The Euphorbiaceae produce a wide range of biologically active diterpenoids. Using castor (Ricinus communis) as a model, this work provides evidence that some of the genes required for the biosynthesis of these diterpenoids are arranged in physical gene clusters and describes a conserved oxidation step required for biosynthesis of a majority of diterpenoids that has been reported in this family.
Abstract
The Euphorbiaceae produce a diverse range of diterpenoids, many of which have pharmacological activities. These diterpenoids include ingenol mebutate, which is licensed for the treatment of a precancerous skin condition (actinic keratosis), and phorbol derivatives such as resiniferatoxin and prostratin, which are undergoing investigation for the treatment of severe pain and HIV, respectively. Despite the interest in these diterpenoids, their biosynthesis is poorly understood at present, with the only characterized step being the conversion of geranylgeranyl pyrophosphate into casbene. Here, we report a physical cluster of diterpenoid biosynthetic genes from castor (Ricinus communis), including casbene synthases and cytochrome P450s from the CYP726A subfamily. CYP726A14, CYP726A17, and CYP726A18 were able to catalyze 5-oxidation of casbene, a conserved oxidation step in the biosynthesis of this family of medicinally important diterpenoids. CYP726A16 catalyzed 7,8-epoxidation of 5-keto-casbene and CYP726A15 catalyzed 5-oxidation of neocembrene. Evidence of similar gene clustering was also found in two other Euphorbiaceae, including Euphorbia peplus, the source organism of ingenol mebutate. These results demonstrate conservation of gene clusters at the higher taxonomic level of the plant family and that this phenomenon could prove useful in further elucidating diterpenoid biosynthetic pathways.
INTRODUCTION
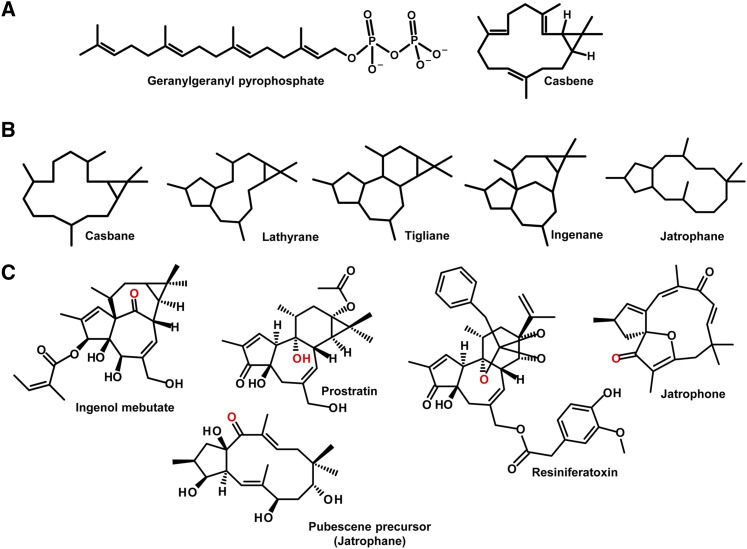
Diterpenoids are 20-carbon compounds derived from the common precursor geranylgeranyl pyrophosphate (Figure 1A). Diterpenoid biosynthesis is an essential process in plants, required for the production of chlorophylls, tocochromenols, and gibberellins. As well as these primary metabolites, some plant families have evolved the ability to produce a diverse range of diterpenoid secondary metabolites (Zerbe et al., 2013). The Euphorbiaceae (spurge family), for example, contains many plant species that produce diterpenoids with casbane, lathyrane, jatrophane, tigliane, or ingenane skeletons (Figure 1B). The Euphorbiaceae family is renowned in ethnomedicine (Mwine and Van Damme, 2011), and analyses of the extracts of a number of species in the Euphorbiaceae have revealed that in many cases the biological activity of these plants can be attributed to particular diterpenoids (Beutler et al., 1989). A number of these diterpenoids are now providing leads in the development of pharmaceutical compounds for the treatment of a wide range of medical conditions (Figure 1C). Ingenol mebutate from Euphorbia peplus has recently been licensed for the treatment of the precancerous skin condition actinic keratosis and has also proven effective in the treatment of superficial basal cell carcinoma in phase IIa clinical trials (Siller et al., 2010). Resiniferatoxin, a tigliane diterpenoid from Euphorbia resinifera, is currently in phase I/II trials for the treatment of intractable pain (Kissin and Szallasi, 2011). Prostratin (a tigliane), which is produced by Homalanthus nutans, Euphorbia fischeriana, and Euphorbia cornigera, has shown potential as an adjuvant therapy for the treatment of latent HIV infection (Miana et al., 1985; Qing-Gao et al., 1997; Kulkosky et al., 2001; Johnson et al., 2008). The jatrophane esters, which are produced by a number of Jatropha and Euphorbia species, have been shown to inhibit p-glycoprotein transporters responsible for efflux of chemotherapeutic agents and may therefore be useful for the treatment of multidrug resistant cancers (Corea et al., 2009; Reis et al., 2012). Jatrophone from Jatropha isabelli has antiplasmodial activity (Hadi et al., 2013).
Figure 1.
Structures of Diterpenoids Produced by Plants of the Euphorbiaceae.
(A) Structure of casbene and geranylgeranyl pyrophosphate, the substrate for casbene and neocembrene synthases.
(B) The casbene, lathyrane, tigliane, ingenane, and jatrophane carbon skeletons.
(C) Diterpenoids isolated from a number of Euphorbiaceae. The oxygen atom highlighted in red in each of these structures corresponds to the position on the casbene molecule that was oxidized by CYP726A14, CYP726A17, and CYP726A18.
Currently, most of these diterpenoids can only be sourced directly from the plant. However, many of these Euphorbiaceae are slow-growing or produce only minute quantities of the desired compound. For example, in the case of prostratin production by H. nutans, the desired compound is produced mainly within the stem of this slow growing plant, at a concentration of 0.2 to 52.6 μg g−1 (Johnson et al., 2008). The amounts of ingenol mebutate present within E. peplus are also low, at around 1.1 mg of ingenol mebutate per kilogram of plant material (Hohmann et al., 2000). To circumvent supply problems of ingenol mebutate, two chemical approaches have been investigated. A semisynthetic route has been developed using ingenol extracted from the seeds of Euphorbia lathyris (Liang et al., 2012), and, recently, a 14-step synthetic route was described (Jørgensen et al., 2013). Both of these routes, however, have limitations. The yield for total synthesis is lower than 1%, and the yield of ingenol from E. lathyris is still low at 275 mg per kg of seeds.
Knowledge of how these diterpenoids are biosynthesized could be used to improve their supply. For example, marker-assisted plant breeding can be used to select higher yielding varieties (Graham et al., 2010). Mutational breeding and heteroduplex mapping could be used to alter flux within metabolic pathways toward desired compounds (Tadele et al., 2009). Alternatively, drugs can be produced in heterologous species such as Escherichia coli or Saccharomyces cerevisiae. The advent of synthetic biology has proven particularly beneficial in this latter approach (Paddon et al., 2013), and this could also be exploited to produce novel compounds by combinatorial biochemistry (Winter and Tang, 2012). However, our understanding of diterpenoid biosynthesis within the Euphorbiaceae is limited at present. The conversion of geranylgeranyl diphosphate into casbene (Figure 1A) is the first committed step in the biosynthesis of terpenoids with jatrophane, tigliane, lathyrane, and ingenane skeletons (Figure 1B). The casbene synthase from castor (Ricinis communis) was one of the first terpene cyclases to be cloned (Mau and West, 1994) and has subsequently been identified in a number of other Euphorbiaceae. (Kirby et al., 2010; Zerbe et al., 2013). However, since the identification of the gene encoding this first committed step, no further steps in the pathways for the biosynthesis of casbene-derived diterpenoids have been identified. The reactions required for the biosynthesis of these diterpenoids include a series of oxidations, ring closures, and, in the case of esters, the addition of acyl groups. In plants, the cytochrome P450s are responsible for the majority of oxidation reactions that occur in secondary metabolism, including terpenoid biosynthesis (Mizutani, 2012). The origin of the intramolecular carbon-carbon bonds present in the jatrophane, tigliane, lathyrane, and ingenane diterpenoids is more obscure. It is possible that cytochrome P450 may be involved in these ring closures; CYP725A4 is able to catalyze the formation of such an intramolecular bond in a taxol molecule (Rontein et al., 2008). Plants have many acyltransferase families that could be responsible for diterpenoid ester biosynthesis, but BAHD acyltransferases, which are also involved in taxol biosynthesis (D’Auria, 2006), are the strongest candidates. Understanding how these diterpenoids are produced by the Euphorbiaceae will provide insights into how these plants have evolved to produce a large chemical diversity of these compounds and could lead to methods of improving the supply of specific diterpenoids for trial purposes or therapeutic use.
