This study shows that 1% of Arabidopsis protein-coding genes are retrogenes (i.e., cDNAs integrated into the genome) and that retrogenes frequently acquire novel promoters at their insertion sites and are transcribed preferentially in male gametes. Thus, RNA-based gene duplication is a common process in plant genome evolution and generates functional genes with specific transcription patterns.
Abstract
Duplications allow for gene functional diversification and accelerate genome evolution. Occasionally, the transposon amplification machinery reverse transcribes the mRNA of a gene, integrates it into the genome, and forms an RNA-duplicated copy: the retrogene. Although retrogenes have been found in plants, their biology and evolution are poorly understood. Here, we identified 251 (216 novel) retrogenes in Arabidopsis thaliana, corresponding to 1% of protein-coding genes. Arabidopsis retrogenes are derived from ubiquitously transcribed parents and reside in gene-rich chromosomal regions. Approximately 25% of retrogenes are cotranscribed with their parents and 3% with head-to-head oriented neighbors. This suggests transcription by novel promoters for 72% of Arabidopsis retrogenes. Many retrogenes reach their transcription maximum in pollen, the tissue analogous to animal spermatocytes, where upregulation of retrogenes has been found previously. This implies an evolutionarily conserved mechanism leading to this transcription pattern of RNA-duplicated genes. During transcriptional repression, retrogenes are depleted of permissive chromatin marks without an obvious enrichment for repressive modifications. However, this pattern is common to many other pollen-transcribed genes independent of their evolutionary origin. Hence, retroposition plays a role in plant genome evolution, and the developmental transcription pattern of retrogenes suggests an analogous regulation of RNA-duplicated genes in plants and animals.
INTRODUCTION
Gene duplications are an important factor in genome evolution, allowing for the functional diversification of genes (Flagel and Wendel, 2009; Innan and Kondrashov, 2010). Duplicated genes are generated by several DNA- and RNA-based mechanisms (Innan and Kondrashov, 2010; Sakai et al., 2011). Whole-genome DNA-based duplication (WGD) by polyploidization has occurred in the evolutionary history of all land plants and many animals (Dehal and Boore, 2005; De Smet et al., 2013). Since WGD amplifies the entire genome, it seems to be a solution toward major evolutionary and/or ecological challenges (Comai, 2005; Fawcett et al., 2009). However, WGDs do not alter protein stoichiometry in most cases; therefore, they may be relatively ineffective in situations where an increased amount of a single or a few specific proteins is required. In such situations, local DNA and RNA duplication mechanisms may be a more sophisticated solution. Local DNA duplications amplify individual genes or short chromosomal regions, presumably by an unequal crossing over mechanism (Zhang, 2003). In RNA-based duplication (retroposition), the mature mRNA of a protein-coding gene is reverse transcribed and integrated at an ectopic position in the genome using retroviral or retrotransposon machinery (Kaessmann et al., 2009). Therefore, retroposition has a high potential to generate evolutionary innovations (e.g., by expressing genes in a new developmental context, generating chimeric genes with new functional domain combinations, or interspecific horizontal gene transfer) (Wang et al., 2006; Yoshida et al., 2010; Sakai et al., 2011). Relatively few studies have searched for retrogenes at the genome-wide scale in plants (Zhang et al., 2005; Wang et al., 2006; Zhu et al., 2009; Sakai et al., 2011). These studies have identified at most 0.38% of protein-coding genes as retrogenes, except for a study in maize (Zea mays) where low-stringency selection criteria were applied (Wang et al., 2006). In human (Homo sapiens), although 19.1% of all genes were identified as retrocopies, 82% of those contain premature stop codons. Therefore, 3.4% of all human genes are retrocopies producing putatively functional proteins (Marques et al., 2005; Pennisi, 2012). In rice (Oryza sativa subsp japonica), transcription was observed for two-thirds of retrogenes, indirectly suggesting that there may be a higher proportion of functional retrogenes in plants (Sakai et al., 2011).
Since retroposition duplicates only transcribed regions, it is expected to cause the loss of promoter sequences. This may represent a major bottleneck to retrogene evolutionary success. However, there are multiple possible mechanisms of retrogene promoter acquisition that have been demonstrated in individual examples (Kaessmann et al., 2009). Nevertheless, it is often not clear how frequent they are at the genome-wide scale. Recent studies in human and rice suggested that retroposition includes parental promoters (Okamura and Nakai, 2008).
Chromatin is an indispensable component that provides regulatory and protective functions to genetic information (reviewed in Li et al., 2007). Transcribed protein-coding genes are associated with permissive chromatin marks. In contrast, transcriptionally repressed genes and repetitive elements are typically labeled by histone H3 Lysine 27 trimethylation (H3K27me3), histone H3 Lysine 9 dimethylation (H3K9me2), and/or high-density DNA methylation in all cytosine sequence contexts in plants (Roudier et al., 2011; Stroud et al., 2013). While H3K27me3 ensures tissue-specific developmental transcription (Lafos et al., 2011), the role of H3K9me2 and promoter DNA methylation is to minimize the activities of repetitive elements, which frequently include retrotransposons (Mosher et al., 2009; Slotkin et al., 2009; Ibarra et al., 2012). Retrogenes are generated by retrotransposon reverse transcriptases and represent duplicated copies. Therefore, they may become targets of epigenetic silencing by repressive chromatin. The association of retrogenes with specific chromatin states has been proposed (Boutanaev et al., 2002; Marques et al., 2005), but only a few have been characterized as to their chromatin states so far (Monk et al., 2011; Pei et al., 2012).
In flies and mammals, many retrogenes show testis-specific transcription (Marques et al., 2005; Vinckenbosch et al., 2006; Bai et al., 2008). This pattern is intriguing, and several explanatory models have been proposed (reviewed in Kaessmann et al., 2009; Kaessmann, 2010). First, it could originate from various chromatin modifications affecting chromosomes and leading to hypertranscription in meiotic and postmeiotic spermatogenic cells. As a consequence of this global chromatin reorganization–induced transcription, some of the testis-transcribed retrogenes could also evolve testis-specific gene functions. The second, not mutually exclusive, hypothesis postulates that retrogenes amplify in the germline tissues and insert preferentially into actively transcribed (open) chromatin. This creates a self-reinforcing loop where the retrogenes insert nearby or into germline-transcribed genes and consequently also would be germline-transcribed. The latter hypothesis is partially supported by observations in Drosophila melanogaster (Bai et al., 2008), but the tissue specificity in the transcription of plant retrogenes has not been clarified.
Here, we developed a search method that we used to identify 251 Arabidopsis thaliana retrogenes, 216 of which are novel. We use this set together with the retrogenes found previously to analyze retrogene and parent-specific features. We show that parents are usually ubiquitously transcribed, while retrogenes are mainly transcribed at low levels and in a stage-specific manner. Most Arabidopsis retrogenes acquired novel cis-regulatory elements at their integration sites, and introns significantly extend retrogene mRNA half-life. Importantly, throughout plant development, retrogenes show peaks of transcription in pollen. This pattern can also be observed for many lowly transcribed genes genome-wide and resembles retrogene transcription in the testis of animals.
RESULTS
Arabidopsis Retrogenes Are Capable of Repeated Retroposition and Occur in Gene-Rich Genomic Regions
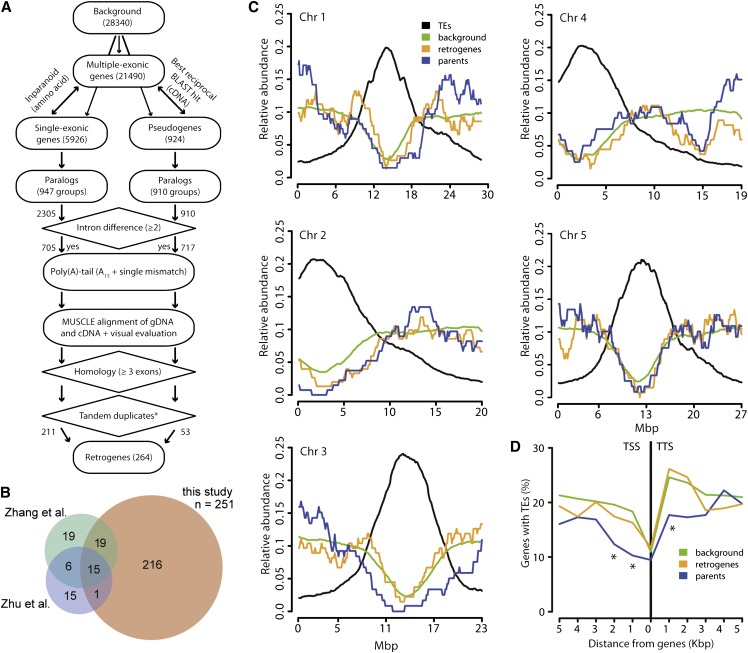
We developed a bioinformatic method to identify retrogenes (Figure 1A). This was based on a genome-wide search for gene paralogy and retrogene-specific characteristics such as differential intron numbers relative to the parental gene and/or the presence of a poly(A) tail. The method was used to screen the genome of Arabidopsis, and in total, 251 retroposition events satisfying stringent quality criteria were identified (Supplemental Data Set 1). Among the retrogenes identified in our list, 36 were shared with two previous Arabidopsis genome-wide retrogene screens and 216 were novel (Figure 1B; Supplemental Data Sets 1 and 2) (Zhang et al., 2005; Zhu et al., 2009). The total number of retrogenes identified in all three studies is 309 (291 were considered for downstream analyses; Supplemental Data Sets 1 and 2), which corresponds to ∼1% of Arabidopsis protein-coding genes and pseudogenes (n = 27,416 and n = 924, respectively).
