Abstract
Drought-induced (Di19) proteins played important roles in plant growth, development, and abiotic stress responses. In the present study, a total of seven Di19 genes were identified in soybean. Each soybean Di19 gene showed specific responses to salt, drought, oxidative, and ABA stresses based on expression profiles. With a relatively higher transcript level among Di19 members under four stress treatments, GmDi19-5 was selected for detailed analysis. Inhibitor assays revealed that ABA inhibitor (Fluridone) or H2O2 inhibitor (DMTU) was involved in the drought- or salt-induced transcription of GmDi19-5. The GUS activity driven by the GmDi19-5 promoter was induced by salt, PEG, ABA, and MV treatments and tended to be accumulated in the vascular bundles and young leaves. A subcellular localization assay showed that GmDi19-5 protein localized in the nucleus. Further investigation showed that GmDi19-5 protein was involved in the interaction with GmLEA3.1. Overexpression of GmDi19-5 increased sensitivity of transgenic Arabidopsis plants to salt, drought, oxidative, and ABA stresses and regulated expression of several ABA/stress-associated genes. This present investigation showed that GmDi19-5 functioned as a negative factor under abiotic stresses and was involved in ABA and SOS signaling pathway by altering transcription of stress-associated genes.
Keywords: Di19 protein, genome-wide analysis, stress response, functional identification, protein interaction, Glycine max
Introduction
Abiotic stress may occur at any stage of plant development and often several types of stresses occur simultaneously (Rizhsky et al., 2004). To reduce the adverse effects of stress, plants have evolved multifaceted strategies, including morphological, physiological, and biochemical adaptations (Ingram and Bartels, 1996; Xiong et al., 2002; Zhu, 2002; Shinozaki et al., 2003; Bohnert et al., 2006). It is also demonstrated that plants have evolved a complex and elaborate signaling network that perceives and responds to continuously changing surroundings by modulating the expression of downstream genes (Xu et al., 2011). It has been demonstrated that the modulation of signaling regulators will be a promising method for improving the stress tolerance of plants. A number of stress-regulated genes encode regulatory proteins, such as transcription factors, that are important in regulating the expression of downstream genes (Seki et al., 2002; Singh et al., 2002; Lee and Lee, 2003; Rabbani et al., 2003). Zinc finger proteins are one of the major families of eukaryotic transcription factors (Klug and Schwabe, 1995; Takatsuji, 1999; Englbrecht et al., 2004). Within this family, the Cys2/His2-type (C2H2) zinc finger proteins contain one or more tandem of C2H2 zinc finger motifs, one of the best-characterized DNA-binding motifs. This motif contains two cysteines and two histidines that serve as zinc ligands and is represented by the signature Cys-X2,4-Cys-X12-His-X3,4,5-His (Brown et al., 1985; Miller et al., 1985; Pabo et al., 2001; Sakamoto et al., 2004). Drought-induced (Di19) proteins are zinc finger transcription factors that play important roles in development, growth, and response to stress (Milla et al., 2006; Li et al., 2010a,b; Liu et al., 2013b; Qin et al., 2014). Di19 proteins are encoded by a small gene family that numbers seven isoforms in Arabidopsis and five in rice (Milla et al., 2006). Di19 proteins are known also in cotton (Li et al., 2010a) and wheat (Li et al., 2010b).
The Di19 family functionally participated in various signaling pathways. Di19s have been found acting as both transcription repressors and activators (Li et al., 2010a,b; Liu et al., 2013b; Qin et al., 2014). In Arabidopsis, AtDi19-1 and AtDi19-3 were rapidly induced by drought stress, whereas transcripts of AtDi19-2 and AtDi19-4 were accumulated at high levels by high salinity stress (Milla et al., 2006). AtDi19-7 has been implicated in regulating light signaling, and did not respond to abiotic stress treatments (Kang et al., 2005). These findings indicate that in the Di19 family, different members may respond to different signal stimuli and accomplish specific functions.
Cys2/His2-type zinc finger proteins can bind to DNA elements (Searles et al., 2000; Wolfe et al., 2000; Liu et al., 2013b; Qin et al., 2014). In Arabidopsis, both AtDi19-1 and AtDi19-3 could bind to the TACA(A/G)T element (Liu et al., 2013b; Qin et al., 2014). Further assays demonstrated AtDi19-1 could directly up-regulate the expressions of PR1, PR2, and PR5 in response to drought stress. In addition to binding DNA elements (Liu et al., 2013b), Cys2/His2-type zinc finger proteins may also participate in protein-protein interactions (Fukamatsu et al., 2005; Milla et al., 2006; Liu et al., 2013b). In Arabidopsis, AtDi19-1 protein interacted with CPK11 (a calcium-binding protein kinase) and its transactivation activity could be enhanced through phosphorylation by CPK11 at the nuclear location signal (NLS)-containing motif (Milla et al., 2006; Liu et al., 2013b). This suggested that posttranslational modification might be important to regulate the function of Di19 protein (Milla et al., 2006). AtDi19-7 interacted with F-box AtLKP2 protein that might be light receptors and function within or very close to the circadian oscillator (Kiyosue and Wada, 2000; Somers et al., 2000; Imaizumi et al., 2003; Fukamatsu et al., 2005). Protein-protein interactions are critically important to many processes that take place in the cell, such as signal transduction, and regulation of gene expression. Therefore, it is significant to identify interacting proteins of Di19s.
In this study, we characterized the Di19 protein family of soybean, by identifying its seven members and their chromosome locations, gene structures, and expression profiles. Moreover, Di19-5 was selected for detailed functional analysis.
Materials and methods
Search and identification of Di19 family members
Arabidopsis Di19 (Milla et al., 2006) sequences were retrieved from the Arabidopsis Information Resource (http://www.arabidopsis.org) and used to search homologous Di19s from the soybean database (http://www.phytozome.org/) (Release 9.1). BLASTN and BLASTP programs were used to identify homologous EST singletons and peptides, respectively. Redundant sequences were removed via the decrease redundancy tool (http://web.expasy.org/decrease_redundancy/). Each non-redundant sequence was checked for the presence of two conserved C2H2 zinc finger domains.
Phylogenetic tree and sequence alignments
The phylogenetic tree of Di19s was constructed using the neighbor-joining method in Molecular Evolutionary Genetics Analysis (MEGA; version 4.1) with the following parameters: Test Neighbor-Joining model and 1000 bootstrap replicates. Multiple sequence alignments were performed using the amino acid sequences of the conserved region and full-length protein by ClustalX2.0 software. Multiple Expectation maximization for Motif Elicitation (MEME) was used to identify the motifs of candidate Di19 proteins. Potential nuclear localization sequences (NLS) and putative nuclear export signal sequences (NES) were predicted by PSORT and NetNES software, respectively (Nakai and Kanehisa, 1992). The subcellular localization was predicted at YLoc (http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi). Phosphorylation sites were predicted at NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/).
Chromosomal distribution, gene structure, and promoter region prediction
Chromosomal distribution was determined by searching the database containing the complete genome sequence of each soybean chromosome (http://www.phytozome.org/). Exon/intron gene structures were constructed by comparing the CDSs with their corresponding genomic DNA sequences and analyzed using the Gene Structure Display Serve tool (http://gsds.cbi.pku.edu.cn/).
To analyze their promoter regions, the 1.8 kb upstream regions of the genes, according to the position of the genes provided by the soybean annotation information, were selected and screened against the PLACE database (Higo et al., 1999).
Soybean stress treatments
Soybean cultivar “Tie feng 8,” with characteristic of salt tolerance, was used in this study. Soybean seeds were grown in pots of peat/vermiculite (1:1 v/v) under conditions of 12 h of light followed 12 h of dark, constant temperature 25°C, and humidity 70%. Salt, drought, H2O2, and abscisic acid (ABA) stresses were applied to 2-week-old soybean seedlings. For salt stress, the roots of seedlings were dipped into solutions of 200 mM NaCl. For dehydration, the root systems of whole plants were washed gently with water to remove soil, and then the plants were put on filter paper for induction of a rapid drought treatment. For H2O2 stress, the roots of seedlings were dipped into solutions of 25 mM H2O2. For ABA treatment, soybean seedlings were sprayed with 100 μM ABA. For inhibitors assay, the plants were pretreated with H2O2 scavenger [10 mM dimethyl thiourea (DMTU)] and ABA scavenger (100 μM fluridone) for 6 h, respectively, and then exposed to dehydration and salt treatments for 0.5, 5, or 12 h. In order to get reliable results for all of the above treatments, the un-treated soybean seedlings with consistent growth were used as control for each series of treatments. At various time points after each treatment soybean seedlings were harvested, frozen in liquid nitrogen, and stored at −80°C until extraction of total RNA for qRT-PCR assays.
Subcellular localization
GmDi19-5 was inserted into the subcellular localization vector p16318 containing the CaMV 35S promoter and green fluorescent protein (GFP) gene. To obtain Di19 coding sequence without the stop codon, Di19-5 cDNA was ligated into the Hind III/Xba I sites of p16318GFP vector (for primer sequences, Supplementary Table S1), upstream to the N-terminal end of GFP under control of the 35S promoter. Subcellular localization of transiently expressed GmDi19-GFP was assessed after transformation by particle bombardment into onion epidermal cells (Xu et al., 2007; Li et al., 2013). Fluorescence was observed by confocal laser scanning microscopy (LSM700; Carl Zeiss) after incubation at 25°C for 24 h on MS medium under dark conditions.