In this article, we report on the characterization of a number of diterpenoid modifying cytochrome P450s present within the genome of castor. These genes were physically clustered with casbene and neocembrene synthases. A BAHD acyltransferase and two alcohol dehydrogenase-like enzymes with homology to terpenoid-modifying enzymes were also present within this cluster. We present evidence of transcriptional coregulation of this gene cluster and demonstrate evidence of similar gene clusters in two other species representative of the main clades of diterpenoid producing Euphorbiaceae: Jatropha curcas and E. peplus.
RESULTS
Diterpene Synthase Genes Are Clustered with Eight Cytochrome P450s, a BAHD Acyltransferase, and Two Short-Chain ADHs in the Castor Genome
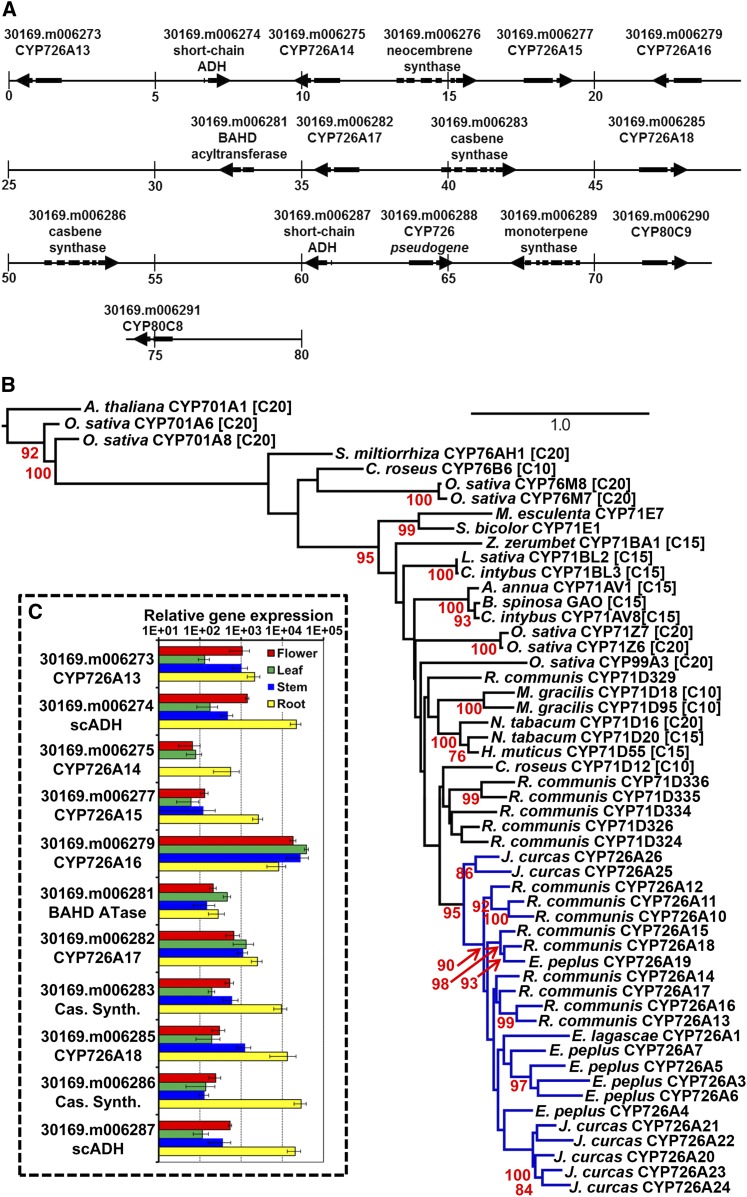
Within the Euphorbiaceae that are known to produce diterpenoids, the species with the most complete genome sequence is R. communis. This species produces casbene (Dueber et al., 1978) and a number of casbane diterpenoids, including (3E,7Z,11E)-19-hydroxycasba-3,7,11-trien-5-one and 6α-hydroxy-10β-methoxy-7α,8α-epoxy-5-oxocasbane-20,10-olide (Tan et al., 2009). Three diterpene synthases have previously been identified from this species, including two casbene synthases and a neocembrene synthase (Mau and West, 1994; Kirby et al., 2010). These diterpene synthases belong to the Tps-a subfamily, which contains mainly sesquiterpene cyclases involved in secondary metabolism (Trapp and Croteau, 2001; Chen et al., 2011). In silico analysis of the castor genome revealed that all three of these diterpene synthase genes were located within the same region of the genome (Figure 2A). Within close proximity to these genes, we also observed eight cytochrome P450 genes, a BAHD acyltransferase, two short-chain alcohol dehydrogenases, and a monoterpene synthase (Figure 2A). Two of the cytochrome P450 genes belonged to the CYP80C subfamily. Phylogenetic analysis revealed the other six were most closely related to CYP726A1, a fatty acid epoxidase from Euphorbia lagascae involved in vernolic acid biosynthesis in seeds (Figure 2B). A similar fatty acid-modifying function in castor seemed unlikely, as the oil of castor seeds contains primarily ricinoleic acid, which is formed by a variant fatty acid desaturase unrelated to the cytochrome P450s (van de Loo et al., 1995). After consulting David Nelson (Nelson, 2009), we were informed that these six cytochrome P450 had been reassigned from their prior CYP71D designations (CYP71D337 to CYP71D342) to CYP726A13 to CYP726A18 due to their homology to CYP726A1. CYP71D331 to CYP71D333 were also reassigned to CYP726A10 to CYP726A12 on this basis. As shown by the phylogenetic analysis (Figure 2B), the CYP726A form a taxon-specific clade within the CYP71D and should be regarded as part of a wider “CYP71D tribe,” which includes the CYP71D and CYP726A subfamilies.
Figure 2.
A Putative Diterpenoid Biosynthetic Gene Cluster from Castor.
(A) An 80-kb section of the R. communis genome containing the proposed diterpenoid biosynthetic gene cluster including genes for diterpene (casbene and neocembrene) synthases, CYP726A proteins, a BAHD acyltransferase, and two short-chain alcohol dehydrogenases. The numbers under the vertical bars represent the position within the gene cluster in kilobases.
(B) Phylogenetic tree obtained for the alignment of peptide sequences of CYP71 clan, including known terpenoid oxidases. Protein sequences are from R. communis CYP726A10 [XP_002532302], CYP726A11 [XP_002532303], CYP726A12 [XP_002532304], CYP71D334 [XP_002523467], CYP71D335 [XP_002514827.1], CYP71D336 [XP_002514826], CYP726A13 [KF986809], CYP726A14 [KF986810], CYP726A15 [KF986811], CYP726A16 [KF986812], CYP726A17 [KF986813], and CYP726A18 [KF986814]); E. lagascae Δ-12 fatty acid epoxidase (CYP726A1 [AAL62063.1]); E. peplus CYP726A3 [KF986822], CYP726A4 [KF986823], CYP726A5 [KF986824], CYP726A6 [KF986825], and CYP726A19 [KF986826]; J. curcas CYP726A20 [KF986815], CYP726A21 [KF986816], CYP726A22 [KF986817], CYP726A23 [KF986818], CYP726A24 [KF986819], CYP726A25 [KF986820], and CYP726A26 [KF986821]; Catharanthus roseus tabersonine-16-hydroxylase (CYP71D12 [ACM92061]) and geraniol-10-hydroxlase (CYP76B6, [Q8VWZ7]; N. tabacum 5-epiaristolochene dihydroxylase (CYP71D20 [Q94FM7]) and α-cembratrienol hydroxylase (CYP71D16 [AAD47832]; Hyoscyamus muticus premnaspirodiene oxidase (CYP71D55 [A6YIH8], Mentha gracilis limonene-6-hydroxylase (CYP71D18 [Q9XHE8] and limonene-3-hydroxylase (CYP71D95 [Q6WKY9]; Zingiber zerumbet α-humulene 10-hydroxlase (CYP71BA1 [E3W9C4]); O. sativa isokaurene C2-hydroxylase (CYP71Z6 [A3A871]), ent-cassadiene C2-hydroxylase (CYP71Z7 [Q6YV88]) ent-cassadiene C11-hydroxylases (CYP76M7 and CYP76M7 [Q69X58 and Q6YTF1] syn-pimaradiene oxidase (CYP99A3 [Q0JF01] and ent-kaurene oxidases (CYP701A6[AAT81230]) and CYP701A8[AAT46567], Lactuca sativa costunolide synthase (CYP71BL2 [F8S1I0]; Cichorium intybus (+)-valencene oxidase (CYP71AV8 [E1B2Z9] and costunolide synthase (CYP71BL3 [G3GBK0]; A. annua amorpha-4,11-diene C-12 oxidase (CYP71AV1 [Q1PS23]; Barnadesia spinosa germacrene A oxidase (BsGAO, D5JBX1.1]; M. esculenta 2-methylbutanal oxime monooxygenase and Sorghum bicolor 4-hydroxyphenylacetaldoxime monooxygenase (CYP71E1 [O48958] and Salvia miltiorrhiza miltiradiene oxidase (CYP76AH1 [AGN04215]), and Arabidopsis ent-kaurene oxidase (CYP701A [NP_197962]). Enzymes known to be involved in mono-, sesqui-, and diterpenoid biosynthesis are indicated by [C10], [C15], and [C20], respectively. CYP726A is highlighted with blue branches. The numbers in red positioned to the left of and below the nodes indicate bootstrap values shown as a percentage based on 1000 replicates. Bootstrap values below 70% are not shown. The scale bar (top right) represents the number of substitutions per site.