Figure 1.
Retrogene Identification and Genomic Features.
(A) Schematic representation of the retrogene identification method developed for our study.
(B) Venn diagrams indicating the numbers of retrogenes identified in three Arabidopsis genome-wide searches (Zhang et al., 2005; Zhu et al., 2009; this study). Note that the Venn diagrams do not include the disputable retrogenes listed in Supplemental Data Set 2.
(C) Relative abundance (y axis) of TEs (black), genes and pseudogenes (background; green), retrogenes (red), and parents (blue) over the five Arabidopsis chromosomes (x axis).
(D) Percentage of genes containing TEs (y axis) in 1-kb intervals from the gene TSS and TTS for all protein-coding genes (background; green), retrogenes (red), and parents (blue). Significant differences (P < 0.05) in the χ2 test relative to background are indicated by asterisks.
Because our method combines multiple retrogene searches within intronless and intronized genes, it allows searching for potential secondary retropositions of retrogene transcripts. This revealed 12 retrogenes that served as templates for another round of retroposition (Supplemental Figure 1 and Supplemental Data Set 3). In these cases, the primary parent gave rise to the primary retrogene, whose mRNA served as the precursor for the secondary retrogene. The model where the primary parent gives rise directly to the secondary retrogene was not supported by the order of protein homologies and suggests retroposition of the retrogene transcript. Hence, 4.3% of Arabidopsis retrogenes underwent repeated retroposition without losing their protein-coding potential. In addition, we identified multiple-retrogene parents. In total, 22 parents gave rise to 54 retrocopies (17 × 2, 3 × 3, 1 × 4, 1 × 7) and a maximum of seven retrocopies derived from a single parent (Supplemental Data Set 1). The observed frequency of multiple retropositions from the same gene is significantly higher than expected at random (Mann-Whitney-Wilcoxon [MWW] test, P < 2.2 × 10−16), strongly arguing that the selection of parental mRNA is not random at least in some cases.
To explore whether retroposition occurs at specific genomic regions, we plotted the densities of all protein-coding genes, transposable elements (TEs), parents, and retrogenes over the five Arabidopsis chromosomes (Figure 1C). In agreement with published data (Arabidopsis Genome Initiative, 2000), TEs were enriched in pericentromeric regions and depleted from chromosome arms, while protein-coding genes showed the opposite pattern. Both retrogenes and parents had profiles similar to that of protein-coding genes, showing that they occur preferentially in gene-rich genomic regions (Figure 1C). To test for the association of retrogenes and/or parents with TEs at the local scale, we estimated the frequency of genes with TEs in 1-kb intervals upstream and downstream of gene transcription start sites (TSSs) and transcription termination sites (TTSs). On average, there are fewer TEs upstream than downstream of genes. The frequency of TEs in TSS upstream regions of the genome-wide genes and retrogenes (17 and 22%, respectively) was not significantly different (Figure 1D). By contrast, parental genes with TEs in the first 2 kb upstream of the TSS were scarce relative to the whole genome (χ2 test, P < 0.05). Similarly, 25% of all genes and retrogenes contained TEs in the first 2 kb of the TTS downstream region, while it was only 17% for parents (χ2 test, P < 0.05 in the first 1 kb). This shows that retrogenes are not enriched for close-lying TEs compared with the genomic average, but parents are depleted of TEs in both upstream and downstream intergenic regions.
Hence, the Arabidopsis genome contains at least 291 retrogenes located predominantly in gene-rich chromosomal regions. About 10% of the parents gave rise to multiple retrogenes, and ∼4.3% of the retrogenes underwent a second retroposition.
Retrogenes Are Derived from Highly Transcribed Parental Genes and Are Transcribed Preferentially by Novel Promoters
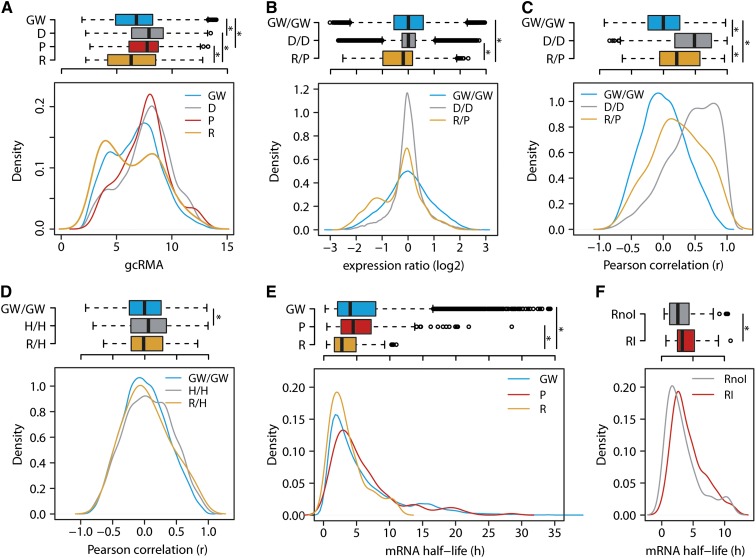
We took advantage of the comprehensive retrogene list assembled in our study and explored the patterns of retrogene transcription in Arabidopsis. The mRNA accumulation was analyzed using microarray data from the 49 Arabidopsis developmental stages assembled by the AtGenExpress consortium (Schmid et al., 2005) and validated for selected tissues by RNA sequencing (Loraine et al., 2013). In total, 209 retrogenes and 245 parents are present on the ATH1 cDNA microarray (Supplemental Data Sets 4 and 5). To compare the effects of RNA- and DNA-based duplications, we also analyzed the set of 3088 Arabidopsis DNA duplicated genes (Blanc and Wolfe, 2004). Plotting the mean log2 robust multiarray averaging (gcRMA; Irizarry et al., 2003) values of all ATH1 probe sets (n = 22,746) revealed a double-peak distribution, with the left peak representing genes with poor mRNA levels and/or background signals (Supplemental Figure 2 and Supplemental Data Set 5). The gcRMA values of some retrogenes and parents overlapped with this region and suggested that some of the candidates may not be transcribed in any of the 49 stages. Therefore, we kept only the genes with gcRMA values of 5 or higher in at least one developmental stage (transcribed genes). In total, 89.4% (n = 20,398) of all genes, 85.2% (n = 178) of retrogenes, 94.7% (n = 232) of parents, and 99.3% (n = 3067) of DNA duplicated genes passed these criteria (Figure 2A; Supplemental Data Set 5). This shows that the majority of Arabidopsis retrogenes are transcribed in at least some developmental stages, and their mean gcRMA values did not differ significantly from the genome-wide gene set (MWW test, P = 0.48; Figure 2A). The parents were significantly enriched for highly transcribed genes relative to both retrogenes and the whole-genome set (MWW test, P = 7.64 × 10−06 and P = 1.86 × 10−11, respectively; Figure 2A). Similarly, DNA duplicated genes were strongly transcribed and therefore similar to parents, but they were strongly different from retrogenes (MWW test, P = 0.16 and P = 1.56 × 10−10, respectively). To reveal the transcription relationships between individual retrogene/parent pairs, we compared their developmental stage–specific gcRMA ratios with the transcription of 5000 randomly selected gene pairs and the 1527 DNA duplicated gene pairs (Figure 2B; Supplemental Data Set 6). Transcript accumulation ratios of random pairs and DNA duplicated genes represented a broad and narrow range of normally distributed values (MWW test, P = 0.85). Although many retrogenes have a comparable degree of transcription relative to their parents, there is a specific group of 2- to 3-fold less transcribed retrogenes, making retrogene/parent pairs significantly different from both the random gene set and DNA duplicated genes (MWW test, both comparisons P < 2.2 × 10−16; Figure 2B). Inspecting the gcRMA values over individual developmental stages for the low-transcribed group revealed that these retrogenes were transcribed above the threshold (gcRMA ≥ 5) in only one or a few tissues, while their parents frequently showed ubiquitous transcription.
Figure 2.
