Transcriptome atlases of Tarenaya hassleriana, a C3 plant, and the C4-species Gynandropsis gynandra were developed. Integrated analysis of RNA-seq and leaf anatomical data indicates altered regulation of the cell cycle and endoreduplication during the establishment of C4 leaf anatomy. C4 genes have been recruited to photosynthetic gene expression regulation from ancestral nonphotosynthetic functions in C3.
Abstract
C4 photosynthesis outperforms the ancestral C3 state in a wide range of natural and agro-ecosystems by affording higher water-use and nitrogen-use efficiencies. It therefore represents a prime target for engineering novel, high-yielding crops by introducing the trait into C3 backgrounds. However, the genetic architecture of C4 photosynthesis remains largely unknown. To define the divergence in gene expression modules between C3 and C4 photosynthesis during leaf ontogeny, we generated comprehensive transcriptome atlases of two Cleomaceae species, Gynandropsis gynandra (C4) and Tarenaya hassleriana (C3), by RNA sequencing. Overall, the gene expression profiles appear remarkably similar between the C3 and C4 species. We found that known C4 genes were recruited to photosynthesis from different expression domains in C3, including typical housekeeping gene expression patterns in various tissues as well as individual heterotrophic tissues. Furthermore, we identified a structure-related module recruited from the C3 root. Comparison of gene expression patterns with anatomy during leaf ontogeny provided insight into genetic features of Kranz anatomy. Altered expression of developmental factors and cell cycle genes is associated with a higher degree of endoreduplication in enlarged C4 bundle sheath cells. A delay in mesophyll differentiation apparent both in the leaf anatomy and the transcriptome allows for extended vein formation in the C4 leaf.
INTRODUCTION
C4 photosynthesis has evolved concurrently and convergently in angiosperms more than 65 times from the ancestral C3 state (Sage et al., 2011) and provides fitness and yield advantages over C3 photosynthesis under permissive conditions, such as high temperatures (Hatch, 1987; Sage, 2004). In brief, C4 photosynthesis represents a biochemical CO2 pump that supercharges photosynthetic carbon assimilation through the Calvin-Benson-Bassham cycle (CBBC) by increasing the concentration of CO2 at the site of its assimilation by the enzyme Rubisco (Andrews and Lorimer, 1987; Furbank and Hatch, 1987). Rubisco is a bifunctional enzyme that catalyzes both the productive carboxylation and the futile oxygenation of ribulose 1,5-bisphosphate. The oxygenation reaction produces a toxic byproduct, 2-phosphoglycolic acid (Anderson, 1971), which is removed by an energy-intensive metabolic repair process called photorespiration. By concentrating CO2 through the C4 cycle, the oxygenation of ribulose 1,5-bisphosphate and thereby photorespiration is massively reduced. However, the C4 cycle requires input of energy to drive the CO2 pump. Photorespiration increases with temperature and above ∼23°C, the energy requirements of metabolic repair become higher than the energy cost of the C4 cycle (Ehleringer and Björkman, 1978; Ehleringer et al., 1991). Hence, operating C4 photosynthesis is beneficial at high leaf temperatures, whereas C3 photosynthesis prevails in cool climates (Ehleringer et al., 1991; Zhu et al., 2008).
With a few exceptions, C4 photosynthesis requires specialized Kranz anatomy (Haberlandt, 1896), in which two distinct cell types share the photosynthetic labor, namely, mesophyll cells (MCs) and bundle sheath cells (BSCs). MCs surround the BSCs in a wreath-like manner and both cell types form concentric rings around the veins. This leads to a stereotypic vein-BSC-MC-MC-BSC-vein pattern (Brown, 1975). MCs serve as carbon pumps that take in CO2 from the leaf intercellular air space, convert it into a C4 carbon compound, and load it into the BSCs. Here, CO2 is released from the C4 compound and assimilated into biomass by the CBBC, and the remaining C3-compound is returned to the MC to be loaded again with CO2. The carbon pump runs at a higher rate than the CBBC (overcycling), which leads to an increased concentration of CO2 in the BSCs. Our understanding of the different elements required for C4 photosynthesis varies, with many components of the metabolic cycle known, while their interplay and regulation remain mostly enigmatic, and very little is known about their anatomical control (Sage and Zhu, 2011).
C4 photosynthesis can be considered a complex trait, since it requires changes to the expression levels of hundreds or perhaps thousands of genes (Bräutigam et al., 2011, 2014; Gowik et al., 2011). While complex traits are typically dissected by measuring the quantitative variation across a polymorphic population, this approach is not promising for C4 photosynthesis, due to lack of known plasticity in “C4-ness” (Sage and McKown, 2006). Historical crosses between C3 and C4 plants (Chapman and Osmond, 1974) are no longer available and would have to be reconstructed before they can be analyzed with molecular tools.
Alternatively, closely related C3 and C4 species provide a platform for studying C4 photosynthesis. In the Cleomaceae and Asteraceae, comparative transcriptomic analyses have identified more than 1000 genes differentially expressed between closely related C3 and C4 species (Bräutigam et al., 2011; Gowik et al., 2011). These studies, however, compared the end points of leaf development, i.e., fully matured photosynthetic leaves. Therefore, they do not provide insight into the dynamics of gene expression during leaf ontogeny, which is important for understanding the establishment of C4 leaf anatomy. Systems analyses of maize (Zea mays) leaf gradients have provided a glimpse into developmental gene expression modules (Li et al., 2010; Pick et al., 2011; Wang et al., 2013); however, maize lacks a close C3 relative and has simple parallel venation making any generalizations to dicot leaf development difficult.
Tarenaya hassleriana, previously known as Cleome hassleriana (Iltis and Cochrane, 2007; Iltis et al., 2011), which is a C3 plant, and Gynandropsis gynandra (previously known as Cleome gynandra), which is a derived C4 plant, represent an ideal pair for a comparative analysis of the complex trait of C4 photosynthesis (Bräutigam et al., 2011). Both species belong to the family of Cleomaceae, are closely related to each other and to the well-annotated C3 plant model species Arabidopsis thaliana (Brown et al., 2005; Marshall et al., 2007; Inda et al., 2008), and both Cleome sister lineages share many traits (Iltis et al., 2011). In addition, the genome of T. hassleriana has been recently sequenced and serves as a reference for expression profiling via RNA sequencing (Cheng et al., 2013).
In this study, we take advantage of the phylogenetic proximity between G. gynandra and T. hassleriana to compare the dynamic changes in gene expression during leaf development (Inda et al., 2008). We generated a transcriptome atlas for each species, consisting of three biological replicates of six different stages of leaf development, three different stages of each seed and seedling development, reproductive organs (carpels, stamen, petals, and sepals), stems, and roots. In parallel, we performed microscopy analysis of the leaf anatomy. Finally, we measured leaf cell ploidy levels by flow cytometry and measurements of nuclear size in different leaf cell types by confocal laser scanning microscopy.
RESULTS
Selection of Tissues Featured in the Comparative Atlases
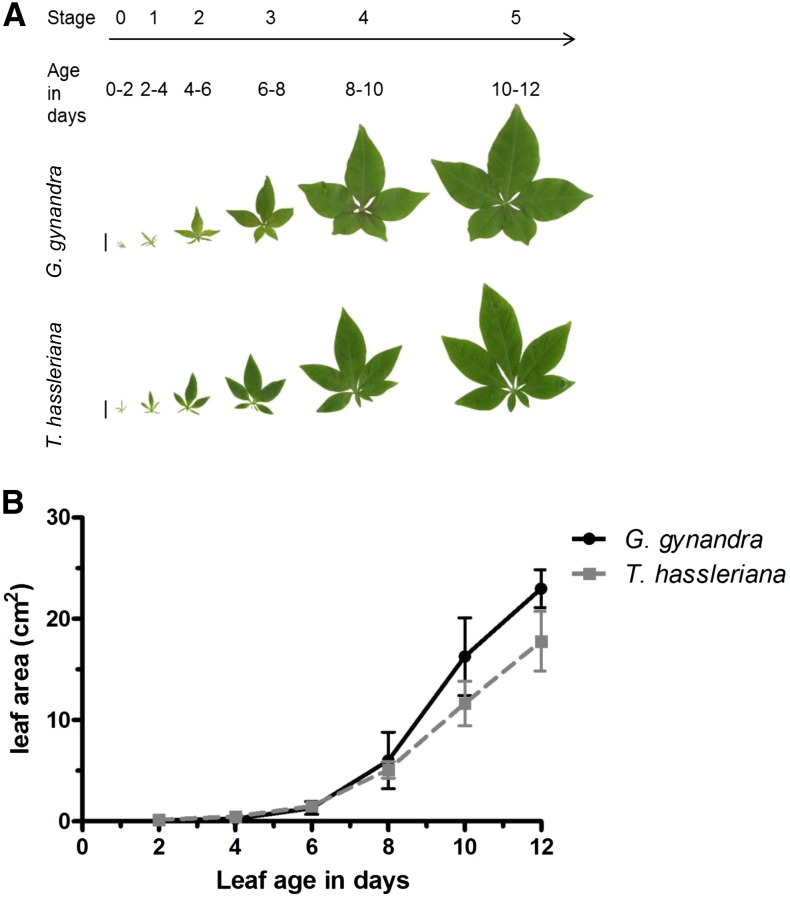
For high-resolution characterization of photosynthetic development between a dicotyledonous C3 and C4 species, a leaf developmental gradient was defined. Stage 0 was the youngest sampled leaf, 2 mm in length, and not yet emerged from the apex. The stage 0 leaves are the first to show a discernible palmate shape and contain the first order vein (midrib vein) in both species (Figure 1A; Supplemental Figure 1A). New leaves emerged from the apex every 2 d (plastochron = 2 d) in both species and were numbered sequentially from the aforementioned stage 0 to stage 5 (Figure 1A). The leaves emerge and initiate secondary vein formation at stage 1 (Supplemental Figure 1B) and fully mature by stages 4 and 5 (Supplemental Figures 1E and 1F). The mature leaf of the C4 species has more minor veins (up to 7°) than that of the C3 species (up to 6°; Supplemental Figure 1F). The leaf expansion rate is initially indistinguishable and never significantly different between the species (Figure 1B). The sampled leaf gradient covered the development from non-light-exposed sink tissues to fully photosynthetic source tissues.
Figure 1.
Overview of Leaf Shape and Expansion Rate in G. gynandra and T. hassleriana.
(A) Image of each leaf category sequenced (bar = 1 cm). Each category is 2 d apart from the other.
(B) Leaf expansion rate of each leaf category in cm2 over 12 d (n = 5; ±sd)
[See online article for color version of this figure.]
Complementary to this and to provide a broader comparison between C3 and C4 plants, seedlings, minor photosynthetic, and heterotrophic tissues were selected for further characterization. The aerial portion of seedlings (cotyledon and hypocotyl) was sampled 2, 4, and 6 d after germination to cover early cotyledon maturation (Supplemental Figure 2). The full root system and stem tissue were sampled from plants after 6 to 8 weeks of growth before inflorescence emergence (Supplemental Figure 3A); floral organs (petals, carpels, stamen, and sepals) were harvested during flowering of 10- to 14-week-old plants as well as three different stages of seed development (Supplemental Figure 3B). In total, 10 phototrophic and 8 heterotrophic tissues per species were included in the atlases (Table 1).