(C) Expression of castor diterpenoid genes. Relative transcript levels were calculated against a housekeeping gene (20-kD nuclear cap binding protein) using real-time PCR. For each tissue sample, three biological and four technical replicates were used. The error bars show the standard deviations.
The CYP71 clan is the largest P450 clan in plants, containing over half of all known cytochrome P450s identified from this kingdom (Nelson and Werck-Reichhart, 2011; Hamberger and Bak, 2013). The function of the vast majority of these enzymes is not known, but many of those that have been characterized function as terpenoid oxidases. The CYP71 subfamily, for example (the largest subfamily of the CYP71 clan), includes monoterpenoid (C10), sesquiterpenoid (C15), and diterpenoid (C20) modifying enzymes, as indicated in Figure 2B. The diterpenoid oxidases include Nicotiana tabacum CYP71D16, an enzyme involved in the biosynthesis of cembratrienediols in trichomes (Wang and Wagner, 2003) and CYP71Z6 and CYP71Z7 from rice (Oryza sativa). Interestingly, these two rice CYP71 genes are located within a physical gene cluster of diterpenoid biosynthetic genes that includes ent-copalyl pyrophosphate synthase (CPS) and kaurene synthase-like (KSL) genes and the CYP76 subfamily genes (also part of the CYP71 clan), CYP76M5, CYP76M6, CYP76M7, and CYP76M8. This cluster of genes is involved in multiple pathways. For example, rice CPS2, KSL7, CYP76M7, CYP76M8, and CYP71Z7 are involved in the conversion of geranylgeranyl pyrophosphate via ent-cassadiene into phytocassanes, whereas CPS2, KSL6, and CYP71Z6 are involved in the biosynthesis of oryzalide A (Swaminathan et al., 2009; Wu et al., 2011; Wang et al., 2012b). CYP99A3 from rice also forms part of a diterpenoid biosynthetic gene cluster involved in momilactone biosynthesis (Wilderman et al., 2004; Shimura et al., 2007; Wang et al., 2011; Wada et al., 2012). More widely, the CYP71 clan contains the CYP701A subfamily. These P450s are mostly ent-kaurene oxidases involved in biosynthesis of the diterpenoid phytohormone gibberellin (a primary metabolite). In rice, however, CYP701A8 has evolved to have a role in secondary metabolism and is involved in the biosynthesis of both oryzalexins and phytocassanes (Wang et al., 2012a).
The colocalization of genes encoding diterpene synthases and CYP726A proteins led us to speculate that these genes may form part of a gene cluster (Osbourn, 2010). To investigate this further, we performed spatial gene expression analysis of the genes present within the putative cluster and determined the ability of the CYP726A to modify casbene and neocembrene.
Coordinate Expression of Genes within the Castor Diterpenoid Cluster
Expression of the genes encoding the three diterpene synthases, six CYP726A, the two short-chain ADH, and the BAHD acyltransferase within roots, stems, leaves, and flowers was analyzed by quantitative PCR (qPCR) (Figure 2C). Transcripts were detected for all genes present within the cluster, with the exception of the putative neocembrene synthase. Expression of CYP726A14 was also not detected in stems. For the majority of the other 11 genes for which transcripts were detected, gene expression was highest in the roots (9 of 11 genes) and lowest in the leaves (7 of 11 genes), with intermediate levels of expression observed in stems (except CYP726A14) and flowers. The most striking differences were for Rc30169.m006274 (short-chain ADH), Rc30169.m006283 (casbene synthase), Rc30169.m006285 (CYP726A18), Rc30169.m006286 (casbene synthase), and Rc30169.m006287 (short-chain ADH). Rc30169.m006279 (CYP726A16) expression was more ubiquitous, with similar levels in leaves, flowers, and stems. Together, these data demonstrate that these genes are not only colocalized within the genome, but share a similar pattern of coexpression.
Modification of Casbene and Neocembrene by CYP726A Enzymes Encoded within the Castor Gene Cluster
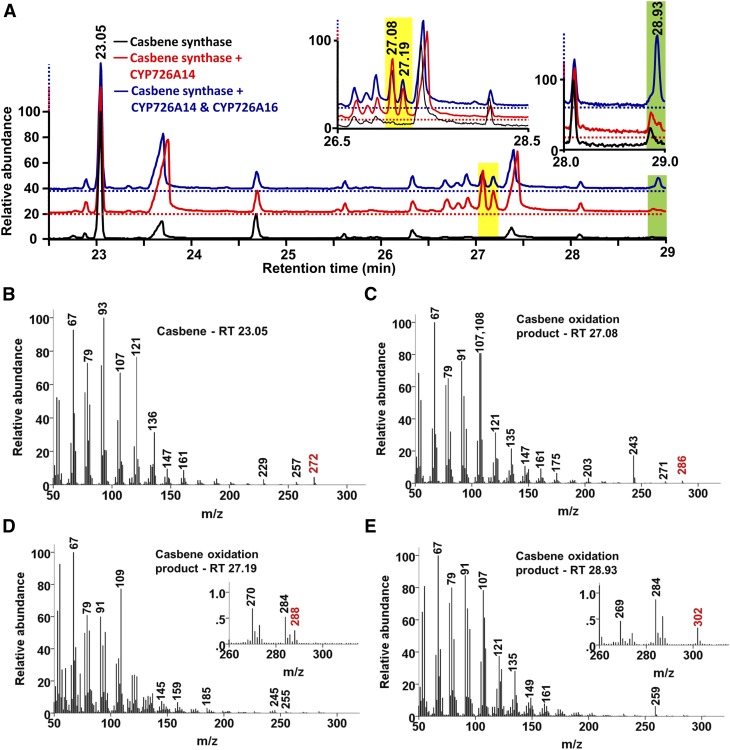
To determine whether any of the CYP726A proteins were capable of modifying casbene or neocembrene, we used the Nicotiana benthamiana transient expression system (Voinnet et al., 2003). A casbene synthase (from J. curcas) or neocembrene synthase (from R. communis) gene and each of the CYP726A genes were coexpressed via infiltration of young N. benthamiana plants by multiple combinations of Agrobacterium tumefaciens strains harboring different expression vectors. Five days after infiltration, chloroform extracts of the leaves were analyzed by gas chromatography-mass spectrometry (GC-MS) and putative casbene-related compounds identified based on mass spectral similarities between the resolved peaks (Figures 3B to 3E). Three of the CYP726A enzymes (CYP726A14, CYP726A17, and CYP726A18) were able to use casbene as a substrate (Figure 3A; Supplemental Figure 1). For these three enzymes, the same two casbene-related product peaks were detected. The apparent molecular weight of the products with retention times of 27.08 and 27.19 min were 286 and 288, respectively, a difference of 14 and 16 atomic mass units compared with casbene (Figures 3C and 3D). CYP726A15 was able to oxidize neocembrene (Supplemental Figure 1).
Figure 3.
GC-MS Analysis of N. benthamiana Leaf Extracts Transiently Expressing Casbene Synthase and Cytochrome P450 Enzymes.
(A) Gas chromatograph obtained with expression of casbene synthase only (lower, black), casbene synthase and CYP726A14 (middle, red), and casbene synthase plus CYP726A14 and CYP726A16 (upper, blue). The two insets display rescaled chromatographs containing the retention times at which the casbene synthase oxidation products eluted from the column.
(B) to (E) Electron ionization mass spectra for casbene (B), casbene oxidation products produced by CYP726A14 ([C] and [D]), and product obtained from coexpression of proteins encoded CYP726A14 and CYP726A16 (E). The molecular ions are indicated in red and are shown in insets for (C) and (E).
To determine whether any of the CYP726A proteins were capable of acting sequentially, we also coinfiltrated N. benthamiana with casbene synthase, CYP726A14, and each of the other CYP726A genes present in the castor gene cluster. Coexpression of casbene synthase with CYP726A14 and CYP726A16 resulted in the formation of an additional product peak (retention time 27.08 min, with an apparent molecular weight of 302). Together, these data confirm that the colocalized diterpene synthase and CYP726A genes of castor represent a bone fide gene cluster.