Retrogenes Are Driven by Novel Promoters and Have Reduced Transcript Stability.
(A) Box and density plots of gcRMA values for genome-wide genes (GW), DNA duplicated genes (D), parents (P), and retrogenes (R) over the 49 Arabidopsis developmental stages.
(B) Log2 transcription ratios of the random genome-wide gene pairs (GW/GW), DNA duplicated pairs (D/D), and retrogene/parent pairs (R/P).
(C) and (D) Pearson correlation of gene cotranscription between random genome-wide gene pairs, DNA duplicated pairs, retrogene/parent pairs, genome-wide head-to-head oriented genes (H/H), and retrogene head-to-head oriented neighboring genes (R/H) in 49 developmental stages.
(E) and (F) mRNA half-lives of genome-wide genes, parents, retrogenes, intronless retrogenes (RnoI), and intronized retrogenes (RI).
Significance values were calculated using the MWW test for all group combinations within each graph, and significant differences (P < 0.05) are indicated by asterisks in box plots. Nonsignificant (P ≥ 0.05) relationships are not shown.
A recent study in rice suggested frequent cotranscription between retrogenes and parents in plants (Sakai et al., 2011). Our retrogene identification criteria and the nature of the Arabidopsis retrogenes (e.g., an absence of retrogenes residing in the introns of other genes) allowed testing three possible mechanisms of retrogene cis-regulatory element origin: (1) carryover of parental promoters, (2) the use of bidirectional promoters, and (3) an acquisition of novel cis-regulatory elements. First, we tested whether the Arabidopsis retrogenes inherit the parental transcription pattern. We calculated the cotranscription of retrogene/parent pairs as Pearson product–moment correlation coefficients (r) across the 49 developmental samples of the AtGenExpress data set. Indeed, cotranscription in the set of retrogene/parent pairs (n = 179) was significantly higher than in the 20,000 randomly selected gene pairs (MWW test, P = 2.30 × 10−6) (Figure 2C; Supplemental Data Set 4). We calculated the frequencies of genes per 0.1 r correlation bins for retrogenes and genome background and used this to calculate the number of highly cotranscribed retrogene/parent pairs. In total, 25% of the retrogene/parent pairs (26 out of 102) were correlated more than random gene pairs. However, the cotranscription of DNA duplicated gene pairs, calculated in the same way, was more prominent (MWW test, P < 2.2 × 10−16; Figure 2C), and 45.6% of them surpassed the random-pairs background.
Second, we tested the possibility for retrogene transcription by bidirectional promoters of head-to-head oriented neighboring genes (Supplemental Data Set 7). The Pearson correlations of random transcribed gene pairs (n = 20,000) and the genome-wide set of transcribed head-to-head oriented genes (n = 2,087; Supplemental Data Set 8) revealed an infrequent but consistent cotranscription between head-to-head oriented gene pairs (MWW test, P = 2.705 × 10−10; Figure 2D). This shows that sharing bidirectional cis-elements is not common in Arabidopsis. Retrogene–head-to-head oriented neighbor pairs (n = 63) displayed an intermediate pattern that was not significantly different from either genome-wide or head-to-head oriented genes (MWW test, both P = 0.60; Figure 2D). Only 2.5% of head-to-head oriented retrogenes had higher correlation than random pairs, illustrating the negligible effect of promoter sharing (Figure 2D).
Most retrocopies are expected to be intronless at the time of integration. However, approximately one-third of retrogenes we found contained introns. This indicated that retrogene intronization has a functional role. We tested whether intronization plays a role in retrogene mRNA stability. First, we compared the mRNA half-life of transcribed retrogenes (n = 100), parents (n = 147), and the genome-wide set of transcribed genes (n = 13,012) included in the publicly available mRNA decay data set (Narsai et al., 2007). The mRNA half-life of the parents and the genome-wide gene set was similar (MWW test, P = 0.21) and significantly longer than that of the retrogene mRNA (MWW test, P = 3.56 × 10−5 and P = 2.54 × 10−5, respectively; Figure 2E). Furthermore, mRNA of intron-containing retrogenes (29%) had a slightly but significantly longer half-life compared with that of intronless retrogenes (MWW test, P = 0.04; Figure 2F).
Hence, retrogenes are transcribed more weakly than their parents and the transcription of most of the retrogene/parent pairs is not correlated, due to the acquisition of novel regulatory elements at their integration sites. Retrogene mRNA half-life is increased by intronization.
Arabidopsis Retrogenes Are Transcribed in Male Gametes
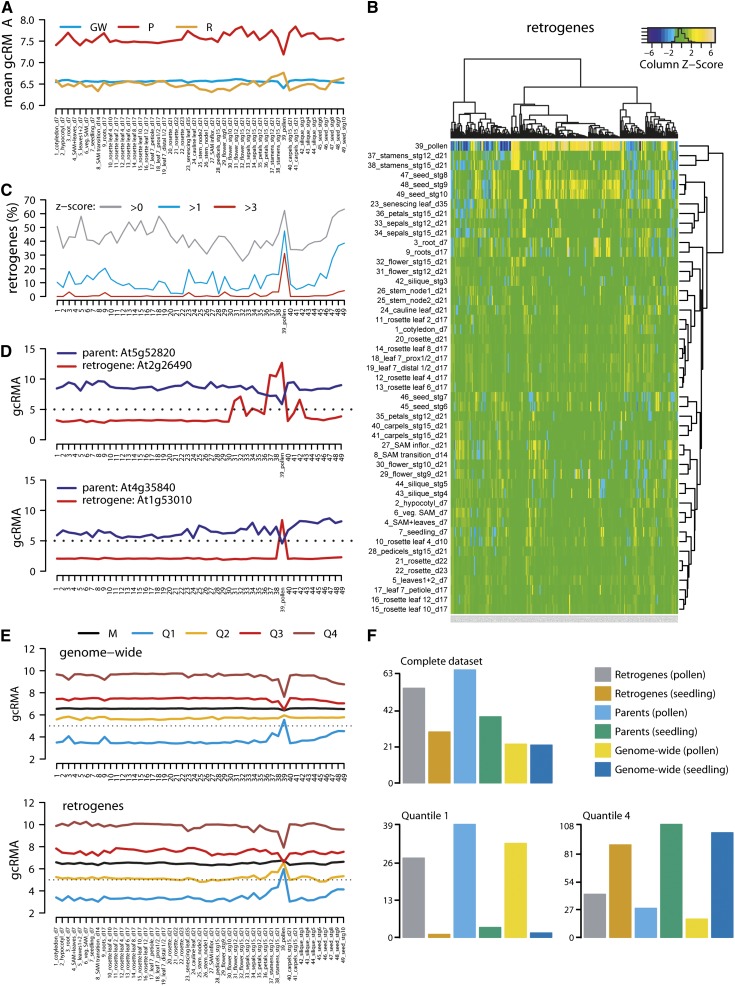
To analyze the developmental regulation of Arabidopsis retrogene transcription, we plotted the mean gcRMA values of genome-wide, parent, and retrogene sets for each of the 49 analyzed developmental stages (Figure 3A). The average mRNA level of parents was higher than that of retrogenes and the genome-wide gene set in all stages. The mean transcription per group was relatively constant, except for pollen, where there was a dip in transcription in the parents and the genome-wide set that contrasted with a peak of retrogene transcription (Figure 3A). To identify relationships between developmental stages and retrogenes, we hierarchically clustered both groups and expressed the result as a heat map of the retrogene transcription z-scores (Figure 3B). This separated stamen and pollen from the rest of the tissues. The highest frequency of retrogenes with positive z-scores (z > 0) was then found in pollen and seeds (62 and 63%, respectively; Figure 3C). However, with more stringent criteria (z > 1 and z > 3), the pollen peak became more prominent relative to other tissues and corresponded to 50 and 30% of retrogenes, respectively (Figure 3C). This shows that many retrogenes reach their transcription maxima in pollen. The pollen-specific transcription pattern has been confirmed by an analysis of individual cases (Figure 3D; Supplemental Figure 3A) and resembles the pattern of retrogene activation in the testis of insects and mammals (reviewed in Kaessmann, 2010).
Figure 3.
Retrogenes Are Transcriptionally Upregulated in Pollen.
(A) Mean gcRMA values for genome-wide genes (GW), parents (P), and retrogenes (R) at each of the 49 Arabidopsis developmental stages.
(B) Hierarchically clustered heat map of retrogene z-scores (y axis) and developmental stages (x axis).
(C) The frequency of retrogenes with row z-scores in (B) >0, >1, and >3 in individual developmental stages.
(D) Examples of retrogenes and parents showing tissue-specific and ubiquitous transcription, respectively, with major transcription changes in pollen (stage 39).
(E) Developmental gcRMA values for the genome-wide set of genes and retrogenes. Transcription is shown for mean (M) and transcription quantiles: lowly transcribed/quantile 1 (Q1), mid-lowly transcribed/quantile 2 (Q2), mid-highly transcribed/quantile 3 (Q3), and highly transcribed/quantile 4 (Q4).
