Abstract
Amygdala and insula hyper-reactivity to threat is implicated in social anxiety disorder (SAD) yet inconsistencies in activation have been reported. One source of variance are individual differences in 5-HTTLPR genotype where the short (S), relative to long (L) allele, corresponds with greater amygdala activation. However, the impact of genotype on insula to threat in SAD is not known. During fMRI, 34 SAD patients and 28 healthy controls completed a perceptual assessment task comprising angry, fear, and happy faces. Results showed no diagnostic group differences in limbic/paralimbic regions but within SAD, greater insula, but not amygdala, activation to fearful faces was observed in subjects with SS genotype compared to LaLa genotype. Findings indicate genotype influenced insula activation to threat in SAD.
Keywords: social anxiety, threat, serotonin, brain imaging, 5-HTTLPR polymorphism, genetics
Introduction
Generalized social anxiety disorder (gSAD) is a common, debilitating disorder characterized by extreme, pervasive fears of situations involving potential negative evaluation by others [1]. The amygdala, which plays a key role in fear responses [2] has been a predominant focus of study. However, as noted in a review of the literature, associations between exaggerated amygdala activity to threat stimuli in SAD relative to healthy volunteers is demonstrated in some emotional face processing studies but not others [3]. A potential source of variance that may contribute to such inconsistent findings are individual differences in the function of serotonin, a neurotransmitter integral in the regulation of anxiety or fear [4]. Causes of alterations in serotonergic function includes the 5-HTTLPR, a polymorphism in the promoter region of the SLC6A4 gene in which the short allele of this variant has lower serotonin transporter availability (“low expressing”) than the long (“high expressing”) allele [5,6].
The relationship between genotype and amygdala has been demonstrated in healthy volunteers where carriers of one or two copies of the short allele compared to subjects homozygous for the long allele display greater amygdala response to threat faces [7]. Regarding social anxiety, positron emission tomography (PET) studies by Furmark and colleagues revealed increased regional cerebral blood flow to the amygdala in response to symptom provocation in SAD patients with at least one short allele relative to those homozygous for the long variant [8]. When comparing SAD to healthy volunteers, heightened amygdala activity to angry faces was attributed to the presence or absence of the short allele, regardless of diagnosis [9]. Collectively, variation in amygdala activation appears to be driven in part by individual differences in the 5-HTTLPR with one meta-analysis estimating that it may account for 10% of the variance in amygdala reactivity [10].
Less clear is the effect of the 5-HTTLPR polymorphism genotype on insula activation in gSAD despite data showing SAD is associated with lower serotonin receptor binding potential in this region, relative to healthy controls, along withlower serotonin binding potential in other areas implicated in the pathophysiology of SAD (e.g., amygdala) [11]. The anterior insula is involved in interoception (i.e., awareness of bodily state) [12], and findings of exaggerated insula reactivity to salient signals in SAD compared to healthy volunteers [3] lends support to the proposal that altered interoception underlies anxiety disorders [13]. Further implicating the role of insula in SAD are negative beliefs regarding somatic cues (e.g., shaking, sweating) [14] and evidence of greater interoceptive accuracy in an analogue sample of SAD participants [15]. Yet, various studies have failed to observe SAD-related insula hyperactivation [3] and a recent meta-analysis of fMRI studies of emotion recognition found that amygdala but not insula activation differentiated individuals with SAD from controls [16]. Given these conflicting findings, individual differences in the 5-HTTLPR genotype may moderate the association of gSAD and insula functioning.
Therefore, the goal of the current study was to examine the effect of genetic variation on activation using a modified version of the Emotional Face Matching Task [17], a task designed to isolate brain response to signals of threat (e.g., angry and fearful faces) against those that do not convey threat (e.g., happy faces). We hypothesized that gSAD patients compared to healthy controls would exhibit greater anterior insula and amygdala response to threat faces and that genotype would have a moderating effect on group differences. Additionally, in light of evidence that individual differences in the 5-HTTLPR genotype impacted amygdala activity to threat more so than a diagnosis of SAD [9], we hypothesized that limbic/paralimbic activity would be heightened in individuals with the short/short relative to longA/longA genotype across diagnostic groups. Lastly, we explored whether symptom severity in gSAD correlated with significant insula and/or amygdala response.