NMR Structural Analysis of Cytochrome P450 Products
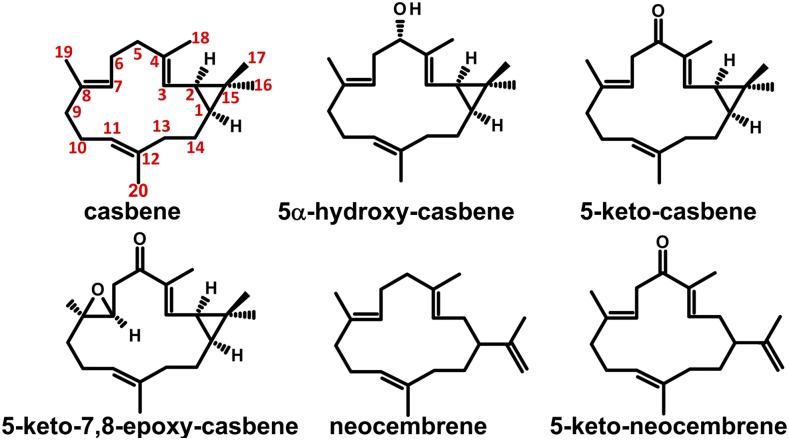
The two casbene oxidation products of CYP726A14 (also equivalent to those from CYP726A17 and CYP726A18) were extracted from 100 g (fresh weight) of infiltrated N. benthamiana leaf material and purified using a combination of normal-phase flash chromatography and semipreparative reversed-phase HPLC. This yielded ∼300 μg of casbene, 15 μg of product 1, and 30 μg of product 2. All three samples were dissolved separately in CDCl3 (500 μL), and NMR data were recorded at 700 MHz. Following acquisition of both 1D-1H and 13C NMR spectra, the 2D experiment and edited-HSQC were used to determine which protons and carbons were directly attached to one another via a single bond. Both HMBC and 1H-1H COSY experiments were then used to connect these fragments of the molecule together through the observation of 2- and 3-bond couplings, respectively, between proton and carbon and (predominantly) 3-bond couplings between protons. The 1H and 13C assignments resulting from these experiments are shown at each position on the structures appearing in Figure 4. Product 1 was identified as 5-keto-casbene and product 2 as 5α-hydroxy-casbene. The perturbations to both 1H and 13C chemical shifts at and around the 5-position of the two oxygenated derivatives of casbene support the two functional groups that are proposed. (However, note that all three structures were established primarily on the basis of connectivities observed in 2D-NMR; these data are presented in Supplemental Figures 2 and 3 and Supplemental Tables 6 to 8). Although it is possible that an endogenous biochemical activity present within N. benthamiana may have been responsible for conversion of 5α-hydroxy-casbene to 5-keto-casbene (or vice versa), it is more likely that both of these products were formed by the castor P450s. Within the CYP71 clan, there are many documented examples of enzymes that are able to catalyze successive oxidation reactions at the same carbon to form hydroxyl, carbonyl, and even carboxyl groups, such as CYP71AV1 from Artemesia annua, which is able to form artemisinic alcohol, artemisinic aldehyde, and artemisinic acid (Ro et al., 2006; Teoh et al., 2006).
Figure 4.
Structures of Casbene, Neocembrene, and the CYP726A Oxidation Products Determined by NMR.
Structures of (-)-casbene (retention time 23.05 in Figure 3A and mass spectrum in Figure 3B), 5α-hydroxy-casbene (retention time 27.08 in Figure 3A and mass spectrum in Figure 3C), 5-keto-casbene (retention time 27.17 in Figure 3A and mass spectrum in Figure 3D), 5-keto-7,8-epoxy-casbene (retention time 28.93 in Figure 3A and mass spectrum in Figure 3E), neocembrene, and 5-keto-neocembrene as established by 1D- and 2D-NMR. NMR data, including 1H and 13C assignments, are provided in Supplemental Figures 2 to 5 and Supplemental Tables 6 to 11. The casbene numbering system (show for casbene only) is based on Tan et al. (2009).
[See online article for color version of this figure.]
In later experiments, we switched from pFGC-derived vectors to pEAQ-HT vectors, which permit a higher level of gene expression (see Methods and Sainsbury et al., 2009). By coexpressing casbene synthase, casbene-5-oxidase, and CYP726A16, we were able to purify ∼500 μg of product from 209 g (fresh weight) of leaves, which was identified as 5-keto-7,8-epoxy-casbene (Figure 4; Supplemental Figure 4 and Supplemental Table 9). CYP726A16 was not able to oxidize casbene (Supplemental Figure 1B). This lack of activity with casbene indicates that the conversion of casbene to 5-keto casbene is a linear step in the diterpenoid biosynthetic pathway.
To characterize the function of CYP726A15, we extracted 84 and 163 g of leaves expressing either neocembrene synthase or neocembrene synthase and CYP726A15, respectively, to obtain ∼1 mg of neocembrene and 1 mg of the oxidation product, which was identified as 5-keto-neocembrene (Figure 4; Supplemental Figure 5 and Supplemental Tables 10 and 11).
Clustering of Diterpenoid Biosynthetic Genes Is Common in the Euphorbiaceae
To investigate whether clustering of diterpenoid biosynthetic genes is a phenomenon widely occurring in the other Euphorbiaceae, we investigated two other species, J. curcas and E. peplus, for the presence of similar clusters. Jatropha and Euphorbia belong to the Crotonoideae and Euphorbioideae subfamilies, respectively, which contain the majority of diterpenoid-producing species within the Euphorbiaceae (Supplemental Figure 6) (Beutler et al., 1989). J. curcas in particular is known to produce a wide variety of diterpenoids, including a number of diterpenoids within roots (Zhang et al., 2012) and phorbol esters within seeds (Haas et al., 2002; He et al., 2011). E. peplus also produces a range of diterpenoids, including the licensed pharmaceutical ingenol mebutate (Hohmann et al., 2000).
Analysis of the genome of J. curcas (Sato et al., 2011; Hirakawa et al., 2012) reveals that it contains three putative full-length diterpene synthases, which are clustered. Prior to the release of the J. curcas genome sequence, we also identified three diterpene synthases using degenerate PCR and determined that these were clustered using long-distance PCR (GenBank accession KJ026361). However, the arrangement of the genes we identified and that reported by the J. curcas genome database differ slightly. This may be due to incorrect assembly of short reads generated by next generation sequencing. Using heterologous gene expression in E. coli, we confirmed that all three of these genes were casbene synthases (Supplemental Figure 7). Using an F2 mapping population we have described previously (King et al., 2013), we were also able to map sequences corresponding to two CYP726A genes and the three casbene synthase genes to a single locus on chromosome 1 (Figure 5A). We also mapped a previously described BAC sequence (GenBank accession AP011970; Sato et al., 2011) to this region, which contains both a casbene synthase and CYP726A pseudogenes. Interestingly, the putative gene cluster in J. curcas is on a separate linkage group to a locus that we have previously identified as being responsible for the loss of phorbol ester biosynthesis in the seed of some naturally occurring provenances of this species (King et al., 2013). However, we observed that varieties of J. curcas that do not produce phorbol esters within seeds are not compromised in their ability to produce diterpenoids within roots, indicating that diterpenoid biosynthesis per se is not blocked in these plants.
Figure 5.
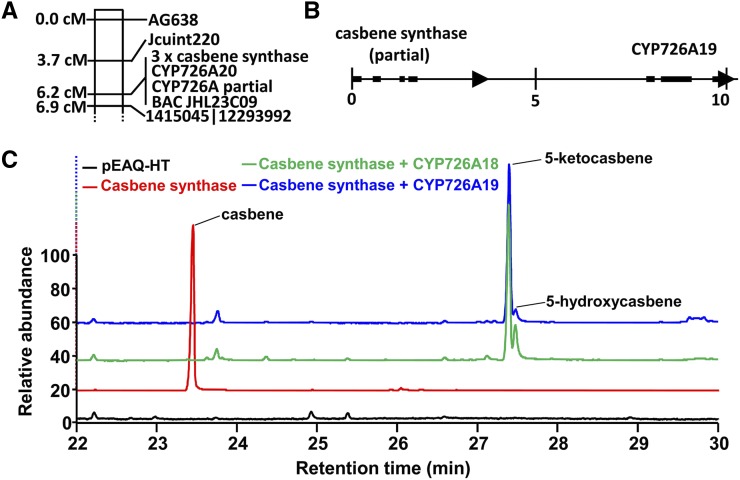
Clustering of Casbene Synthases and CYP726A Genes in J. curcas and E. peplus.
(A) Physical linkage of three casbene synthase genes (GenBank accession KJ026361), two cytochrome P450 genes (GenBank accession KF986815 and J. curcas genome database accession Jcr4S16860.10) and casbene synthase and CYP726A pseudogenes (AP011970) on chromosome 1 of J. curcas determined by linkage mapping in a population of 220 F2 individuals (47).
(B) Physical linkage of a casbene synthase homolog and CYP721A19 in E. peplus determined by long-distance PCR (GenBank accession KJ026362). The numbers under the vertical bars represent the position within the gene cluster in kilobases.
(C) Casbene 5-oxidase activity of CYP726A18 (R. communis) and CYP726A19 (E. peplus). The CYP726A genes were co-expressed with a casbene synthase gene from J. curcas (CAS1).