(F) Mean RNA sequencing reads per kilobase per million reads (RPKM) values for all genes (genome-wide), parents, and retrogenes in vegetative rosettes and pollen as complete data sets, quantile 1 (lowly transcribed genes), and quantile 4 (highly transcribed genes).
However, plotting the transcription quantiles (Supplemental Data Set 9) of retrogene gcRMA revealed that not all retrogenes followed this simple trend and that the retrogenes with a negative z-score (pollen downregulated) were usually derived from the group of developmentally highly transcribed genes (Figure 3E, bottom). Remarkably, this distribution also held true for the genome-wide gene set (Figure 3D, top). The parents and the DNA duplicated genes showed more prominent downregulation of the highly transcribed genes (quantile 4) and less obvious upregulation of lowly transcribed genes (quantile 1), while TEs showed upregulation for all quantiles (Supplemental Figure 3B). Hence, we found a pollen-specific activation of retrogenes that is a part of the global pollen-specific transcriptional reprogramming.
Pollen development includes several steps (Honys and Twell, 2003). To find out whether retrogenes are transcribed in specific pollen developmental stages, we compared their transcription in unicellular microspores, bicellular pollen, tricellular pollen, and two highly correlated (r = 0.92) samples of mature pollen grains (Honys and Twell, 2004; Schmid et al., 2005). This revealed a continuous increase of mean retrogene transcription throughout pollen development that contrasted with the downregulation of parental genes in tricellular pollen and mature pollen grains (Supplemental Figure 3C). Next, we asked whether there is an enrichment for retrogene transcripts in vegetative and sperm cells (Honys and Twell, 2003). We used TEs as the control for vegetative cell–specific transcription based on the recently proposed model (Slotkin et al., 2009). Although we observed strong TE upregulation in pollen relative to leaves (MWW test, P < 2.2 × 10−16), there was a significantly higher amount of TE transcripts in sperm cells relative to the entire pollen (MWW test, P = 0.013; Supplemental Figure 3D). This indicates that there is a higher amount of TE transcripts in both pollen cell types. The parents were significantly more highly transcribed in sperm cells relative to seedlings (MWW test, P = 0.001) and were underrepresented for the lowly transcribed genes in this tissue relative to entire pollen (Figure 3E). Therefore, retrogene parents are transcribed preferentially in sperm cells. The median of retrogene transcription was higher than that of TEs and increased in both pollen samples relative to seedlings, but only the entire pollen differed significantly (MWW test, P = 0.008; Figure 3E). In combination with pollen developmental stage data, this shows that retrogenes are transcribed in both pollen cell types.
In order to validate our results by independent experiment, we tested whether our findings hold true in data sets generated by RNA sequencing. Gene transcription in mature pollen grains was compared with that in seedling tissues (Loraine et al., 2013). Plotting the mean reads per kilobase per million reads values for the entire set, quantile 1 (lowest transcribed), and quantile 4 (highest transcribed) of all genes, retrogenes, and parents confirmed the microarray data (Figures 3A, 3E, and 3F; Supplemental Figure 3B). The only exception was a higher transcription of parents in pollen relative to seedlings in RNA sequencing (mean and quantile 1 samples; absent in the quantile 4 sample), while the opposite results were obtained using microarrays (Figures 3A and 3F). This difference is due to the higher sensitivity of RNA sequencing technology to quantify transcripts from lowly transcribed genes (Mooney et al., 2013; Zhao et al., 2014). This partially applies also to retrogenes, as the upregulation in pollen versus seedling is more pronounced in RNA sequencing compared with microarrays (Figure 3F).
From this, we conclude that retrogene activation starts prior to pollen maturation and later occurs in both terminal pollen cell types.
Retrogenes Are Deficient for Transcription-Permissive Chromatin Marks in Leaf Tissues
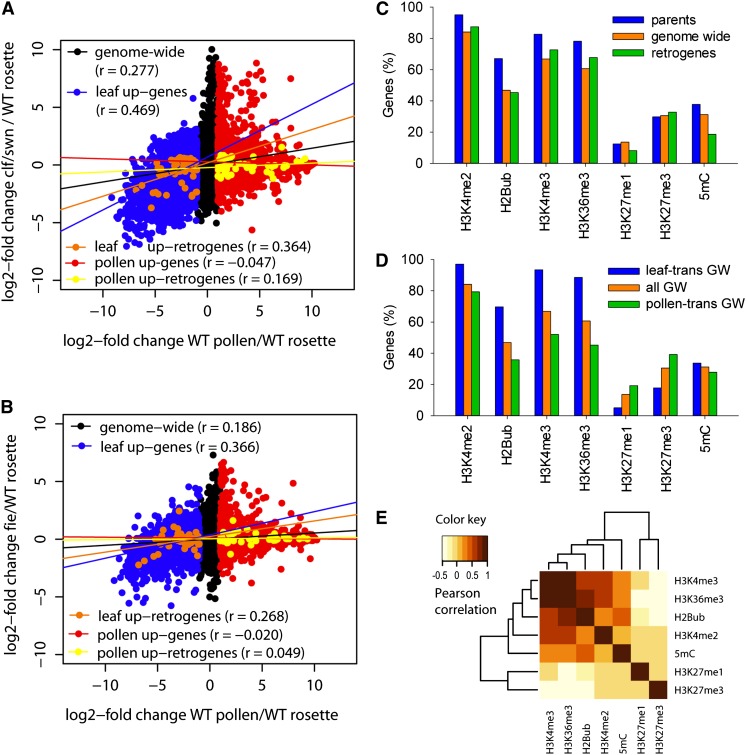
Analysis of transcription quantiles suggests that global transcriptional changes in pollen have a major effect on retrogene transcription. This may be achieved by a global chromatin reprogramming (Kaessmann et al., 2009). Therefore, we calculated log2-fold transcription changes between pollen and 21-d-old rosettes (ATGE_73/ATGE_22; Schmid et al., 2005) and correlated those with transcriptional changes induced by chromatin mutants (mutant rosettes/wild-type rosettes). Five groups were compared (Supplemental Data Set 4): all genes (n = 22,746), pollen upregulated genes (n = 5171), leaf upregulated genes (n = 6057), pollen upregulated retrogenes (n = 51), and leaf upregulated retrogenes (n = 53). Tissue upregulated genes were defined as having log2-fold change ≥ 1 in one versus the other tissue. First, we estimated the effects of the transposon-silencing machinery by testing mutants for DECREASED DNA METHYLATION1 (DDM1), KRYPTONITE (KYP), and HISTONE DEACETYLASE6 (HDA6) (Baubec et al., 2010; Inagaki et al., 2010; Popova et al., 2013), which lead to a loss of DNA methylation, a loss of H3K9me2, and a gain of histone acetylation at heterochromatic loci, respectively. There was no clear correlation (maximum r = 0.040) between transcription in pollen relative to leaves and transcriptional changes induced by ddm1, kyp, and hda6 for all tested groups (Supplemental Figures 4A to 4C). This demonstrates that TE silencing components do not determine the global gene transcription pattern in pollen or affect retrogenes. Next, we tested the effects of the H3K27me3 mark by analyzing mutants of the Polycomb group repressive complex factors CURLY LEAF (CLF) and SWINGER (SWN), which have been shown to regulate transcription during development (Farrona et al., 2011; Lafos et al., 2011). The correlation between clf and swn single mutants with pollen-specific transcriptional changes was low (r < 0.20; Supplemental Figures 4D and 4E). Because CLF and SWN are partially functionally redundant (Lafos et al., 2011), we tested for effects in the clf swn double mutant. The correlation between pollen and clf swn transcription profiles for the set of all genes was higher (r = 0.277) than for the clf and swn single mutants (Figure 4A; Supplemental Figures 4D and 4E). Surprisingly, the high correlation was mainly due to leaf upregulated genes and retrogenes (r = 0.469 and 0.364, respectively) that were coordinately downregulated in both pollen and clf swn (Figure 4A). By contrast, pollen upregulated genes showed generally uncorrelated transcription with clf swn (r = −0.047). To further test the connection with H3K27me3 changes, we analyzed transcription in a mutant for FERTILIZATION INDEPENDENT ENDOSPERM (FIE), another key gene of the Polycomb repressive complex (Bouyer et al., 2011). Although the correlations between fie and pollen transcription profiles were weaker (r = 0.186, 0.366, and 0.268 for all genes, leaf upregulated genes, and retrogenes, respectively; Figure 4B), they perfectly recapitulated trends observed in the comparison between clf swn and pollen. Hence, loss of key components of the Polycomb repressive complex correlates with pollen-specific gene downregulation of leaf transcribed genes but does not explain pollen-specific gene upregulation.
Figure 4.