Table 1. Sequencing and Mapping Stats for Each Averaged Tissue Sample in T. hassleriana and G. gynandra.
|
T. hassleriana |
G. gynandra |
||||||
|---|---|---|---|---|---|---|---|
| Total No. of Reads in Three Replicates | No. of Genes Expressed > 1 RPKM | No. of Genes Expressed > 1000 RPKM | Total No. of Reads in Three Replicates | No. of Genes Expressed > 1 RPKM | No. of Genes Expressed > 1000 RPKM | ||
| Leaf gradient | 0 | 58,874,878 | 23,238 | 64 | 75,895,556 | 22,357 | 104 |
| 1 | 59,389,701 | 23,134 | 74 | 66,822,298 | 22,021 | 133 | |
| 2 | 63,590,283 | 23,104 | 81 | 55,247,053 | 22,143 | 129 | |
| 3 | 90,654,684 | 23,004 | 90 | 75,944,275 | 21,854 | 144 | |
| 4 | 36,572,303 | 22,844 | 106 | 69,951,930 | 21,734 | 119 | |
| 5 | 102,018,867 | 22,905 | 106 | 69,639,670 | 21,039 | 119 | |
| Floral organs | Sepal | 103,721,357 | 23,656 | 74 | 77,430,418 | 23,145 | 83 |
| Petal | 21,754,853 | 21,379 | 86 | 10,872,686 | 21,322 | 77 | |
| Stamen | 57,929,412 | 22,642 | 140 | 55,748,506 | 22,489 | 133 | |
| Carpel | 28,021,839 | 23,910 | 67 | 4,929,824 | 23,577 | 76 | |
| Stem | 30,932,633 | 23,292 | 75 | 59,516,389 | 22,508 | 98 | |
| Root | 88,911,824 | 24,255 | 68 | 86,879,963 | 23,430 | 89 | |
| Seedling | 2 DAG | 90,777,012 | 23,306 | 120 | 89,262,140 | 21,960 | 130 |
| 4 DAG | 89,517,055 | 23,041 | 116 | 112,658,149 | 22,036 | 130 | |
| 6 DAG | 71,271,739 | 22,877 | 138 | 64,470,699 | 21,910 | 136 | |
| Seed maturation | 1 | 52,229,844 | 23,708 | 118 | 32,763,383 | 22,991 | 118 |
| 2 | 31,872,067 | 22,969 | 145 | 29,958,720 | 22,262 | 148 | |
| 3 | 53,271,349 | 21,737 | 138 | 56,453,325 | 20,082 | 152 | |
Reads were normalized as RPKM (n = 3). DAG, days after germination.
The C3 and C4 Transcriptomes Are of High Quality and Comparable between Species
Cross-species mapping provided a more reliable data set than de novo transcriptome assembly. Between 1.4 and 67 million high-quality reads were generated per replicate (Supplemental Data Set 2). Initially, paired-end reads from each tissue were assembled by VELVET/OASES (Supplemental Table 1). Comparing the resulting contigs to reference data, including the T. hassleriana genome (Cheng et al., 2013), revealed several quality issues. These include excessive numbers of contigs mapping to single loci, fused and fragmented contigs, and the absence of C4 transcripts known to be highly expressed in G. gynandra (Supplemental Figures 4A to 4C and Supplemental Data Set 3). As an alternative, we aligned single-end reads from both species to the recently sequenced T. hassleriana genome (Cheng et al., 2013). Albeit slightly lower, the mapping efficiency and specificity remained comparable between both species with 60 to 70% of reads mapped for both leaf gradients (Supplemental Data Set 1). To define an upper boundary for any artifacts caused by cross-species mapping, three T. hassleriana samples (mature leaf stage 5, stamen, and young seed) were mapped to Arabidopsis. The correlation between replicates was equivalent in reads mapped to the cognate genome and across species with an average r = 0.98. Furthermore, there was a strong correlation between both mappings, reaching an average Pearson correlation of r = 0.86 after collapsing expression data to Arabidopsis identifiers to minimize bias from different genome duplication histories (Supplemental Table 2 and Supplemental Figure 5). Cross-species mapping has been successfully used for inter species comparisons before (Bräutigam et al., 2011, 2014; Gowik et al., 2011), and in this study mapping of both species to the T. hassleriana genome provided a quality data set with a limited degree of artifacts.
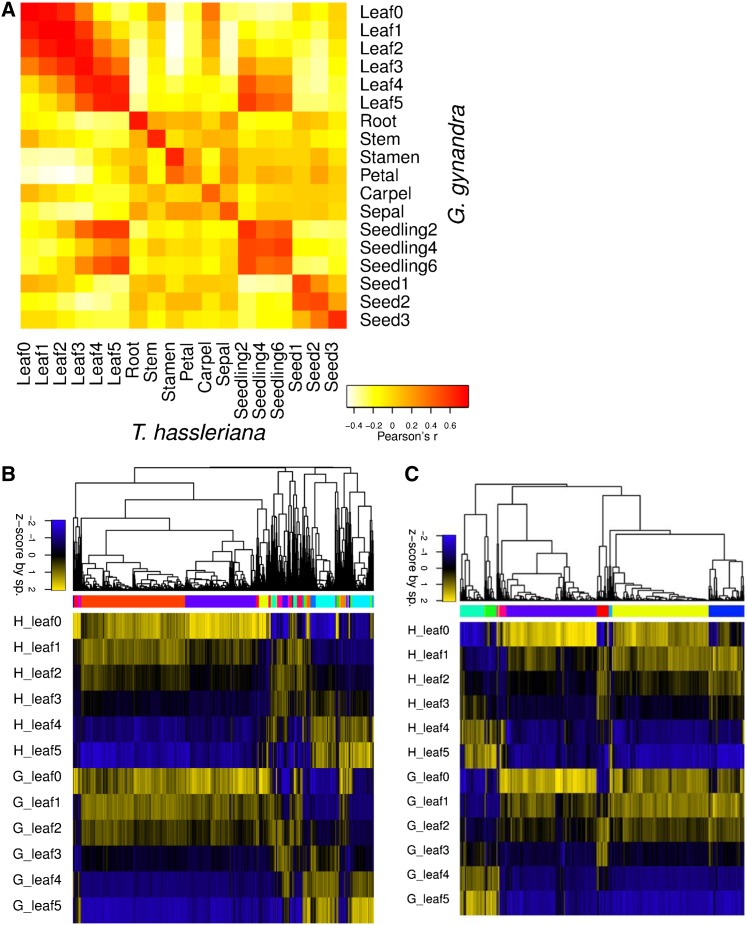
The generated transcriptome atlases were reproducible and comparable between species. To reduce noise, downstream analyses focused on genes expressed above 20 reads per mappable million (RPKM; Supplemental Figure 6), unless otherwise noted. Biological replicates of each tissue clustered closely together and were highly correlated (mean r = 0.92, median r = 0.97; Figure 2A; Supplemental Figures 7A and 7B and Supplemental Table 3). On average, 4686 and 5308 genes displayed significantly higher expression values in G. gynandra and T. hassleriana, respectively, with the greatest differences observed in seed and stem tissue (Supplemental Table 4). In contrast, the transcriptome patterns were highly similar between the sister species (Figure 2A; Supplemental Figure 7C). Principle component analysis (PCA) showed that the first component separated the species and accounted for only 15% of the total variation (Supplemental Figure 8A).
Figure 2.
Comparative Tissue Dynamics and Gene Expression Pattern between G. gynandra and T. hassleriana.
(A) Pearson’s correlation heat map of the expression of tissue-specific signature genes (RPKM) of all leaf gradient sample averages (n = 3) per species. Yellow, low expression; red, high expression. G, G. gynandra; H, T. hassleriana.
(B) Pearson’s correlation hierarchical cluster of all leaf gradient sample averages as Z-scores. Blue is the lowest expression and yellow the highest expression.
(C) Expression patterns of transcriptional regulators in both species within the leaf gradient. Pearson’s correlation hierarchical cluster of all sample averages as Z-scores. Blue is the lowest expression and yellow the highest expression.
Gene expression patterns and dynamics are conserved between species. The number of genes expressed above 20 RPKM varied by tissue from 6900 to 12,000, with the fewest in the mature leaf and most in the stem and youngest leaf in both species (Table 1; Supplemental Data Set 2). Hierarchical clustering revealed major modules with increasing and decreasing expression along the leaf gradient (Figure 2B), a large overlap of peak expression between seedlings and mature tissue, and distinct gene sets for the other sampled tissues (Supplemental Figure 9A). In leaves, the genes with decreasing expression split into two primary clusters, of which the smaller cluster maintained higher expression longer in the C4 than the C3 species (Figure 2B). Clustering of the tissues with 10,000 bootstrap replications confirmed the visual similarity of mature leaves and seedlings and showed further major branches consisting of (1) carpel, stem, and root; (2) a seed gradient and remaining floral organs; and (3) young leaves (Supplemental Figure 9A). Limiting the clustering to transcription factors (TFs) showed equivalent results (Supplemental Figure 9B; Figure 2C), except that in leaves, a higher proportion of the TFs with decreasing expression maintained expression longer in the C4 species. Notably, this delay impacted the clustering of the tissues and older C4 leaves tended to cluster with younger C3 leaves by TF expression (Supplemental Figures 9A and 9B). The delay was further reflected in a PCA of the leaf gradient where stage 0 and 1 show much less separation in G. gynandra than in T. hassleriana (Supplemental Figure 8B).
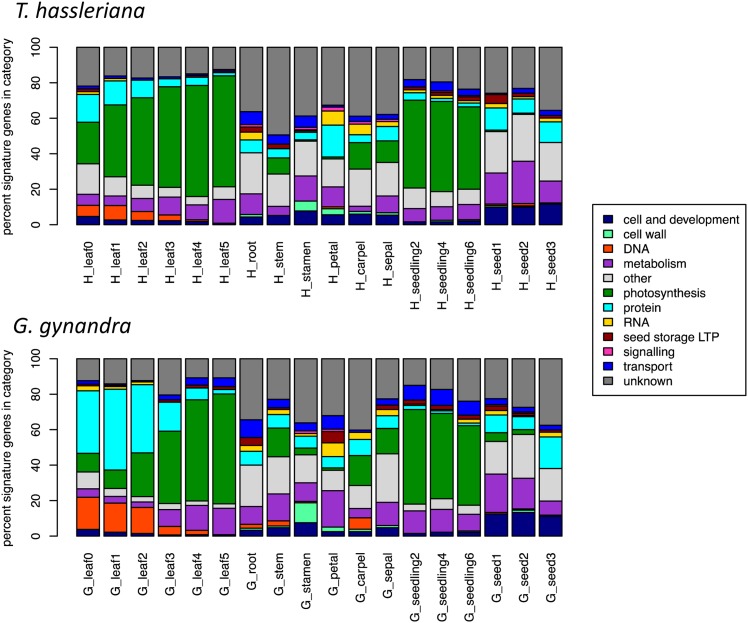
The functional categories with dominant expression showed distinct patterns across the tissues and high conservation between the species. As in the hierarchical clustering, the species showed similar profiles when examining the number of signature genes (expressed over 1000 RPKM; Figure 3) or the total RPKM (Supplemental Figure 9) in each functional category. As expected, in mature leaves and seedlings, transcriptional activity is dominated by photosynthesis, which is almost entirely lacking from roots, seeds, stamens, and petals (Figure 3; Supplemental Figure 9). Younger leaf tissues of the C3 species show higher expression of genes in the photosynthetic category, displayed as signature genes (Figure 3) or as cumulative RPKM per category (Supplemental Figure 9). In all floral tissues, roots, and stems, transcriptional activity is comparatively balanced between categories. In seeds, a major portion of the total expression is allocated to a few, extremely highly expressed lipid transfer protein type seed storage proteins (Supplemental Figure 9). The differences between the two species lie in the details, especially within the developmental leaf gradient. In young G. gynandra leaves, more signature genes encode DNA and protein-associated MapMan terms than in T. hassleriana (Figure 3). A close examination of secondary MapMan categories shows that specifically histone proteins (34 genes with P < 0.05 in stage 1, enriched with Fisher's exact test P = 2.6⋅10−13) and protein synthesis (222 genes with P < 0.05 in stage 1, enriched with Fisher's exact test P = 1.8⋅10−17) are upregulated in G. gynandra and that these categories have a larger dynamic range in G. gynandra than T. hassleriana (Supplemental Figure 10).
Figure 3.
Distribution of Signature Genes in Each Tissue in G. gynandra and T. hassleriana.
Percentage of signature genes expressed over 1000 RPKM falling in each basal MapMan category for every averaged tissue.
In summary, transcriptomic analysis indicates the tissues are well paired and comparable between species and although there are differences in expression level, there is conservation of expression patterns between species. Within the leaf gradient, there is a subset of genes that shows a delay in the onset of expression changes in G. gynandra.