Methods
Ethics statement
All participants provided written informed consent and all procedures were approved by the Institutional Review Boards of the University of Chicago and Michigan Medical School.
Participants comprised 34 unmedicated gSAD patients not in treatment and 28 healthy controls (HC). All participants completed the Structured Clinical Interview for DSM-IV [18] conducted by licensed clinicians as well as the Liebowitz Social Anxiety Scale (LSAS), a clinician-rated measure of symptom severity [19]. GSAD was the primary diagnosis for all patients and we did not exclude patients with co-morbid disorders, which were generalized anxiety disorder (n=4), specific phobia (n=3), panic disorder (n=1), somatoform disorder (n=1), and eating disorder (n=1). HC had no history of any Axis I disorder. Exclusion criteria for gSAD patients additionally included major depressive disorder and substance abuse/dependence (within 6 months of study), or any history of other major psychiatric illness (e.g., bipolar disorder, psychosis).
Participants were between 18 and 55 years of age, right-handed, and free of current and past major medical or neurologic illness, as confirmed by a Board Certified psychiatrist. None of the participants tested positive for alcohol or illegal substances at the time of fMRI scanning.
5-HTTLPR Genotyping
DNA was extracted from either saliva (n=40) or blood (n=22) samples. After extraction, DNA was quantified with Quant-iTPicoGreen dsDNA Assay (Invitrogen, Carlsbad, CA) and normalized to a concentration of 10ng/uL. DNA was amplified with FAM –labeled forward primer 5′-FAM-CTG AAT GCC AGC ACC TAA CCC CTA ATG T-3′ (Applied Biosystems, Foster City, CA) and reverse primer (with pigtail sequence in parentheses) 5′(GTTTCT) TGG GGA ATA CTG GTA GGG TGC AAG GAG AA-3′ (Applied Biosystems, Foster City, CA) using Dynazyme EXT Polymerase (Finnzymes, Espoo, Finland) with an initial denaturation step of 96°C for 12 min. followed by 45 cycles of 96°C for 30sec, 68°C for 45sec, 72°C for 3min, one hold at 72°C for 10 min., and a final hold at 10°C. Products were digested using Msp I restriction enzyme (Promega, Madison, WI) to genotype rs25531 since the G allele is cut and the A allele is uncut. Both cut and uncut products were separated on a 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA) in the UIC Research Resources Center DNA Services Facility. Genotypes were called blind to phenotype data using Genemapper v 3.7. 5HTTLPR, including the SNP contained within the length variant, was coded into the following three categories: low (Sa/Sa, Sg/Sa) (“SS”), intermediate (Sa/Lg, Sa/La, Lg/La, Sa/XLa, Lg/XXLa), and high (“LaLa”) expressing. A priori, we decided to compare the homozygous low (SS; n=40), and homozygous high (LaLa; n=22) expressing genotype between and across gSAD and HC groups.
fMRI Task
This task has been previously described in detail [20]. In brief, in each trial, three photographs from a validated set of face stimuli [21] were presented and participants selected whether one of two faces (bottom) expressed the same emotion as the target face (top). The target and congruent probe faces displayed one of three expressions (angry, fearful, or happy), and the other (incongruent) probe face always displayed a neutral/non-emotional expression. Trials were presented in blocks and the paradigm consisted of 18 blocks in total, specifically, 9 blocks of matching emotional faces with each target expression of fearful, angry, or happy interleaved with 9 blocks of matching shapes (i.e., “baseline” condition). Participants used a right-handed button press to record responses.