Chromatin Regulation of Pollen-Specific Gene Transcription.
(A) and (B) Dot plots of log2-fold changes in wild-type pollen/rosettes (x axis) and clf swn double mutants (y axis) (A) or fie/wild-type rosettes (y axis) (B). Specific gene sets were superimposed on the genome-wide set in different colors. The lines indicate transcription correlation (r) between the x and y axes for specific gene sets. The r values are given in parentheses.
(C) The frequency of seven chromatin modifications at gene-coding sequences for all genes, parents, and retrogenes in young leaf tissues.
(D) The same as (C) for all genes (all GW), leaf transcribed genes (leaf-trans GW), and pollen transcribed genes (pollen-trans GW).
(E) Hierarchical clustering and heat map of Pearson correlation values of colocalization between seven chromatin modifications for all Arabidopsis genes.
Therefore, we used publicly available chromatin data from young Arabidopsis leaves (Roudier et al., 2011) to test which chromatin modification(s) is associated with retrogenes and pollen upregulated genes in somatic tissues. We extracted information on chromatin marks for every gene and compared the full sets of retrogenes, parents, and all genes (Figure 4C). In accordance with high and ubiquitous transcription, the parents were enriched for the permissive chromatin marks histone H3 Lysine 4 dimethylation and histone H3 Lysine 4 trimethylation (H3K4me3), histone H3 Lysine 36 trimethylation (H3K36me3), and histone H2B ubiquitination (H2Bub), followed by retrogenes and the genome-wide set. None of these groups was enriched for the repressive histone H3 Lysine 27 modifications. The enrichment for gene body DNA methylation in highly expressed genes is consistent with the currently proposed function of this modification (Coleman-Derr and Zilberman, 2012). Next, we compared previously defined groups of pollen upregulated and leaf upregulated genes (Supplemental Data Set 10). The pattern of distribution of chromatin marks for each individual group (retrogenes, parents, and all genes) was relatively similar (Figure 4D; Supplemental Figures 5A and 5B). There were no changes in gene body DNA methylation. While the histone H3 Lysine 27 modifications were enriched in pollen upregulated genes of the genome-wide set, this mark does not seem to play a major role in the somatic silencing of pollen upregulated retrogenes (Supplemental Figure 5A). By contrast, all analyzed transcription-permissive marks (histone H3 Lysine 4 dimethylation, H3K4me3, H2Bub, and H3K36me3) were underrepresented in pollen upregulated genes in leaf tissues (Figure 4D; Supplemental Figures 5A and 5B). The presence or absence of these marks was strongly correlated in pair-wise comparisons of individual modifications (Figure 4E; Supplemental Figure 5C).
This suggests that in leaf tissues, retrogenes and other pollen upregulated genes are depleted of permissive chromatin marks without enrichment for repressive marks. By contrast, leaf upregulated genes are downregulated in pollen by a mechanism involving the Polycomb repressive complex components CLF, SWN, and FIE.
Gain of Transcription Factor Binding Sites Facilitates PCR11 Retrogene Sperm-Specific Transcription
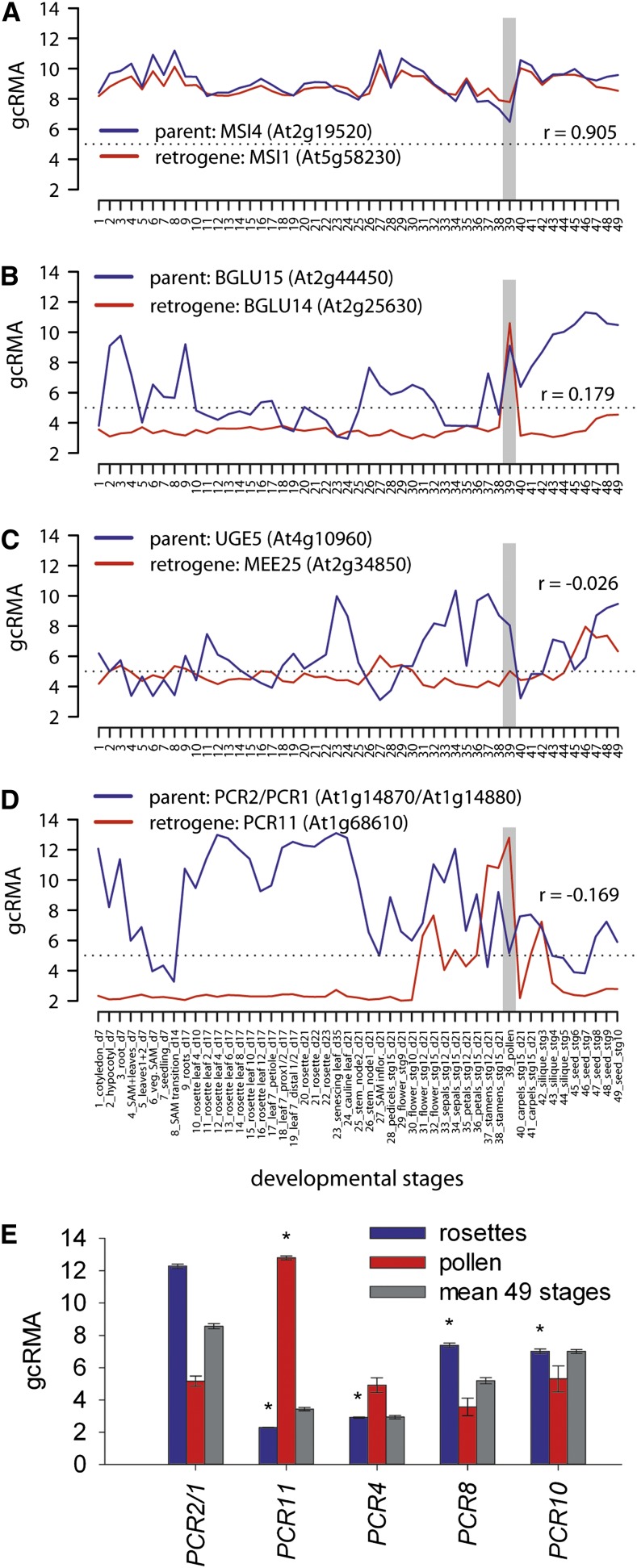
Next, we tested whether the gamete-specific transcription of retrogenes has evolved into gamete-specific developmental functions. Five retrogenes found in our screen, MULTICOPY SUPRESSOR OF IRA1 (MSI1), PLANT CADMIUM RESISTANCE11 (PCR11), BETA-GLUCOSIDASE14 (BGLU14), MATERNAL EFFECT EMBRYO ARREST25 (MEE25), and PEROXIDASE, are associated with pollen development, sperm cell differentiation, pollen tube growth, and development (TAIR10). To investigate the relationship between the transcription of these retrogenes and their parents, we plotted their mean developmental gcRMA values and calculated transcription Pearson correlations (Figures 5A to 5D; Supplemental Data Set 4). The parental gene of PEROXIDASE was not included on the ATH1 array; therefore, we did not continue its analysis. The transcription of MSI1 was strongly correlated (r = 0.905) with its parent MSI4, and both were ubiquitously transcribed throughout development (Figure 5A). BGLU14 and its parent BGLU15 were both upregulated in pollen (Figure 5B). The MEE25 retrogene was transcribed at low levels throughout development, and higher transcription was found only in embryonic tissues (Figure 5C). However, its parent, At4g10960, was transcribed mainly in floral tissues, seeds, and pollen, where it greatly surpassed MEE25 transcription. Hence, these three retrogenes did not provide evidence for the development of parent-independent pollen-specific transcription. In contrast, PCR11 was transcribed at low levels almost throughout the entire process of development but was activated in floral tissues, stamen, and pollen. This pattern was opposite to that of its parent, PCR2, which was active mainly in the photosynthetically active tissues and downregulated in stamen and pollen (Figure 5D). PCR11 has been shown to be transcribed specifically in pollen sperm cells by the MYB transcription factor DUO1 (Borg et al., 2011). Therefore, we compared the promoter regions of PCR11 and PCR2 and looked for previously described DUO1 binding motifs (Borg et al., 2011). There are three binding regions in the 500-bp region upstream of the PCR11 TSSs (TAACCGTC at −47 to −54 bp and AAACCG at −153 to −158 and −452 to −457 bp). However, only a single DUO1 binding motif (AAACCGT at −100 to −106 bp from the TSS) is found in the promoter of PCR2. To test whether this represents a gain of function in PCR11 or a loss of function in PCR2, we compared the promoter regions of several other PCR family members representing both the PCR2 clade (PCR1 and PCR3) and the outgroups (PCR4, PCR8, and PCR10) (Song et al., 2010). None of these genes contained a single DUO1 binding motif in the 500-bp region upstream of the TSS. Furthermore, comparing their transcript levels revealed that only PCR11 is significantly upregulated in pollen relative to PCR2 (Figure 5E).
