This study found that IPUT1 is an enzyme required for the biosynthesis of glycosyl inositol phosphorylceramide sphingolipids in plants. These glycolipids are highly abundant and have many important cellular roles. IPUT1 adds glucuronic acid residues to the lipids. When the glucuronic acid residues are not incorporated in the sphingolipids, the pollen is unable to mediate fertilization.
Abstract
Glycosyl inositol phosphorylceramide (GIPC) sphingolipids are a major class of lipids in fungi, protozoans, and plants. GIPCs are abundant in the plasma membrane in plants, comprising around a quarter of the total lipids in these membranes. Plant GIPCs contain unique glycan decorations that include a conserved glucuronic acid (GlcA) residue and various additional sugars; however, no proteins responsible for glycosylating GIPCs have been identified to date. Here, we show that the Arabidopsis thaliana protein INOSITOL PHOSPHORYLCERAMIDE GLUCURONOSYLTRANSFERASE1 (IPUT1) transfers GlcA from UDP-GlcA to GIPCs. To demonstrate IPUT1 activity, we introduced the IPUT1 gene together with genes for a UDP-glucose dehydrogenase from Arabidopsis and a human UDP-GlcA transporter into a yeast mutant deficient in the endogenous inositol phosphorylceramide (IPC) mannosyltransferase. In this engineered yeast strain, IPUT1 transferred GlcA to IPC. Overexpression or silencing of IPUT1 in Nicotiana benthamiana resulted in an increase or a decrease, respectively, in IPC glucuronosyltransferase activity in vitro. Plants in which IPUT1 was silenced accumulated IPC, the immediate precursor, as well as ceramides and glucosylceramides. Plants overexpressing IPUT1 showed an increased content of GIPCs. Mutations in IPUT1 are not transmitted through pollen, indicating that these sphingolipids are essential in plants.
INTRODUCTION
Glycosyl inositol phosphorylceramide (GIPC) sphingolipids are a major class of lipids in fungi, protozoans, and plants. These lipids consist of a phosphorylceramide backbone linked to an inositol and additional sugar residues. In plants, GIPCs are highly glycosylated and thus have limited solubility in typical lipid extraction solvents (Sperling and Heinz, 2003); as a result, they have not received as much attention as other major types of lipids. However, recent reports indicate that they make up 25 to 50% of the plasma and tonoplast membranes (Sperling et al., 2005; Markham et al., 2006, 2013) and that they are involved in many essential processes, including pathogen defense (Wang et al., 2008; Mortimer et al., 2013), symbiosis (Perotto et al., 1995), and membrane organization such as formation of lipid rafts (Borner et al., 2005).
Sphingolipid biosynthesis has been discussed in several recent reviews (Zäuner et al., 2010; Markham et al., 2013). In brief, synthesis begins with the condensation of serine and palmitoyl-CoA by the enzyme serine palmitoyltransferase to form the long-chain sphingobase (LCB) 3-ketosphinganine (Dietrich et al., 2008). Mutations in genes encoding subunits of serine palmitoyltransferase are pollen lethal, indicating that sphingolipids are essential in plants (Chen et al., 2006; Kimberlin et al., 2013). 3-Ketosphinganine reductase then converts 3-ketosphinganine to sphinganine (Chao et al., 2011), which can be modified by hydroxylation and unsaturation to create up to nine different LCBs with variations in structure. Free LCBs can be converted to ceramide by ceramide synthases, also known as Lag 1 Homolog (LOH) enzymes, which acylate LCBs with fatty acids. Different LOH enzymes acylate trihydroxy and dihydroxy LCBs, implicating these enzymes in control of sphingolipid flux through different biosynthetic pathways (Markham et al., 2011; Ternes et al., 2011). Ceramides may then be phosphorylated by ceramide kinases such as ACCELERATED CELL DEATH5 (Liang et al., 2003), glucosylated by glucosylceramide synthases to synthesize glucosylceramides (Hillig et al., 2003), or substituted with inositol phosphate by inositol phosphorylceramide (IPC) synthases (IPCSs) (Wang et al., 2008). IPC may then be glycosylated by glycosyltransferases in the Golgi to produce GIPCs.
Identification of genes encoding sphingolipid biosynthetic proteins has implicated sphingolipids in many important processes including ion transport (Chao et al., 2011), endomembrane trafficking (Markham et al., 2011), programmed cell death (Liang et al., 2003), cold tolerance (Chen et al., 2012), stomatal closure (Coursol et al., 2005), and pollen development (Chen et al., 2006). However, little information is available about the roles of sphingolipid glycosylation. Identifying glycosyltransferases involved in GIPC synthesis is challenging because while IPC structure is largely conserved, glycosylation patterns vary more widely between kingdoms. Plant GIPCs contain a core α(1,4)-linked GlcA that can be modified by addition of (N-acetyl)glucosamine, mannose, galactose, arabinose, and fucose residues (Carter et al., 1969; Kaul and Lester, 1978; Sperling and Heinz, 2003; Buré et al., 2011; Cacas et al., 2013; Mortimer et al., 2013). These glycan structures are unique to plants; thus, finding the enzymes responsible for synthesizing the glycosyl portion of these glycolipids cannot rely on homology, and no enzymes have yet been identified in this pathway. Only one protein involved in GIPC glycosylation has been identified: GOLGI-LOCALIZED NUCLEOTIDE SUGAR TRANSPORTER1, a transporter that supplies GDP-mannose to the Golgi lumen for mannosylation of GIPCs (Mortimer et al., 2013). gonst1 GIPCs have reduced mannose content, and mutant plants are dwarfed and develop spontaneous lesions on leaves.