The Comparative Transcriptome Atlases Revealed Diverse Recruitment Patterns from the C3 Plant T. hassleriana to C4 Photosynthesis
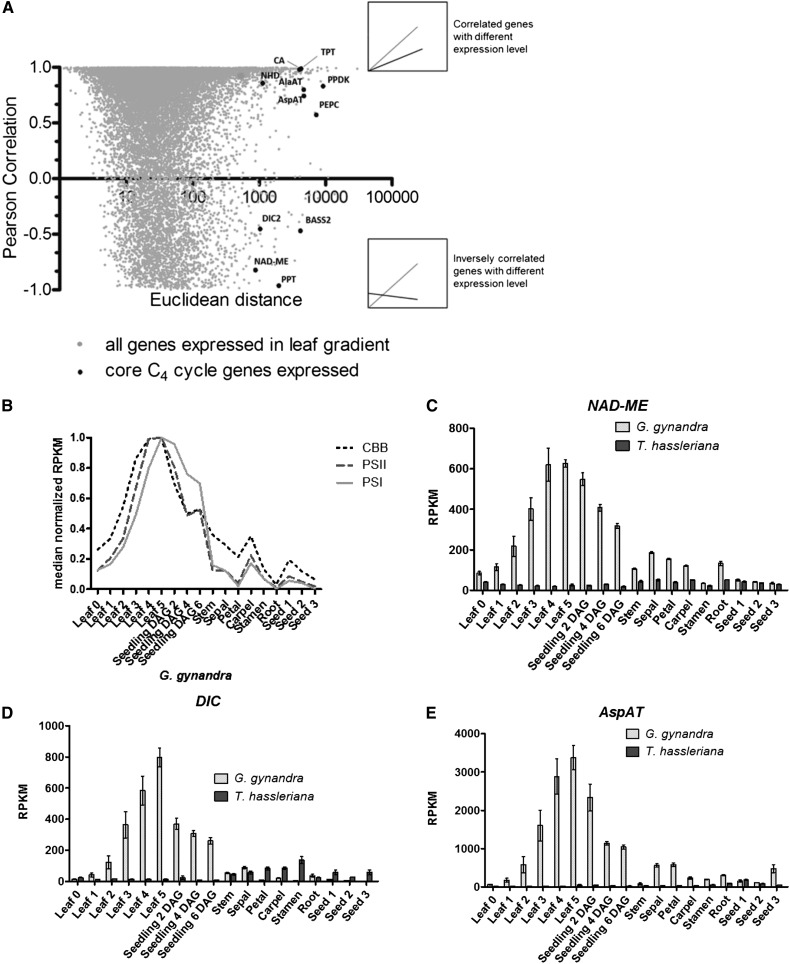
The expression patterns of the core C4 cycle genes were compared in G. gynandra and T. hassleriana to gain insight into the evolutionary recruitment of C4 cycle genes to photosynthesis. During convergent evolution of C4 photosynthesis, these genes were recruited from ancestral C3 genes (Sage, 2004; Edwards et al., 2010; Sage et al., 2011). To contextualize the change in expression of the C4 cycle genes, the between species Euclidean (absolute) and Pearson (pattern) distances were calculated and compared from the leaf developmental gradients (Figure 4A). All known C4 cycle genes showed a large Euclidean distance (844 to 9156 RPKM), while they split between a correlated and an inversely correlated pattern. In addition to the known C4 genes, histones, lipid transfer proteins, protein synthesis, and DNA synthesis are functional categories found among genes with greater than 844 RPKM differences in absolute expression (Supplemental Data Set 6).
Figure 4.
Comparison of Gene Expression Dynamics within the Leaf Gradient of Both Species.
(A) Euclidean distance versus Pearson’s correlation of average RPKM (n = 3) of genes expressed (>20 RPKM) in both leaf developmental gradients. Comparison of gene expression by similarity of expression pattern and expression level in T. hassleriana and G. gynandra. Relevant highly expressed C4 cycle genes are marked in plot. Above inset shows an example of two highly correlated genes by expression trend and strength. Lower inset shows an example of two genes inversely correlated with different expression level.
(B) Expression pattern across the atlas of averaged relative expression of transcripts encoding for photosystem I (PSI), photosystem II (PSII), and soluble enzymes of the Calvin-Benson-Bassham (CBB) cycle in G. gynandra.
(C) to (E) Average expression pattern of highest abundant ortholog of C4 cycle genes (NAD-ME, DIC, and AspAT) in photo- and heterotrophic tissues in G. gynandra (light gray) and T. hassleriana (dark gray); ±se, n = 3.
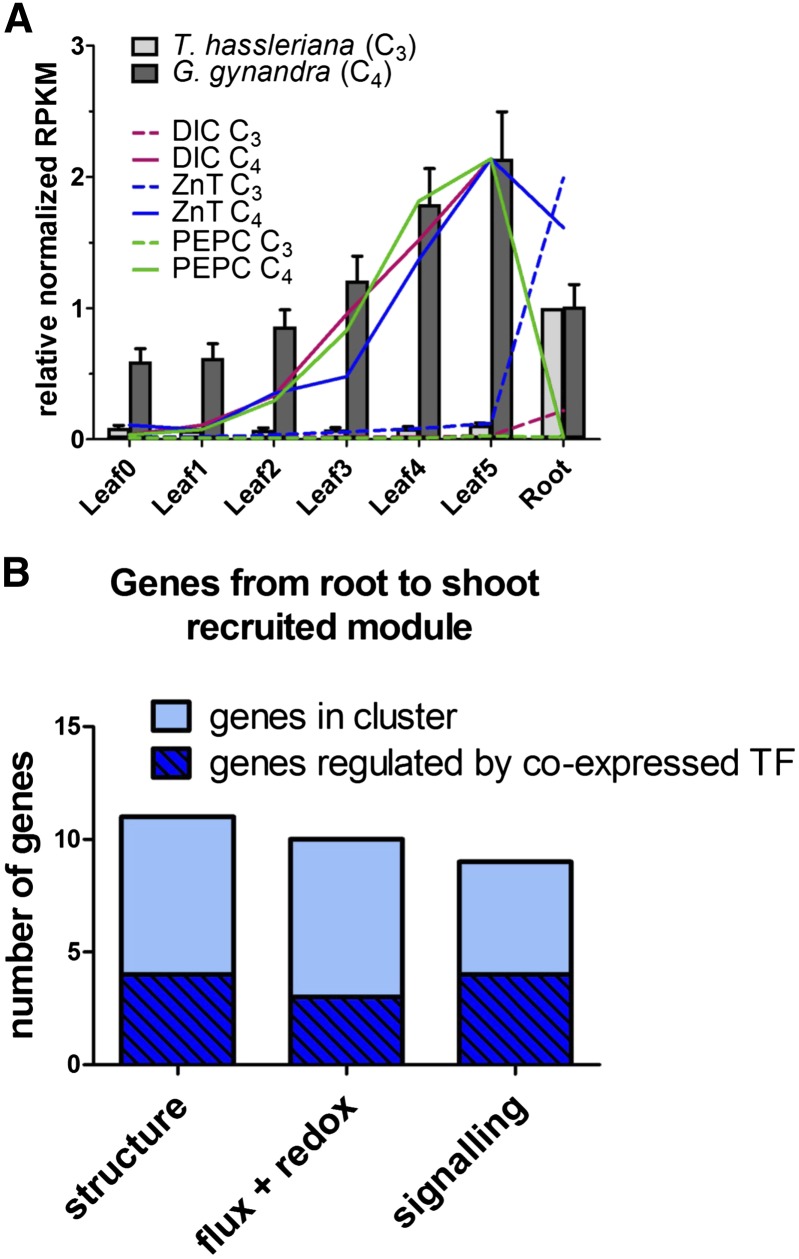
To identify ancestral C3 expression domains from which C4 genes were recruited, the expression of the core C4 cycle genes was compared between species. In G. gynandra, all core C4 cycle genes increase in expression along the leaf gradient and are high in seedlings (Figures 4C and 4D; Supplemental Figures 12A to 12F); this pattern matches that of other photosynthetic genes (Figure 4B). For each C4 cycle gene, the T. hassleriana sequence to which most G. gynandra reads mapped was taken as the most likely closest putative ortholog (Supplemental Figures 13 and 14). The putative orthologs of core C4 genes are expressed at comparatively low levels in C3 (Supplemental Figures 13 and 14). Activity measurements of the core C4 cycle enzymes match the observed gene expression profiles (Supplemental Figure 15). In contrast to leaves and seedlings, the remaining tissues show a variety of expression patterns of C4 cycle genes in both species (Figures 4C to 4E; Supplemental Figures 12A to 12G). Of the C4 cycle genes, NAD-MALIC ENZYME (NAD-ME) and the SODIUM:HYDROGEN ANTIPORTER (NHD) show a fairly constitutive expression pattern in C3, while the others have a small number of tissues where the expression peaks (Figure 4C; Supplemental Figure 12A). The expression of PYRUVATE PHOSPHATE DIKINASE (PPDK), the PHOSPHOENOLPYRUVATE TRANSLOCATOR (PPT), and DICARBOXYLATE CARRIER (DIC) peaks in floral organs (Supplemental Figures 12B and 12C; Figure 4D); the expression of ASPARTATE AMINO TRANSFERASE (AspAT) and ALANINE AMINOTRANSFERASE (AlaAT) peaks in seed (Figure 4E; Supplemental Figure 12D); and the expression of the pyruvate transporter BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN2 (BASS2) peaks in the young leaf (Supplemental Figure 12E). Albeit erroneous identification of the closest C3 ortholog in some cases (e.g., BASS2 and PHOSPHOENOLPYRUVATE CARBOXYLASE [PEPC]) impedes identification of the ancestral C3 expression domain (Supplemental Figures 12 and 13), the majority of known C4 cycle genes were recruited to a photosynthetic expression pattern from a variety of expression domains (Figure 4B).
To assess the possibility of small modular recruitment from other tissues to the C4 leaf, we searched for evidence of an expression shift between the C3 root and the C4 leaf. This shift is expected, if the bundle sheath tissue is partially derived from the regulatory networks of root endodermis, as proposed previously (Slewinski, 2013). Expression pattern filters were used to identify 37 genes that were expressed primarily in the C3 root and the C4 leaf (C3 leaf/root < 0.3; C4/C3 leaf > 1; C4 leaf4-5/root > 0.5; C4 leaf5 > 30 RPKM; leaf5/root enrichment 6-fold greater in C4), significantly more than in a randomized data set (P value < 10−29; Supplemental Table 5). This set of genes showed a very similar expression pattern to photosynthetic genes along the C4 leaf gradient (Figure 5A).
Figure 5.
Recruitment of Genes from the Root to Leaf Expression Domain in the C4 Plant G. gynandra.
(A) Relative average RPKM normalized to expression in G. gynandra leaf 5 (gray bars). Bars represent the arithmetic means of all 37 genes; lines show expression patterns of a reference C4 cycle gene (PEPC) and of two genes found in the shifted module.
(B) Genes in the module displayed as functional groups. Light blue: absolute number of genes in the group. Dark blue overlay: portion of genes controlled by a transcription factor of the module.
The functions encoded by the genes that were apparently recruited to the leaf from a root expression domain were consistent with structural modifications and C4 photosynthesis. In Arabidopsis, 29 of the corresponding homologs are heterogeneously expressed across different root tissues with their highest expression in either the endodermis or cortex, analogous to bundle sheath and mesophyll cells, respectively (Slewinski, 2013). Three functional groups could be identified in the cluster. The first is related to tissue structure, i.e., cell wall modification and plasmodesmata, the second to metabolic flux and redox balance, and the third to signaling (Figure 5B). Among these genes are two C4 cycle genes, namely, DIC1, and a carbonic anhydrase. The group contains three TFs, one of which is involved in auxin response stimulation. Coexpression network analysis of the Arabidopsis homologs (ATTED-II) shows 11 genes from the cluster occur in a shared regulatory network. In summary, a set of genes related to cell wall, metabolic/redox flux, and signaling was recruited from the C3 root to the C4 leaf, many of which are coexpressed in Arabidopsis and found in leaf tissues analogous to BSC and MC.