fMRI Data Acquisition
This study was conducted on two separate 3 Tesla GE Signa System (General Electric; Milwaukee, WI) scanners using the same standard radiofrequency coil –one at the University of Chicago (gSAD n=14; HC n=8) and another at the University of Michigan (gSAD n=20; HC n=20). Of note (see below) our planned analyses used scanner site as a covariate. All scanning was performed with blood oxygen-level dependent (BOLD)-sensitive whole-brain fMRI using the same GE software (LX 8.3, Neuro-optimized gradients) and acquired using the exact same T2*-weighted reverse spiral gradient-recall echo sequence (echo time=25ms, repetition time=2000ms, 64×64 matrix, flip angle=77°, field of view=24cm, 3.75mm2 inplane voxels, 30 contiguous 5mm axial slices/volume) optimized to minimize susceptibility artifacts in the regions of interest (i.e., amygdala and insula). A high-resolution T1 scan was also acquired for anatomical localization.
fMRI data preprocessing and analysis
All the participants included in this analysis met inclusion criteria for minimal head movement (<3mm or <3 degrees of displacement) during scans. The first four volumes from each run were discarded to allow for T1 equilibration effects. Data were preprocessed and analyzed using statistical parametric mapping (SPM8; Wellcome Department of Cognitive Neurology, London; www.fil.ion.ucl.ac.uk/spm). Functional scans were preprocessed using the following steps: 1) temporally/slice-time corrected; 2) spatial realignment for motion correction; 3) coregistered to each individual's T1 image; 4) anatomical images were segmented into grey and white matter and normalized to match a canonical template brain in Montreal Neurologic Institute (MNI) space using the VBM8 toolbox; 5) functional images were normalized based on the VBM8 deformation field and resampled to 2mm3 voxels; and 6) smoothing was applied using an 8 mm3 kernel. A general linear model was applied to the time series, convolved with the canonical hemodynamic response function and with a 128 s high-pass filter.
Using a box-car model, contrasts of interest (Angry vs. Shapes, Fear vs. Shapes, and Happy vs. Shapes) were generated for each subject, and then entered into a second-level general linear model treating subject as a random effect (i.e., a random effects analysis). A 2 (Group: gSAD, HC)×2 (Genotype: SS, LaLa)×3 (Emotion: Angry, Fear, Happy) Analysis of Variance (ANOVA), controlling for scanner site, was conducted in SPM8.
To test our a priori hypotheses, we used a region of interest (ROI) approach localized to anatomically-based bilateral amygdala and insula masks [22]. To search for reactivity within the anterior insula (aINS), the anterior portion was demarcated as y-axis=0 and forward. The F-statistical map was set at p<0.001 with at least 20 contiguous voxels. To correct for multiple comparisons in anatomically-based ROIs, a small volume correction (svc) was applied [23]. The amygdala search volume comprised 1760 voxels on the left and 1984 voxels on the right, and for anterior insula, the search volume consisted of 9552 voxels on the left and 8776 voxels on the right.
To clarify results, parameter estimates (β weights, arbitrary units [a.u.]) of brain activation based on spherical 10 mm-diameter ROIs centered on peak task-related activations depicting interactions with group or group main effects was extracted from each participant and analyzed in the Statistical Package for the Social Sciences (SPSS; Chicago, IL version 20). To examine potential influences of genotype and diagnostic group on significant a priori activity, β weights were submitted to simple slopes analysis in SPSS. To illustrate the direction and magnitude of significant simple slopes findings, follow-up two-tailed t-tests were conducted in SPSS. Additionally, two-tailed Pearson correlations were used to examine relationships between symptom severity (i.e., LSAS scores) and significant neural activity.