Generation and stress treatments of transgenic arabidopsis plants
To generate Arabidopsis transgenic plants constitutively overexpressing the GmDi19-5 gene, the coding sequence of GmDi19-5 was cloned into the pBI121 vector with Sma I/Sac I sites to replace the GUS gene under the control of the CaMV 35S promoter (for primer sequences, Supplementary Table S1). To develop Arabidopsis transgenic plants expressing the GUS reporter gene under the control of the GmDi19-5 promoter, the promoter sequence of GmDi19-5 was cloned into the pBI121 vector with Hind III /Xba I sites to replace CaMV 35S promoter and fused to the N-terminal end of GUS (for primer sequences, Supplementary Table S1). Each construct was then transferred into Arabidopsis by the floral dip method. Arabidopsis lines carrying promoter CaMV 35S-GmDi19-5 gene were used for phenotypic analysis (Zhang et al., 2004). Arabidopsis lines (proDi19-5) carrying promoter Di19-5-GUS gene were used for expression analysis.
Seeds of transgenic overexpressing Arabidopsis and WT plants were grown on 10 × 10 cm MS agar plates. They were routinely kept for 3 d in darkness at 4°C to break dormancy and transferred in a tissue culture room under a day/night 16/8 h cycle at 22°C. For seed germination, a total of 50 seeds of transgenic lines or WT were kept on MS media supplemented with 50 mM NaCl, 2% PEG, 1.0 μM methylviologen (MV), or 1.5 μM ABA for 5 d. Seeds were considered germinated when radicles had emerged from the seed coat. For root growth, 30 5-d-old seedlings with consistent growth state of transgenic lines or WT were transferred to MS agar plates containing 100 mM NaCl, 4% PEG, 5 μM MV, or 10 μM ABA for 5 d. Seed germination rates and root lengths were analyzed. Each treatment contained three independent replicates.
Screening of cDNA libraries and yeast two-hybrid interaction assay
The soybean seedling cDNA library was constructed in a pGADT7-Rec2 vector containing a GAL4 activation domain using Matchmaker Library Construction (Clontech) and then transformed into the yeast strain AH109 (Clontech). GmDi19-5 was cloned into pGBKT7 bait vector (for primer sequences, Supplementary Table S1) and transformed into yeast strain Y187 (Clontech). Yeast two-hybrid screening was performed using the MATCHMAKER two-hybrid system (Clontech) as previously described (Liu et al., 2013a).
Candidates were retransformed with the bait vector into the yeast strain AH109 for two-hybrid analysis. Transformants were selected by growing on SD-Trp-Leu- at 30°C for 4 d. Surviving clones were retransferred to SD-Trp-Leu-His-Ade- medium according to the manufacturer's instructions (Clontech).
Bimolecular fluorescence complementation (BiFC) assay
For BiFC analysis, the full-length coding sequence of GmDi19-5 was cloned into PUC-pSPYNE vector and fused with the N-terminal fragment of YFP to form YFPN-Di19-5 construct (for primer sequences, Supplementary Table S1). The full-length coding sequence of GmLEA3.1 was cloned into PUC-pSPYCE vector as a fusion with the C-terminal fragment of YFP to form YFPC-LEA3 construct (for primer sequences, Supplementary Table S1). For transient expression, plasmids of YFPN-Di19-5 and YFPC-LEA3 were co-transformed into onion epidermal cells by a particle gun and monitored by confocal microscopy as previously described (Liu et al., 2013a).
Detection of β-glucuronidase (GUS) activity assay
For GUS activity assay, proDi19 transgenic seeds were germinated and grown on MS media for 5 d, and then exposed to MS media containing 100 mM NaCl, 4% PEG, 5 μM MV, or 10 μM ABA for 5 d to a tissue culture room under a day/night 16/8 h cycle at 22°C. Seedlings without any stress treatment were used as controls. Histochemical localization of GUS activities were analyzed after incubating the transgenic plants in 10 ml tubes with 1 mg/mL 5-bromo-4-chloro-3-indolyl-glucuronic acid, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.03% Triton X-100 and 0.1 M sodium phosphate buffer, pH 7.0 overnight at 37°C. Seedlings were cleared with 70% ethanol to remove chlorophyll from green tissue. GUS-stained plants were examined with a light microscope (Leica) at a low magnification and photographed with a digital camera.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA template was extracted from isolated different plant materials using Trizol reagent (Takara, Dalian, China) and reverse transcription was performed with 2 μg of total RNA for the first strand cDNA synthesis with a PrimeScript 1st Strand cDNA Synthesis kit (Takara, Dalian, China), according to the manufacturer's instructions. The qRT-PCR primers were designed from non-conserved regions of the genes (Supplementary Table S1). Soybean Actin (U60506) or Arabidopsis Tublin was used as internal controls for normalization of the template cDNA. qRT-PCR analyses were performed on an ABI7300 system with SYBR Premix ExTaq II (Takara, Dalian, China). The amount of transcript accumulated for GmDi19 genes or Arabidopsis stress-related genes normalized to the internal control gene was analyzed using 2−ΔΔCT method (Livak and Schmittgen, 2001). Three independent experiments were accomplished and for each sample three technical replicates were analyzed.
Statistical analysis
Statistical analyses were performed using the software in Excel. Analysis of variance was used to compare the statistical difference based on Student's t-test, at a significant level of P < 0.05, P < 0.01.
Results
Identification of the soybean Di19 family
Seven Di19s were identified in the soybean genome. They contained conserved two conserved C2H2 zinc finger domains which were consistent with Arabidopsis Di19 homologs (Supplementary Dataset S1). Based on suggested Arabidopsis Di19 nomenclature, each gene was named with a two-letter code corresponding to G. max (Gm), followed the family designation (Di19), and finally a number (Table 1). The soybean Di19s encode proteins with predicted molecular mass of ~25 kD and isoelectric point (pI) < 7.
Table 1.
Nomenclature for Di19s in soybean.
| GmDi19 gene name | GmDi19 gene model | Amino acid | Isoelectronic point (pI) | Molecular mass (kD) | Chromosome |
|---|---|---|---|---|---|
| GmDi19-1 | Glyma03g37530 | 218 | 4.66 | 24.35 | 3 |
| GmDi19-2 | Glyma19g40150 | 216 | 4.83 | 24.26 | 19 |
| GmDi19-3 | Glyma10g29030 | 219 | 5.52 | 24.35 | 10 |
| GmDi19-4 | Glyma15g15560 | 215 | 5.51 | 24.04 | 15 |
| GmDi19-5 | Glyma09g04490 | 215 | 5.38 | 24.07 | 9 |
| GmDi19-6 | Glyma07g32120 | 233 | 5.98 | 26.13 | 7 |
| GmDi19-7 | Glyma13g24420 | 237 | 6.12 | 26.40 | 13 |
Phylogenetic tree and domain analysis of the Di19 family
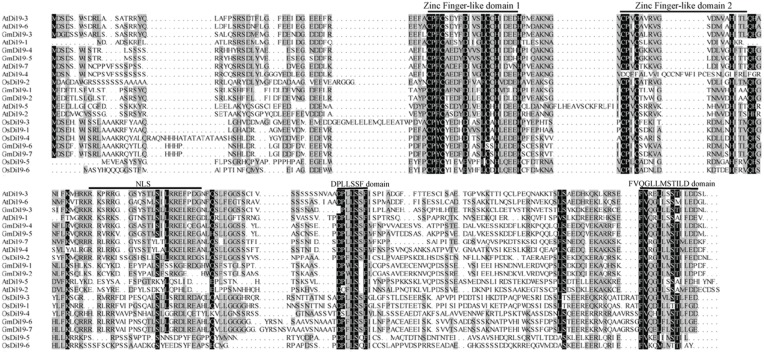
Multiple alignments were performed using the ClustalX program to examine sequence features of Di19 proteins. All soybean Di19 proteins contain highly conserved region with two unusual C2H2 zinc finger-like domains at the N-terminus, which span approximate about 60 amino acids (Figure 1). The two motifs differ slightly from the canonical C2H2 domain sequence (Cys–X2−5–Cys–X12–His–X2−5–His) (Klug and Schwabe, 1995). In soybean Di19 proteins, the spacing between the two Cys and two His amino acids in the first and second finger domain was 11 and 10 amino acids, respectively, as in Arabidopsis and rice Di19 proteins (Figure 1; Supplementary Dataset S2). Outside the putative zinc finger-like domain, a Leu-rich motif and two short regions in the C-terminus half of the proteins (consensus sequences “DPLLSSF” and “FVQGLLMSTILD”) were conserved among all members of the family (Figure 1). Members of the soybean Di19 family (except GmDi19-1 and GmDi19-2) contained NLS (Supplementary Table S2). Potential NES was also present in several soybean Di19 proteins (Supplementary Table S2). Subcellular localization predictions revealed that all soybean Di19 proteins were located in the nucleus (Supplementary Table S2). Moreover, a BLAST search against the NCBI translated database resulted in identification of homology proteins with the same features in Arabidopsis, rice, maize, and Brachypodium (Supplementary Table S3). MEME analysis identified 10 motifs (Supplementary Figure S1), of which motifs 1 and 2 were located in the conserved Di19 domain region of all Di19 proteins (namely the two C2H2 zinc finger-like domains). Domain analysis indicated that Di19 proteins were present and well conserved in both dicotyledonous and monocotyledonous plants (Supplementary Figures S1, S2; Supplementary Table S4). Different logos of these 10 motifs were shown in Supplementary Figure S2.
Figure 1.
Multiple alignments of soybean Di19s, AtDi19s, and OsDi19s.