Changes in the Leaf Transcriptomes Reveal Differences in Cellular Architecture and Leaf Development in the C4 Species
Altered expression of cell cycle genes and enlarged BSC nuclei in G. gynandra suggest the occurrence of endoreduplication within this cell type. During early leaf development, G. gynandra leaf samples clustered together with younger samples in T. hassleriana (Supplemental Figures 8A and 8B), indicating a delay in leaf maturation. We hypothesized this delay in G. gynandra leaf maturation is manifested through alterations of cell cycle gene expression during leaf development. Hierarchical clustering of absolute expression values showed that the majority of known core cell cycle genes (Vandepoele et al., 2002; Beemster et al., 2005) have comparable expression patterns between both species (Supplemental Figure 16 and Supplemental Data Set 7). However, two distinct groups of genes were identified, which are either upregulated in G. gynandra between stage 0 to 2 (group 1: 9 of 18 genes with P value < 0.05) or show a delayed decrease during C4 leaf development (group 2: 9 of 12 genes with P value < 0.05 between stage 0 and 3; Supplemental Figure 16 and Supplemental Data Set 7). Interestingly, GT-2-LIKE1 (GTL1), a key cell cycle regulator, was not correlated between G. gynandra and T. hassleriana during leaf development. GTL1 is upregulated in later stages of leaf development in T. hassleriana but not in G. gynandra (P value < 0.001 in stage 5; Supplemental Figure 16 and Supplemental Data Set 7).
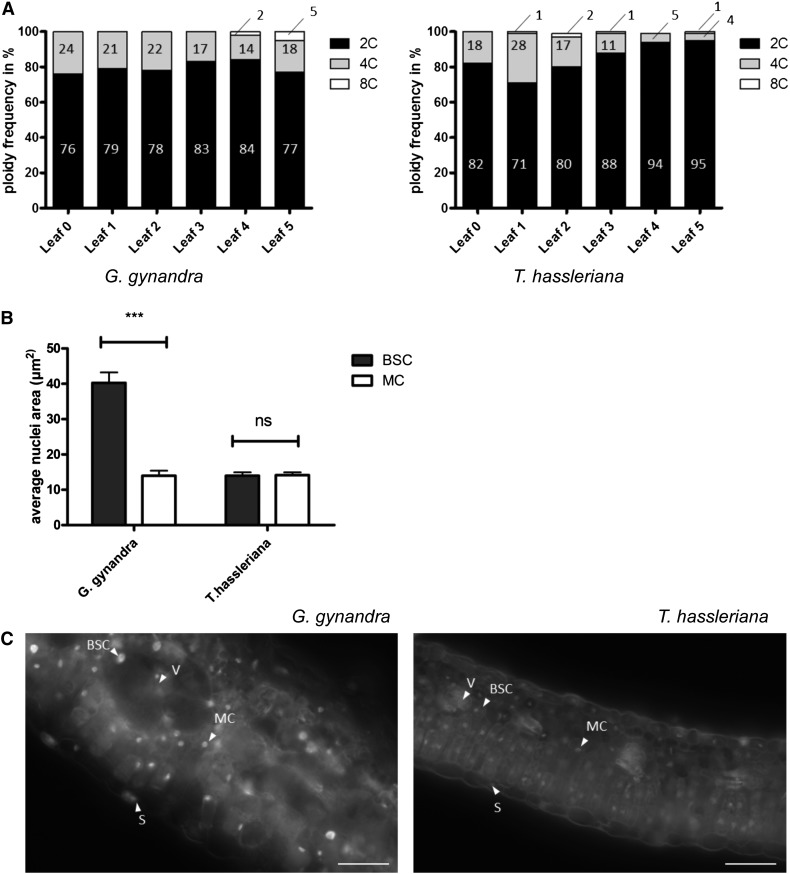
As GTL1 has been demonstrated to operate as an inhibitor of endoreduplication and ploidy-dependent cell growth (Breuer et al., 2009, 2012), we examined whether nuclei were enlarged in any G. gynandra leaf tissues. First, both leaf developmental gradients were subjected to flow cytometry. Polyploidy (DNA content > 2C) was observed in both species, but clearly enriched in C4 compared with C3, especially in the more mature leaves (5% versus 1% ≥ 8C, 16% versus 4% ≥ 4C; Figure 6A). In the G. gynandra C4 leaf, the BSC nuclei were 2.9-fold larger than those in the MC (P < 0.001; Figures 6B and 6C). In contrast, the C3 T. hassleriana nuclei of both cell types were similar sizes with a size ratio of 1.0 (Figures 6B and 6C). The proportion of BSC in the leaf was estimated from transversal sections as 15% in G. gynandra and 6% in T. hassleriana (Figures 7A to 7L). This number fits with the subpopulation of cells with higher ploidy observed in G. gynandra in the mature leaf. In summary, the extended expression of a subgroup of cell cycle genes and downregulation of GTL1 correlate with higher ploidy levels in the G. gynandra mature leaf based on BSC nuclei area and flow cytometry measurements.
Figure 6.
Distribution of Ploidy Levels during Leaf Development and Nuclei Area of BSC and MC between G. gynandra and T. hassleriana.
(A) Ploidy distribution of developing leaf (category 0 till 5) in percentage in G. gynandra and T. hassleriana. Measurements performed in n = 3 (except G0 = 1 replicate). For each replicate, at least 2000 nuclei were measured by flow cytometry.
(B) Quantification of BSC and MC nuclei area in cross sections (n = 3 ± se) of mature G. gynandra and T. hassleriana leaves (stage 5). Area of nuclei in µm2 with at least 150 nuclei analyzed per cell type per species per replicate. Asterisks indicate statistically significant differences between BSC and MC (***P value < 0.001); n.s., not significant.
(C) Fluorescence microscopy images of propidium iodide-stained leaf cross sections (stage 5) of T. hassleriana (left) and G. gynandra (right). Arrowheads point to nuclei of the indicated cell type. V, vein; S, stomata. Bar = 50 μm.
Figure 7.
Analysis of Shifted Gene Expression Pattern and Leaf Anatomy during Leaf Ontogeny.
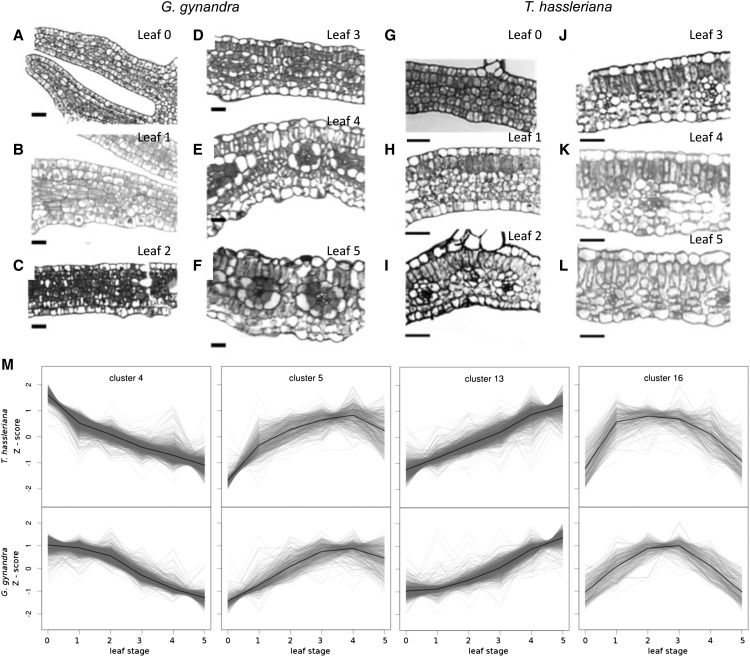
(A) to (L) Leaf anatomy development along the gradient in G. gynandra and T. hassleriana depicted by cross sections stained with toluidine blue. Bar = 20 μm.
(M) Selected clusters from K-means clustering of gene expression shown as Z-scores, which show a phase shift between G. gynandra and T. hassleriana during leaf development.
The C4 Species Shows Delayed Differentiation of Mesophyll Tissue, Coinciding with Increased Vein Formation
The transcriptional delay in a large subset of G. gynandra genes (Figures 2B, 2C, and 3) reflects a later differentiation of the C4 leaf. The delayed pattern of this large subset of genes indicated that there might be a delay in the differentiation of leaf internal anatomy, although leaf growth rates and shape are similar between species (Figure 1A). Thus, the leaves were examined microscopically. Since dicotyledonous leaves differentiate in a wave from tip toward petiole (Andriankaja et al., 2012), leaves were cross-sectioned at the midpoint (50% leaf length) for comparison. The cross sections revealed that in C4 leaves, cell differentiation was delayed in the transition from undifferentiated ground tissue toward fully established palisade parenchyma (Figures 7A to 7L). Both species start undifferentiated at leaf stage 0 with only the primary vein distinctly visible in cleared leaves (Figures 7A and 7G; Supplemental Figure 1A). In stage 1, the C3 leaf starts to differentiate its palisade parenchyma, while the C4 leaf shows dividing undifferentiated cells (Figures 7B and 7H). Mesophyll differentiation has finished by stage 2 in the C3 leaf (Figure 7I), but not until stage 4 in the C4 leaf (Figure 7D). Classical mature C4 leaf architecture appears in stage 4 in G. gynandra (Figure 7E). C4 leaves ultimately develop more veins and open veinlets leading to Kranz anatomy (Supplemental Figure 1). Leaf mesophyll tissue of the C3 species differentiates faster and develops fewer veins than the C4 species.
The expression of genes related to vein development was consistent with greater venation in the C4 leaf but failed to explain the larger delay in expression patterns and mesophyll differentiation in the C4 leaf. Hierarchical clustering indicated that most known leaf and vasculature developmental factors (reviewed in Ohashi-Ito and Fukuda, 2010) showed similar expression patterns in the two species (Supplemental Figure 17 and Supplemental Table 6). However, two clusters with distinct expression patterns were detected. In the C4 species, seven genes were upregulated (P value < 0.05), including vasculature facilitators PIN-FORMED (PIN1), HOMEOBOX GENE8 (HB8), and XYLOGEN PROTEIN1 (XYP1) (Motose et al., 2004; Scarpella et al., 2006; Donner et al., 2009), while five genes were downregulated (P value < 0.05), among those the negative regulators KANADI1 and 2, as well as HOMEOBOX GENE15 (Supplemental Figure 17 and Supplemental Table 6; Ilegems et al., 2010).
To further elucidate the magnitude and nature of the delayed expression changes on the transcriptional level, the leaf gradient data were clustered with the K-means algorithm (Supplemental Figures 17A and 17B and Supplemental Data Set 9). Of 16 clusters, six were divergent (1 to 3, 8, 9, and 15; 1270 genes). The remaining clusters were similar; however, four showed a transcriptional delay (4, 5, 13, and 16; 3361 genes), while six did not (6, 7, 10 to 12, and 14; 5162 genes). Of all clustered genes, 87% belonged to highly conserved clusters, 34% with a delay and 53% without. Thus, the transcriptional delay cannot be explained by general slower development.
All of the K-means clusters were functionally characterized by testing for enrichment in MapMan categories (Supplemental Data Set 10). The visually “shifted” patterns were: later onset of increase in clusters 13 and 5 (1058 and 395 genes, respectively), delayed decrease in cluster 4 (1644 genes), and a later peak in cluster 16 (264 genes; Figure 7M). The “late decrease” cluster 4 is enriched in genes related to mitochondrial electron transfer, CONSTITUTIVE PHOTOMORPHOGENESIS9 (COP9) signalosome, and protein degradation by the proteasome (Figure 7M; Supplemental Data Set 10). The “late onset” cluster 13 is enriched in all major photosynthetic categories: N-metabolism, and chlorophyll, isoprenoid, and tetrapyrrole biosynthesis (P value < 0.05; Supplemental Figures 17C and 17D and Supplemental Data Sets 9 and 10). The smaller “late onset” cluster 5 is enriched in the categories protein synthesis, tetrapyrrole synthesis, carotenoids, and peroxiredoxin. Cluster 16 peaks earlier in T. hassleriana than G. gynandra and is enriched in lipid metabolism (e.g., ACYL CARRIER PROTEIN4, CHLOROPLASTIC ACETYLCOA CARBOXYLASE1, 3-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE1, and 3-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III) and plastid division genes, such as the FILAMENTATION TEMPERATURE-SENSITIVE genes FtsZ2, FtsH, and FtsZ, as well as ACCUMULATION AND REPLICATION OF CHLOROPLASTS11 (Figure 7M; Supplemental Data Sets 9 and 10).