Results
Study participants
GSAD and HC groups were similar in age [t(60)=1.42, p=0.16], gender [χ2(1, N=62)=0.14, p=0.71], and race/ethnicity (Caucasian 77%, African American 10%, Asian 10%, Other 3%) [χ2(3, N=62)=4.88, p=0.18]. The frequency distribution for genotype was comparable between gSAD (SS=35.5%; LaLa=19.4%) and HC (SS=29%; LaLa=16.1%) groups [χ2(3, N=62)=5.87, p=0.12]. A 2 (Group)×2 (Genotype) ANOVA revealed a significant main effect of Group for LSAS[F(1, 58)=405.23, p<0.001] but there was no main effect for Genotype (p=0.76) or Group × Genotype interaction (p=0.96). Post-hoc analyses showed the gSAD patients exhibited greater symptom severity (LSAS mean±SD: 81.5±15.8) than the HC group (9.4±9.2) (p<0.001).
fMRI results
The ANOVA findings revealed a Group × Genotype × Emotion interaction for bilateral anterior insula (aINS) [right (42, 16, -14), F=9.02, volume=264 mm3, svc p<0.02; left (-34, 10, -12), F=9.04, volume=264 mm3, svc p<0.02] (Fig 1). There were no main effects for a priori ROIs or other interactions with insula or amygdala.
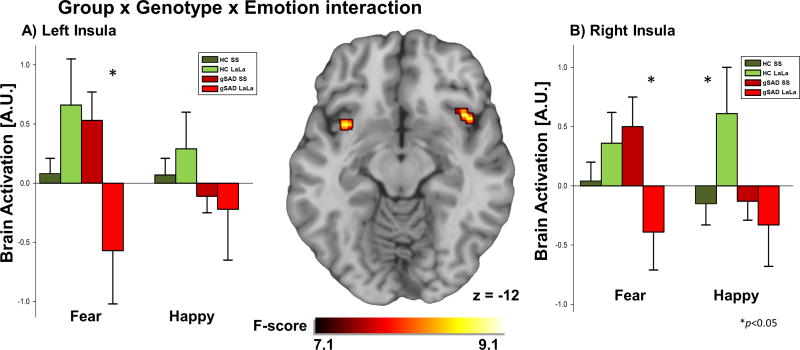
Figure 1.
A) Brain map depicting whole-brain voxel-wise ANOVA F-map showing a significant Group × Genotype × Emotion interaction in the insula in response to fearful and happy faces. B) Bar graph depicts extracted BOLD signal change from insula region of interest showing right and left insula reactivity to fearful faces is greater in gSAD patients with SS than patients with LaLa, who showed reduced insula activation (gSAD SS >gSAD LaLa; right p<0.04, left p<0.02); in the HC group, right insula response to happy faces was greater in controls with LaLa than for controls with the SS genotype (HC LaLa> HC SS, p<0.05).
gSAD, Generalized Social Anxiety Disorder; Healthy Control (HC); SS, short allele for the serotonin transporter gene; LaLa, long allele for the serotonin transporter gene
Regarding right aINS, simple slopes analysis showed a significant Genotype × Group interaction for fearful faces (vs. shapes) (p<0.03) where genotype significantly modulated insula response in gSAD patients (B=0.42, p<0.02) but not in controls (B=-0.15, p=0.42) (Fig 2). Specifically, within the gSAD group, patients with SS exhibited greater insula activity compared to patients with LaLa who showed reduced insula activation [t(32)=2.14, p<0.04]. Insula response to fearful faces within gSAD SS and gSAD LaLa groups did not correlate with symptom severity. Furthermore, no significant results were observed for fearful faces between gSAD patients and controls for the SS (p=0.15) or LaLa (p=0.09) genotype.