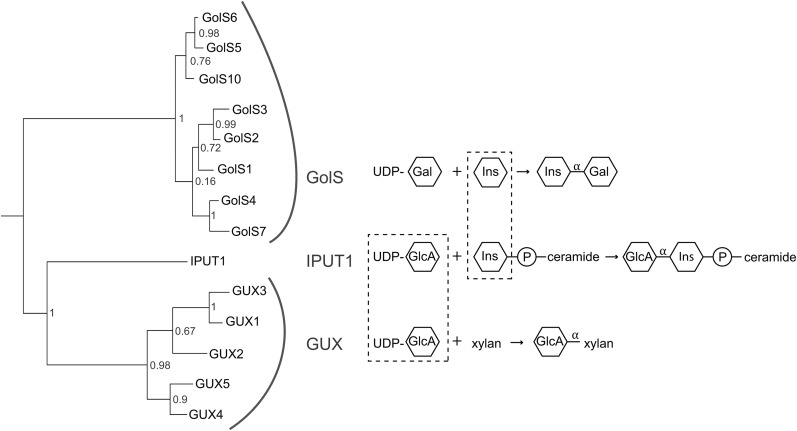
We focused our investigation on an Arabidopsis thaliana protein that we have named INOSITOL PHOSPHORYLCERAMIDE GLUCURONOSYLTRANSFERASE1 (IPUT1) based on the results described in this report. IPUT1 is a member of Glycosyltransferase Family 8 and was formerly named PLANT GLYCOGENIN-LIKE STARCH INITIATION PROTEIN6 because it is distantly related to glycogenin and has been suggested to catalyze the initiation of starch synthesis (Chatterjee et al., 2005). However, several of lines of evidence led us to suspect that IPUT1 is actually an IPC glucuronosyltransferase. First, IPUT1 is closely related to both the GLUCURONIC ACID SUBSTITUTION OF XYLAN (GUX) proteins, which transfer GlcA onto the cell wall polymer xylan (Mortimer et al., 2010; Oikawa et al., 2010; Rennie et al., 2012), and the Galactinol Synthase (GolS) proteins, which transfer galactose onto inositol to synthesize galactinol (Taji et al., 2002). We reasoned that IPUT1 might use the same substrate, UDP-GlcA, as the GUX proteins while using the same acceptor, inositol (in the form of inositol phosphorylceramide), as the GolS proteins (Figure 1; Supplemental Data Set 1). Second, IPUT1 belongs to a glycosyltransferase family that transfers sugars with a retaining mechanism resulting in α-linked sugars, consistent with the α conformation of the GlcA in GIPCs. Finally, both fluorescent protein fusions and proteomic studies have shown that IPUT1 is localized to the Golgi apparatus (Dunkley et al., 2004; Parsons et al., 2012; Rennie et al., 2012), where glycosylation of sphingolipids is expected to occur (Markham et al., 2013).
Figure 1.
Phylogenetic Relationship of the GolS, GUX, and IPUT1 Proteins.
IPUT1 is related to both the GolS and GUX proteins, which transfer sugars with a retaining mechanism that creates α glycosidic linkages. IPUT1 uses the same substrate, UDP-GlcA, as the GUX proteins, while using the same acceptor, inositol (Ins) or inositol phosphorylceramide, as the GolS proteins (dashed lines). Likelihood values are given at nodes.
We used several approaches to investigate the role of IPUT1 in GIPC synthesis. Our results show that IPUT1 does in fact possess IPC glucuronosyltransferase activity and that expression of IPUT1 is correlated with this activity in plants. In addition, we found that IPUT1 is required in Arabidopsis, highlighting the importance of these abundant, yet poorly understood, sphingolipids.