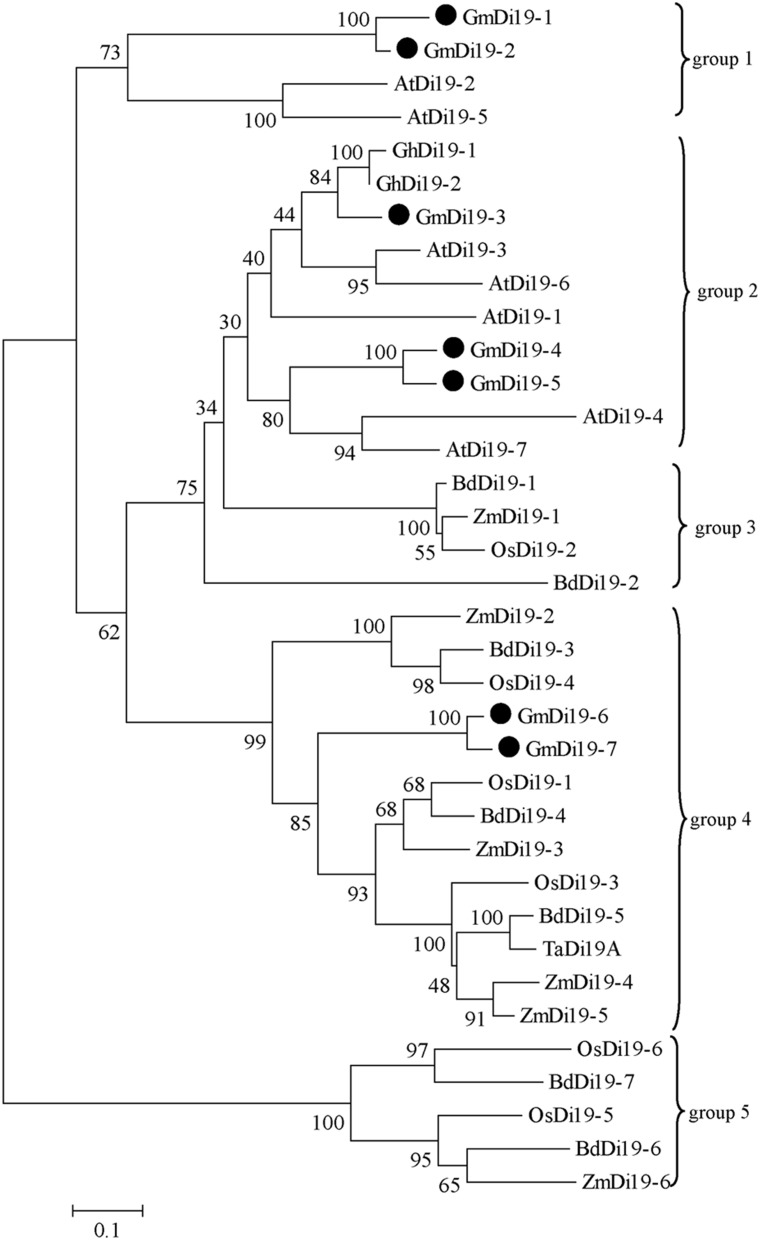
Due to the high similarity of Di19 proteins, a phylogenetic tree was built on the basis of the full amino acid sequence of soybean, Arabidopsis, rice, maize, Brachypodium, cotton, and wheat Di19 proteins. Three pairs of closely related proteins were found: (i) GmDi19-1 and GmDi19-2, (ii) GmDi19-4 and GmDi19-5, and (iii) GmDi19-6 and GmDi19-7 (Figure 2). Phylogenetic tree showed all Di19s were divided into five groups (Figure 2). Dicotyledonous Di19s formed two groups 1 and 2. Monophyletic clade formed three groups 3, 4, and 5. The best orthology matches of the GmDi19-1, GmDi19-2, GmDi19-4, and GmDi19-5 proteins were AtDi19-2, AtDi19-5, AtDi19-4, and AtDi19-7, respectively. The best orthology matches of the GmDi19-3 protein were AtDi19-1, AtDi19-3, AtDi19-6, GhDi19-1, and GhDi19-2. The best orthology matches of the GmDi19-6 and GmDi19-7 proteins were ZmDi19-3, OsDi19-1, and BdDi19-4.
Figure 2.
Phylogenetic relationship of the soybean Di19s with those of Arabidopsis, rice, maize, Brachypodium, cotton, and wheat.
Additionally, the reliability of the phylogeny was further evidenced by parameters like motif compositions of individual subfamilies. Motif sequence conservation or variation between the proteins might specify the functional equivalence or diversification, respectively, with respect to various aspects of biological function. Members of a particular subfamily showed a tendency to have similar motif compositions and certain motifs were deleted or duplicated within particular clades. For example, motifs 7, 9, and 10 were absent from almost all members of Di19-group 1, and instead, Di19-group 2 included motif 9. Motif 7 only existed in Di19-group 4. Motifs 4, 5, 7, 8, and 10 were absent from almost all members of Di19-group 5 (Figure 2; Supplementary Figure S1). Specific motif deletion or duplication within a protein might be crucial for dispensing undesirable regions and maintaining or developing only those regions needed to develop a particular phenotype.
Chromosomal location and gene structure analysis
The seven genes were located on different soybean chromosomes (Supplementary Table S3). However, two Di19 genes were found on each of chromosomes 3 and 5 in Arabidopsis, three Di19 genes were present on chromosome 5 in rice, and five were located on chromosome 2 in maize (Supplementary Table S3). Clearly, certain chromosomes in some species had a relatively high density of Di19 genes (Supplementary Table S3).
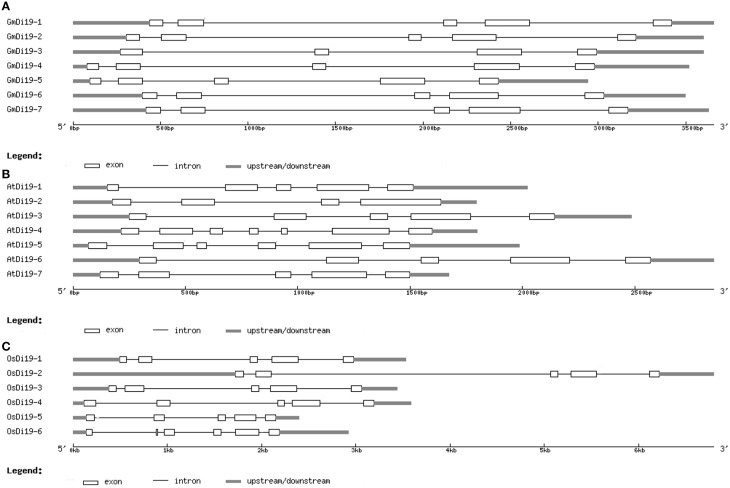
To obtain some insight into the gene structures of the soybean Di19 family genes, their exon/intron organizations were analyzed. All soybean Di19 genes were disrupted by four or five introns (Figure 3A). Interestingly, each of the putative zinc finger domains in soybean Di19 proteins was encoded by the same adjacent exons (second and third exons). We also compared the gene structures of Di19 genes in Arabidopsis and rice. The exon-intron structures were highly conserved in all cases (Figures 3B,C). These findings suggested that each subgroup of Di19 genes was conserved in a relatively constant exon-intron composition during evolution.
Figure 3.
Gene structures of soybean Di19s (A), AtDi19s (B), and OsDi19s (C). Exons and introns are represented by white boxes and blank lines, respectively.
