Solovyova NI. ICOM Committee for Conservation, 8th Triennial Meeting. Vol. 3. Sydney, Australia: 1987. Electron microscopic and biochemical investigation of parchment; pp. 1197–1200. [Google Scholar]
- Reed R. Ancient Skins Parchments and Leathers. London – New York: Seminar Press; 1972. [Google Scholar]
- Reed R. The Nature and Making of Parchment. Leeds, England: The Elmete Press; 1975. [Google Scholar]
- Ryder ML. Remains derived from skin. In: Higgs ES, editor; Brothwell DR, editor. Science in Archaeology. London, UK: Thames and Hudson; 1969. pp. 529–544. and (eds).. (revised chap. in 2nd edn. 1970) [Google Scholar]
- Sambrook J, Fritsch EF. Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbour, New York, USA: Cold Spring Harbour Laboratory; 1989. [Google Scholar]
- Samson RA. Von der Lustgraaf B. Aspergillus penicilloidesEurotium halophilicum. Vol. 64. 1978. pp. 13–16. and and in association with house- dust mites. Mycopathologia. [DOI] [PubMed] [Google Scholar]
- Samson RA, Hoekstra ES, Frisvad JC. Filtenborg O. Introduction to food- and airborne fungi. 6th edn. Utrecht, The Netherlands: ‘Centraalbureau Voor Schimmelculture’; 2002. [Google Scholar]
- Schabereiter-Gurtner C, Piñar G, Lubitz W. Rölleke S. An advanced molecular strategy to identify bacterial communities on art objects. J Microbiol Methods. 2001;45:77–87. doi: 10.1016/s0167-7012(01)00227-5. [DOI] [PubMed] [Google Scholar]
- Schäfer S, Jäckel U. Kämpfer P. Analysis of Actinobacteria from mould-colonized water damaged building material. Syst Appl Microbiol. 2010;33:260–268. doi: 10.1016/j.syapm.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Sigler L, Sutton DA, Gibas CF, Summerbe RC, Noel RK. Iwen PC. Phialosimplex, a new anamorphic genus associated with infections in dogs and having phylogenetic affinity to the Trichocomaceae. Med Mycol. 2010;48:35–45. doi: 10.3109/13693780903225805. [DOI] [PubMed] [Google Scholar]
- Szabó Z. Szabó IM. Mikroorganizmusok részvételének igazolása a Korvinak pergamen anyagának biodeteriorációjában (Verification of the participation of microorganisms in biodeterioration of parchment material of Corvina codices) Vol. 13. Budapest, Hungary: Múzeumi mutárgyvédelem; 1984. pp. 87–96. and, Vol.. (AATA Nos.:1986–26946 and 23–647) [Google Scholar]
- Tang SK, Wang Y, Wu JY, Cao LL, Lou K, Xu LH, et al. Saccharopolyspora qijiaojingensis sp. nov., a halophilic actinomycete isolated from a salt lake. Int J Syst Evol Microbiol. 2009;59:2166–2170. doi: 10.1099/ijs.0.009860-0. [DOI] [PubMed] [Google Scholar]
- Tardieux M, Moussin MH, Flieder F. Pochon MJ. Altération Biologique des Parchemins. Paris, France Comptes Rendus de l'Academie des Sciences. 1971;272:1817–1818. [PubMed] [Google Scholar]
- Trüper HG. Galinski EA. Concentrated brines as habitats for microorganisms. Experientia. 1986;42:1182–1187. [Google Scholar]
- Voronina LI, Nazarova ON. Petushkova PYU. Disinfection and straightening of parchment damaged by microorganisms. Restaurator. 1980;4:91–97. and. Denmark. [Google Scholar]
- White TJ, Bruns T, Lee S. Taylor J. PCR Protocols: A Guide to Methods and Applications, Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Woods DR, Rawlings DE, Cooper DR. Galloway AC. Collagenolytic activity of hide bacteria and leather decay. J Appl Bacteriol. 1973;36:289–295. [Google Scholar]
- Zhang E, Tanaka T, Tajima M, Tsuboi R, Nishikawa A. Sugita T. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol. 2011;55:625–632. doi: 10.1111/j.1348-0421.2011.00364.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. DGGE fingerprints derived from seven different parchment samples showing:
A. Bacterial communities.
B. Fungal communities.
The linear chemical gradient of denaturants used was 35–60% for bacteria and 20–50% for fungi. Lane 1: 148D; lane 2: MXIV; lane 3: MS-492; lane 5: To800; lane 6: Bo-Arch; lane 7: Ve-3; lane 8: Ve-21; lanes + : positive controls for bacteria and fungi respectively.
Table S1. Bacteria on parchment: phylogenetic affinities of partial 16S rRNA coding sequences.
Table S2. Fungi on parchment: phylogenetic affinities of the fungal ITS coding sequences.