RESULTS
Introducing the Plant-Type Sphingolipid Pathway into Yeast Confirms That IPUT1 Is an Inositol Phosphorylceramide Glucuronosyltransferase
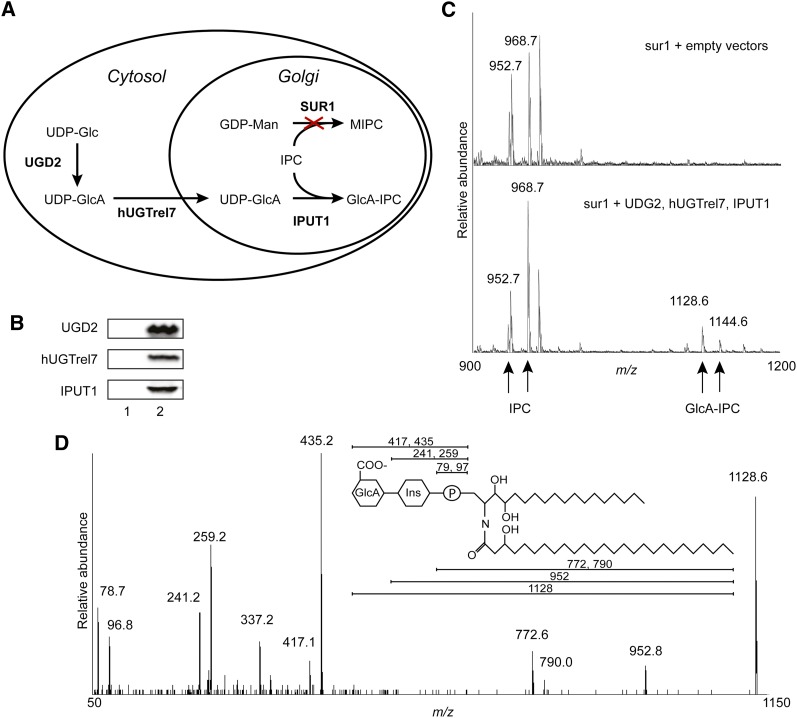
Because GIPCs from Saccharomyces cerevisiae do not contain GlcA, we used this system to test whether IPUT1 can transfer GlcA onto IPC. The major GIPC species in yeast are mannosyl inositol phosphorylceramide (MIPC) and mannosyl-diinositolphosphorylceramide (Obeid et al., 2002); however, the yeast mutant sur1Δ is defective in the mannosyltransferase that transfers mannose to IPC to synthesize MIPC and therefore accumulates IPC (Beeler et al., 1997), the predicted acceptor substrate for IPUT1. Because S. cerevisiae does not naturally synthesize UDP-GlcA, the predicted donor substrate for IPUT1, we introduced the UDP-glucose dehydrogenase gene UGD2 from Arabidopsis (Klinghammer and Tenhaken, 2007) into the sur1Δ mutant yeast strain to produce this substrate. We also introduced the human UDP-GlcA transporter, hUGTrel7, to ensure that UDP-GlcA is present in the Golgi lumen (Muraoka et al., 2001). This strategy is outlined in Figure 2A. Lysates from sur1Δ yeast expressing IPUT1, UGD2, and hUGTrel7 were analyzed by immunoblotting to ensure that proteins were expressed (Figure 2B).
Figure 2.
Production of GlcA-IPC in Yeast.
(A) Diagram of strategy for expressing proteins to produce GlcA-IPC. UDP-Glucose Dehydrogenase 2 (UGD2), human UDP-Galactose Transporter Relative 7 (hUGTrel7), and IPUT1 were transformed into sur1Δ yeast. Cross indicates deletion of sur1.
(B) Immunoblot showing expression of heterologous proteins in yeast with empty vectors (1) or vectors driving expression of UGD2, hUGTrel7, and IPUT1 (2).
(C) Mass spectrometry analysis of lipids from yeast transformed with control (empty) or UGD2, hUGTrel7, and IPUT1 vectors. Major IPC species at m/z 952.7 and 968.7 containing trihydroxy C18 saturated long-chain sphingobases acylated with mono- or dihydroxy C26 saturated fatty acids, respectively, and corresponding GlcA-IPC species at m/z 1128.6 and 1144.6 are indicated.
(D) Tandem mass spectrometry confirming identification of the GlcA-IPC peak at m/z 1128.6. Expected m/z for each fragment ion is indicated in the diagram.
[See online article for color version of this figure.]
Lipids from sur1Δ yeast expressing UGD2, hUGTrel7, and IPUT1 were extracted and analyzed by mass spectrometry (Figure 2C). The major IPC species in these yeast contained trihydroxy C18 saturated (t18:0) LCBs acylated to mono- or dihydroxy C26 fatty acids, with [M-H]− peaks at m/z 952.7 and 968.7, respectively (Guan and Wenk, 2006) (Figure 2C). These IPC species are relatively well characterized in yeast and are referred to as IPC-C and IPC-D (Dunn et al., 1998). In wild-type yeast, additional peaks at m/z 1114.8 and 1130.8 represent MIPC-C and MIPC-D with the addition of a mannose residue (162 atomic mass units) (Guan and Wenk, 2006). However, these peaks were reduced in sur1Δ yeast as expected, and when UGD2, hUGTrel7, and IPUT1 were expressed, peaks appeared at m/z 1128.6 and 1144.6 that represent the addition of a GlcA residue (176 atomic mass units) (Figure 2C). Tandem mass spectrometry confirmed the identity of the compound giving rise to the peak at m/z 1128.6 as GlcA-IPC-C with a C26 fatty acid (Figure 2D). Because the compound represented by the peak at m/z 1144.6 appeared to be present in only a small amount, we were not able to collect tandem mass spectrometry data on this compound; however, because it was present only in yeast expressing both UDG2 and IPUT1 (Supplemental Figure 1), it is very likely that this compound is GlcA-IPC-D containing a C26 fatty acid.
Lipids from transformed yeast were also visualized by thin-layer chromatography (TLC). Several bands representing polar lipids including IPC were visible near the solvent front, and expression of IPUT1 led to the appearance of an additional lipid band (arrowhead, Figure 3A). The slower migration of this band compared with IPC in the solvent system used is consistent with the addition of a charged group such as GlcA (Kaul and Lester, 1975), and when the band was excised and analyzed with mass spectrometry it was determined to contain the peak at m/z 1128.6 representing GlcA-IPC.