Abiotic stress responses
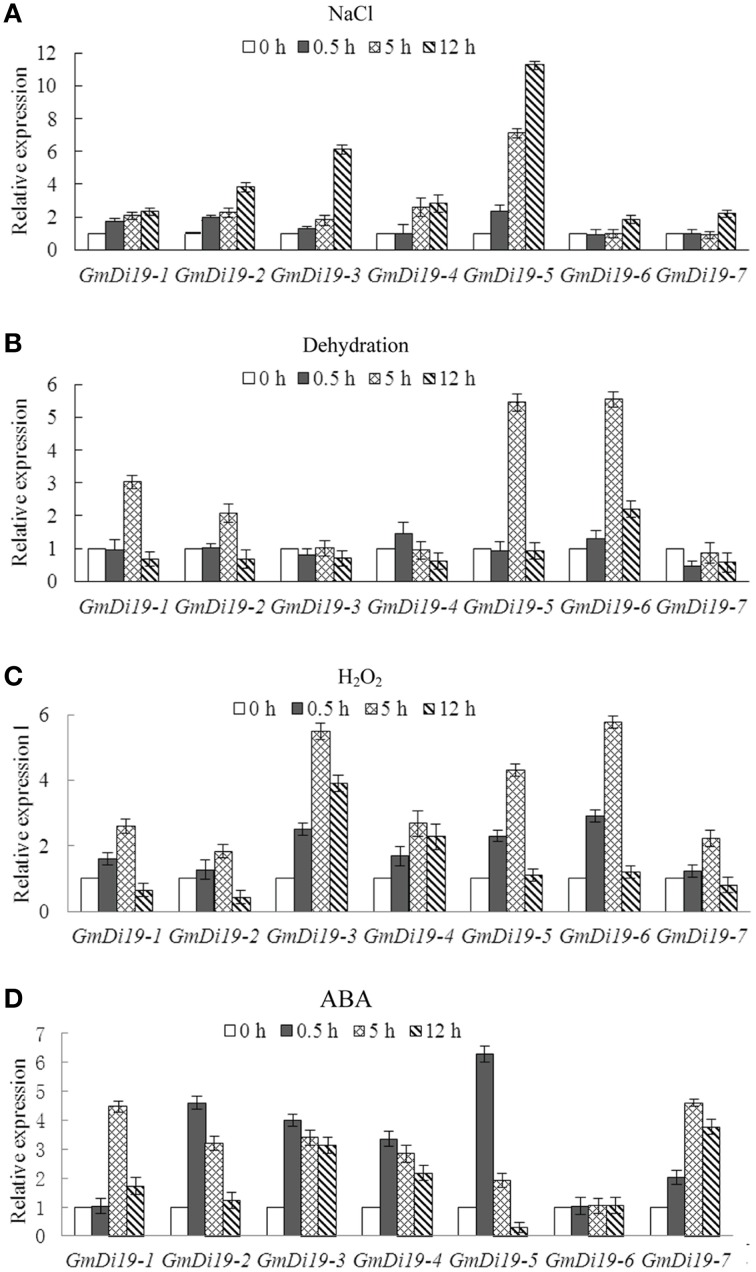
To investigate the effect of abiotic stresses on Di19s, expression pattern was measured by qRT-PCR in soybean seedlings subjected to salt, drought, H2O2,and ABA treatments. As shown in Figure 4, there were wide variations in accumulation of different mRNAs following different types of stress. For salt treatment most of the transcript levels of Di19 genes increased and remained constant during the first 12 h of treatment. GmDi19-5 was upregulated more than 10-fold after 12 h of salt treatment (Figure 4A). Drought and H2O2 stresses also significantly modulated the expressions of most Di19 genes. As shown in Figure 4B, transcripts of GmDi19-5 and GmDi19-6 were increased by more than 5-fold after 5 h of drought stress. Figure 4C showed that transcript levels of Di19 genes, except GmDi19-2, instantaneously increased by more than 2-fold with H2O2 treatment; after 5 h GmDi19-3 and GmDi19-6 were upregulated by more than 5-fold. With ABA treatment, Di19 transcript levels were increased more than 3-fold, except for GmDi19-6 (Figure 4D). The transcript of GmDi19-5 was increased more than 6-fold at 0.5 h of ABA stress.
Figure 4.
Relative expression profiles of the Di19 genes in the 2-week-old soybean seedlings subjected to NaCl (A), dehydration (B), H2O2 (C), and ABA (D) stress treatments. The transcript levels of each Di19 in the stress-treated plants were plotted as the relative expression (fold) of the non-stressed control plants for 0.5, 5, and 12 h. The transcript level of Actin was used as a reference. Mean values and standard errors (bar) were shown from three independent experiments.
Due to relatively high up-regulated transcript levels under stress treatments (Figure 4), GmDi19-5 was selected for investigation. The full-length cDNA of GmDi19-5 was comprised of 1252 bp with a 648 bp open reading frame and the deduced protein contained 215 amino acid residues with a predicted molecular mass of 24.07 kD (Table 1; Supplementary Dataset S3). Serine, threonine, and tyrosine phosphorylation sites were all found in GmDi19-5 protein (Supplementary Table S5).
ABA and H2O2 were involved in induction of GmDi19-5 under stress treatments
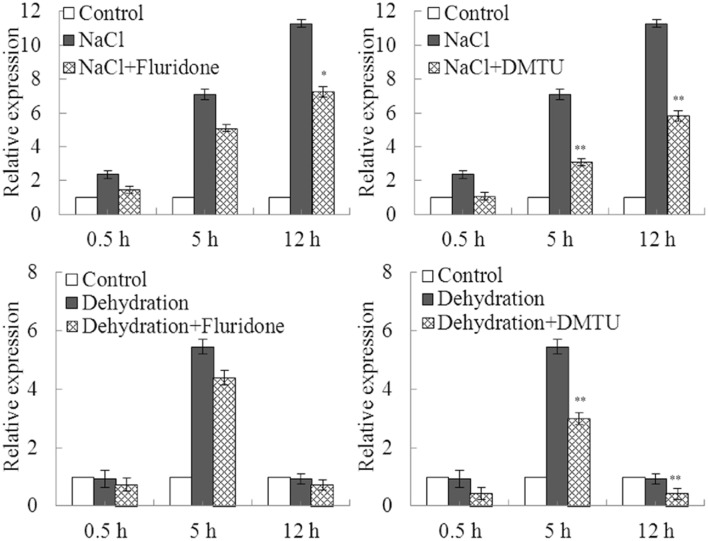
To explore whether ABA and H2O2 were involved in up-regulation of GmDi19-5 under drought and salt stresses, fluridone, and DMTU were chosen as ABA and H2O2 inhibitors, respectively (Hu et al., 2012; Ma et al., 2014; Yoshida et al., 2014; You et al., 2014; Zhang et al., 2014). Treatment with fluridone or DMTU had no effect on transcript of GmDi19-5 under normal treatments (Figure 5). Pretreatment with the fluridone inhibitor partially prevented up-regulation of GmDi19-5 under NaCl, while fluridone was not so effective in block of up-regulation of GmDi19-5 under drought stress in soybean. Pretreatment with the DMTU inhibitor partially prevented up-regulation of GmDi19-5 under NaCl or drought stresses in soybean. Therefore, ABA was involved in up-regulation of GmDi19-5 under salt stresses, and H2O2 was involved in up-regulation of GmDi19-5 under both salt and drought stresses.
Figure 5.
Effects of pretreatment with inhibitor of ABA and H2O2 on the expression of GmDi19-5 under abiotic treatments. ABA inhibitor (Fluridone) and H2O2 inhibitor (DMTU) on expression of GmDi19-5 in soybean seedlings exposed to dehydration and salt stresses. Mean values and standard errors (bar) were shown from three independent experiments. * and ** indicated significant expression differences in comparison with expression without inhibitor pretreatment at 0.01 < P < 0.05 and P < 0.01, respectively (t-test).
The activity of GmDi19-5 promoter was driven by various stresses
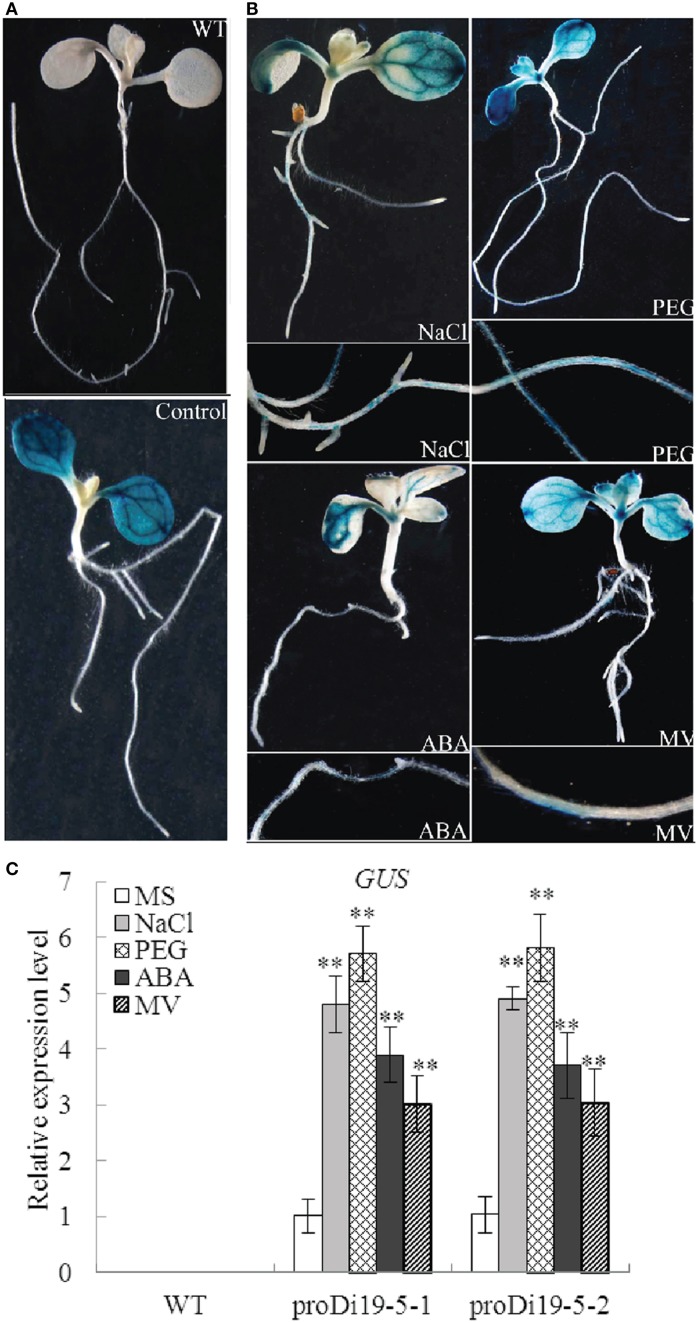
To investigate the activity of GmDi19-5 promoter, two homozygous transgenic Arabidopsis lines (proDi19-5-1 and proDi19-5-2) were selected for phenotypic analysis under stress treatments. Under normal growth conditions, GUS staining revealed that activity of GUS gene driven by GmDi19-5 promoter was detected throughout cotyledons, especially in the vascular bundles of cotyledons (Figure 6A). After NaCl, PEG, ABA, and MV treatments, GUS activities significantly increased in roots, leaf primordium, and young leaves (Figure 6B). In addition, the expression level of GUS gene in the transgenic plants with NaCl, PEG, ABA, and MV treatments was remarkably stronger than those without NaCl, mannitol, or ABA treatment (Figure 6C).