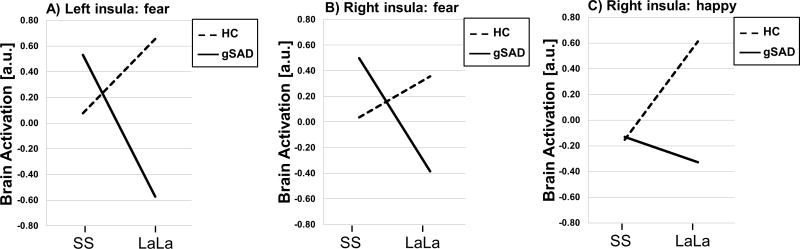
Figure 2.
A) Slopes depicting genotype (i.e., SS vs. LaLa) modulated left insula to fearful faces in gSAD patients (p<0.02) but not in controls (p=0.42). B) Slopes showing genotype impacted right insula to fearful faces in gSAD (p<0.01) but not controls (p=0.19). C) Slopes illustrating genotype modulated right insula to happy faces in HC (p<0.04) but not gSAD (p=0.56)
gSAD, Generalized Social Anxiety Disorder; Healthy Control (HC); SS, short allele for the serotonin transporter gene; LaLa, long allele for the serotonin transporter gene
In the control group, genotype impacted right aINS activity to happy faces though the interaction was a non-significant trend (p=0.06). In exploring this trend, we observed a significant influence of genotype in controls (B=-0.38, p<0.04) with no evidence of a moderational effect in gSAD (B=0.10, p=0.56). Controls with the high expressing (LaLa) genotype exhibited an enhanced response to happy faces whereas SS was associated with less activation [t(26)=2.03, p<0.05]. No significant results emerged for happy faces between HC and gSAD groups for SS (p=0.93) or LaLa (p=0.09). With regard to angry faces, there was no evidence genotype interacted with group (p>0.10).
For left aINS, simple slopes analysis revealed a similar outcome for fearful faces (vs. shapes) as the interaction was significant (p<0.01) and activity was modulated by genotype in gSAD patients (B=0.46, p<0.01) but not controls (B=-0.24, p=0.19) (Fig 2). Again, within the gSAD group, SS corresponded with increased insula and LaLa with decreased activation [t(32)=2.39, p<0.02]. The differential activation did not, however, correlate with symptom severity. With regard to LaLa participants only, there was a non-significant trend toward gSAD patients exhibiting greater activation to fearful faces than controls with LaLa (p=0.06). No effect between diagnostic group regarding the SS genotype was evident (p=0.12), and no interactions were observed for happy (p=0.51) or angry faces (p=0.13).
Discussion
In this fMRI study we examined the impact of 5-HTTLPR genotype on anterior insula (aINS) and amygdala response to fearful, angry, and happy faces in patients with generalized social anxiety disorder (gSAD) and healthy controls. We observed bilateral aINS activation was moderated by genotype within the gSAD group; specifically, patients with the homozygous low expressing (i.e., short “SS”) allele for the serotonin transporter gene exhibited greater aINS activation to fear (vs. shapes) stimuli compared to patients homozygous for the high expressing (i.e., LaLa) allele, who demonstrated reduced aINS activation. In healthy controls right aINS response to happy faces (vs. shapes) was impacted by geneotype in that LaLa was associated with enhanced activation compared to SS.
The finding that genotype's effect on bilateral aINS response to fear-evoking stimuli was specific to patients and not controls points to the importance of individual differences in the 5-HTTLPR polymorphism. Indeed, there was no main effect of diagnostic group for insula response. The greater aINS activation in gSAD indicates a hyper-sensitivity to fear signals and suggests patients with the SS relative to the LaLa genotype may be more vulnerable to make negative interpretations about internal state prompted by salient external stimuli and/or more likely to be vigilant to perceived threat due to an amplified interoceptive system [13].