Figure 5.
Gain of Pollen-Specific Transcription by the PCR11 Retrogene.
(A) to (D) Developmental gcRMA transcription profiles of retrogenes associated with pollen growth and development and their parents. Pollen stage is highlighted by the vertical gray bars.
(E) gcRMA transcription values of PCR family genes in rosettes, pollen, and mean of 49 developmental stages and tissues. PCR2 and PCR1 correspond to a single microarray element and therefore are shown together. Transcription values were compared with PCR2/PCR1 transcription in the same tissue, and statistically significant differences (P < 0.05) in t tests are labeled with asterisks. Error bars denote sd of three biological replicates.
Next, we tested these results in an independent experiment by analyzing retrogene and parent transcription in Arabidopsis lines carrying somatically inducible DUO1 (Borg et al., 2011). Upon 6, 12, and 24 h of DUO1 induction, we observed 36, 131, and 125 significantly upregulated genes and 47, 124, and 121 significantly downregulated genes, respectively. The number of upregulated and downregulated retrogenes (2 and 1, respectively) was small (Supplemental Data Set 11), showing that DUO1 regulates the transcription of only a few specific retrogenes. Importantly, the set of significantly upregulated retrogenes included the PCR11 retrogene (log2-fold changes in 6, 12, and 24 h: 0.26, 2.21, and 4.03; t test P values: 0.010, 3.3 × 10−5, and 5.4 × 10−5, respectively). This has been reflected by significant downregulation of its parent PCR2 in two out of three experimental points (log2-fold changes in 6, 12, and 24 h: −1.18, −1.60, and −0.74; t test P values: 0.003, 0.007, and 0.19, respectively). Therefore, we conclude that the PCR11 retrogene gained sperm cell–specific DUO1-dependent transcription relative to its parent PCR2.
DISCUSSION
Multiple and Repeated Retropositions in Arabidopsis
We found 251 retrogenes in Arabidopsis, 216 of which are newly identified. The limited overlap of our set with the previous Arabidopsis retrogene lists was most likely due to partly different search criteria and thresholds of individual methods (Zhang et al., 2005; Zhu et al., 2009). We detected ∼50% of the retrogenes found in the study of Zhang et al. (2005). A specific subset of the remaining retrogenes was not accepted by our method, owing to different thresholds for selection or a lack of positive evidence for retroposition, such as missing information on the parental gene or insufficient difference in intron number (Supplemental Data Set 2). The smaller (43.2%) overlap with the set identified by Zhu et al. (2009) is due to their use of very specific criteria to identify chimeric retrogenes. These criteria apparently hamper the identification of structurally simple retrogenes; conversely, our method does not allow the identification of chimeric retrogenes. The higher number of retrogenes detected with our analysis is due to several factors: (1) search among Arabidopsis pseudogenes, (2) allowing intronized retrogenes, and (3) accepting multiple retrocopies derived from a single parent (applied also in Zhang et al., 2005). Although we increased the number of retrogenes in Arabidopsis 3-fold, our selection criteria were conservative and the current number is most likely an underestimate based on two facts. First, we omitted several hundred candidates that had at least one paralog within the Arabidopsis genome but did not show evidence of retroposition [i.e. did not differ by two or more introns or did not have a poly(A) tail]. Second, none of the plant genome-wide retrogene screens detected retrogenes of the Ser-Glu-Thr domain protein group (Zhang et al., 2005; Zhu et al., 2009; this study), which were identified in studies focusing specifically on the evolution of this gene family (Baumbusch et al., 2001; Zhu et al., 2011). Hence, 1% of Arabidopsis genes estimated to be retrogenes is most likely an underestimation.
Although we have found 3-fold more retrogenes in Arabidopsis than were previously found in rice (Sakai et al., 2011), the number of conservatively estimated retrogenes per plant genome is much smaller compared with metazoans (e.g., 19.1% in human) (Marques et al., 2005; Pennisi, 2012). This difference may have multiple reasons. Since most of the retrogenes are identified based on intron loss, greater intron numbers in parents would simplify retrogene identification. This may partially explain the difference between the genomes of Arabidopsis and human, which have average numbers of 4.2 and 7.8 introns per gene, respectively (Arabidopsis Genome Initiative, 2000; Sakharkar et al., 2004). Another possibility, which is not mutually exclusive, builds on the scarcity of WGDs in many groups of higher animals compared with plants (Gregory and Mable, 2005). This may favor local gene duplication mechanisms, including retroposition, in metazoa versus plants. Finally, the higher activity of LONG INTERSPERSED ELEMENT (LINE) element reverse transcriptases may be responsible for an increased retroposition rate in animals (Beck et al., 2010).
In contrast with animals, where 82% of retrocopies contain premature stop codons (Marques et al., 2005), only 17.4% of Arabidopsis retrogenes are annotated as pseudogenes. This suggests a higher retrogene success rate in plants relative to the total number of retrocopies. Further support comes from our observation that several retrogenes served as parents and produced secondary retrocopies. Therefore, retroposition contributes to functional plant genome evolution.
One of the unresolved questions in retrogene biology is how transcripts are selected for retroposition. Although retroposition in animals has been associated with LINE element amplification machinery, this link has not been firmly proven in plants (Ohshima, 2013). We describe 22 parents that produced up to seven retrogenes each, which suggests one or more common features or a signal for retroposition in Arabidopsis. Additional support comes from the 13 cases where a repeated retroposition has been found. Since retrotransposon reverse transcriptases favor specific sequences in combination with transcript folding (Ohshima, 2013), it is possible that such structures exist also in transcripts of some protein genes. Similar to other plant and animal studies (Marques et al., 2005; Potrzebowski et al., 2008; Sakai et al., 2011), we have confirmed that parents are generally strongly and ubiquitously transcribed, indicating that higher amounts of transcript may increase the probability of retroposition. Although produced by the retrotransposon amplification machinery, they are located in gene-rich chromosome arms in Arabidopsis and thus fundamentally differ in their genomic distribution from repetitive elements. This also holds true for their upstream and downstream intergenic regions that are not enriched for repetitive DNA.
Arabidopsis Retrogenes Are Transcribed via Newly Acquired Promoters
One of the major limitations to the establishment of retrogenes as functional genes is the loss of cis-regulatory sequences (Kaessmann et al., 2009). Hence, we analyzed the retrogene transcription in Arabidopsis using genome-wide transcription data of 49 different Arabidopsis developmental stages by microarrays. In agreement with the observations in rice (Sakai et al., 2011), we found that retrogenes are transcribed less compared with their parents. However, retrogene transcription resembles the whole-genome average, suggesting that they are not “dead on arrival” in Arabidopsis. The parents are mostly recruited from highly and ubiquitously transcribed genes, indirectly supporting the hypothesis that transcript abundance is an important prerequisite for retroposition.
In human, it has been shown that retrogenes and parents may share promoter sequences, implying a carryover of the parental promoter by retroposition of transcripts from an upstream TSS (Okamura and Nakai, 2008). Furthermore, a recent study in rice revealed a number of retrogene/parent pairs with positively correlated transcription profiles among seven developmental stages (Sakai et al., 2011). However, this analysis did not include correction for the cotranscription of random gene pairs (Sakai et al., 2011); therefore, the extent of correlation may be partially overestimated. Our data show that ∼25% of retrogene/parent pairs and 3% of retrogene head-to-head oriented neighboring genes are cotranscribed beyond the genome background in Arabidopsis. Hence, rice and Arabidopsis data support the mechanism of cis-regulatory element carryover in plants. However, DNA sequence analysis of parent and retrogene promoters did not reveal significant homology in rice (Sakai et al., 2011). Therefore, it remains unclear whether retrogenes retropose including parental upstream regulatory sequences that mutate rapidly afterward or they carry cryptic exonic regulatory sequences. In Arabidopsis, the majority (72%) of retrogenes are transcribed in a pattern that is not correlated to that of parents and neighboring genes, suggesting the acquisition of novel cis-regulatory elements in most cases. Currently, it is unknown whether this pattern is the result of postintegration selection or whether the compact Arabidopsis genome offers a sufficient density of cryptic promoters.
Previous studies in Arabidopsis showed that transcripts of single-exon genes are relatively short-lived (Narsai et al., 2007). We observed that this also holds true for single-exon retrogenes and that retrogene intronization significantly increases their mRNA half-life. Hence, intron retention in the retrogene parent mRNAs and/or retrogene neointronization may help establish retrogenes as mature genes.