The core of the phase-shifted clusters, defined as genes with Pearson's correlation coefficient of r > 0.99 to the cluster center, contained candidate regulators for the observed delayed patterns. The core of cluster 13 contained 17 TFs and genes involved in chloroplast maintenance (Supplemental Data Set 11). The core of cluster 4 contained 30 transcriptional regulators, including PROPORZ1 (PRZ1), and eight other chromatin-remodeling genes. Nineteen cell cycle genes were found in the core of cluster 4 (Supplemental Figures 19A and 19B), including CELL DIVISION CYCLE20 (CDC20), CDC27, and CELL CYCLE SWITCH PROTEIN52 (CCS52), which are key components of cell cycle progression from M-phase to S-phase (Pérez-Pérez et al., 2008; Mathieu-Rivet et al., 2010b).
Our data were quantitatively compared with data from Arabidopsis leaf development to test if the observed phase shift related to a switch from proliferation to differentiation (Andriankaja et al., 2012). This study identified genes that were significantly up- or downregulated during the shift from proliferation to expansion (Andriankaja et al., 2012). Putative orthologs of these genes were clustered by the K-means algorithm (without prior expression filtering), producing seven clusters for the upregulated genes (containing 483 genes in total) and five clusters for the downregulated genes (1112 genes in total; Supplemental Figure 20). The trend was well conserved across species, with 75% of the upregulated and 96% of the downregulated genes falling into clusters with a matching trend. The genes showed a higher proportion of delay in G. gynandra than in the total data set, with 60 and 68% falling in delayed up- and downregulated clusters, respectively (Supplemental Figure 20).
In summary, about a third of all gene expression patterns show a delay in the G. gynandra leaf (Figure 7M; Supplemental Figure 18). Delayed genes include major markers of leaf maturity such as the upregulation of photosynthetic gene expression and downregulation of mitochondrial electron transport (Supplemental Figures 19C and 19D and Supplemental Data Set 10). This delay was more common in putative orthologs of genes differentially regulated during the shift from cell proliferation to expansion (Supplemental Figure 19; Andriankaja et al., 2012). The slow maturation can be seen on the anatomical level as a delayed differentiation that coincides with increased vein formation in the C4 species (Figures 7A to 7L).
DISCUSSION
Comparative Transcriptome Atlases Provide a Powerful Tool for Understanding C4 Photosynthesis
Two transcriptome atlases were generated to allow the analysis of gene recruitment to photosynthesis and to detect differences related to C4 leaf anatomy. Two Cleomaceae species were chosen for this study due to their phylogenetic proximity to the model species Arabidopsis (Marshall et al., 2007). The sampled leaf tissues covered development from sink tissue to fully mature source tissue (Figures 1 and 3), and all higher order vein development (Supplemental Figure 1). Since C4 genes are recruited from genes already present in C3 ancestors, where they carry out housekeeping functions (Sage, 2004; Besnard et al., 2009; Christin and Besnard, 2009; Christin et al., 2009), seed, stem, floral, and root tissues were included in the atlases in addition to leaves and seedlings.
The high similarity in expression pattern between the species maximizes our ability to detect differences related to C4 photosynthesis. While PCA analysis showed that the first principle component separated the data set by species, this accounted for only 15% of the variation (Supplemental Figure 8A). Excluding floral organs and stem, all tissues correlated with r > 0.7 between species (Supplemental Figure 7C and Supplemental Table 3). Hierarchical and K-means clustering showed the vast majority of genes had a similar pattern between species, and tissue types clustered closely with the same tissue in the other species. Specific groups of highly expressed genes exclusively expressed in one tissue type, such as root, stamen, and petal, are shared between G. gynandra and T. hassleriana, suggesting that these genes might represent drivers for the respective tissue identity (Supplemental Figure 9). A subset of genes showed a consistent adjustment to their expression pattern, namely, a delay in the leaf gradient of G. gynandra relative to T. hassleriana (Figure 7M). Thus, organ identity is highly conserved between G. gynandra and T. hassleriana, but the rate at which organ identity, especially the leaf, is established can differ.
Expression Patterns of C3 Putative Orthologs Support Small-Scale or Modular Recruitment to Photosynthesis, Implying That a General C4 Master Regulator Is Unlikely
Ancestral expression patterns can be compared with assess whether a master regulator could have facilitated recruitment of genes to C4 photosynthesis. The patterns of gene expression in T. hassleriana provide a good proxy for the ancestral C3 expression pattern due to its phylogenetic proximity to G. gynandra (Inda et al., 2008; Cheng et al., 2013). Genes active in the C4 cycle were recruited from previously existing metabolism (Matsuoka, 1995; Chollet et al., 1996; Streatfield et al., 1999; Wheeler et al., 2005; Tronconi et al., 2010). Expression patterns in T. hassleriana reflect known metabolism and expression; for instance, PPDK is expressed in seeds, stamens, and petals (Supplemental Figure 12B), which is similar to the expression domain reported by Chastain et al. (2011). Furthermore, PPT is highly expressed in stamens and during seed development (Supplemental Figure 12C; Knappe et al., 2003a, 2003b), since it is required for fatty acid production (Hay and Schwender, 2011).
The C3 putative orthologs of C4 cycle genes show a variety of expression patterns within the atlas, providing strong evidence they could not have been recruited by a single master regulator. All C4 cycle genes are expressed to a low degree in T. hassleriana, either constitutively or in defined tissues such as stamens, seeds, or young leaves (Figures 4C to 4E). Expression of NHD, AlaAT, AspAT, and PPDK increased along the leaf gradient in both C3 and C4 species, but in C3, the expression was highest in tissues other than the leaf (Figure 4E; Supplemental Figures 12A, 12B, and 12D). In contrast, DIC, BASS2, NAD-ME, and PPT are expressed in inverse patterns between C3 and C4 along the leaf gradient (Figures 4C and 4D; Supplemental Figures 12C and 12E), and PEPC is expressed only in mature leaves in the C3 species (Supplemental Figure 12F). Except for DIC and PPDK, the expression level of the C4 cycle genes was higher in G. gynandra across all tissues (Figure 4; Supplemental Figures 12 to 14). Thus, most of the C4 cycle genes may still maintain their ancestral functions in addition to the acquired C4 function. The correct ortholog in C3 may not have been conclusively determined by cross species read mapping in all cases reported here. However, the main conclusion—that C4 cycle genes are recruited from a variety of C3 expression patterns—holds regardless of which putative C3 paralog is selected (Supplemental Figures 13 and 14).
A set of genes shifted from a root to leaf expression domain during C4 evolution provides an example of small-scale modular recruitment. The proposed analogy between root endodermis and bundle sheath and between root cortex and mesophyll (Slewinski, 2013) has been linked to cooption of the SCARECROW (SCR) and SHORTROOT (SHR) regulatory networks into developing leaves (Slewinski et al., 2012; Wang et al., 2013). A set of 37 genes consistent with such a recruitment module was identified. For this gene set, the C3 species T. hassleriana (Figure 5; Supplemental Table 5) and Arabidopsis (Brady and Provart, 2009) showed conserved root expression, while the C4 species showed an expression pattern similar to photosynthesis. Much of the root to leaf gene set was coregulated in Arabidopsis, and it contained TFs, including ETHYLENE RESPONSE FACTOR1 (Mantiri et al., 2008), as well as an AUX/IAA regulator (Pérez-Pérez et al., 2010) and VND-INTERACTING2 (Yamaguchi et al., 2010). Functionally, the majority of the gene set is involved in processes related to cell wall synthesis and modification. The set contains the cell wall-plasma membrane linker protein (Stein et al., 2011) and the xyloglucan endotransglycosylase TOUCH4 (Xu et al., 1995), the tonoplast intrinsic protein involved in cell elongation (Beebo et al., 2009), and a plasmodesmata-located protein (Bayer et al., 2008). The observed coregulation and structural functions support an underlying structural relationship between the root tissues endodermis and cortex, and the leaf tissues bundle sheath and mesophyll.
It is still unresolved whether expression level recruitment of genes to the C4 cycle was facilitated by the action of one or a few master switches controlling C4 cycle gene expression and/or by changes to promoter sequences of C4 genes (Westhoff and Gowik, 2010). The diverse transcriptional patterns of the core C4 cycle genes in T. hassleriana provide strong evidence that they were not recruited as a single transcriptional module facilitated by one or a few master regulators. However, the identified root to leaf module indicates that small-scale corecruitment occurs, and this may help bring about the 3 to 4% overall transcriptional changes occurring during C4 evolution (Bräutigam et al., 2011, Gowik et al., 2011). The similarities in expression pattern between photosynthetic genes and C4 cycle genes are evident (Figure 4B), and light-dependent induction of C4 genes has been reported (Christin et al., 2013), leading us to hypothesize that C4 cycle genes may use the same light-induced regulatory circuits employed for the photosynthetic genes, possibly through acquisition of cis-regulatory elements or modification of chromatin structure, as has been shown for the PEPC gene promoter in maize (Tolley et al., 2012).
Cell Size in G. gynandra Coincides with Nuclei Size and Ploidy
In addition to the biochemical C4 cycle genes, transcriptional changes related to cell and tissue architecture are required for C4 leaf development (Westhoff and Gowik, 2010). The comparative atlases were contextualized with anatomical data to better understand BSC size.
G. gynandra has generally larger cells (Figures 7A to 7L), which might be attributed to a larger genome. After divergence from T. hassleriana, the G. gynandra lineage has undergone a putative whole-genome duplication (Inda et al., 2008). Cell size has been tied to genome ploidy status previously (Sugimoto-Shirasu and Roberts, 2003; Lee et al., 2009b; Chevalier et al., 2011). A relationship between ploidy and cell size could explain the generally larger cells in G. gynandra leaves (Figures 7A to 7L) or relate to the upregulation of DNA and histone-associated genes in developing leaves (Figure 3; Supplemental Figures 10 and 11).
Changes in the expression of key cell cycle genes indicated endoreduplication may be increased in G. gynandra, and follow-up nuclear size measurements indeed indicate BSCs have undergone endoreduplication. Enlargement of BSC is a common feature of C4 plants (Sage, 2004; Christin et al., 2013) including G. gynandra (Figures 7D to 7F), but the genetic mechanism is unknown. During leaf development, key cell cycle genes showed changes in expression pattern and expression level between G. gynandra and T. hassleriana (Supplemental Figure 16). CDC20 and CCS52A, which are closely linked with cell cycle M-to-S-phase progression or endocycle onset (Lammens et al., 2008; Larson-Rabin et al., 2009; Kasili et al., 2010; Mathieu-Rivet et al., 2010a), exhibit prolonged expression during C4 leaf development, whereas the expression of the master endoreduplication regulator GTL1 (Breuer et al., 2009, 2012; Caro et al., 2012) is suppressed in the older leaf stages (Supplemental Figure 16). Although a comparison of the more distantly related species Arabidopsis and G. gynandra discounted endoreduplication as a factor in bundle sheath cell size (Aubry et al., 2013), the BSC and MC nuclei area measurements of mature G. gynandra and T. hassleriana leaves revealed that the BSC nuclei are 2.9-fold enlarged compared with MC nuclei in G. gynandra (Figures 6B and 6C). At the same time, T. hassleriana BSC and MC cells do not differ significantly in nuclei size (Figures 6A and 6C).These results are supported by a flow cytometry analysis of both leaf developmental gradients, where the proportion of endoreplicated cells in the mature C4 leaf (Figures 6A) matches the number of BSCs present in G. gynandra (Figures 6A and 7A to F). Interestingly, we also find significant (P > 0.001) enlarged BSC nuclei in other C4 species (e.g., Flaveria trinervia, Megathyrsus maximum, and maize; Supplemental Figure 22), indicating that larger nuclei size in BSC compared with the MC could be a general phenomenon in C4 plants conserved across mono- and dicotyledons. Whether endoreplication is the cause of increased cell size in C4 BSC, as found for trichomes and tomato (Solanum lycopersicum) karyoplasm (Traas et al., 1998; Chevalier et al., 2011) or whether endoreplication only occurs to support the high metabolic activity and large size of the BSCs (Sugimoto-Shirasu and Roberts, 2003) remains to be determined.