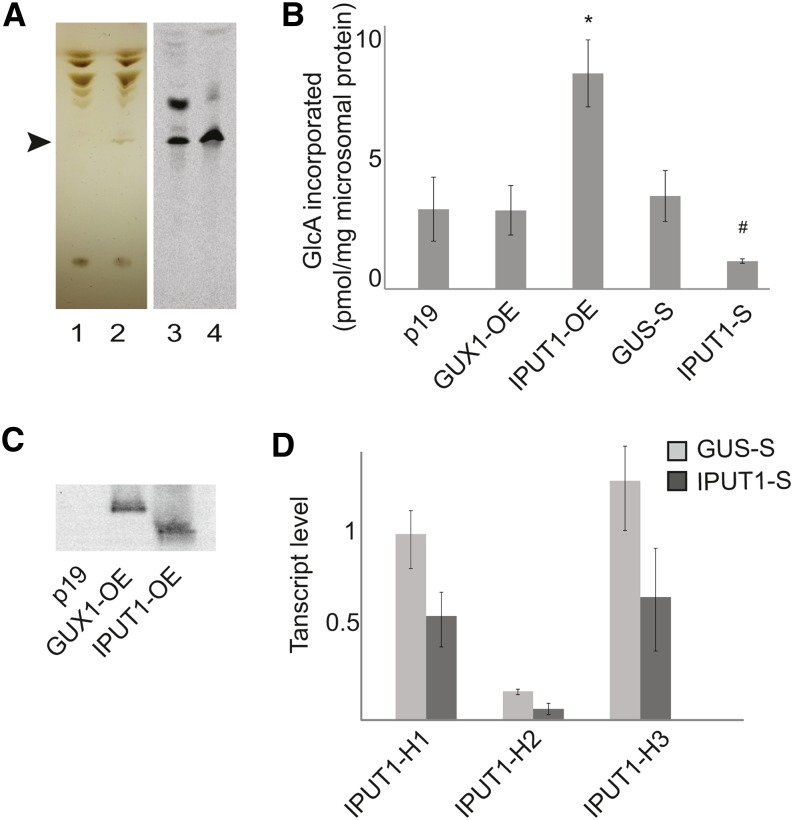
Figure 3.
Assay for IPC Glucuronosyltransferase Activity in N. benthamiana.
(A) TLC of lipids extracted from sur1Δ yeast with empty vectors (1) or vectors with UGD2, hUGTrel7, and IPUT1 (2). GlcA-IPC is indicated by the arrowhead. Radiolabeled lipids from N. benthamiana microsomes incubated with UDP-[14C]GlcA followed by mock (3) or mild alkaline hydrolysis treatment (4) were developed on the same TLC plate.
(B) Incorporation of radiolabel into GlcA-IPC bands produced from microsomes prepared from N. benthamiana plants expressing p19 (control), overexpressing GUX1 (GUX-OE) or IPUT1 (IPUT1-OE), or plants silencing the control gene GUS (GUS-S) or IPUT1 (IPUT1-S). *, Significantly different from p19 (t test, P < 0.05); #, significantly different from GUS-S (t test, P < 0.05).
(C) Immunoblot of microsomal proteins from N. benthamiana plants expressing p19 or overexpressing GUX1 (GUX-OE) or IPUT1 (IPUT1-OE).
(D) Expression levels of N. benthamiana IPUT1 homologs H1, H2, and H3 in IPUT1-silenced plants. All transcript levels were normalized to ACT2, UBI, and EF1α. Values shown are normalized to the transcript level of IPUT1-H1 in GUS-silenced plants. All values are the mean ± se of three biological replicates.
[See online article for color version of this figure.]
As additional controls, sur1Δ yeast were transformed with all combinations of either empty vectors or vectors containing UGD2, hUGTrel7, and/or IPUT1 cDNA. Both UGD2 and IPUT1 were necessary for production of GlcA-IPC (Supplemental Figure 1). The requirement for UGD2 indicates that the uronic acid in the compound with m/z 1128.6 is GlcA and that UDP-GlcA is the substrate used in glucuronosylation. GlcA-IPC production was more pronounced when the UDP-GlcA transporter hUGTrel7 was expressed. The detectable GlcA-IPC production in the absence of hUGTrel7 indicates that there is background transport of UDP-GlcA into the Golgi lumen in yeast; this is consistent with previous observations (Muraoka et al., 2001).
Overexpressing IPUT1 Leads to Increased IPC Glucuronosyltransferase Activity, While Silencing IPUT1 Leads to Decreased Activity in Nicotiana benthamiana
We developed an enzyme assay for IPC glucuronosyltransferase activity in microsomes from N. benthamiana. Although the acceptor IPC is not commercially available, it is present in plant microsomal fractions (Bromley et al., 2003). Microsomes were incubated with UDP-[14C]GlcA to allow [14C]GlcA to be transferred onto endogenous IPC, and the reaction was then stopped and lipids were extracted and separated by TLC. The position of the radiolabeled lipids was visualized using a phosphor imager (Figure 3A). Radiolabel was incorporated into several bands; however, only one band was resistant to mild alkaline hydrolysis, which cleaves ester-linked glycerolipid but not amide-linked sphingolipid fatty acids (Pinto et al., 1992). In addition, this band migrated to the same position as the GlcA-IPC produced in transgenic yeast, which was used as a standard. The band was therefore designated as [14C]GlcA-IPC.