Data signifying decreased aINS response to fearful faces in gSAD patients with the homozygous long genotype is generally consistent with evidence that homozygous high expressing individuals exhibit reduced limbic activation [7] and less attention [24] to aversive stimuli. Interestingly, there was a non-significant trend towards less left insula activation to fearful faces in gSAD patients with LaLa than healthy controls with LaLa indicating this allele may moderate insula response relative to normative function. Furthermore, individuals homozygous for the long allele have been shown to exhibit attentional bias to positive information [24] and in keeping with a positivity bias, LaLa in the healthy control group was linked to enhanced right aINS activation to happy faces compared to controls with the SS genotype. However, a comparable response was not observed in patients suggesting LaLa did not aid in eliciting activation to a socio-emotional signal that conveys acceptance in gSAD.
In contrast to our hypothesis, we did not observe a main effect of genotype in gSAD or a group by genotype interaction for amygdala activation. The results are fairly surprising given evidence of exaggerated amygdala response to threat in gSAD relative to controls [3] and influence of genotype on amygdala activation in healthy volunteers [7]. Nonetheless, the study of genetics on brain response in gSAD is at a nascent stage. The failure to replicate amygdala findings include methodological differences between our study and previous ones such as neuroimaging approach (e.g., PET vs. fMRI) and distribution of allele frequencies in participants [8,9], which differ between Caucasian and Asian groups [25], for example, together with our small sample size. The sample size may have also limited power to detect angry face effects or correlations with symptom severity. Therefore, these preliminary data need to be interpreted with caution as further study is necessary to estimate the effect size to ensure power is adequate to examine the impact of genotype on limbic/paralimbic response to threat stimuli in gSAD.
Conclusion
Preliminary results indicate bilateral aINS activity to threat-relevant stimuli (i.e., fearful faces) was modulated by the 5-HTTLPR genotype such that patients homozygous for the short variant (SS) exhibited greater bilateral activation to fearful faces relative to patients homozygous for the long variant (LaLa), who showed reduced aINS activation. In healthy controls, a right aINS response to happy faces was influenced by genotype where homozygotes for the short allele demonstrated reduced activation and those who were homozygous for the long allele, a heightened response to positive stimuli. Given the small sample size, interpretations should be made with caution. Nevertheless, our observations underscore the value of taking into account genotype of the 5-HTTLPR polymorphism when examining neural mechanisms of anxiety.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Institute of Mental Health (MH076198 to KLP and MH093679 to HK).
Footnotes
Conflicts of Interest: None.
References
- 1.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 2.LeDoux JE. Emotion circuits in the brain. Annu Rev Neurosci. 2000;23:155–184. doi: 10.1146/annurev.neuro.23.1.155. [DOI] [PubMed] [Google Scholar]
- 3.Freitas-Ferrari MC, Hallak JEC, Trzesniak C, Filho AS, Machado-de-Sousa JP, Chagas MHN, et al. Neuroimaging in social anxiety disorder: A systematic review of the literature. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34(4):565–580. doi: 10.1016/j.pnpbp.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Charney DS, Woods SW, Krystal JH, Heninger GR. Serotonin function and human anxiety disorders. Ann N Y Acad Sci. 1990;600(1):558–572. doi: 10.1111/j.1749-6632.1990.tb16910.x. [DOI] [PubMed] [Google Scholar]
- 5.Heils A, Teufel A, Petri S, Stöber G, Riederer P, Bengel D, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66(6):2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 6.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274(5292):1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 7.Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297(5580):400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 8.Furmark T, Tillfors M, Garpenstrand H, Marteinsdottir I, Långström B, Oreland L, et al. Serotonin transporter polymorphism related to amygdala excitability and symptom severity in patients with social phobia. Neurosci Lett. 2004;362(3):189–192. doi: 10.1016/j.neulet.2004.02.070. [DOI] [PubMed] [Google Scholar]