For E. peplus, there are currently limited resources available for the elucidation of a potential biosynthetic gene cluster. However, transcriptome data have been released for this species (Xiao et al., 2013), and a casbene synthase from this plant has already been cloned (Zerbe et al., 2013). Long-distance PCR revealed that this casbene synthase gene is adjacent to the CYP726A19 gene (Figure 5B; GenBank accession KJ026362). Phylogenetic analysis of the CYP726A19 protein (Figure 2B) indicated that it was most closely related to CYP726A18 from castor. The positioning and orientation of the gene relative to casbene synthase also suggested that CYP726A19 may be an ortholog of CYP726A18. To determine whether CYP726A19 was also a casbene 5-oxidase, we coinfiltrated N. benthamiana plants with constructs containing a casbene synthase gene and CYP726A19 (Figure 5C). For these experiments, we used the pEAQ-HT vector (Sainsbury et al., 2009). As well as having a copy of the P19 suppressor of RNA silencing present on the vector, eliminating the need for coinfiltration with a separate P19 vector, pEAQ-HT contains a modified cowpea mosaic virus 35S promoter and 5′ untranslated region that can drive very high levels of gene expression (Sainsbury and Lomonossoff, 2008). Coexpression of casbene synthase and CYP726A19 resulted in the formation of both 5-keto- and 5-hydroxy-casbene. Interestingly, unlike previous experiments using the pFGC5941 vector (Figure 3A), we observed almost complete conversion of casbene into oxidated products. The relative amounts of 5-keto- and 5-hydroxy-casbene were also altered, with vastly more of the ketone being produced. Although this may be due to enzyme specificity of the E. peplus enzymes compared with those of castor, we obtained a similar result when casbene synthase and CYP726A18 were expressed in plants using the pEAQ-HT vector. We therefore attributed this shift in the balance of product from the hydroxy- to the keto- product as being a consequence of higher gene expression.
DISCUSSION
Significance of the Diterpenoid Biosynthetic Gene Clusters in Castor
A number of studies have shown that the clustering of genes for the biosynthesis of secondary metabolites occurs with some frequency in plants (Nützmann and Osbourn, 2014). Many of the gene clusters described to date are involved in terpenoid biosynthesis. Triterpenoid biosynthetic gene clusters have been described in Arabidopsis thaliana and oat (Avena sativa; Qi et al., 2004; Swaminathan et al., 2009; Field et al., 2011). As mentioned previously, work in rice has described two diterpenoid biosynthetic gene clusters involved in the production of phytocassanes, oryzalides, and momilactones (Wilderman et al., 2004; Shimura et al., 2007; Swaminathan et al., 2009; Wu et al., 2011). Both of these clusters produce labdane diterpenoids, which are structurally similar to the phytohormone gibberellin and the diterpene synthase genes present within these gene clusters and are related to the copalyl diphosphate and kaurene synthase genes of the TPS-c and TPS-e/f clades responsible for the production of this primary metabolite. Despite the role of the Tps-a clade of terpene synthases in producing the large diversity of sequiterpenoid and diterpenoid secondary metabolites (Trapp and Croteau, 2001; Chen et al., 2011), whether there are other gene clusters containing terpene cyclases from this clade remains unclear. As members of the Tps-a clade, casbene, and neocembrene synthases are unusual in that they contain a chloroplast transit peptide and use geranylgeranyl pyrophosphate as a substrate rather than 15-carbon farnesyl pyrophosphate substrate used by most of the other members within this family, which are sesquiterpene cyclases. However, in silico analysis of plant genomes has indicated that other gene clusters carrying Tps-a genes may occur in other plant species. For example, the TPS12 and TPS13 genes of Arabidopsis, which have both been characterized as (Z)-γ-bisabolene synthases, are colocated on the genome with the uncharacterized CYP71A19 and CYP71A20 genes (Tholl and Lee, 2011).
The Evolution of Diterpenoid Biosynthetic Gene Clusters in the Euphorbiaceae
The Euphorbiaceae is one of the largest families of plants, containing 229 known genera and over 6500 species (www.theplantlist.org). It is also recognized as being one of the most morphologically and phytochemically diverse families of the Malpighiales order (Wurdack et al., 2005), which is thought to have evolved around 100 million years ago (Bell et al., 2010). The majority of the diterpenoid producing Euphorbiaceae reported belong to two of the three major subfamilies, the Euphorboideae and the Crotonoideae. However, the third major subfamily, the Acalyphoideae, also contains a number of species that produce diterpenoids, including castor (Supplemental Figure 6). The wide distribution of diterpenoid producing capability within the Euphorbiaceae and the evidence we presented for gene clustering from a member of all three major subfamilies indicate that the ability to produce diterpenoids is likely to have evolved early in the evolution of the Euphorbiaceae. Strikingly, although the overall structural diversity reported for diterpenoids is very large, molecules with similar structures can be observed across all three subfamilies. For example, phorbols have been found in representatives of the Crotonoideae (e.g., Jatropha and Croton), Euphorboideae (e.g., Euphorbia, Homulanthus, and Hura), and Acalyphoidae (Pycnocoma). Although this may be an example of convergent evolution, it also suggests that the ancestor of these three subfamilies may have already contained a functional biosynthetic pathway for the production of phorbols.
One of the plant species assigned to the Crotonoideae is cassava (Manihot esculenta), a staple crop in the diet of an estimated 800 million people in Africa, Asia, and the Americas (Prochnik et al., 2012). Thus far, there have been no reports of casbene or neocembrene derived diterpenoids in this species. We performed a tBLASTn search of the M. esculenta genome and could not find any TpsA family genes containing a chloroplast transit peptides (i.e., potential homologs of casbene and neocembrene synthase) or a CYP726A family gene (data not shown). A number of molecular phylogenetic studies have shown that Manihot does not form a single monophyletic group with the other Crotonoideae (Wurdack et al., 2005; Tokuoka and Tobe, 2006). Therefore, it is possible that this genus may have either evolved separately from the other diterpenoid producing Euphorbiaceae or may have coevolved and then subsequently lost the diterpenoid biosynthetic gene cluster.
Casbene-5-Oxidation as a Conserved Key Step in the Biosynthesis of Biologically Active Diterpenoids in the Euphorbiaceae
Four of the CYP726A proteins described in this study (CYP726A14, CYP726A17, and CYP726A18 from castor and CYP726A19 from E. peplus) were casbene-5-oxidases. We compared the structures from a number of biologically active diterpenoids produced by the Euphorbiaceae and observed that the oxidation of casbene at this 5-position is conserved in almost all of these diterpenoids described so far. For ingenol mebutate, prostratin, resiniferatoxin, jatrophane, and a pubescene precursor, the carbon atom at this position was oxidized (see oxygen atoms highlighted in red on Figure 1C). A number of other casbane, lathyrane, tigliane, jatrophane, and ingenane diterpenoids that are also oxidized at this position are shown in Supplemental Figure 8. This strongly indicates that casbene-5-oxidation is likely to be a conserved step in the biosynthesis of the majority of the bioactive diterpenoids produced by the Euphorbiaceae. 5-Keto-casbene has not been reported in castor. However, Tan et al. (2009) previously reported two diterpenoids obtained from extracting large quantities of the aerial parts of the castor plant: (3E,7Z,11E)-19-hydroxycasba-3,7,11-trien-5-one and 6α-hydroxy-10β-methoxy-7α,8α-epoxy-5-oxocasbane-20,10-olide. Both these structures contain a keto-group at the 5-position, and the latter contains an epoxide group at the 7,8-position. The casbene derivatives we reported in this article are therefore likely precursors for these molecules. Neocembranes have not previously been reported in castor, and the transcript for neocembrene synthase was not detected within this study. Nonetheless, the genome of castor contains both functional neocembrene synthase and neocembrene oxidase, suggesting that these genes may be functional under certain conditions. As well as 5-oxidation, a number of other oxygenation steps also appeared to be highly conserved. For example, in both phorbol (prostratin and resiniferatoxin) and ingenol molecules shown in Figure 1C, equivalent casbene positions of 9, 10, 11, and 20 have all been oxidized. This suggests that many of the cytochrome P450 genes are likely to be conserved between species.
Toward the Exploitation of the Euphorbiaceae Gene Clusters
At this stage in our studies, it is not clear whether the diterpenoid biosynthetic gene clusters that appear in other Euphorbiaceae will be sufficient for the production of the many biologically active end products that have been reported, such as ingenol mebutate and prostratin, or whether these gene clusters only contain a partial pathway, with the remaining steps being located at other loci. Nevertheless, the presence of clusters in the Euphorbiaceae could be exploited to facilitate the more rapid identification of genes involved in the biosynthesis of particular metabolites. For example, by screening BAC libraries, it may be possible to simultaneously clone multiple genes within a pathway. For most species, whole-genome sequences are either not available or the available drafts are not sufficiently assembled to permit the identification of putative gene clusters using bioinformatic techniques. However, BAC library screening, particularly where a PCR-based screening approach can be adopted, provides a much more rapid method of selecting targeted regions of the genome for sequencing. This BAC sequencing approach has already been used for the identification of triterpenoid biosynthetic genes in oat (Mugford et al., 2013) and alkaloid biosynthetic genes in the opium poppy (Papaver somniferum; Winzer et al., 2012). Another advantage of this approach compared with the identification of genes based on transcriptomic studies is sequencing of genomic DNA also provides cis-acting sequences such as promoter regions, which could provide insight into the regulation of genes within a cluster.