To test whether IPUT1 is responsible for the production of [14C]GlcA-IPC in planta, we transiently expressed both IPUT1-YFP-HA and GUX1-YFP-HA fusion proteins in N. benthamiana leaves by infiltration with Agrobacterium tumefaciens. GUX1 is a glucuronosyltransferase that acts on the cell wall polymer xylan and was used as a control to determine whether IPC glucuronosyltransferase activity was specific to IPUT1. Both constructs were coexpressed with p19, a viral protein that is used as a suppressor of gene silencing, to ensure that proteins were highly expressed. GUX1-YFP-HA and IPUT1-YFP-HA proteins were expressed at similar levels, as monitored by immunoblot (Figure 3C). Radiolabeled standards were spotted onto TLC plates to ensure that images were collected in the linear range of detection and to allow the amount of radiolabel incorporated into GlcA-IPC to be quantified. Overexpressing IPUT1-YFP-HA caused a 170% increase in the amount of [14C]GlcA transferred to IPC compared with that from the p19 control (Figure 3B). This difference was significant (P < 0.05, t test). In comparison, for microsomes from plants overexpressing GUX1-YFP-HA, the amount of [14C]GlcA transferred was similar to that in microsomes from plants infiltrated with p19 alone.
We used virus-induced gene silencing (VIGS) to reduce expression of the N. benthamiana homologs of IPUT1. Plants were also infiltrated with a control construct designed to silence the β-glucuronidase (GUS) transcript, which is not endogenous to plants. The N. benthamiana genome encodes three IPUT1 homologs (Supplemental Figure 2 and Supplemental Data Set 2). We used three different housekeeping reference genes to confirm that IPUT1 genes were silenced: ACT2, UBI, and EF1α. Expression of the VIGS construct reduced expression of all three homologs to between 40 and 50% of levels in GUS-silenced control plants (Figure 3D). Silencing of the N. benthamiana IPUT1 homologs resulted in a reduction of the [14C]GlcA-IPC band to ∼35% of that produced by GUS-silenced control microsomes (significant at P < 0.05, t test) (Figure 3B). These results indicate that the amount of IPC glucuronosyltransferase activity in plants correlates with IPUT1 expression levels.
Sphingolipid Levels Are Altered in N. benthamiana Plants with Changes in IPUT1 Expression
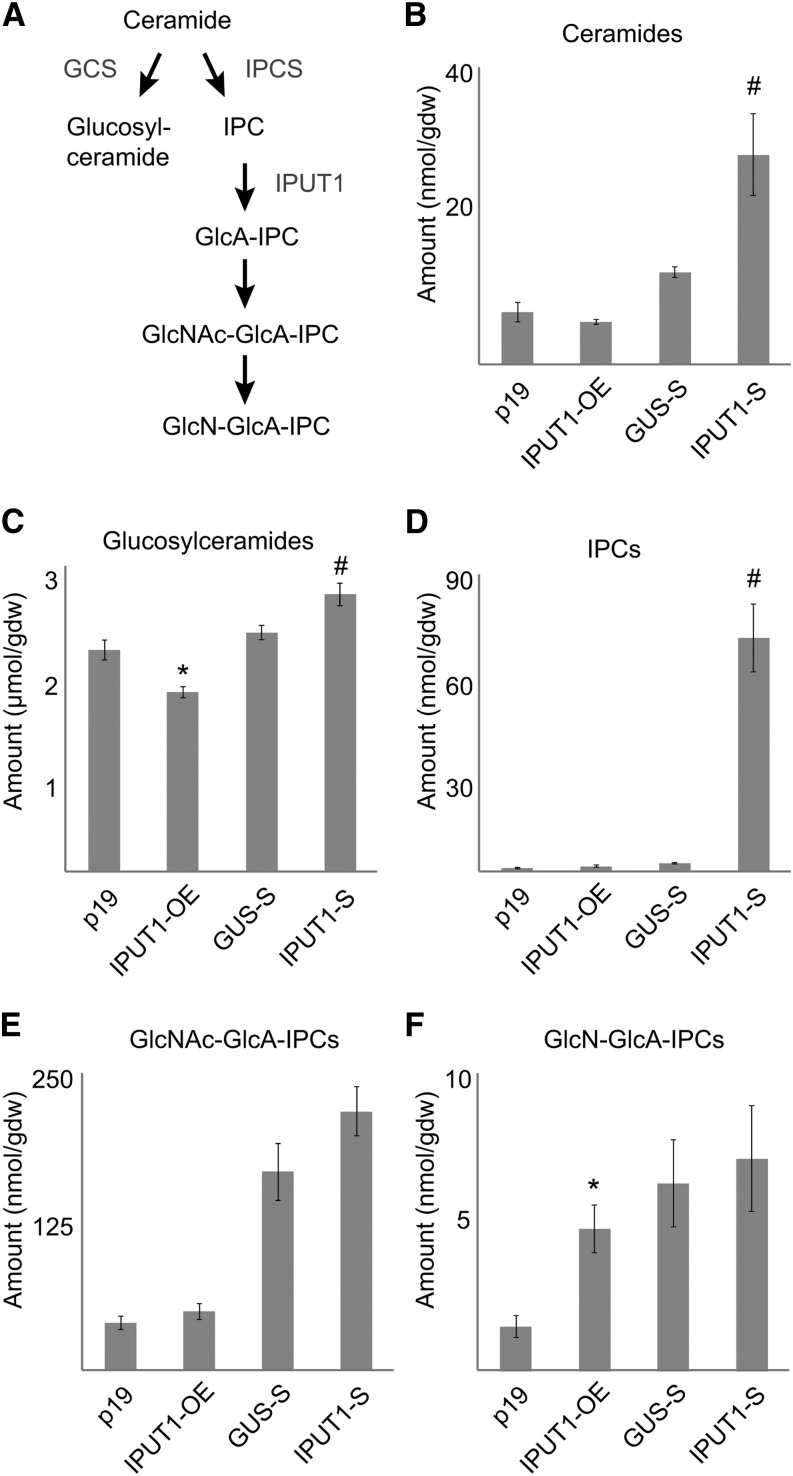
We used mass spectrometry to profile sphingolipids in the IPUT1-overexpressing and IPUT1-silencing N. benthamiana plants (Figure 4). In IPUT1-overexpressing plants, total glucosylceramide levels decreased slightly compared with the p19 control, while levels of GlcN-GlcA-IPC increased (P < 0.05, t test). Ceramide, IPC, and GlcNAc-GlcA-IPC levels did not change significantly. In contrast, plants in which IPUT1 homologs were silenced showed a nearly 30-fold increase in IPC compared with the GUS-silenced control. Ceramide levels more than doubled in these plants, while glucosylceramide levels were moderately increased. Levels of GlcN(Ac)-GlcA-IPC did not change significantly. The relative proportions of different sphingobases and fatty acids in the GIPCs were not affected in overexpressing or silencing plants (Supplemental Data Set 3). However, the ceramide pool in the silenced plants had an overrepresentation of the t18:0/h24:0 structure, which is the most abundant ceramide in tobacco GIPCs.