Figure 6.
Analysis of GmDi19-5 promoter activities under abiotic treatments. (A) 10-d-old seedlings of WT and transgenic plants. (B) Transgenic seedlings treated with NaCl, PEG, ABA, and MV. (C) qRT-PCR analysis of expression of GUS gene in proDi19-5-GUS transgenic Arabidopsis plants under NaCl, PEG, ABA, and MV stress treatments. ** indicated significant differences in comparison with the WT lines at P < 0.01 (t-test).
The GmDi19-5 protein was localized in the nucleus
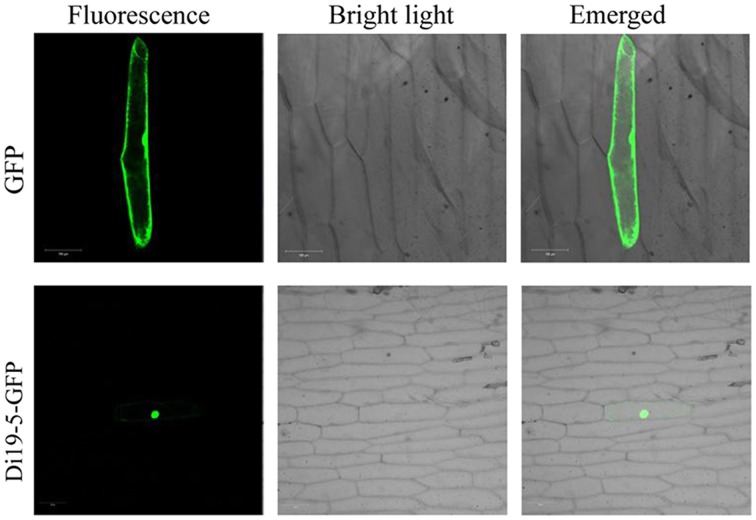
The predicted GmDi19-5 protein contained one conserved NLS (97–118 amino acids) and one conserved NES (117–125 amino acids) (Supplementary Table S2). To determine the cellular localization of GmDi19-5 protein, the GmDi19-5 gene was cloned into the p16318GFP vector downstream of the constitutive CaMV 35S promoter and upstream of the GFP gene to create the GmDi19-GFP fusion construct, which was then transformed into onion epidermal cells. Green fluorescence of GmDi19-5-GFP was mainly in the nucleus, whereas GFP in the control was uniformly distributed throughout the cell (Figure 7).
Figure 7.
Subcellular localization of GmDi19-5 protein. GFP and GmDi19-5-GFP represented subcellular localization of the control 16318GFP and GmDi19-5 in onion epidermal cells, respectively.
Gmdi19-5 might be involved in the interaction with GmLEA3.1
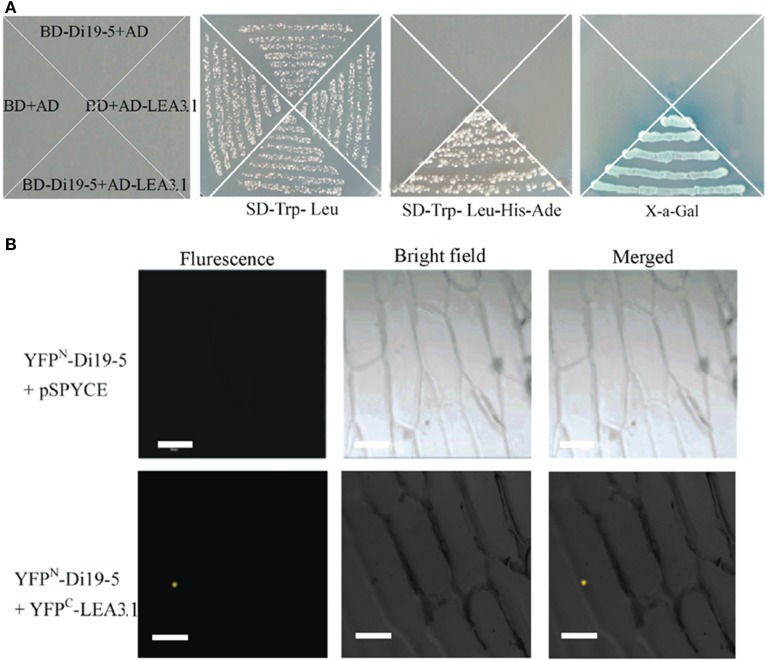
Interactions of proteins with other proteins are important for most biological functions. Search for interacting partners is necessary to understand the function of GmDi19-5. One positive interactor late embryogenesis abundant (LEA) protein (Glyma10g02210) was screened and identified using the yeast two-hybrid system. BLASTN and BLASTP analysis in soybean database revealed that this protein possessed the full ORF and the conserved LEA3 protein domain (Pfam 02987). Therefore, it was named as GmLEA3.1 protein. GmLEA3.1 encoded a predicted product of 95 amino acid residues with a molecular mass of 10.01 kD. In yeast two-hybrid screening, strong growth on SD-Trp-Leu-Ade-His medium and activity of the reporter gene were observed only in yeast cells co-transformed with pGBKT7-GmDi19-5 and pGADT7-GmLEA3.1 vectors (Figure 8A), indicating interaction of GmDi19-5 with GmLEA3.1 in yeast cells.
Figure 8.
Interaction of GmDi19-5 with GmLEA3.1. (A) Yeast two-hybrid interaction assay. Vectors were co-introduced into yeast in different combinations: 1, pGADT7 and pGBKT7; 2, pGBKT7-GmDi19-5 and pGADT7; 3, pGBKT7 and pGADT7-GmLEA3.1; 4, pGBKT7-GmDi19-5 and pGADT7-GmLEA3.1. Transformants were placed on selection medium and grown for 4 d. (B) In vivo BiFC assay. The expression of GmDi19-5 alone (YFPN-GmDi19-5 + pSPYCE) was used as the control.
Further, we used the BiFC technology to investigate the interaction in plant cells. In contrast to the almost total absence of fluorescence in the negative control (GmDi19-5-YFPN and empty vector pSPYCE), interaction of GmDi19-5-YFPN (N-terminal fragment of yellow fluorescent protein) and GmLEA3.1-YFPC (C-terminal fragment of yellow fluorescent protein) was observed mainly in the nucleus of onion cells (Figure 8B).
Phenotypes of GmDi19-5 transgenic arabidopsis
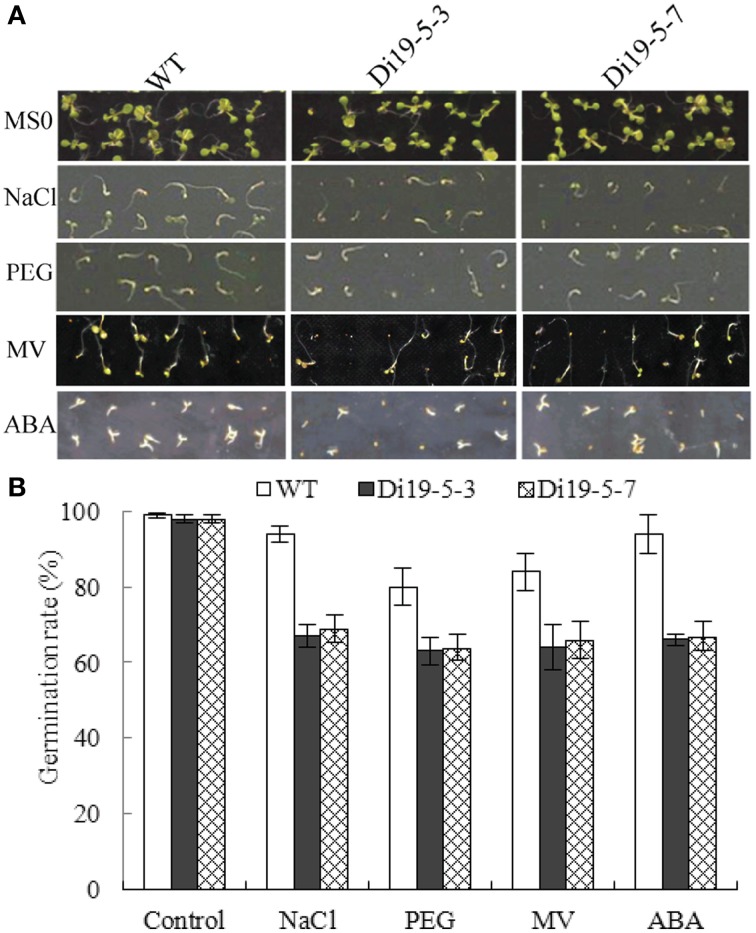
To investigate the function of GmDi19-5, two homozygous constitutively overexpressing Arabidopsis lines (Di19-5-3 and Di19-5-7) with higher GmDi19-5 expression were selected for phenotypic analysis under NaCl, PEG, MV, and ABA stress treatments. On MS medium alone, no obvious difference was observed between the transgenic and wide type (WT) seeds. When sown on MS medium containing 50 mM NaCl, GmDi19-5 transgenic seeds germinated much later than WT seeds. After 5 d, approximately 94% of WT seeds germinated compared to 67% for transgenic seeds. After sowing on MS medium containing 2% PEG for 5 d, approximately 80% of the WT seeds germinated, compared to about 64% for transgenic seeds. The ultimate germination rate of GmDi19-5 transgenic seeds was slightly lower than that of WT seeds. The germination rate on MS medium containing 1.0 μM MV was also analyzed. Germination of GmDi19-5 transgenic seeds was approximately 64% compared to 84% for WT. The germination rate on MS medium containing 1.5 μM ABA was again similar to levels observed with other stress treatment experiments. Treatment with ABA decreased the germination of GmDi19-5 transgenic plants to approximately 66%, whereas WT retained a 94% germination rate under the same ABA concentration (Figure 9).