The identification of the complement of genes responsible for a biochemical pathway would provide the opportunity to improve production through engineering of the natural host or transfer of production to a heterologous host. The use of alternative host systems such as yeasts to produce either a final bioactive product or a precursor for semisynthesis is another approach that has been used to increase the supply of medicinal compounds (Paddon et al., 2013). This synthetic biology approach could also be used to combine diterpenoid biosynthesis from a number of different source organisms, which could allow libraries of novel diterpenoid structures to be produced for bioactivity screens. Our future work will concentrate on characterizing the functions of other cytochrome P450 genes from the Euphorbiaceae, as well as the BAHD acyltransferases and short-chain ADH genes. As well as determining the functions of other genes in diterpenoid biosynthetic pathways, it is envisaged that this could provide insights into the evolution of gene clusters and chemical diversity within this large family of plants and the extent of their involvement in the production of more complex molecules such as phorbols and ingenols.
METHODS
In Silico Genome Analysis and Phylogenetic Tree Construction
The location of Ricinus communis genes was determined using the TIGR Castor Bean Genome Database (Chan et al., 2010). Jatropha genome data were obtained from release 4.5 of the Jatropha curcas genome database (Sato et al., 2011; Hirakawa et al., 2012). Partial and full-length cDNA sequences for Euphorbia peplus homologs of diterpene synthase and cytochrome P450s were obtained from the PhytoMetaSyn database (Xiao et al., 2013). Peptide sequence alignments were performed using the tools provided at www.phylogeny.fr (Dereeper et al., 2008); sequence alignments were first constructed using MUSCLE in full mode with 16 iterations (Edgar, 2004). Nonconserved regions were then removed from the alignment using GBLOCKS (Talavera and Castresana, 2007). The resulting 296 amino acid sequence alignment (Supplemental Data Set 1) was used to construct a phylogenetic tree using PhyML (Guindon and Gascuel, 2003) with the LG amino acid replacement matrix (Le and Gascuel, 2008). The phylogenetic tree was drawn using FigTree version 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/).
qPCR Analysis of Gene Expression of the Castor Gene Cluster
For each tissue, the analysis was performed using three biological and four technical replicates. RNA was extracted from plants using the CTAB-lithium chloride method (Gasic et al., 2004). RNA samples were DNase treated and further purified using the on-column digestion protocol for the Qiagen RNeasy miniprep kit. cDNA was synthesized from 5 μg of total RNA using random hexamers using Superscript II reverse transcriptase (Life Technologies). qPCR primers (Supplemental Table 1) were designed using QuantPrime (Arvidsson et al., 2008) or Primer3Plus (Untergasser et al., 2007). Due to the similarity of many of the target genes, optimal primer annealing temperatures were obtained by gradient PCR and then primer specificity was confirmed using template for both target gene and genes of the same family.
Real-time PCR was performed on Bio-Rad MyIQ iCycler (Bio-Rad Laboratories) using iQ SybrGreen supermix. Each 20-μL reaction contained 5 μL of a 25-fold dilution of the cDNA synthesis reaction, 10 μL of 2× supermix, and primers at a final concentration of 150 nM. The cycling conditions included an initial activation step for 10 min at 95°C followed by 40 cycles of 95°C for 10 s, annealing (58 to 63°C; Supplemental Table 1) for 15 s, and extension at 72°C for 30 s. Fluorescence data were acquired during the extension phase. A melt curve was obtained at the end of the amplification to allow confirmation of product specificity. Relative transcript abundance was determined using the delta-delta CT method (Pfaffl, 2001) with correction for amplification efficiencies obtained using LinReg PCR (Ruijter et al., 2009). Gene expression was normalized against the 20-kD nuclear cap binding protein (GenBank accession XM_002517818). The expression levels of this housekeeping gene were found to be stable against both an actin 1 homolog (XM_002522148) and a ubiquitin 1 homolog (XM_002513332).
Transient Expression of Genes in Nicotiana benthamiana
cDNA was prepared from total RNA samples using Superscript II reverse transcriptase and a 5′-T(18)VN-3′ primer. The open reading frame of casbene synthase, neocembrene synthase, and cytochrome P450 genes were amplified from this cDNA using Phusion Pfu polymerase (New England Biolabs) using the primers detailed in Supplemental Table 2 (R. communis genes), Supplemental Table 3 (J. curcus genes), or Supplemental Table 4 (E. peplus genes). The open reading frames (ORFs) were then cloned into the pJET1.2 vector (Thermo Scientific). Dye-terminator sequencing was used to confirm the sequence of each cDNA. Restriction sites were added to the 5′ and 3′ ends of the ORF by PCR using the primers detailed in Supplemental Tables 2 to 4. For each ORF, a 5′-AAAA-3′ Kozak sequence was included immediately before each start codon. After restriction digestion, the ORF was then inserted into the 10-kb fragment obtained from digestion of pFGC5941 with AscI and PacI positioning each gene under the control of cauliflower mosaic virus 35S promoter (Kerschen et al., 2004) or between the AgeI and XhoI sites of the pEAQ-HT vector (Sainsbury et al., 2009). The expression vectors were then transformed into Agrobacterium tumefaciens GV3101:pMP90 using the freeze-thaw method (Höfgen and Willmitzer, 1988). Infiltration of N. benthamiana plants was performed as described previously (Voinnet et al., 2003). Where the pFGC5941 was used, coinfiltration with a vector carrying the p19 protein of tomato bushy stunt virus was performed. Five days after Agrobacterium infiltration, three leaves were collected from each plant, ground in liquid nitrogen, and then extracted with 10 mL of chloroform. The extracts were dried over anhydrous sodium sulfate, concentrated to <500 μL under a stream of nitrogen, and then 5 μL of the extract was analyzed by GC-MS using a Thermofinnigan GCQ or a Leco Pegasus IV GC-TOF instrument. In both cases, the GC oven was fitted with a Restek RTX-5SIL MS capillary column (30 m, 0.25-mm ID, 0.25 μm df). The oven temperature was set at 100°C for 2 min and then increased to 300°C at a rate of 5°C min−1. Mass spectral data was acquired over the m/z range of 50 to 450 in positive electron ionization mode at −70 eV.
Purification of Diterpenes and Diterpenoids
To isolate casbene and the two oxidation products of CYP726A14, N. benthamiana plants were coinfiltrated with Agrobacterium carrying pFGC vectors conferring the expression of casbene synthase, CYP726A14, and a separate vector containing the p19 sequence. Five days after infiltration, 100 g of leaves was dried by lyophilization and then extracted twice with 200 mL of 60/40 hexane/isopropanol. The extract was dried over anhydrous sodium sulfate and the solvent was then removed by rotary evaporation to yield 520 mg of a green oily residue. This extract was dissolved in 10 mL of hexane and then fractionated by flash chromatography using 25 g of silica gel 60 (particle size 35 to 70 μm, 220 to 440 mesh) as a stationary phase. The mobile phases were (1) 250 mL of hexane, (2) 250 mL of 2% ethyl acetate in hexane, (3) 250 mL 10% ethyl acetate in hexane, and (4) 50% ethyl acetate in hexane. Fractions of 25 mL were collected, and an aliquot from each was analyzed for the presence of casbene or casbene oxidation products by GC-MS (as above). Fractions containing the desired compounds were pooled then concentrated via rotary evaporation. The casbene fraction did not require further purification, but the two casbene oxidation products were further purified using nonaqueous reverse phased HPLC. The fractions were evaporated to dryness and then dissolved in 250 μL of methanol. Ten-microliter aliquots were separated on a Develosil C30-UG-S column (3-mm ID, 5-μm particle size; Nomura Chemical Co.) using a three-solvent gradient. Solvent A was 20 mM ammonium formate, 0.2% formic acid, and 20% water in methanol. Solvent B was 0.2% formic acid in methanol. Solvent C was 0.2% formic acid in tetrahydrofuran. The gradient used was as follows; at injection, 80% solvent A and 20% solvent B ramping via a linear gradient to 40% solvent A and 60% solvent B over 16 min. After holding at this ratio for a further 1 min, the solvents were switched to 40% solvent B and 60% solvent C for a further 8 min. The flow rate was 1 mL min−1. Detection was performed using atmospheric pressure chemical ionization (+ve). The fractions were then pooled, evaporated to dryness in a GeneVac EZ-2 plus, and dissolved in 500 μL of CDCl3. NMR analysis was then performed as detailed in Supplemental Methods. In subsequent experiments to isolate the second casbene oxidation product (i.e., of CYP726A16), neocembrene, and the neocembrene oxidation product (CYP726A15), a similar strategy was employed, but instead using the pEAQ-HT expression vectors and omitting the separate p19 vector. The initial flash chromatography was performed using a Biotage Isolera Spektra LS system fitted with GraceResolve 40 g silica columns. Extractions from 209 g (casbene synthase + CYP726A14 + CYP726A16), 84 g (neocembrene synthase), and 163 g (neocembrene synthase + CYP726A15) of N. benthamiana leaves yielded approximately 500 μg, 1 mg, and 1 mg of metabolite, respectively.