Figure 4.
Sphingolipid Profiles in N. benthamiana Plants with Altered Expression of IPUT1.
(A) Outline of complex sphingolipid synthesis. Known enzymes in the pathway are shown in gray. GCS, glucosylceramide synthase.
(B) to (F) Profiles of sphingolipids extracted from leaves of plants expressing p19 (control), overexpressing IPUT (IPUT1-OE), silencing the control gene (GUS-S), or silencing IPUT1 (IPUT1-S).
*, Significantly different from p19 (t test, P < 0.05); #, significantly different from GUS-S (t test, P < 0.05). gdw, gram dry weight. Values are the mean ± se of five biological replicates.
Transmission of iput1 Alleles through Pollen Is Significantly Reduced
We identified two insertional mutants in Arabidopsis IPUT1, named iput1-1 and iput1-2. Both insertions are located within the gene; iput1-1 is found in the third intron, while iput1-2 is found in the eighth exon (Supplemental Figure 3). We were unable to obtain homozygotes for either mutant allele. To investigate the transmission of iput1 alleles, progeny from heterozygous iput1-1/+ and iput1-2/+ plants were scored for resistance to the Basta or sulfadiazine markers encoded on T-DNA insertions. Self-fertilization of iput1-1/+ and iput1-2/+ plants resulted in an ∼1:1 ratio of resistant to susceptible progeny, indicating a deficiency in transmission through either the male or female gametophyte (Table 1). Reciprocal crosses with wild-type plants showed that while transmission of iput1 alleles through female gametophytes was not significantly affected, transmission through pollen was significantly reduced (P < 0.01, χ2 test).
Table 1. Transmission Efficiency of iput1 Alleles.
| Parent Genotype | Self-Fertilized | TEFemale | TEMale |
|---|---|---|---|
| iput1-1/+ | 0.98:1 (672) | 95% (455) | 1% (527) |
| iput1-2/+ | 0.93:1 (349) | 92% (349) | 2% (519) |
Transmission efficiency of the iput1 T-DNA alleles was determined using the ratio of Basta-resistant to Basta-sensitive progeny (iput1-1/+) or the ratio of Sulfadiazine-resistant to Sulfadiazine-sensitive progeny (iput1-2/+) after self-fertilization and after performing reciprocal crosses with wild-type plants. Self-fertilized, ratio of number of progeny with T-DNA insertion to number of progeny without T-DNA insertion. TE, number of progeny with T-DNA insertion/number of progeny without T-DNA insertion × 100. Total numbers of seedlings scored are indicated in parentheses. Pooled data represented in this table are from multiple generations.