Late Differentiation of Mesophyll Tissue Allows Denser Venation
General regulators of leaf anatomy and shape (reviewed in Byrne, 2012) are expressed in very similar patterns between the two species (Supplemental Figure 17), reflecting the very similar palmate five-fingered leaf shape and speed of leaf expansion (Figures 1A and 1B). However, anatomical studies of leaf development show that differentiated palisade parenchyma is already observed at the midpoint of stage 1 leaves in T. hassleriana (Figure 7H) but can only be detected in the middle of the leaf in stages 3 and 4 in G. gynandra (Figures 7D to 7F). Hierarchical clustering of transcriptome data indicates a similarity between younger T. hassleriana and older G. gynandra tissues (Supplemental Figure 9), which we attribute to a delay in G. gynandra leaf expression changes observed in the hierarchical clusters (Figures 2B and 2C) and observed for K-means clustering involving about a third of clustered genes (Figure 7M; Supplemental Figure 18). Analysis of the delayed clusters for significant enrichment of functional categories indicated that the metabolic shift from sink to source tissue was delayed (Figures 3 and 7M; Supplemental Figure 18 and Supplemental Data Set 10). Furthermore, the “delayed decrease” cluster 4 was enriched in COP9 signalosome and marker genes of the still developing heterotrophic leaf.
Cell cycle and cell differentiation regulators show a delayed expression pattern in G. gynandra. The expression of PRZ1, which switches development from cell proliferation to differentiation in Arabidopsis (Sieberer et al., 2003; Anzola et al., 2010), is prolonged in the C4 leaf (Figure 7M, cluster 4), as is the expression of chromatin remodeling factor GRF1-INTERACTING FACTOR3 implicated in the control of cell proliferation upstream of cell cycle regulation (Lee et al., 2009a). Plastid division genes peak around leaf stage 1 in T. hassleriana and leaf stage 2 in G. gynandra (Figure 7M, cluster 16). It has recently been shown that chloroplast development and division precedes photosynthetic maturity in Arabidopsis leaves and retrograde signaling from the chloroplasts affects cell cycle exit from proliferation (Andriankaja et al., 2012). Quantitative comparison of differentially regulated genes during the shift from cell proliferation to cell expansion found in Arabidopsis (Supplemental Figure 20; Andriankaja et al., 2012) to the expression patterns of the putatively orthologous genes along leaf developmental gradients in Cleome, reveals a strong conservation of expression pattern between Arabidopsis and Cleome during development. A higher proportion of delay of G. gynandra genes is observed in this gene set. This supports the idea that the transcriptional delay is directly linked to the anatomical delay in differentiation observed in G. gynandra (Supplemental Figure 19).
The delay in cell differentiation allows for increased vein formation in the C4 leaf. Mesophyll differentiation has already been shown to limit minor vein formation in Arabidopsis (Scarpella et al., 2004; Kang et al., 2007). G. gynandra and T. hassleriana have altered vein densities, which result from more minor vein orders in G. gynandra (Supplemental Figure 1), similar to results for the dicot Flaveria species (McKown and Dengler, 2009). Given that differentiation of photosynthetic mesophyll cells limits minor vein formation in Arabidopsis (Scarpella et al., 2004; Kang et al., 2007) and that mesophyll differentiation is delayed in the C4 species compared with the C3 species (Figure 7), dense venation may indeed be achieved by delaying mesophyll differentiation.
Genes related to vascular patterning are expressed in a manner consistent with higher venation in the C4 leaf. The high expression of vascular pattern genes such as PIN1, HB8, ARF3, and XYP1 in the C4 leaf (Supplemental Figure 17) is similar to that observed for Kranz patterned leaves in maize (Wang et al., 2013). However, these genes may be a consequence, rather than a cause, of higher venation, especially since some of these markers are only expressed after pre-procambial or procambial identity is introduced (Ohashi-Ito and Fukuda, 2010). Once procambial fate is established, cellular differentiation of vein tissues proceeds through positional cues and localized signaling, possibly via the SCR/SHR pathway (Langdale and Nelson, 1991; Nelson and Langdale, 1992; Nelson and Dengler, 1997; Griffiths et al., 2013; Wang et al., 2013; Lundquist et al., 2014). Interestingly, in accordance with the delay in leaf differentiation in G. gynandra, we could monitor a delay in higher expression for SHR peaking around leaf stage 1 to 3 (Supplemental Figure 21A). SCR transcript abundance is clearly divided in both G. gynandra and T. hassleriana between two homologs, one of which is more abundant in the C4 leaf and the other in the C3 leaf (Supplemental Figure 21B). SCR expression in G. gynandra follows the SHR pattern with a delayed upregulation. This is in accordance with earlier studies conducted in maize, where SHR transcript highly accumulates in the BSC to activate SCR expression (reviewed in Slewinski et al., 2012)
The identification of mesophyll differentiation as the proximate cause for fewer minor vein orders in T. hassleriana raises the question of how mesophyll differentiation is controlled. In both C4 and C3 species, vascular patterning precedes photosynthetic tissue differentiation (Sud and Dengler, 2000; Scarpella et al., 2004; McKown and Dengler, 2010). Light is one of the most important environmental cues that regulate leaf development, including its cellular differentiation and onset of photosynthesis (Tobin and Silverthorne, 1985; Nelson and Langdale, 1992; Fankhauser and Chory, 1997). The COP9 signalosome, which plays a central role in repression of photomorphogenesis and G2/M cell cycle progression (Chamovitz et al., 1996; Dohmann et al., 2008), showed a delayed decrease in G. gynandra compared with T. hassleriana (Supplemental Figure 19B). The delay and earlier vein formation termination induced by excess light in Arabidopsis (Scarpella et al., 2004) suggest that light perception and its signal transduction may be differentially regulated in species with denser venation patterns.
Conclusions
In this study, we report a detailed comparison of the transcriptomes and the leaf development of two Cleomaceae species with different modes of photosynthetic carbon assimilation, i.e., C3 and C4 photosynthesis. The gene expression patterns are quite similar between both species, which facilitates the identification of differences related to C4 photosynthesis. We could link two key features of Kranz anatomy to developmental processes through integration of expression and anatomical data. First, we show that the larger size of the bundle sheath cells in the C4 species is associated with a higher ploidy in these cells, which might be controlled by delayed repression of the endocycle via the transcription factor GTL1. Second, a prominent difference between C3 and C4 leaf development is the delayed differentiation of the leaf cells in C4, which is associated with a delayed onset of photosynthetic gene expression, chloroplast proliferation and development, and altered expression of a few distinct cell cycle genes. Delayed mesophyll differentiation allows for increased initiation of vascular tissue and thus contributes to the higher vein density in C4. We hypothesize that delayed onset of mesophyll and chloroplast differentiation is a consequence of the prolonged expression of the COP9 signalosome and, hence, a delayed derepression of photomorphogenesis.
METHODS
Plant Material and Growth Conditions
Gynandropsis gynandra and Tarenaya hassleriana plants for transcriptome profiling by Illumina Sequencing were grown in standard potting mix in a greenhouse between April and August 2011. Internal transcribed spacer sequences of G. gynandra and T. hassleriana were analyzed and plant identity confirmed according to Inda et al. (2008). Leaves were harvested from 4- to 6-week-old plants, prior to inflorescence initiation. All samples were harvested during midday. Flowers, stamens, sepals, and carpels were harvested after induction of flowering. Green tissues from seedlings were harvested 2, 4, and 6 d after germination. Root material was harvested from plants grown in vermiculite for 6 weeks and supplemented with Hoagland solution. Leaf material for the ontogeny analysis was selected by the order of leaf emergence from the apex in leaf stages from 0 to 5. Up to 40 plants were pooled for each biological replicate.
Leaf Expansion Rate
Leaves from stage 0 to 5 were analyzed in five biological replicates for each G. gynandra and T. hassleriana. Leaves were scanned on a flat bed scanner (V700 Photo; Epson), and the area was analyzed with free image analysis software ImageJ.
Leaf Cross Sections for Anatomical Studies
Leaves from stage 0 to 5 were analyzed in biological triplicates. Leaf material (2 × 2 mm) was cut next to the major first order vein at 50% of the whole leaf length. Leaf material was fixed in 4% paraformaldehyde solution overnight at 4°C, transferred to 0.1% glutaraldehyde in phosphate buffer, and vacuum infiltrated three times for 5 min. The leaf material was then dehydrated with an ascending ethanol series (70, 80, 90, and 96%) with a 1-h incubation in each solution. Samples were incubated twice in 100% ethanol and twice in 100% acetone, each for 20 min, and infiltrated with an acetone:araldite (1:1) mixture overnight at 4°C. After acetone evaporation, fresh araldite was added to the leaf samples until samples were covered and incubated for 3 to 4 h. Samples were transferred to fresh araldite in molds and polymerized at 65°C for 48 h. Cross sections were stained with toluidine blue for 15 s and washed with H2Odest. Cross sections were imaged with bright-field settings using an Eclipse Ti-U microscope (Nikon).
Flow Cytometry
Three biological replicate samples were chopped with a razor blade in 200 μL of Cystain UV Precise P Nuclei extraction buffer followed by the addition of 800 μL of staining buffer (buffers from Partec). The chopped leaves in buffer were filtered through a 50-μm mesh. The distribution of the nuclear DNA content was analyzed using a CytoFlow ML flow cytometer and FLOMAX software (Partec) as described (Zhiponova et al., 2013).
Measurement of Nuclei from Mature Leaves
Fresh mature leaves (leaf stage 5, three biological replicates) of G. gynandra and T. hassleriana were cut transversally, fixed in 1× PBS buffer (1% Tween 20 and 3% glutaraldehyde) overnight at room temperature, and stained with propidium iodide solution directly on the microscopic slide. Cross sections were imaged by fluorescence microscopy using an Axio Imager M2M fluorescence microscope (Zeiss) with an HE DS-Red Filter. Images were processed with ZEN10 software (Zeiss), and the nuclear area of at least 200 nuclei per cell type per species was measured with ImageJ.
RNA Extraction, Library Construction, and Sequencing
Plant material was extracted using the Plant RNeasy extraction kit (Qiagen). RNA was treated on-column (Qiagen) and in solution with RNA-free DNase (New England Biolabs). RNA integrity, sequencing library quality, and fragment size were checked on a 2100 Bioanalyzer (Agilent). Libraries were prepared using the TruSeq RNA Sample Prep Kit v2 (Illumina), and library quantification was performed with a Qubit 2.0 (Invitrogen). Single-end sequenced samples were multiplexed with six libraries per lane with ∼20 million reads per library. For paired-end sequencing, RNA of all photosynthetic and nonphotosynthetic samples was pooled equally for each species and prepared as one library per species. Paired end libraries were run on one lane with ∼175 million clean reads for T. hassleriana and 220 million clean reads for G. gynandra. All libraries were sequenced on the HISEQ2000 Illumina platform. Libraries were sequenced in the single-end or paired-end mode with length ranging from 80 to 100 nucleotides. The paired-end library of G. gynandra had an average fragment size of 304 bp; T. hassleriana had an average fragment size of 301 bp.
Gene Expression Profiling
Reads were checked for quality with FASTQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/), subsequently cleaned and filtered for quality scores greater than 20 and read length greater than 50 nucleotides using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit). Expression abundances were determined by mapping the single-end read libraries (each replicate for each tissue) independently against T. hassleriana representative coding sequences (Cheng et al., 2013) using BLAT V35 (Kent, 2002) in protein space and counting the best mapping hit based on e-value for each read uniquely. Default BLAT parameters were used for mapping both species. Expression was normalized to reads per kilobase T. hassleriana coding sequence per million mappable reads (RPKM). T. hassleriana coding sequences were annotated using BLASTX searches (cutoff 1e−10) against the TAIR10 proteome database. The best BLAST hit per read was filtered by the highest bit score. A threshold of 20 RPKM per coding sequence in at least one species present in at least one tissue was chosen to discriminate background transcription (Supplemental Figure 14). Differential expression between T. hassleriana and G. gynandra was determined by EdgeR (Robinson et al., 2010) in R (R Development Core Team, 2009). A significance threshold of 0.05 was applied after the P value was adjusted with false discovery rate via Bonferroni-Holms correction (Holm, 1979).