Figure 9.
Effect of salt, drought, oxidative and ABA stresses on seed germination of GmDi19-5 transgenic and WT seeds. (A) Germination rate of WT and transgenic seeds on MS medium with or without NaCl, PEG, MV, and ABA. (B) Statistics analysis of germination rate. Mean values and standard errors (bar) were shown from three independent experiments.
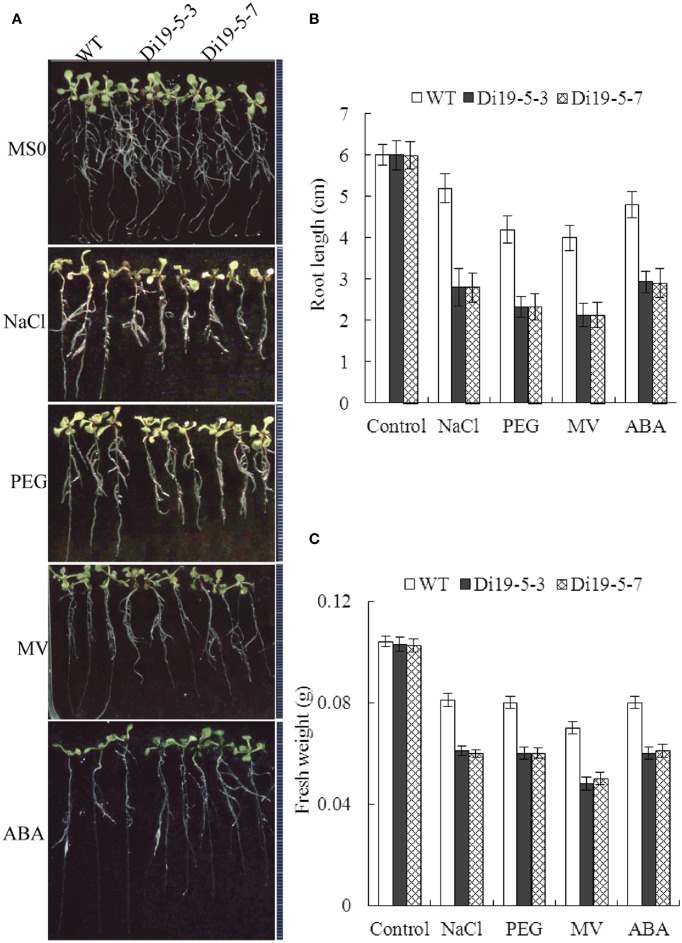
Root growth of GmDi19-5 transgenic Arabidopsis seedlings was also investigated on MS media containing NaCl, PEG, MV, and ABA. As shown in Figure 10, when the seedlings were grown on MS medium supplemented with 100 mM NaCl or 4% PEG for 5 d, primary root growth was significantly retarded compared to WT. When the seedlings were grown on MS medium supplemented with 5 μM MV or 10 μM ABA for 5 d, the root lengths and fresh weight of the transgenic seedlings were less than those of WT. Seed germination and post-germination assays revealed increased sensitivity of the transgenic plants overexpressing GmDi19-5 under NaCl, PEG, MV, and ABA stress conditions.
Figure 10.
Effect of salt, drought, oxidative, and ABA stresses on root length of GmDi19-5 transgenic and WT seedlings. (A) Root length of seedlings were transferred to a medium with or without NaCl, PEG, MV, and ABA before the images were taken. Statistics analysis of root length (B) and fresh weight (C). Mean values and standard errors (bar) were shown from three independent experiments.
GmDi19-5 regulated stress-responsive gene expression
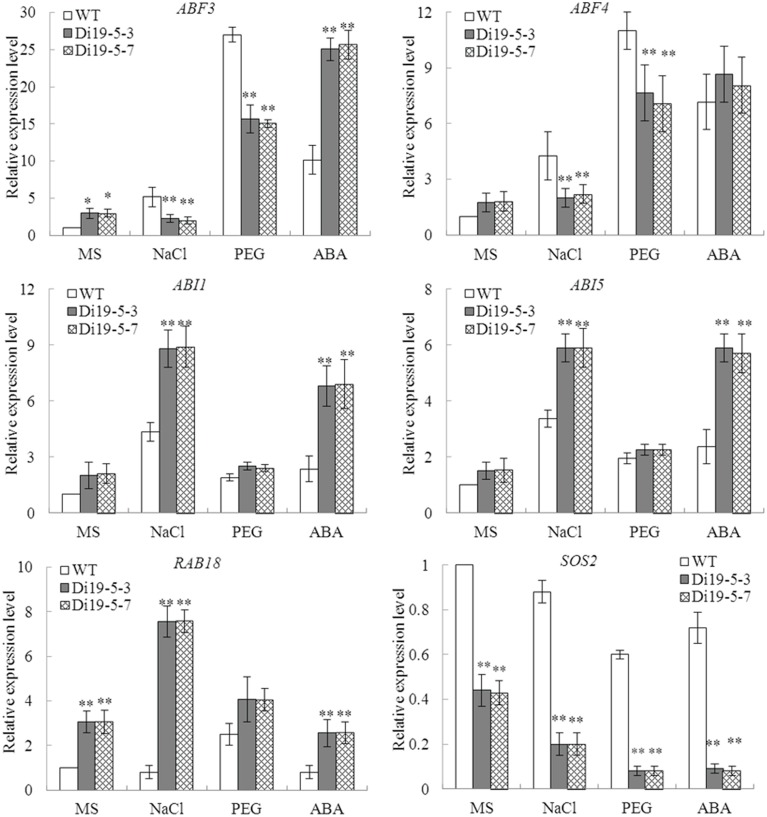
To elucidate the possible molecular mechanism in stress response, the expressions of six stress-response genes (ABF3, ABF4, ABI1, ABI5, RAB18, and SOS2) were investigated in constitutively overexpressing GmDi19-5 Arabidopsis lines (Di19-5-3 and Di19-5-7) and WT plants under normal and stress conditions. Under normal conditions, transcript levels of the ABF3 and RAB18 accumulated to much higher level in the transgenic lines than WT plants, while transcript level of SOS2 in transgenic lines were remarkably lower than WT plants. Under NaCl and PEG treatments, expression levels of ABF3, ABF4, and SOS2 in the transgenic lines were lower than those in WT. Under ABA stress conditions, ABI1, ABI5, and RAB18 showed increased expression in the transgenic plants relative to WT plants and SOS2 was substantially lower in transgenic lines than in WT plants (Figure 11).
Figure 11.
Expression of stress-responsive genes in GmDi19-5 transgenic Arabidopsis. * and ** indicated significant differences in comparison with the WT lines at 0.01 < P < 0.05 and P < 0.01, respectively (t-test).
Discussion
Di19 family was involved in plant stress responses and development (Kang et al., 2005; Milla et al., 2006; Parkinson et al., 2009; Li et al., 2010a,b; Liu et al., 2013b). However, further investigation was still needed to elaborate Di19 cascades and functions. This provided an impetus for investigation of the biological roles and interacting proteins of Di19 protein family in soybean.
Functional divergence of Di19 genes
Di19 proteins, encoded by small multigenes, were hydrophilic, low molecular weight, and stress-responsive proteins. Domain compositions and gene structure of individual group tended to preserve similar motif compositions and relatively constant exon-intron compositions during evolution (Figures 2, 3; Supplementary Figures S1, 2). Phosphorylation sites and conserved cysteine, leu, aspartic acid, and N-myristoylation sites were found in soybean Di19 proteins, which may play important role in keeping the protein structure (Figure 1; Supplementary Table S5). Despite evolutionarily well conserved, Di19 proteins presented amino acid diversity outside the three conserved regions (Figure 1).
Di19 was involved in multiple stress responses in plants (Milla et al., 2006; Parkinson et al., 2009; Li et al., 2010a,b; Liu et al., 2013b). Arabidopsis Di19 family genes displayed various transcriptional regulations in response to specific abiotic stress, including cold, drought, osmotic, oxidative, and salt stresses, in roots or shoots (Supplementary Figure S3; Supplementary Table S7). Promoter analysis showed there were many abiotic stress-related elements in the upstream region of soybean Di19 family genes (Supplementary Table S6). Expression profiles analysis revealed that transcripts of soybean Di19 genes were increased by salt, drought, oxidative, and ABA stresses (Figure 4). Meanwhile, GUS activity driven by the GmDi19-5 promoter tended to increase under NaCl, PEG, ABA, and MV treatments (Figure 6). It was reported that several genes responsive to salt and drought might be involved in ABA and H2O2 signaling pathways that were conserved components of these stress signal pathways (Hu et al., 2012; Ma et al., 2014; Yoshida et al., 2014; You et al., 2014; Zhang et al., 2014). Pretreatment with the inhibitors of these pathways prevented up-regulation of GmDi19-5 in NaCl- and drought-treated soybean seedlings (Figure 5). Thus, it is plausible that GmDi19-5 could be part of the ABA- and ROS-mediation process. However, more research is needed to confirm this mechanism through other biological technologies.