Retrogenes Are Preferentially Upregulated in Pollen
The separation of gametes from somatic cells is very much delayed in plants compared with animals. Therefore, somatic retroposition events in the shoot apical meristems may also be transmitted to the next generations. Thus, we tested for tissue-specific transcription of retrogenes in Arabidopsis using a developmental transcription data series (Schmid et al., 2005) and validated these findings using RNA sequencing data sets (Loraine et al., 2013). Surprisingly, this revealed that retrogenes are overtranscribed in pollen while overall transcription was not increased at this stage. However, this pattern was not uniform for the whole group, as lowly transcribed retrogenes became upregulated in pollen while highly transcribed ones were downregulated. In addition, the set of all Arabidopsis genes showed a similar trend. Hence, this transcription pattern is not restricted to retrogenes. More likely, many retrogenes are part of global cellular reprogramming in male gametes. So far, chromatin changes in male gametes have been associated mainly with DNA methylation changes (Slotkin et al., 2009; Ibarra et al., 2012), but there is emerging evidence that histone modifications may also contribute to pollen-specific gene reprogramming (Hoffmann and Palmgren, 2013). In order to identify possible causes of the observed pollen-specific transcription, we explored available data on tissue- and mutant-specific transcription and the distribution of chromatin modifications. By comparing transcriptional profiles of pollen and mutants defective in transcriptional gene silencing, we excluded the loss of DNA methylation and H3K9me2 or heterochromatin-specific histone hyperacetylation as the factors leading to global transcription changes in pollen. The analysis of chromatin profiles in leaves revealed that pollen upregulated genes (and retrogenes) are depleted of the transcription-permissive marks (H2Bub, H3K4me3, and H3K36me3) in these tissues. Recently, it was reported that pollen-specific genes are regulated by histone H3 Lysine 27 methylation in Arabidopsis (Hoffmann and Palmgren, 2013), but this trend was much less pronounced in our data set. This is due to the different selection criteria of candidate genes in both studies. Our set of pollen upregulated genes (n = 5171) included the entire (99.1%) set of pollen-specific genes (n = 584; Hoffmann and Palmgren, 2013). This is most likely masking the enrichment for histone H3 Lysine 27 methylation modifications of a specific subset of pollen-transcribed genes in leaves. However, it has to be noted that H3K27me3 modification may regulate pollen-specific transcription indirectly, as suggested by our transcription analysis of the clf swn and fie mutants. This also holds true for the group of pollen-specific genes associated with histone H3 Lysine 27 monomethylation and H3K27me3 in leaf tissues (Hoffmann and Palmgren, 2013), as only a few of those genes are upregulated in clf swn (Supplemental Figure 5D). Unexpectedly, we found correlated downregulation of similar sets of genes (and retrogenes) in pollen and leaves of clf swn and fie (r = 0.462 and 0.366, respectively). Gene downregulation in response to the loss of a repressive mark is counterintuitive and suggests that the effect is indirect and may be achieved by the activation of specific H3K27me3-regulated suppressors such as microRNAs (Lafos et al., 2011). Based on this, we propose that it is most likely a temporary absence of permissive marks (without strong enrichment for repressive marks) that causes the upregulation of specific genes in pollen relative to somatic tissues.
Pollen-specific transcription of Arabidopsis retrogenes was unanticipated and is analogous to retrogene transcription in animal spermatocytes (Marques et al., 2005; Vinckenbosch et al., 2006; Bai et al., 2008). Although the molecular nature of this specific transcription is so far unknown, two explanatory models have been proposed in animals (Kaessmann et al., 2009). The first suggests sperm-specific retroposition and integration into open (and thus more likely to be transcribed) chromatin that allows transcription and perpetuates this behavior. However, our data do not support this model in two respects. First, integration into active chromatin would most likely be reflected by cotranscription between neighboring genes, which was rare in Arabidopsis. Second, we observed many nonretrogene genes with pollen-specific transcription. The second model proposes spermatocyte-specific transcriptional reprogramming by global chromatin changes and transcriptional activation of retrogenes and their subsequent functionalization specific to spermatocytes (Marques et al., 2005; Potrzebowski et al., 2008). In plants, pollen has been identified as the hotspot of chromatin reprogramming (Slotkin et al., 2009; Ibarra et al., 2012; Hoffmann and Palmgren, 2013), and we have shown that pollen upregulated genes are depleted from transcription-permissive chromatin marks in somatic tissues. Furthermore, we found several retrogenes that are associated with pollen growth and development and the PCR11 retrogene, which is transcribed in pollen, contrary to its parent. This is due to the presence of multiple pollen-specific DUO1 transcription factor binding motifs in its promoter. Hence, our data support the second model and suggest that a small number of retrogenes have developed or retained male gamete–specific functions in Arabidopsis.
The activation of many normally lowly transcribed genes and the subsequent downregulation of highly transcribed genes just prior to the onset of the next generation is an intriguing pattern with no known molecular function. However, it seems to be present in both plant and animal lineages and suggests evolutionarily conserved or analogous mechanisms that regulate gene transcription during this critical stage of development.
METHODS
Retrogene Identification
The principal steps in retrogene identification in Arabidopsis thaliana are given in Figure 1A. First, the paralogy groups between sets of intronless (n = 5923) and intron-containing (n = 21,481) protein-coding genes according to TAIR10 were established using protein homologies in InParanoid 4.1 (Remm et al., 2001). When the paralogy group had multiple intron-containing “inparalogs” with different intron numbers, they were also considered for downstream analysis. Similarly, paralogy groups between pseudogenes (n = 924) and intron-containing protein-coding genes were identified as the best reciprocal BLAST hits using cDNA sequences (Altschul et al., 1990). Accepted retrogene–parent candidate pairs had a minimum homology score of 10−10 and a minimum difference in intron number of two. Intronless genes were also considered as candidates when differing by only a single intron, if the poly(A) tail was detected within 150 or 250 bp downstream of the retrogene candidate stop codon with or without an annotated 3′ untranslated region, respectively. The poly(A) tail was defined as ≥15 consecutive adenines with a single mismatch. We determined poly(A) tail length as the shortest stretch of adenines present significantly above random (Supplemental Figure 6). Since the absence of introns can be due to a loss of splicing signals (intron retention), the homology of exonic and intronic sequences was validated visually. A retrogene was accepted when a minimum of three consecutive homologous exons, spanning two lost introns, were observed (Edgar, 2004). If multiple parents were predicted for a retrogene, we accepted the candidate with the highest pair-wise alignment score in multiple (cDNA) sequence alignment (Larkin et al., 2007). The protocol was executed with customized bioperl and awk scripts (Stajich et al., 2002).
Genome-Wide Transcription and mRNA Half-Life Analysis
All microarray analyses were based on publicly available data sets. Throughout the study, we used the following ATH1 cDNA microarrays (Affymetrix): wild-type Arabidopsis development data produced by the AtGenExpress consortium (Schmid et al., 2005), Arabidopsis pollen development and sperm cell data sets NASCARRAYS-48 (Honys and Twell, 2003, 2004), the ddm1-12 data set deposited at the Gene Expression Omnibus (GEO) as GSE18977 (Baubec et al., 2010), the kyp GEO data set GSE22957 (Inagaki et al., 2010), the clf, swn, and clf swn GEO data set GSE20256, and the hda6 (rts1-1) data set NASCARRAYS-538 (Popova et al., 2013). The raw data were processed and normalized using the robust multiarray averaging method (Irizarry et al., 2003) in R software (www.R-project.org) using Bioconductor (www.bioconductor.org) and the affy package. The fie transcription values were retrieved from the GEO data set GSE19851 (Bouyer et al., 2011) as the normalized transcription values. Retrogene and parent probes that corresponded to multiple gene models were excluded from genome-wide analysis. The transcription borderline for transcribed genes (gcRMA ≥ 5) was based on the minimal density of genes between peaks indicating absent or background signals versus high transcription signals (Supplemental Figure 2). The Arabidopsis mRNA half-life data and rosette- and pollen-specific RNA sequencing data were extracted from previously published data sets (Narsai et al., 2007; Loraine et al., 2013). Randomized sets of genes or gene pairs were generated, plots drawn, and statistical tests calculated in R. The significance of density distributions was tested using the MWW rank-sum test with correction and cotranscription correlation by the Pearson product–moment correlation coefficient (r).
Chromatin Analysis
Chromatin data of 10-d-old Arabidopsis seedlings were retrieved from the publicly available genome-wide atlas of chromatin modifications (Roudier et al., 2011). The frequencies for individual groups were compared. Pearson correlations were calculated in Excel (Microsoft), and heat maps were built in R.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MSI1, At5g58230; PCR1, At1g14880; PCR2, At1g14870; PCR3, At5g35525; PCR4, At3g18460; PCR8, At1g52200; PCR10, At2g40935; PCR11, At1g68610; BGLU14, At2g25630; BGLU15, At2g44450; MEE25, At2g34850; parent of MEE25 retrogene, At4g10960; and PEROXIDASE, At4g17690. Other genes listed in the supplemental data sets include Arabidopsis Genome Initiative codes.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Examples of Repeated Retroposition in Arabidopsis.