- 9.Furmark T, Henningsson S, Appel L, Ahs F, Linnman C, Pissiota A, et al. Genotype over-diagnosis in amygdala responsiveness: Affective processing in social anxiety disorder. J Psychiatry Neurosci. 2009;34(1):30–40. [PMC free article] [PubMed] [Google Scholar]
- 10.Munafò MR, Brown SM, Hariri AR. Serotonin transporter (5-HTTLPR) genotype and amygdala activation: A meta-analysis. Biol Psychiatry. 2008;63(9):852–857. doi: 10.1016/j.biopsych.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanzenberger RR, Mitterhauser M, Spindelegger C, Wadsak W, Klein N, Mien LK, et al. Reduced serotonin-1A receptor binding in social anxiety disorder. Biol Psychiatry. 2007;61(9):1081–1089. doi: 10.1016/j.biopsych.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Critchley HD, Wiens S, Rotshtein P, Öhman A, Dolan RJ. Neural systems supporting interoceptive awareness. Nat Neurosci. 2004;7(2):189–195. doi: 10.1038/nn1176. [DOI] [PubMed] [Google Scholar]
- 13.Paulus MP, Stein MB. Interoception in anxiety and depression. Brain Struct Funct. 2010;214(5-6):451–463. doi: 10.1007/s00429-010-0258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark DM, Wells A. A cognitive model of social phobia. In: Heimberg RG, Liebowitz MR, Hope DA, Schneier FR, editors. Soc Phobia Diagn Assess Treat. New York, NY: US: Guilford Press; 1995. pp. 69–93. [Google Scholar]
- 15.Stevens S, Gerlach AL, Cludius B, Silkens A, Craske MG, Hermann C. Heartbeat perception in social anxiety before and during speech anticipation. Behav Res Ther. 2011;49(2):138–143. doi: 10.1016/j.brat.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Hattingh CJ, Ipser J, Tromp S, Syal S, Lochner C, Brooks SJB, et al. Functional magnetic resonance imaging during emotion recognition in social anxiety disorder: An activation likelihood meta-analysis. Front Hum Neurosci. 2013;6:347. doi: 10.3389/fnhum.2012.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hariri AR, Tessitore A, Mattay VS, Fera F, Weinberger DR. The amygdala response to emotional stimuli: A comparison of faces and scenes. NeuroImage. 2002;17(1):317–323. doi: 10.1006/nimg.2002.1179. [DOI] [PubMed] [Google Scholar]
- 18.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured clinical interview for DSM-IV Axis I disorders, clinician version (SCID-CV) Washington, D.C.: American Psychiatric Association; 1996. [Google Scholar]
- 19.Liebowitz MR. Social phobia. Mod Probl Pharmacopsychiatry. 1987;22:141–173. doi: 10.1159/000414022. [DOI] [PubMed] [Google Scholar]
- 20.Phan KL, Coccaro EF, Angstadt M, Kreger KJ, Mayberg HS, Liberzon I, et al. Corticolimbic brain reactivity to social signals of threat before and after sertraline treatment in generalized social phobia. Biol Psychiatry. 2013;73(4):329–336. doi: 10.1016/j.biopsych.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gur RC, Schroeder L, Turner T, McGrath C, Chan RM, Turetsky BI, et al. Brain activation during facial emotion processing. NeuroImage. 2002;16(3, Part A):651–662. doi: 10.1006/nimg.2002.1097. [DOI] [PubMed] [Google Scholar]
- 22.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 23.Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC. A unified statistical approach for determining significant signals in images of cerebral activation. Hum Brain Mapp. 1996;4(1):58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 24.Fox E, Ridgewell A, Ashwin C. Looking on the bright side: Biased attention and the human serotonin transporter gene. Proc R Soc B Biol Sci. 2009;276(1663):1747–1751. doi: 10.1098/rspb.2008.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smits KM, Smits LJM, Schouten JSAG, Stelma FF, Nelemans P, Prins MH. Influence of SERTPR and STin2 in the serotonin transporter gene on the effect of selective serotonin reuptake inhibitors in depression: A systematic review. Mol Psychiatry. 2004;9(5):433–441. doi: 10.1038/sj.mp.4001488. [DOI] [PubMed] [Google Scholar]