It was demonstrated that salt and drought stresses were able to induce ABA biosynthesis and trigger ABA-dependent signaling pathways, and ABA could maintain seed dormancy, prevented germination and inhibited seedling growth (Finkelstein et al., 1998, 2002; Leung and Giraudat, 1998; Achard et al., 2006; Piskurewicz et al., 2008; Bari and Jones, 2009). In cotton, Di19-like genes GhDi19-1 and GhDi19-2 were identified to be positive nuclear regulators in the ABA signaling pathway (Li et al., 2010a). Wheat TaDi19A was positively regulated by ABA signaling and was responsive to NaCl and high osmotic stress (Li et al., 2010b). GmDi19-5 affected the expression of genes related to ABA/stress signaling in transgenic plants, including ABF3, ABF4, ABI1, ABI5, RAB18, and SOS2 genes (Figure 11). ABF3 and ABF4 were two bZIP transcription factors which positively regulated ABA signaling. The constitutive overexpression of ABF3 or ABF4 in Arabidopsis exhibited enhanced drought tolerance and ABA hypersensitivity (Kang et al., 2002). The relative expression levels of these two genes were higher after ABA treatment. ABI1 encoded one ser/thr protein phosphatase of the PP2C family that acted as negative regulator of ABA response (Koornneef et al., 1989). ABI5 was one bZIP transcription factor, positively regulating ABA-mediated control of seed germination and early seedling development (Leung and Giraudat, 1998). The level of ABI1 and ABI5 were significantly increased under ABA treatment. The transcription level of known ABA up-regulated gene RAB18 was also elevated in GmDi19-5 transgenic plants. These findings suggested that ABA was more effectively synthesized in transgenic plants than in WT plants. GmDi19-5 might act a positive regulator in ABA response. In addition, ABF3, ABF4, and SOS2 were significantly low in GmDi19-5 transgenic plants after NaCl and PEG treatments. Overexpression of GmDi19-5 displayed increased sensitivities to salt, drought, and oxidative stress during the germination and seedling stages in Arabidopsis (Figures 7, 8). Recently, it was reported that AtDi19-3 displayed drought- and salt-sensitive phenotype in overexpression lines (Qin et al., 2014). Similar to AtDi19-3, GmDi19-5 might act as a negative regulator in response to salt and drought stresses (Qin et al., 2014).
Di19s might be involved in the interactions with various proteins
Zinc-finger domain is involved in protein-DNA interactions, protein-protein interactions between cytoskeleton dystrophin and calmodulin, and between transcriptional adapters (such as CREB-binding proteins and p300) and transcriptional activators (Dure, 1993; Davies et al., 1996; Takatsuji, 1999; Searles et al., 2000; Wolfe et al., 2000). The Cys2/His2 zinc finger-like domain may have a similar function in protein-protein interaction. LEA proteins possessed high hydrophilicity and chaperone-like activity and involved in stress resistance in plants, such as chilling, drought, and high salinity (Battaglia et al., 2008; Bies-Etheve et al., 2008; Battaglia and Covarrubias, 2013). Physical interaction with other proteins was one of the important features of LEAs, such as between hydrophilic proteins and LEA proteins (Hanin et al., 2011; Cuevas-Velazquez et al., 2014). Maize LEA protein Rab17 interacted with and was phosphorylated protein kinase CK2 (Riera et al., 2004). Opuntia streptacantha LEA proteins were shown to form dimer (Hernández-Sánchez et al., 2014). LEAs belonged to a multigene family, which was classified into seven groups based on expression patterns and sequences. Recently, there were extensive correlative data linking the expression of Group 3 LEA (LEA3) proteins with tolerance suggested various biochemical mechanisms for LEA3s, such as the repair of improperly folded proteins as a chaperone, the binding of metal ions, the stabilization of membrane structure and the increase of cellular mechanical strength through the generation of filaments (Browne et al., 2002; Tolleter et al., 2007; Tunnacliffe and Wise, 2007). We found that GmDi19-5 interacted with GmLEA3.1 (Figure 8), thus we might suppose that GmLEA3.1 protein might improve the stability of GmDi19-5 protein.
In addition, GmDi19-5 contained a conserved NLS in which putative kinase phosphorylation sites (Serine and Tyrosine) were located (Supplementary Table S5). Arabidopsis AtDi19-1 interacted with and was phosphorylated by AtCPK11 at the NLS-containing motif (Milla et al., 2006). AtDi19-4 was phosphorylated by AtCPK3 at the NLS-containing motif with Serine 116 being identified as one of the putative phosphorylated sites (Milla et al., 2006). It was possible that GmDi19-5 protein might interact with other proteins or be modulated at the post-translational level by changing their phosphorylation states to enhance the abiotic stress in plants. Interactions with different factors may be essential for activation of Di19 proteins or one mechanism distinguishing the different functions of the Di19 members. Therefore, their function and interacting proteins still need to be studied in detail, which will help to elaborate Di19 cascades.
Author contributions
ZSX coordinated the project, conceived and designed experiments, and edited the manuscript. ZJF conducted the bioinformatic work, generated and analyzed data, and wrote the first draft. XYC and XYC performed experiments and analyzed the data. MC and GXY managed reagents and provided analytical tools. YZM and GYH contributed with valuable discussions. All authors have read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was financially supported by the National Transgenic Key Project of MOA (2014ZX08009-016B and 2014ZX08002-002). We are grateful to Dr. Lijuan Qiu of the Institute of Crop Science, Chinese Academy of Agricultural Sciences for kindly providing soybean seeds.
Glossary
Abbreviations
- ABA
abscisic acid
- Di19
drought-induced
- DMTU
dimethyl thiourea
- GFP
green fluorescent protein
- MV
methylviologen
- qRT-PCR
quantitative real-time PCR
- WT
wild-type.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00179/abstract
Fasta file of soybean Di19 protein and conserved domain sequences.
Fasta file of all Arabidopsis and rice Di19 protein conserved regions.
Fasta files of soybean Di19-5 gene and protein sequences.
List of primers used in this paper.
NLS and NES sequences identified in soybean Di19 family proteins by the PSORT and NetNES softwares.
Nomenclature for Di19s in soybean, Arabidopsis, rice, maize, and Brachypodium.
Conserved motifs identified for the Di19 proteins from soybean, Arabidopsis, rice, maize, and Brachypodium by MEME software.
Phosphorylation sites predicted in soybean Di19 family proteins by NetPhos 2.0 Server softwares.
Prediction cis-elements of soybean Di19 family genes.
Expression profiles of Arabidopsis Di19 genes in response to various stress treatments. The data was downloaded from http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi.
Variation in motif clades of Di19 proteins from soybean, Arabidopsis, rice, maize, and Brachypodium. The MEME motifs were shown as different-colored boxes.
Logos of the conserved Di19 domain alignments of Arabidopsis, rice, maize, and Brachypodium.