Supplemental Figure 2. Defining Lowly and Highly Transcribed Genes in Arabidopsis.
Supplemental Figure 3. Arabidopsis Retrogenes Are Transcribed in Pollen.
Supplemental Figure 4. Transcription Correlations between Pollen and Chromatin Mutants.
Supplemental Figure 5. Chromatin Regulation of Pollen-Specific Gene Transcription.
Supplemental Figure 6. Defining the Minimum Length of Nonrandom Poly(A) Tail for the Arabidopsis Genome.
Supplemental Data Set 1. List of Retrogenes and Parents Identified by Genome-Wide Searches in Arabidopsis.
Supplemental Data Set 2. Putative Arabidopsis Retrogenes and Parents Not Considered in This Study.
Supplemental Data Set 3. Repeated Retroposition Events in Arabidopsis.
Supplemental Data Set 4. Retrogene and Parent Transcription in 49 Arabidopsis Developmental Stages and Tissues.
Supplemental Data Set 5. Transcription of Specific Gene Sets in the Wild Type and Chromatin Mutants.
Supplemental Data Set 6. Transcription Analysis of DNA Duplicated Genes (Blanc and Wolfe, 2004).
Supplemental Data Set 7. List of Genome-Wide Head-to-Head Oriented Genes and Transposable Elements.
Supplemental Data Set 8. Transcription of Retrogene and Head-to-Head Oriented Neighboring Genes in 49 Arabidopsis Developmental Stages and Tissues.
Supplemental Data Set 9. Transcription Quantiles of Specific Sets of Genes.
Supplemental Data Set 10. The Frequency of Chromatin Modifications at Specific Groups of Genes.
Supplemental Data Set 11. Genes Significantly Upregulated and Downregulated in Arabidopsis pMDC7-DUO1 Lines after 6, 12, and 24 h of DUO1 Induction.
Supplementary Material
Acknowledgments
We thank A. Srinivasan, N. Müller, T. Baubec, and D. Twell for discussions on data analysis, K. Schneeberger, D. Schubert, and M. Koornneef for careful reading of the article, and T. Harrop for language editing. This work was supported by the Max Planck Society and by the German Research Foundation (Grant 1853/2) to A.P.
AUTHOR CONTRIBUTIONS
Both authors designed and conceived the experiments. A.P. wrote the article, and A.A. contributed to its final version.
Glossary
- WGD
whole-genome DNA-based duplication
- H3K27me3
histone H3 Lysine 27 trimethylation
- H3K9me2
histone H3 Lysine 9 dimethylation
- MWW
Mann-Whitney-Wilcoxon
- TE
transposable element
- TSS
transcription start site
- TTS
transcription termination site
- gcRMA
log2 robust multiarray averaging
- H3K4me3
histone H3 Lysine 4 trimethylation
- H3K36me3
histone H3 Lysine 36 trimethylation
- H2Bub
histone H2B ubiquitination
- GEO
Gene Expression Omnibus
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Bai Y., Casola C., Betrán E. (2008). Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics 9: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T., Dinh H.Q., Pecinka A., Rakic B., Rozhon W., Wohlrab B., von Haeseler A., Mittelsten Scheid O. (2010). Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states in Arabidopsis. Plant Cell 22: 34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch L.O., Thorstensen T., Krauss V., Fischer A., Naumann K., Assalkhou R., Schulz I., Reuter G., Aalen R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29: 4319–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V. (2010). LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M., Brownfield L., Khatab H., Sidorova A., Lingaya M., Twell D. (2011). The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell 23: 534–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev A.M., Kalmykova A.I., Shevelyov Y.Y., Nurminsky D.I. (2002). Large clusters of co-expressed genes in the Drosophila genome. Nature 420: 666–669. [DOI] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E.D., Gey D., Nowack M.K., Goodrich J., Renou J.-P., Grini P.E., Colot V., Schnittger A. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D., Zilberman D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Dehal P., Boore J.L. (2005). Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R., Adams K.L., Vandepoele K., Van Montagu M.C.E., Maere S., Van de Peer Y. (2013). Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. USA 110: 2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Thorpe F.L., Engelhorn J., Adrian J., Dong X., Sarid-Krebs L., Goodrich J., Turck F. (2011). Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 23: 3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J.A., Maere S., Van de Peer Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 106: 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Wendel J.F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183: 557–564. [DOI] [PubMed] [Google Scholar]
- Gregory, R.T., and Mable, B.K. (2005). Polyploidy in animals. In The Evolution of the Genome, R.T. Gregory, ed (New York: Elsevier), pp. 427–483. [Google Scholar]
- Hoffmann R.D., Palmgren M.G. (2013). Epigenetic repression of male gametophyte-specific genes in the Arabidopsis sporophyte. Mol. Plant 6: 1176–1186. [DOI] [PubMed] [Google Scholar]
- Honys D., Twell D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132: 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D., Twell D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra C.A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S., Miura-Kamio A., Nakamura Y., Lu F., Cui X., Cao X., Kimura H., Saze H., Kakutani T. (2010). Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome. EMBO J. 29: 3496–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kondrashov F. (2010). The evolution of gene duplications: Classifying and distinguishing between models. Nat. Rev. Genet. 11: 97–108. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Kaessmann H. (2010). Origins, evolution, and phenotypic impact of new genes. Genome Res. 20: 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H., Vinckenbosch N., Long M. (2009). RNA-based gene duplication: Mechanistic and evolutionary insights. Nat. Rev. Genet. 10: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M., Kroll P., Hohenstatt M.L., Thorpe F.L., Clarenz O., Schubert D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719. [DOI] [PubMed] [Google Scholar]
- Loraine A.E., McCormick S., Estrada A., Patel K., Qin P. (2013). RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 162: 1092–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A.C., Dupanloup I., Vinckenbosch N., Reymond A., Kaessmann H. (2005). Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 3: e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D., et al. (2011). Human imprinted retrogenes exhibit non-canonical imprint chromatin signatures and reside in non-imprinted host genes. Nucleic Acids Res. 39: 4577–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney M., et al. (2013). Comparative RNA-Seq and microarray analysis of gene expression changes in B-cell lymphomas of Canis familiaris. PLoS ONE 8: e61088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286. [DOI] [PubMed] [Google Scholar]
- Narsai R., Howell K.A., Millar A.H., O’Toole N., Small I., Whelan J. (2007). Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K. (2013). RNA-mediated gene duplication and retroposons: Retrogenes, LINEs, SINEs, and sequence specificity. Int. J. Evol. Biol. 2013: 424726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Nakai K. (2008). Retrotransposition as a source of new promoters. Mol. Biol. Evol. 25: 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei B., et al. (2012). The GENCODE pseudogene resource. Genome Biol. 13: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. (2012). Genomics. ENCODE project writes eulogy for junk DNA. Science 337: 1159–1161, 1161. [DOI] [PubMed] [Google Scholar]
- Popova O.V., Dinh H.Q., Aufsatz W., Jonak C. (2013). The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 6: 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L., Vinckenbosch N., Marques A.C., Chalmel F., Jégou B., Kaessmann H. (2008). Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 6: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M., Storm C.E., Sonnhammer E.L. (2001). Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Mizuno H., Kawahara Y., Wakimoto H., Ikawa H., Kawahigashi H., Kanamori H., Matsumoto T., Itoh T., Gaut B.S. (2011). Retrogenes in rice (Oryza sativa L. ssp. japonica) exhibit correlated expression with their source genes. Genome Biol. Evol. 3: 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar M.K., Chow V.T.K., Kangueane P. (2004). Distributions of exons and introns in the human genome. In Silico Biol. 4: 387–393. [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.-Y., Choi K.S., Kim Y., Geisler M., Park J., Vincenzetti V., Schellenberg M., Kim S.H., Lim Y.P., Noh E.W., Lee Y., Martinoia E. (2010). Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. Plant Cell 22: 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J.E., et al. (2002). The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N., Dupanloup I., Kaessmann H. (2006). Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. USA 103: 3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2006). High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18: 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Maruyama S., Nozaki H., Shirasu K. (2010). Horizontal gene transfer by the parasitic plant Striga hermonthica. Science 328: 1128. [DOI] [PubMed] [Google Scholar]
- Zhang J. (2003). Evolution by gene duplication: An update. Trends Ecol. Evol. 18: 292–298. [Google Scholar]
- Zhang Y., Wu Y., Liu Y., Han B. (2005). Computational identification of 69 retroposons in Arabidopsis. Plant Physiol. 138: 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Fung-Leung W.-P., Bittner A., Ngo K., Liu X. (2014). Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 9: e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Ma H., Chen Z. (2011). Phylogenetics and evolution of Su(var)3-9 SET genes in land plants: Rapid diversification in structure and function. BMC Evol. Biol. 11: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Zhang Y., Long M. (2009). Extensive structural renovation of retrogenes in the evolution of the Populus genome. Plant Physiol. 151: 1943–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.