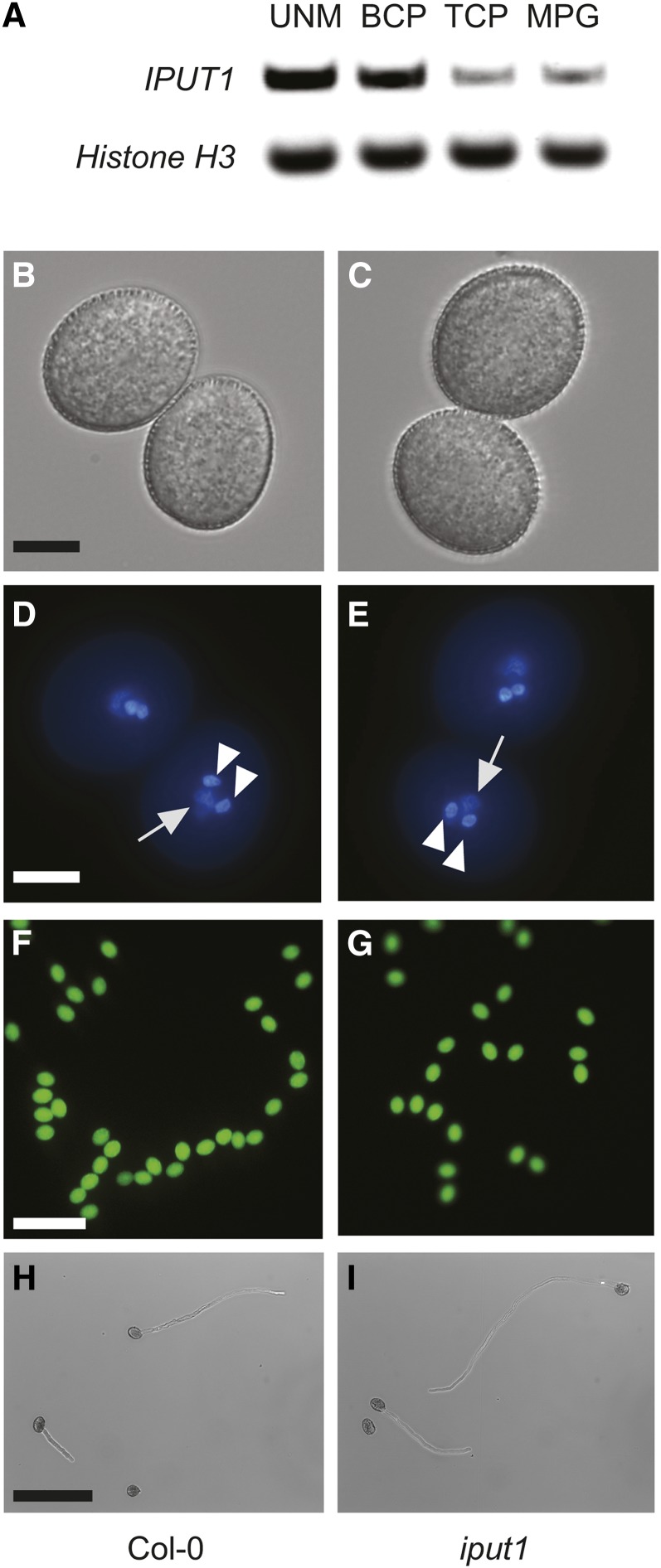
We tested expression of IPUT1 by quantitative PCR and found that it is expressed throughout pollen development (Figure 5A). These results indicate that IPUT1 could be required for normal pollen development. However, examination of pollen from both iput1-1/+ and iput1-2/+ plants showed no difference from wild-type pollen grains when viewed under a light microscope (Figures 5B and 5C), indicating that there are no gross morphological pollen defects. Staining with 4′,6-diamidino-2-phenylindole (DAPI) (Figures 5D and 5E) or fluorescein diacetate (Figures 5F and 5G) further indicated that pollen nuclei develop normally and that pollen grains are viable. Pollen germination rates were only slightly lower in both iput1-1/+and iput1-2/+ than in wild-type plants, indicating that iput1 pollen is able to germinate (Table 2), and iput1 pollen tubes appeared to grow normally (Figures 5H and 5I). Tube growth rates of pollen from iput1-1/+ and iput1-2/+ plants, based on measurement of tube length 6-8 h after germination, were only slightly lower than those of pollen from wild-type plants (Supplemental Figure 5). Thus, the defect in transmission through the male gametophyte is not due to improper pollen development or inability of pollen tubes to germinate and grow.
Figure 5.
iput1 Pollen Develops Normally.
(A) RT-PCR of IPUT1 and Histone H3 (control) in the uninucleate microspore (UNM), bicellular pollen (BCP), tricellular pollen (TCP), and mature pollen grains (MPG). RT-PCRs used pooled spores from two biological replicates.
(B), (D), (F), and (H) Pollen from wild-type plants.
(C), (E), (G), and (I) Pollen from iput1-1/+ plants. Phenotype of wild type and iput1 pollen is indistinguishable.
Light microscopy of mature pollen grains ([A] and [B]); fluorescence microscopy after DAPI staining ([D] and [E]) and fluorescein diacetate staining ([F] and [G]). Arrows indicate the diffusely stained vegetative nucleus; arrowheads indicate the densely stained sperm cell nuclei. Light microscopy of germinated pollen tubes, 6 h after germination ([H] and [I]). Bars = 10 μm in (B) to (F) and 100 μm in (G) to (I).
Table 2. Germination Rate of Pollen from iput1/+ Heterozygotes.
| Parent Genotype | Pollen Germination Rate |
|---|---|
| Col-0 | 74.6% (2372) |
| iput1-1/+ | 71.5% (3078) |
| iput1-2/+ | 70.0% (3427) |
Pollen germination rates of iput1 heterozygotes. Total numbers of pollen grains scored are indicated in parentheses.
Quantitative PCR indicated that IPUT1 is expressed in all Arabidopsis tissues and at all stages tested (Supplemental Figure 5). This ubiquitous expression is similar to that of other sphingolipid biosynthetic proteins (Chen et al., 2006; Tsegaye et al., 2007) and is expected for a protein required for synthesis of a major lipid class.