Expression profiles of Arabidopsis Di19 genes in response to various stress treatments. Hierarchical clustering of differential gene expression in response to cold (A), drought (B), heat (C), osmotic (D), oxidative (E), and salinity (F) stresses.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. 10.1126/science.1118642 [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Battaglia M., Covarrubias A. A. (2013). Late Embryogenesis Abundant (LEA) proteins in legumes. Front. Plant Sci. 4:190. 10.3389/fpls.2013.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A. (2008). The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24. 10.1104/pp.108.120725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies-Etheve N., Gaubier-Comella P., Debures A., Lasserre E., Jobet E Raynal M., Cooke R., et al. (2008). Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 67, 107–124. 10.1007/s11103-008-9304-x [DOI] [PubMed] [Google Scholar]
- Bohnert H. J., Gong Q., Li P., Ma S. (2006). Unraveling abiotic stress tolerance mechanism, getting genomics going. Curr. Opin. Plant Biol. 9, 180–188. 10.1016/j.pbi.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Brown R. S., Sander C., Argos P. (1985). The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 186, 271–274 10.1016/0014-5793(85)80723-7 [DOI] [PubMed] [Google Scholar]
- Browne J., Tunnacliffe A., Burnell A. (2002). A hydrobiosis, plant desiccation gene found in a nematode. Nature 416:38. 10.1038/416038a [DOI] [PubMed] [Google Scholar]
- Cuevas-Velazquez C. L., Rendón-Luna D. F., Covarrubias A. A. (2014). Dissecting the cryoprotection mechanisms for dehydrins. Front. Plant Sci. 5:583. 10.3389/fpls.2014.00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K. E., Blake D. J., Ponting C. P., Winder S. J., Kendrick-Jones J. (1996). ZZ and TAZ, New putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 21, 11–13. 10.1016/0968-0004(96)80878-4 [DOI] [PubMed] [Google Scholar]
- Dure L. (1993). A repeating 11-mer amino acid motif and plant desiccation. Plant J. 3, 363–369. 10.1046/j.1365-313X.1993.t01-19-00999.x [DOI] [PubMed] [Google Scholar]
- Englbrecht C. C., Schoof H., Böhm S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC genomics 5:39. 10.1186/1471-2164-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Gampala S. S., Rock C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, S15–S45. 10.1105/tpc.010441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Wang M. L., Lynch T. J., Rao S., Goodman H. M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukamatsu Y., Mitsui S., Yasuhara M., Tokioka Y., Ihara N., Fujita S., et al. (2005). Identification of LOV KELCH PROTEIN2 (LKP2)-interacting factors that can recruit LKP2 to nuclear bodies. Plant Cell Physiol. 46, 1340–1349. 10.1093/pcp/pci144 [DOI] [PubMed] [Google Scholar]
- Hanin M., Brini F., Ebel C., Toda Y., Takeda S., Masmoudi K. (2011). Plant dehydrins and stress tolerance: versatile proteins for complex mechanisms. Plant Signal. Behav. 6, 1503–1509. 10.4161/psb.6.10.17088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Sánchez I. E., Martynowicz D. M., Rodríguez-Hernández A. A., Pérez-Morales M. B., Graether S. P., Jiménez-Bremont J. F. (2014). A dehydrin-dehydrin interaction: the case of SK3 from Opuntia streptacantha. Front. Plant Sci. 5:520 10.3389/fpls.2014.00520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. 10.1093/nar/27.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yuan Q., Wang Y., Cai R., Deng X., Wang J., et al. (2012). Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol. 53, 2127–2141. 10.1093/pcp/pcs154 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Tran H. G., Swartz T. E., Briggs W. R., Kay S. A. (2003). FKF1 is essential for photoperiodic-specific light signaling in Arabidopsis. Nature 426, 302–306. 10.1038/nature02090 [DOI] [PubMed] [Google Scholar]
- Ingram J., Bartels D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403. 10.1146/annurev.arplant.47.1.377 [DOI] [PubMed] [Google Scholar]
- Kang J. Y., Choi H. I., Im M. Y., Kim S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. 10.1105/tpc.010362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Chong J., Ni M. (2005). HYPERSENSITIVE TO RED AND BLUE 1, a ZZ-type zinc finger protein, regulates phytochrome B-mediated red and cryptochrome-mediated blue light responses. Plant Cell 17, 822–835. 10.1105/tpc.104.029165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T., Wada M. (2000). LKP1 (LOV kelch protein 1) a factor involved in the regulation of flowering time in Arabidopsis. Plant J. 23, 807–815. 10.1046/j.1365-313x.2000.00850.x [DOI] [PubMed] [Google Scholar]
- Klug A., Schwabe J. W. (1995). Protein motifs 5. Zinc fingers. FASEB J. 9, 597–560. [PubMed] [Google Scholar]
- Koornneef M., Hanhart C., Hilhorst H., Karssen C. (1989). In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 90, 463–469. 10.1104/pp.90.2.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Lee D. H. (2003). Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiol. 132, 517–529. 10.1104/pp.103.020511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Giraudat J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. 10.1146/annurev.arplant.49.1.199 [DOI] [PubMed] [Google Scholar]
- Li G., Tai F. J., Zheng Y., Luo J., Gong S. Y., Zhang Z. T., et al. (2010a). Two cotton Cys2/His2-type zinc-finger proteins, GhDi19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling. Plant Mol. Biol. 74, 437–452. 10.1007/s11103-010-9684-6 [DOI] [PubMed] [Google Scholar]
- Li S., Xu C., Yang Y., Xia G. (2010b). Functional analysis of TaDi19A, a salt-responsive gene in wheat. Plant Cell Environ. 33, 117–129. 10.1111/j.1365-3040.2009.02063.x [DOI] [PubMed] [Google Scholar]
- Li Z. Y., Xu Z. S., Chen Y., He G. Y., Yang G. X., Li L. C., et al. (2013). A novel role for Arabidopsis CBL1 in affecting plant responses to glucose and gibberellin during germination and seedling development. PLoS ONE 8:e56412. 10.1371/journal.pone.0056412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Xu Z. S., Lu P. P., Hu D., Chen M., Li L. C., et al. (2013a). A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J. Exp. Bot. 64, 2915–2927. 10.1093/jxb/ert133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. X., Zhang F. C., Zhang W. Z., Song L. F., Wu W. H., Chen Y. F. (2013b). Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 6, 1487–1502. 10.1093/mp/sst031 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma F., Wang L., Li J., Samma M. K., Xie Y., Wang R., et al. (2014). Interaction between HY1 and H2O2 in auxin-induced lateral root formation in Arabidopsis. Plant Mol. Biol. 85, 49–61. 10.1007/s11103-013-0168-3 [DOI] [PubMed] [Google Scholar]
- Milla M. A., Townsend J., Chang I. F., Cushman J. C. (2006). The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways. Plant Mol. Biol. 61, 13–30. 10.1007/s11103-005-5798-7 [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K., Kanehisa M. (1992). A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14, 897–911. 10.1016/S0888-7543(05)80111-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Peisach E., Grant R. A. (2001). Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 70, 313–340 10.1146/annurev.biochem.70.1.313 [DOI] [PubMed] [Google Scholar]
- Parkinson H., Kapushesky M., Kolesnikov N., Rustici G., Shojatalab M., Abeygunawardena N., et al. (2009). ArrayExpress update–from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 37, D868–D872. 10.1093/nar/gkn889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. 10.1105/tpc.108.061515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L. X., Li Y., Li D. D., Xu W. L., Zheng Y., Li X. B. (2014). Arabidopsis drought-induced protein Di19-3 participates in plant response to drought and high salinity stresses. Plant Mol. Biol. 86, 609–625. 10.1007/s11103-014-0251-4 [DOI] [PubMed] [Google Scholar]
- Rabbani M. A., Maruyama K., Abe H., Khan M. A., Katsura K., Ito Y., et al. (2003). Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133, 1755–1767. 10.1104/pp.103.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera M., Figueras M., López C., Goday A., Pagès M. (2004). Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proc. Natl. Acad. Sci. U.S.A. 101, 9879–9884. 10.1073/pnas.0306154101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L., Liang H., Shuman J., Shulaev V., Davletova S., et al. (2004). When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134, 1683–1696. 10.1104/pp.103.033431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H., Maruyama K., Sakuma Y., Meshi T., Iwabuchi M., Mittler R. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136, 2734–2746. 10.1104/pp.104.046599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles M. A., Lu D., Klug A. (2000). The role of the central zinc fingers of transcription factor IIIA in binding to 5 s RNA. J. Mol. Biol. 301, 47–60. 10.1006/jmbi.2000.3946 [DOI] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292. 10.1046/j.1365-313X.2002.01359.x [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shionzaki K., Seki M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. 10.1016/S1369-5266(03)00092-X [DOI] [PubMed] [Google Scholar]
- Singh K. B., Foley R. C., Oñate-Sánchez L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. 10.1016/S1369-5266(02)00289-3 [DOI] [PubMed] [Google Scholar]
- Somers D. E., Schultz T. F., Milnamow M., Kay S. A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. 10.1016/S0092-8674(00)80841-7 [DOI] [PubMed] [Google Scholar]
- Takatsuji H. (1999). Zinc-finger proteins, the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078. 10.1023/A:1006184519697 [DOI] [PubMed] [Google Scholar]
- Tolleter D., Jaquinod M., Mangavel C., Passirani C., Saulnier P., Manon S., et al. (2007). Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell 19, 1580–1589. 10.1105/tpc.107.050104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A., Wise M. J. (2007). The continuing conundrum of the LEA proteins. Naturwissenschaften 94, 791–812. 10.1007/s00114-007-0254-y [DOI] [PubMed] [Google Scholar]
- Wolfe S. A., Nekludova L., Pabo C. O. (2000). DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29, 183–212. 10.1146/annurev.biophys.29.1.183 [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K. S., Zhu J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183. 10.1105/tpc.000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. S., Chen M., Li L. C., Ma Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. 10.1111/j.1744-7909.2011.01062.x [DOI] [PubMed] [Google Scholar]
- Xu Z. S., Xia L. Q., Chen M., Cheng X. G., Zhang R. Y., Li L. C., et al. (2007). Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65, 719–732. 10.1007/s11103-007-9237-9 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Mogami J., Yamaguchi-Shinozaki K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21C, 133–139 10.1016/j.pbi.2014.07.009 [DOI] [PubMed] [Google Scholar]
- You J., Zong W., Hu H., Li X., Xiao J., Xiong L. (2014). A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 166, 2100–2114. 10.1104/pp.114.251116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Chen L., Li D., Lv B., Chen Y., Chen J., et al. (2014). OsRACK1 is involved in abscisic acid- and H2O2-mediated signaling to regulate seed germination in rice (Oryza sativa, L.). PLoS ONE 9:e97120. 10.1371/journal.pone.0097120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Z., Creelman R. A., Zhu J. K. (2004). From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 135, 615–621. 10.1104/pp.104.040295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fasta file of soybean Di19 protein and conserved domain sequences.
Fasta file of all Arabidopsis and rice Di19 protein conserved regions.
Fasta files of soybean Di19-5 gene and protein sequences.
List of primers used in this paper.
NLS and NES sequences identified in soybean Di19 family proteins by the PSORT and NetNES softwares.
Nomenclature for Di19s in soybean, Arabidopsis, rice, maize, and Brachypodium.
Conserved motifs identified for the Di19 proteins from soybean, Arabidopsis, rice, maize, and Brachypodium by MEME software.
Phosphorylation sites predicted in soybean Di19 family proteins by NetPhos 2.0 Server softwares.
Prediction cis-elements of soybean Di19 family genes.
Expression profiles of Arabidopsis Di19 genes in response to various stress treatments. The data was downloaded from http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi.
Variation in motif clades of Di19 proteins from soybean, Arabidopsis, rice, maize, and Brachypodium. The MEME motifs were shown as different-colored boxes.
Logos of the conserved Di19 domain alignments of Arabidopsis, rice, maize, and Brachypodium.
Expression profiles of Arabidopsis Di19 genes in response to various stress treatments. Hierarchical clustering of differential gene expression in response to cold (A), drought (B), heat (C), osmotic (D), oxidative (E), and salinity (F) stresses.