Data Analysis
Data analysis was performed with the R statistical package (R Development Core Team, 2009) unless stated otherwise. For Pearson’s correlation and PCA analysis, Z-scores were calculated by gene across both species. For all other analyses, Z-scores were calculated by gene within each species, to focus on comparing expression patterns. For K-means and hierarchical clustering, genes were filtered to those with more than 20 RPKM in at least one of the samples used in each species. To determine the number of centers for K-means clustering, the sum of se within clusters was plotted against cluster number and compared with randomized data (Supplemental Figures 18B, 20C, and 20D). A total of 16 centers was chosen, and K-means clustering was performed 10,000 times and the best solution, as defined by the minimum sum of se of genes in the cluster, was taken for downstream analyses (Peeples, 2011). Multiscale bootstrap resampling of the hierarchical clustering was performed for samples with 10,000 repetitions using the pvclust R package (Suzuki and Shimodaira, 2006).
Stage enrichment was tested for all K-means clusters and for tissue “signature genes” with expression of over 1000 RPKM in each tissue using TAIR10 MapMan categories (from http://mapman.gabipd.org) for the best Arabidopsis thaliana homolog. Categories with more than five members in the filtered (K-means) or complete (signature genes) data set were tested for enrichment by Fisher’s exact test, and P values were adjusted to false discovery rates via Benjamini-Yekutieli correction, which is tolerant of dependencies (Yekutieli and Benjamini, 1999).
Accession Numbers
Sequence data from this article can be found in NCBI GenBank under the following accession numbers: SRP036637 for G. gynandra and SRP036837 for T. hassleriana.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Venation Patterning during Leaf Development of G. gynandra and T. hassleriana.
Supplemental Figure 2. G. gynandra Cotyledon Anatomy 2, 4, and 6 d after germination (DAG).
Supplemental Figure 3. Images of Tissues Harvested for Atlases in G. gynandra and T. hassleriana.
Supplemental Figure 4. Quality Assessment of Velvet/OASES Assembled T. hassleriana Contigs against Predicted Corresponding CDS from T. hassleriana Genome
Supplemental Figure 5. Quality Assessment of the Biological Replicates of T. hassleriana Libraries Mapped to A. thaliana and Mapping Similarity of T. hassleriana Libraries Mapped to A. thaliana and to Its Own CDS.
Supplemental Figure 6. Determination of Baseline Gene Expression via a Histogram of Photosystem (PS) I and II Transcript Abundances (RPKM) in the G. gynandra Root.
Supplemental Figure 7. Quality Assessment of the Biological Replicates within Each Species and Tissue Similarity between G. gynandra and T. hassleriana.
Supplemental Figure 8. Principle Component Analysis between G. gynandra and T. hassleriana.
Supplemental Figure 9. Hierarchical Cluster Analysis with Bootstrapped Samples of G. gynandra and T. hassleriana.
Supplemental Figure 10. Transcriptional Investment of Each Tissue Compared in Both Species.
Supplemental Figure 11. Transcriptional Investment at Secondary MapMan Category Level of Each Tissue Compared in Both Species.
Supplemental Figure 12. Comparison of Gene Expression Dynamics within the Leaf Gradient of Both Species.
Supplemental Figure 13. Plot of the Expression Pattern (RPKM) of all C4 Gene Orthologs Expression Pattern in G. gynandra.
Supplemental Figure 14. Plot of the Expression Pattern of all C4 Gene Putative Orthologs Expression Pattern (RPKM) in T. hassleriana.
Supplemental Figure 15. Enzyme Activity Measurement of Soluble C4 Cycle Enzymes.
Supplemental Figure 16. Hierarchical Clustering of Average RPKM with Euclidean Distance of Core Cell Cycle Genes.
Supplemental Figure 17. Hierarchical Clustering with Pearson’s Correlation of Leaf Developmental Factors.
Supplemental Figure 18. K-Means Clustering of Leaf Gradient Expression Data and Quality Assessment.
Supplemental Figure 19. Z-Score Plots of Enriched MapMan Categories in the Shifted Clusters.
Supplemental Figure 20. K-Means Clustering of Genes Differentially Regulated during the Transition from Proliferation to Enlargement.
Supplemental Figure 21. Transcript Abundances of SCARECROW and SHORTROOT Homologs in G. gynandra and T. hassleriana Leaf and Root.
Supplemental Figure 22. Nuclei Area and Images of C4 and C3 Species.
Supplemental Table 1. Velvet/OASES Assembly Stats from G. gynandra and T. hassleriana Paired-End Reads.
Supplemental Table 2. Cross-Species Mapping Results.
Supplemental Table 3. Pearson's Correlation between G. gynandra and T. hassleriana Individual Tissues.
Supplemental Table 4. Number of Significantly Up- or Downregulated Genes in G. gynandra Compared with T. hassleriana within the Different Tissues.
Supplemental Table 5. List of Genes Present in Root-to-Shoot Recruitment Module.
Supplemental Table 6. List of Clustered General Leaf Developmental and Vasculature Regulating Genes along Both Leaf Gradients.
Supplemental Methods.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.8v0v6.
Supplemental Data Set 1. Annotated Transcriptome Expression Data of Both Atlases in RPKM.
Supplemental Data Set 2. Sequencing and Mapping Statistics for All Single-End Libraries Sequenced.
Supplemental Data Set 3. Quality Assessment of Representative Contigs against Predicted CDS within T. hassleriana.
Supplemental Data Set 4. MapMan Categories of Highly Expressed Genes in Each Tissue.
Supplemental Data Set 5. Transcriptional Investment of Each Enriched Basal MapMan Categories in Percentage for Each Tissue.
Supplemental Data Set 6. List of All Genes with Euclidean Distance over 800 RPKM Expressed within Both Leaf Gradients.
Supplemental Data Set 7. List of Core Cell Cycle Genes Selected for Clustering.
Supplemental Data Set 8. Statistical Analysis of Differential Transcript Abundances between G. gynandra and T. hassleriana for Each Tissue.
Supplemental Data Set 9. Genes Assigned by K-Means Clustering to Each Cluster.
Supplemental Data Set 10. MapMan Enrichment Analysis of K-Means Clustering.
Supplemental Data Set 11. List of Genes Highly Correlated with Cluster Centers of Shifted Clusters.
Supplementary Material
Acknowledgments
Work in our laboratory was supported by grants from the Deutsche Forschungsgemeinschaft (EXC 1028, IRTG 1525, and WE 2231/9-1 to A.P.M.W.). We thank the HHU Biomedical Research Center (BMFZ) for support with RNA-seq analysis and the MSU High Performance Computing Cluster for support with computational analysis of RNA-seq data. We thank Stefanie Weidtkamp-Peters and the HHU Center for Advanced Imaging for expert advice and support with confocal microscopy and image analysis.
AUTHOR CONTRIBUTIONS
C.K. performed experimental work, analyzed data, and wrote the article. A.K.D. analyzed data and cowrote the article. M.S. assisted in data analysis, identified the root-to-shoot shift, and cowrote the article. J.M., S.S., T.J.W., and E.G.-C. assisted in data analysis. B.B. assisted in design of ploidy experiments. C.R.B assisted in data analysis and experimental design. R.S. assisted in data discussion. L.D.V. assisted in ploidy determination. A.B. analyzed data and wrote the article. A.P.M.W. designed the study and wrote the article.
Glossary
- CBBC
Calvin-Benson-Bassham cycle
- MC
mesophyll cell
- BSC
bundle sheath cell
- RPKM
reads per mappable million
- PCA
principle component analysis
- TF
transcription factor
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Anderson L.E. (1971). Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases. Biochim. Biophys. Acta 235: 237–244. [DOI] [PubMed] [Google Scholar]
- Andrews T.J., Lorimer G.H. (1987). Rubisco: Structure, mechanisms, and prospects for improvement. In The Biochemistry of Plants, Vol. 10, Photosynthesis, M.D. Hatch and N.K. Boardman, eds (San Diego, CA: Academic Press; ), pp. 131–218. [Google Scholar]
- Andriankaja M., Dhondt S., De Bodt S., Vanhaeren H., Coppens F., De Milde L., Mühlenbock P., Skirycz A., Gonzalez N., Beemster G.T.S., Inzé D. (2012). Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22: 64–78. [DOI] [PubMed] [Google Scholar]
- Anzola J.M., Sieberer T., Ortbauer M., Butt H., Korbei B., Weinhofer I., Müllner A.E., Luschnig C. (2010). Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. USA 107: 10308–10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S., Knerová J., Hibberd J.M. (2013). Endoreduplication is not involved in bundle-sheath formation in the C4 species Cleome gynandra. J. Exp. Bot. 65: 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer E., Thomas C., Maule A. (2008). Symplastic domains in the Arabidopsis shoot apical meristem correlate with PDLP1 expression patterns. Plant Signal. Behav. 3: 853–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebo A., et al. (2009). Life with and without AtTIP1;1, an Arabidopsis aquaporin preferentially localized in the apposing tonoplasts of adjacent vacuoles. Plant Mol. Biol. 70: 193–209. [DOI] [PubMed] [Google Scholar]
- Beemster G.T.S., De Veylder L., Vercruysse S., West G., Rombaut D., Van Hummelen P., Galichet A., Gruissem W., Inzé D., Vuylsteke M. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138: 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G., Baali-Cherif D., Bettinelli-Riccardi S., Parietti D., Bouguedoura N. (2009). Pollen-mediated gene flow in a highly fragmented landscape: consequences for defining a conservation strategy of the relict Laperrine’s olive. C. R. Biol. 332: 662–672. [DOI] [PubMed] [Google Scholar]
- Brady S.M., Provart N.J. (2009). Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell 21: 1034–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., et al. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155: 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Schliesky S., Külahoglu C., Osborne C.P., Weber A.P.M. (2014). Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. J. Exp. Bot. 65: 3579–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C., Morohashi K., Kawamura A., Takahashi N., Ishida T., Umeda M., Grotewold E., Sugimoto K. (2012). Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J. 31: 4488–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C., Kawamura A., Ichikawa T., Tominaga-Wada R., Wada T., Kondou Y., Muto S., Matsui M., Sugimoto K. (2009). The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21: 2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.J., Parsley K., Hibberd J.M. (2005). The future of C4 research—maize, Flaveria or Cleome? Trends Plant Sci. 10: 215–221. [DOI] [PubMed] [Google Scholar]
- Brown W.V. (1975). Variations in anatomy, associations, and origins of Kranz tissue. Am. J. Bot. 62: 395–402. [Google Scholar]
- Byrne M.E. (2012). Making leaves. Curr. Opin. Plant Biol. 15: 24–30. [DOI] [PubMed] [Google Scholar]
- Caro E., Desvoyes B., Gutierrez C. (2012). GTL1 keeps cell growth and nuclear ploidy under control. EMBO J. 31: 4483–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz D.A., Wei N., Osterlund M.T., von Arnim A.G., Staub J.M., Matsui M., Deng X.W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121. [DOI] [PubMed] [Google Scholar]
- Chapman E.A., Osmond C.B. (1974). The effect of light on the tricarboxylic acid cycle in green leaves: III. A Comparison between some C(3) and C(4) plants. Plant Physiol. 53: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain C.J., Failing C.J., Manandhar L., Zimmerman M.A., Lakner M.M., Nguyen T.H.T. (2011). Functional evolution of C(4) pyruvate, orthophosphate dikinase. J. Exp. Bot. 62: 3083–3091. [DOI] [PubMed] [Google Scholar]
- Cheng S., et al. (2013). The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 25: 2813–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C., Nafati M., Mathieu-Rivet E., Bourdon M., Frangne N., Cheniclet C., Renaudin J.-P., Gévaudant F., Hernould M. (2011). Elucidating the functional role of endoreduplication in tomato fruit development. Ann. Bot. (Lond.) 107: 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R., Vidal J., O’Leary M.H. (1996). Phosphoenolpyruvate carboxylase: A ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 273–298. [DOI] [PubMed] [Google Scholar]
- Christin P.-A., Besnard G. (2009). Two independent C4 origins in Aristidoideae (Poaceae) revealed by the recruitment of distinct phosphoenolpyruvate carboxylase genes. Am. J. Bot. 96: 2234–2239. [DOI] [PubMed] [Google Scholar]
- Christin P.-A., Osborne C.P., Chatelet D.S., Columbus J.T., Besnard G., Hodkinson T.R., Garrison L.M., Vorontsova M.S., Edwards E.J. (2013). Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proc. Natl. Acad. Sci. USA 110: 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P.A., Salamin N., Kellogg E.A., Vicentini A., Besnard G. (2009). Integrating phylogeny into studies of C4 variation in the grasses. Plant Physiol. 149: 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann E.M.N., Levesque M.P., De Veylder L., Reichardt I., Jürgens G., Schmid M., Schwechheimer C. (2008). The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135: 2013–2022. [DOI] [PubMed] [Google Scholar]
- Donner T.J., Sherr I., Scarpella E. (2009). Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246. [DOI] [PubMed] [Google Scholar]
- Edwards E.J., et al. C4 Grasses Consortium (2010). The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328: 587–591. [DOI] [PubMed] [Google Scholar]
- Ehleringer J.R., Björkman O. (1978). A comparison of photosynthetic characteristics of encelia species possessing glabrous and pubescent leaves. Plant Physiol. 62: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehleringer J.R., Sage R.F., Flanagan L.B., Pearcy R.W. (1991). Climate change and the evolution of C(4) photosynthesis. Trends Ecol. Evol. (Amst.) 6: 95–99. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13: 203–229. [DOI] [PubMed] [Google Scholar]
- Furbank R.T., Hatch M.D. (1987). Mechanism of c(4) photosynthesis: the size and composition of the inorganic carbon pool in bundle sheath cells. Plant Physiol. 85: 958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U., Bräutigam A., Weber K.L., Weber A.P.M., Westhoff P. (2011). Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23: 2087–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths H., Weller G., Toy L.F., Dennis R.J. (2013). You’re so vein: bundle sheath physiology, phylogeny and evolution in C3 and C4 plants. Plant Cell Environ. 36: 249–261. [DOI] [PubMed] [Google Scholar]
- Haberlandt G. (1896). Physiologische Pflanzenanatomie. (Leipzig, Germany: Verlag von Wilhelm Engelmann).