Linkage Mapping in J. curcas
Linkage mapping was performed in a mapping population described previously (King et al., 2013), using either simple sequence repeat markers or single nucleotide polymorphism markers detailed in Supplemental Table 5. The genetic linkage map was constructed using CRI-MAP software (www.animalgenome.org).
Partial Reconstruction of the E. peplus Gene Cluster Using Long-Distance PCR
Genomic DNA was extracted from E. peplus plants using the Qiagen DNeasy plant mini kit. Long-distance PCR was performed using NEB LongAmp Taq DNA polymerase using primers detailed in Supplemental Table 4. The PCR product was then subcloned and sequenced using dye-terminator sequencing.
Accession Numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: R. communis cytochrome P450s CYP726A13 (KF986809), CYP726A14 (KF986810), CYP726A15 (KF986811), CYP726A16 (KF986812), CYP726A17 (KF986813), CYP726A18 (KF986814), CYP80C9 (XM_002513301), and CYP80C8 (XM_002513348); short-chain alcohol dehydrogenase-like (XM_002513286 and XM_002513298), casbene synthases (XM_002513294 and XM_002513297), neocembrene synthase (XM_002513288), BAHD acyltransferase (XM_002513292), monoterpene synthase (XM_002513300), 20-kD nuclear cap binding protein (XM_002517818), actin 1 homolog (XM_002522148), ubiquitin 1 homolog (XM_002513332); J. curcas cytochrome P450s CYP726A20 (KF986815), CYP726A21 (KF986816), CYP726A22 (KF986817), CYP726A23 (KF986818), CYP726A24 (KF986819), CYP726A25 (KF986820), and CYP726A26 (KF986821); and genomic sequence containing three casbene synthases (KJ026361). E. peplus cytochrome P450s CYP726A3 (KF986822), CYP726A4 (KF986823), CYP726A5 (KF986824), CYP726A6 (KF986825), CYP726A19 (KF986826), and genomic sequence containing casbene synthase and CYP726A19 (KJ026362). The castor diterpenoid biosynthetic gene cluster is located on scaffold 30169 of the castor genome database, which can be accessed at www.plantgdb.org.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GC Analysis of Leaf Extracts Expressing Genes from the Castor Gene Cluster.
Supplemental Figure 2. Critical NOESY Correlations Observed for Casbene.
Supplemental Figure 3. Critical NOESY Correlations Used in Establishing the Relative Configuration at the 5-Position of 5α-Hydroxy-Casbene.
Supplemental Figure 4. Critical NOESY Correlations Observed for 5-Keto-7,8-Epoxy-Casbene.
Supplemental Figure 5. Critical NOESY Correlations Observed for 5-Keto-Neocembrene.
Supplemental Figure 6. Phylogenetic Relationship and Diterpenoid Production in the Euphorbiaceae.
Supplemental Figure 7. Conversion of Geranylgeranyl Diphosphate into Casbene by R. communis and J. curcas Casbene Synthases.
Supplemental Figure 8. Structures and Biological Activities of Euphorbiaceae Diterpenoids.
Supplemental Table 1. Primers Used for qPCR Analysis of R. communis Genes.
Supplemental Table 2. Primers Used for Cloning and Subcloning of R. communis Genes.
Supplemental Table 3. Primers Used for Cloning and Subcloning of J. curcas Genes.
Supplemental Table 4. Primers Used for Cloning and Subcloning of E. peplus Genes.
Supplemental Table 5. Primers Used for Linkage Mapping in J. curcas.
Supplemental Table 6. NMR Analysis of Casbene.
Supplemental Table 7. NMR Analysis of 5-Hydroxy-Casbene.
Supplemental Table 8. NMR Analysis of 5-Keto-Casbene.
Supplemental Table 9. NMR Analysis of 5-Keto-7,8-Epoxy-Casbene.
Supplemental Table 10. NMR Analysis of Neocembrene.
Supplemental Table 11. NMR Analysis of 5-Keto-Neocembrene.
Supplemental Data Set 1. FASTA Formatted Text File Containing the Edited Peptide Sequence Alignment of CYP71 Clan Proteins Used to Construct the Phylogenetic Tree Shown in Figure 2B.
Supplemental Data Set 2. FASTA Formatted Text File Containing the Edited DNA Sequence Alignment of Plastidial Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase Large Subunit (rbcL) Gene Used to Construct the Phylogenetic Tree Shown in Supplemental Figure 6.
Supplementary Material
Acknowledgments
We thank David Nelson (University of Tennessee) for providing assignments for the cytochrome P450 sequences, George Lomonossoff for providing the pEAQ-HT vector, and Quinvita for proving the mapping populations used in this study.
AUTHOR CONTRIBUTIONS
A.J.K. and I.A.G. designed research. A.J.K., A.D.G., and G.D.B. performed research. A.J.K., G.D.B., and T.R.L. analyzed data. A.J.K., G.D.B., and I.A.G. wrote the article.
Glossary
- qPCR
quantitative PCR
- GC-MS
gas chromatography-mass spectrometry
- ORF
open reading frame
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Arvidsson S., Kwasniewski M., Riaño-Pachón D.M., Mueller-Roeber B. (2008). QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.D., Soltis D.E., Soltis P.S. (2010). The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97: 1296–1303. [DOI] [PubMed] [Google Scholar]
- Beutler J.A., Alvarado A.B., McCloud T.G., Cragg G.M. (1989). Distribution of phorbol ester bioactivity in the Euphorbiaceae. Phytother. Res. 3: 188–192. [Google Scholar]
- Chan A.P., et al. (2010). Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 28: 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tholl D., Bohlmann J., Pichersky E. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66: 212–229. [DOI] [PubMed] [Google Scholar]
- Corea G., Di Pietro A., Dumontet C., Fattorusso E., Lanzotti V. (2009). Jatrophane diterpenes from Euphorbia spp. as modulators of multidrug resistance in cancer therapy. Phytochem. Rev. 8: 431–447. [Google Scholar]
- D’Auria J.C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9: 331–340. [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber M.T., Adolf W., West C.A. (1978). Biosynthesis of the diterpene phytoalexin casbene: partial purification and characterization of casbene synthetase from Ricinis communis. Plant Physiol. 62: 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B., Fiston-Lavier A.-S., Kemen A., Geisler K., Quesneville H., Osbourn A.E. (2011). Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. USA 108: 16116–16121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic K., Hernandez A., Korban S.S. (2004). RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Rep. 22: 437a–437g. [Google Scholar]
- Graham I.A., et al. (2010). The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 327: 328–331. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Haas W., Sterk H., Mittelbach M. (2002). Novel 12-deoxy-16-hydroxyphorbol diesters isolated from the seed oil of Jatropha curcas. J. Nat. Prod. 65: 1434–1440. [DOI] [PubMed] [Google Scholar]
- Hadi V., Hotard M., Ling T., Salinas Y.G., Palacios G., Connelly M., Rivas F. (2013). Evaluation of Jatropha isabelli natural products and their synthetic analogs as potential antimalarial therapeutic agents. Eur. J. Med. Chem. 65: 376–380. [DOI] [PubMed] [Google Scholar]
- Hamberger B., Bak S. (2013). Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., King A.J., Khan M.A., Cuevas J.A., Ramiaramanana D., Graham I.A. (2011). Analysis of seed phorbol-ester and curcin content together with genetic diversity in multiple provenances of Jatropha curcas L. from Madagascar and Mexico. Plant Physiol. Biochem. 49: 1183–1190. [DOI] [PubMed] [Google Scholar]
- Hirakawa H., et al. (2012). Upgraded genomic information of Jatropha curcas L. Plant Biotechnol. 29: 123–130. [Google Scholar]
- Höfgen R., Willmitzer L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann J., Evanics F., Berta L., Bartók T. (2000). Diterpenoids from Euphorbia peplus. Planta Med. 66: 291–294. [DOI] [PubMed] [Google Scholar]
- Johnson H.E., Banack S.A., Cox P.A. (2008). Variability in content of the anti-AIDS drug candidate prostratin in Samoan populations of Homalanthus nutans. J. Nat. Prod. 71: 2041–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen L., McKerrall S.J., Kuttruff C.A., Ungeheuer F., Felding J., Baran P.S. (2013). 14-step synthesis of (+)-ingenol from (+)-3-carene. Science 341: 878–882. [DOI] [PubMed] [Google Scholar]
- Kerschen A., Napoli C.A., Jorgensen R.A., Müller A.E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566: 223–228. [DOI] [PubMed] [Google Scholar]
- King A.J., et al. (2013). Linkage mapping in the oilseed crop Jatropha curcas L. reveals a locus controlling the biosynthesis of phorbol esters which cause seed toxicity. Plant Biotechnol. J. 11: 986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J., Nishimoto M., Park J.G., Withers S.T., Nowroozi F., Behrendt D., Rutledge E.J.G., Fortman J.L., Johnson H.E., Anderson J.V., Keasling J.D. (2010). Cloning of casbene and neocembrene synthases from Euphorbiaceae plants and expression in Saccharomyces cerevisiae. Phytochemistry 71: 1466–1473. [DOI] [PubMed] [Google Scholar]