- Hatch M.D. (1987). C-4 photosynthesis - a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 895: 81–106. [Google Scholar]
- Hay J., Schwender J. (2011). Computational analysis of storage synthesis in developing Brassica napus L. (oilseed rape) embryos: flux variability analysis in relation to ¹³C metabolic flux analysis. Plant J. 67: 513–525. [DOI] [PubMed] [Google Scholar]
- Holm S. (1979). A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6: 65–70. [Google Scholar]
- Ilegems M., Douet V., Meylan-Bettex M., Uyttewaal M., Brand L., Bowman J.L., Stieger P.A. (2010). Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 137: 975–984. [DOI] [PubMed] [Google Scholar]
- Iltis H.H., Cochrane T.S. (2007). Studies in the Cleomaceae V: A new genus and ten new combinations for the flora of North America. Novon 17: 447–451. [Google Scholar]
- Iltis H.H., Hall J.C., Cochrane T.S., Sytsma K.J. (2011). Studies in the Cloemaceae I. On the separate recognition of Capparaceae, Cleomaceae, and Brassicaceae. Annals Miss. Bot. Gard. 98: 28–36. [Google Scholar]
- Inda L.A., Torrecilla P., Catalán P., Ruiz-Zapata T. (2008). Phylogeny of Cleome L. and its close relatives Podandrogyne Ducke and Polanisia Raf. (Cleomoideae, Cleomaceae) based on analysis of nuclear ITS sequences and morphology. Plant Sys. Evol. 274: 111–126. [Google Scholar]
- Kang J., Mizukami Y., Wang H., Fowke L., Dengler N.G. (2007). Modification of cell proliferation patterns alters leaf vein architecture in Arabidopsis thaliana. Planta 226: 1207–1218. [DOI] [PubMed] [Google Scholar]
- Kasili R., Walker J.D., Simmons L.A., Zhou J., De Veylder L., Larkin J.C. (2010). SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 185: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. (2002). BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S., Flügge U.I., Fischer K. (2003a). Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S., Löttgert T., Schneider A., Voll L., Flügge U.I., Fischer K. (2003b). Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis—AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 36: 411–420. [DOI] [PubMed] [Google Scholar]
- Lammens T., Boudolf V., Kheibarshekan L., Zalmas L.P., Gaamouche T., Maes S., Vanstraelen M., Kondorosi E., La Thangue N.B., Govaerts W., Inzé D., De Veylder L. (2008). Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. USA 105: 14721–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale J.A., Nelson T. (1991). Spatial regulation of photosynthetic development in C4 plants. Trends Genet. 7: 191–196. [DOI] [PubMed] [Google Scholar]
- Larson-Rabin Z., Li Z., Masson P.H., Day C.D. (2009). FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 149: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Ko J.-H., Lee S., Lee Y., Pak J.-H., Kim J.H. (2009a). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151: 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.O., Davidson J.M., Duronio R.J. (2009b). Endoreplication: polyploidy with purpose. Genes Dev. 23: 2461–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Lundquist P.K., Rosar C., Bräutigam A., Weber A.P. (2014). Plastid signals and the bundle sheath: mesophyll development in reticulate mutants. Mol. Plant 7: 14–29. [DOI] [PubMed] [Google Scholar]
- Mantiri F.R., Kurdyukov S., Lohar D.P., Sharopova N., Saeed N.A., Wang X.-D., Vandenbosch K.A., Rose R.J. (2008). The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 146: 1622–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D.M., Muhaidat R., Brown N.J., Liu Z., Stanley S., Griffiths H., Sage R.F., Hibberd J.M. (2007). Cleome, a genus closely related to Arabidopsis, contains species spanning a developmental progression from C(3) to C(4) photosynthesis. Plant J. 51: 886–896. [DOI] [PubMed] [Google Scholar]
- Mathieu-Rivet E., Gévaudant F., Cheniclet C., Hernould M., Chevalier C. (2010a). The anaphase promoting complex activator CCS52A, a key factor for fruit growth and endoreduplication in tomato. Plant Signal. Behav. 5: 985–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu-Rivet E., Gévaudant F., Sicard A., Salar S., Do P.T., Mouras A., Fernie A.R., Gibon Y., Rothan C., Chevalier C., Hernould M. (2010b). Functional analysis of the anaphase promoting complex activator CCS52A highlights the crucial role of endo-reduplication for fruit growth in tomato. Plant J. 62: 727–741. [DOI] [PubMed] [Google Scholar]
- Matsuoka M. (1995). The gene for pyruvate, orthophosphate dikinase in C4 plants: structure, regulation and evolution. Plant Cell Physiol. 36: 937–943. [DOI] [PubMed] [Google Scholar]
- McKown A.D., Dengler N.G. (2009). Shifts in leaf vein density through accelerated vein formation in C4 Flaveria (Asteraceae). Ann. Bot. (Lond.) 104: 1085–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown A.D., Dengler N.G. (2010). Vein patterning and evolution in C-4 plants. Botany 88: 775–786. [Google Scholar]
- Motose H., Sugiyama M., Fukuda H. (2004). A proteoglycan mediates inductive interaction during plant vascular development. Nature 429: 873–878. [DOI] [PubMed] [Google Scholar]
- Nelson T., Langdale J.A. (1992). Developmental genetics of C-4 photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 25–47. [Google Scholar]
- Nelson T., Dengler N. (1997). Leaf vascular pattern formation. Plant Cell 9: 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Fukuda H. (2010). Transcriptional regulation of vascular cell fates. Curr. Opin. Plant Biol. 13: 670–676. [DOI] [PubMed] [Google Scholar]
- Peeples M.A. (2011). R Script for K-Means Cluster Analysis. http://www.mattpeeples.net/kmeans.html.
- Pérez-Pérez J.M., Candela H., Robles P., López-Torrejón G., del Pozo J.C., Micol J.L. (2010). A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 51: 1661–1673. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Serralbo O., Vanstraelen M., González C., Criqui M.C., Genschik P., Kondorosi E., Scheres B. (2008). Specialization of CDC27 function in the Arabidopsis thaliana anaphase-promoting complex (APC/C). Plant J. 53: 78–89. [DOI] [PubMed] [Google Scholar]
- Pick T.R., Bräutigam A., Schlüter U., Denton A.K., Colmsee C., Scholz U., Fahnenstich H., Pieruschka R., Rascher U., Sonnewald U., Weber A.P.M. (2011). Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23: 4208–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing).
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage R.F. (2004). The evolution of C-4 photosynthesis. New Phytol. 161: 341–370. [DOI] [PubMed] [Google Scholar]
- Sage R.F., McKown A.D. (2006). Is C4 photosynthesis less phenotypically plastic than C3 photosynthesis? J. Exp. Bot. 57: 303–317. [DOI] [PubMed] [Google Scholar]
- Sage R.F., Christin P.A., Edwards E.J. (2011). The C(4) plant lineages of planet Earth. J. Exp. Bot. 62: 3155–3169. [DOI] [PubMed] [Google Scholar]
- Sage R.F., Zhu X.G. (2011). Exploiting the engine of C(4) photosynthesis. J. Exp. Bot. 62: 2989–3000. [DOI] [PubMed] [Google Scholar]
- Scarpella E., Francis P., Berleth T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131: 3445–3455. [DOI] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20: 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer T., Hauser M.T., Seifert G.J., Luschnig C. (2003). PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr. Biol. 13: 837–842. [DOI] [PubMed] [Google Scholar]
- Slewinski T.L. (2013). Using evolution as a guide to engineer kranz-type c4 photosynthesis. Front. Plant Sci. 4: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski T.L., Anderson A.A., Zhang C., Turgeon R. (2012). Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant Cell Physiol. 53: 2030–2037. [DOI] [PubMed] [Google Scholar]
- Stein H., Honig A., Miller G., Erster O., Eilenberg H., Csonka L.N., Szabados L., Koncz C., Zilberstein A. (2011). Elevation of free proline and proline-rich protein levels by simultaneous manipulations of proline biosynthesis and degradation in plants. Plant Sci. 181: 140–150. [DOI] [PubMed] [Google Scholar]
- Streatfield S.J., Weber A., Kinsman E.A., Häusler R.E., Li J., Post-Beittenmiller D., Kaiser W.M., Pyke K.A., Flügge U.I., Chory J. (1999). The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell 11: 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud R.M., Dengler N.G. (2000). Cell lineage of vein formation in variegated leaves of the C-4 grass Stenotaphrum secundatum. Ann. Bot. (Lond.) 86: 99–112. [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. (2003). “Big it up”: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6: 544–553. [DOI] [PubMed] [Google Scholar]
- Suzuki R., Shimodaira H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542. [DOI] [PubMed] [Google Scholar]
- Tobin E.M., Silverthorne J. (1985). Light regulation of gene-expression in higher plants. Annu. Rev. Plant Biol. 36: 569–593. [Google Scholar]
- Tolley B.J., Woodfield H., Wanchana S., Bruskiewich R., Hibberd J.M. (2012). Light-regulated and cell-specific methylation of the maize PEPC promoter. J. Exp. Bot. 63: 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J., Hülskamp M., Gendreau E., Höfte H. (1998). Endoreduplication and development: rule without dividing? Curr. Opin. Plant Biol. 1: 498–503. [DOI] [PubMed] [Google Scholar]
- Tronconi M.A., Gerrard Wheeler M.C., Maurino V.G., Drincovich M.F., Andreo C.S. (2010). NAD-malic enzymes of Arabidopsis thaliana display distinct kinetic mechanisms that support differences in physiological control. Biochem. J. 430: 295–303. [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Kelly S., Fouracre J.P., Langdale J.A. (2013). Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J. 75: 656–670. [DOI] [PubMed] [Google Scholar]
- Westhoff P., Gowik U. (2010). Evolution of C4 photosynthesis—looking for the master switch. Plant Physiol. 154: 598–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.C.G., Tronconi M.A., Drincovich M.F., Andreo C.S., Flügge U.I., Maurino V.G. (2005). A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol. 139: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Purugganan M.M., Polisensky D.H., Antosiewicz D.M., Fry S.C., Braam J. (1995). Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Ohtani M., Mitsuda N., Kubo M., Ohme-Takagi M., Fukuda H., Demura T. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekutieli D., Benjamini Y. (1999). Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J. Stat. Plan. Inference 82: 171–196. [Google Scholar]
- Zhiponova M.K., et al. (2013). Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 197: 490–502. [DOI] [PubMed] [Google Scholar]
- Zhu X.G., Long S.P., Ort D.R. (2008). What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr. Opin. Biotechnol. 19: 153–159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.