- Kissin I., Szallasi A. (2011). Therapeutic targeting of TRPV1 by resiniferatoxin, from preclinical studies to clinical trials. Curr. Top. Med. Chem. 11: 2159–2170. [DOI] [PubMed] [Google Scholar]
- Kulkosky J., Culnan D.M., Roman J., Dornadula G., Schnell M., Boyd M.R., Pomerantz R.J. (2001). Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98: 3006–3015. [DOI] [PubMed] [Google Scholar]
- Le S.Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Liang X., Grue-Sørensen G., Klarskov Petersen A., Högberg T. (2012). Semisynthesis of ingenol 3-angelate (PEP005): efficient stereoconservative angeloylation of alcohols. Synlett 23: 2647–2652. [Google Scholar]
- Mau C.J., West C.A. (1994). Cloning of casbene synthase cDNA: evidence for conserved structural features among terpenoid cyclases in plants. Proc. Natl. Acad. Sci. USA 91: 8497–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miana G.A., Bashir M., Evans F.J. (1985). Isolation of prostratin from Euphorbia cornigera. Planta Med. 51: 353–354. [DOI] [PubMed] [Google Scholar]
- Mizutani M. (2012). Impacts of diversification of cytochrome P450 on plant metabolism. Biol. Pharm. Bull. 35: 824–832. [DOI] [PubMed] [Google Scholar]
- Mugford S.T., Louveau T., Melton R., Qi X., Bakht S., Hill L., Tsurushima T., Honkanen S., Rosser S.J., Lomonossoff G.P., Osbourn A. (2013). Modularity of plant metabolic gene clusters: a trio of linked genes that are collectively required for acylation of triterpenes in oat. Plant Cell 25: 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwine J.T., Van Damme P. (2011). Why do Euphorbiaceae tick as medicinal plants? A review of Euphorbiaceae family and its medicinal features. J. Med. Plants Res. 5: 652–662. [Google Scholar]
- Nelson D.R. (2009). The cytochrome p450 homepage. Hum. Genomics 4: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D., Werck-Reichhart D. (2011). A P450-centric view of plant evolution. Plant J. 66: 194–211. [DOI] [PubMed] [Google Scholar]
- Nützmann H.-W., Osbourn A. (2014). Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 26: 91–99. [DOI] [PubMed] [Google Scholar]
- Osbourn A. (2010). Gene clusters for secondary metabolic pathways: an emerging theme in plant biology. Plant Physiol. 154: 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C.J., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496: 528–532. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik S., Marri P.R., Desany B., Rabinowicz P.D., Kodira C., Mohiuddin M., Rodriguez F., Fauquet C., Tohme J., Harkins T., Rokhsar D.S., Rounsley S. (2012). The Cassava genome: Current progress, future directions. Trop. Plant Biol. 5: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004). A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101: 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing-Gao M., Wen-Zi L., Xiao-Yun W., Tian-Xi Z., Guo-Wei Q. (1997). Diterpenoids from Euphorbia fischeriana. Phytochemistry 44: 663–666. [Google Scholar]
- Reis M., Ferreira R.J., Serly J., Duarte N., Madureira A.M., Santos D.J., Molnár J., Ferreira M.J.U. (2012). Colon adenocarcinoma multidrug resistance reverted by Euphorbia diterpenes: structure-activity relationships and pharmacophore modeling. Anticancer. Agents Med. Chem. 12: 1015–1024. [DOI] [PubMed] [Google Scholar]
- Ro D.-K., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943. [DOI] [PubMed] [Google Scholar]
- Rontein D., Onillon S., Herbette G., Lesot A., Werck-Reichhart D., Sallaud C., Tissier A. (2008). CYP725A4 from yew catalyzes complex structural rearrangement of taxa-4(5),11(12)-diene into the cyclic ether 5(12)-oxa-3(11)-cyclotaxane. J. Biol. Chem. 283: 6067–6075. [DOI] [PubMed] [Google Scholar]
- Ruijter J.M., Ramakers C., Hoogaars W.M.H., Karlen Y., Bakker O., van den Hoff M.J.B., Moorman A.F.M. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F., Lomonossoff G.P. (2008). Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148: 1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F., Thuenemann E.C., Lomonossoff G.P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7: 682–693. [DOI] [PubMed] [Google Scholar]
- Sato S., et al. (2011). Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 18: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282: 34013–34018. [DOI] [PubMed] [Google Scholar]
- Siller G., Rosen R., Freeman M., Welburn P., Katsamas J., Ogbourne S.M. (2010). PEP005 (ingenol mebutate) gel for the topical treatment of superficial basal cell carcinoma: results of a randomized phase IIa trial. Australas. J. Dermatol. 51: 99–105. [DOI] [PubMed] [Google Scholar]
- Swaminathan S., Morrone D., Wang Q., Fulton D.B., Peters R.J. (2009). CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadele Z., Mba C., Till B.J. (2009). TILLING for mutations in model plants and crops. [Google Scholar]
- Talavera G., Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56: 564–577. [DOI] [PubMed] [Google Scholar]
- Tan Q.-G., Cai X.-H., Du Z.-Z., Luo X.-D. (2009). Three terpenoids and a tocopherol-related compound from Ricinus communis. Helv. Chim. Acta 92: 2762–2768. [Google Scholar]
- Teoh K.H., Polichuk D.R., Reed D.W., Nowak G., Covello P.S. (2006). Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580: 1411–1416. [DOI] [PubMed] [Google Scholar]
- Tholl D., Lee S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. Arabidopsis Book 9: e0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuoka T., Tobe H. (2006). Phylogenetic analyses of Malpighiales using plastid and nuclear DNA sequences, with particular reference to the embryology of Euphorbiaceae sens. str. J. Plant Res. 119: 599–616. [DOI] [PubMed] [Google Scholar]
- Trapp S.C., Croteau R.B. (2001). Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158: 811–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35: W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Loo F.J., Broun P., Turner S., Somerville C. (1995). An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc. Natl. Acad. Sci. USA 92: 6743–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wada M., Takahashi H., Altaf-Ul-Amin M., Nakamura K., Hirai M.Y., Ohta D., Kanaya S. (2012). Prediction of operon-like gene clusters in the Arabidopsis thaliana genome based on co-expression analysis of neighboring genes. Gene 503: 56–64. [DOI] [PubMed] [Google Scholar]
- Wang E., Wagner G.J. (2003). Elucidation of the functions of genes central to diterpene metabolism in tobacco trichomes using posttranscriptional gene silencing. Planta 216: 686–691. [DOI] [PubMed] [Google Scholar]
- Wang Q., Hillwig M.L., Peters R.J. (2011). CYP99A3: functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J. 65: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Hillwig M.L., Wu Y., Peters R.J. (2012a). CYP701A8: a rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 158: 1418–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Hillwig M.L., Okada K., Yamazaki K., Wu Y., Swaminathan S., Yamane H., Peters R.J. (2012b). Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J. Biol. Chem. 287: 6159–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman P.R., Xu M., Jin Y., Coates R.M., Peters R.J. (2004). Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135: 2098–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J.M., Tang Y. (2012). Synthetic biological approaches to natural product biosynthesis. Curr. Opin. Biotechnol. 23: 736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T., et al. (2012). A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708. [DOI] [PubMed] [Google Scholar]
- Wu Y., Hillwig M.L., Wang Q., Peters R.J. (2011). Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization of CYP71Z6 & 7. FEBS Lett. 585: 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdack K.J., Hoffmann P., Chase M.W. (2005). Molecular phylogenetic analysis of uniovulate Euphorbiaceae (Euphorbiaceae sensu stricto) using plastid RBCL and TRNL-F DNA sequences. Am. J. Bot. 92: 1397–1420. [DOI] [PubMed] [Google Scholar]
- Xiao M., et al. (2013). Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 166: 122–134. [DOI] [PubMed] [Google Scholar]
- Zerbe P., Hamberger B., Yuen M.M.S., Chiang A., Sandhu H.K., Madilao L.L., Nguyen A., Hamberger B., Bach S.S., Bohlmann J. (2013). Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 162: 1073–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-Q., Li F., Zhao Z.-G., Liu X.-L., Tang Y.-X., Wang M.-K. (2012). Diterpenoids from the root bark of Jatropha curcas and their cytotoxic activities. Phytochem. Lett. 5: 721–724. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.