DISCUSSION
In this study, we provide evidence showing that IPUT1 is an inositol phosphorylceramide glucuronosyltransferase. The expression of IPUT1 in yeast led to the production of GlcA-IPC, indicating that IPUT1 is capable of transferring a GlcA residue from UDP-GlcA onto IPC in yeast. In addition, assays using microsomes from N. benthamiana plants transiently overexpressing IPUT1 confirmed that IPUT1 is an active IPC:glucuronosyltransferase in plants. Plants in which IPUT1 homologs were silenced showed a decrease in IPC:glucuronosyltransferase activity.
Plants produce several different ceramide species with variations in long-chain sphingobase and fatty acid structure. The makeup of ceramides differs between sphingolipid pools such as free ceramides, ceramide phosphates, glucosylceramides, and GIPCs (Markham et al., 2006). We therefore undertook a sphingolipidomic approach to determine how changes in IPUT1 expression affected various sphingolipid pools. Altering IPUT1 expression in N. benthamiana led to changes in sphingolipids including ceramides, glucosylceramides, IPC, and the glycosylated sphingolipids GlcN-GlcA-IPC and GlcNAc-GlcA-IPC (Figure 4). Most notably, silencing the N. benthamiana homologs of IPUT1 caused a nearly 30-fold increase in levels of IPC, the substrate for IPUT1. Given that production of IPC by IPCS is the first committed step in GIPC synthesis, it is not surprising that this large increase in IPC levels was accompanied by changes in other sphingolipid pools. Ceramide levels were also significantly increased in IPUT1-silenced plants, probably because the accumulation of IPC prevented ceramide from being utilized by IPCS. More detailed analysis of the ceramide pool supports the notion that the major ceramide structure in the IPCs and GIPCs we examined was t18:0/h24:0, and this ceramide was preferentially enriched in IPUT1-silenced plants (Supplemental Figure 3). When the flux toward GIPCs was decreased, ceramide precursors may instead have been redirected toward glucosylceramide synthesis. The small increase in glucosylceramide levels in IPUT1-silenced plants is consistent with this scenario. Conversely, glucosylceramide levels decreased in IPUT1-overexpressing plants, indicating that sphingolipids that may have otherwise been used in glucosylceramide synthesis were instead used to synthesize GIPCs.
Due to the lack of available acceptors, we were not able to investigate the in vitro acceptor specificity of IPUT1 in detail; however, expression of IPUT1 in yeast caused glucuronosylation of IPCs containing at least two different ceramide species (Figure 2B), indicating that IPUT1 is capable of somewhat relaxed acceptor specificity at least with respect to ceramide fatty acid hydroxylation and length. Further investigation of IPUT1 specificity, as well as of ceramide specificities for the three IPCS orthologs that catalyze the committed step of GIPC synthesis (Mina et al., 2010), may help clarify how GIPC synthesis is controlled.
IPUT1 is essential in Arabidopsis. We identified two allelic mutants in IPUT1. iput1 alleles were transmitted through pollen at 1 to 2% of wild-type levels, indicating a role for IPUT1 in pollen function; however, iput1 pollen and pollen tubes appeared to develop normally (Figure 5). Although germination and tube growth rates were decreased slightly in pollen from iput1-1/+ and iput1-2/+ plants (Table 2; Supplemental Figure 4), the decreases were not large enough to indicate that iput1 pollen are unable to germinate or grow properly or to explain why only 1 to 2% of iput1 alleles were transmitted through pollen. These results indicate that the major defect in iput1 pollen is manifested during tube guidance or ovule fertilization. Several scenarios could explain how GIPCs are involved in these processes. Many plasma membrane-localized transporters and signaling components are required for tube guidance and fertilization and are located only at the pollen tube tips, indicating that precise spatial organization of the membrane is necessary for these processes (Cheung and Wu, 2008). Lipid rafts have been suggested to play a role in positioning membrane proteins during pollen tube growth (Lalanne et al., 2004; Liu et al., 2009), and there is some evidence that GIPCs are enriched in lipid rafts (Borner et al., 2005). Lipid rafts may also be directly involved in signal transduction, for example, by organizing proteins such as receptors in microenvironments that allow specific receptor proteins to interact (Simons and Toomre, 2000) or by regulating receptors allosterically (Coskun et al., 2011). It is possible that membrane composition influences how pollen tubes perceive signaling molecules produced by maternal tissues. Further work with IPUT1 may help to determine more precisely how GIPCs are involved in pollen tube guidance and fertilization.
IPUT1 function is not limited to pollen. We found that IPUT1 is expressed at all developmental stages tested (Figure 5). IPUT1 has also been found in Golgi proteomes from several studies using tissue cultures, indicating that it is expressed under cell culture conditions (Dunkley et al., 2004; Parsons et al., 2012). This widespread expression is not unexpected, since recent reports have indicated that GIPCs are a major component of plant plasma membranes (Sperling et al., 2005; Markham et al., 2006). Sphingolipids are also concentrated in the outer leaflet of the membrane, indicating that they are subject to additional levels of organization (Markham et al., 2013). It is becoming increasingly evident that GIPCs are quantitatively important and probably have functions in the secretory system and plasma membrane that have not yet been determined.
GIPCs have also recently been implicated in salicylic acid-dependent signaling and the hypersensitive response during defense against pathogens. inositol phosphorylceramide synthase1 (ipcs1) mutants expressing the resistance gene RPW8 have elevated salicylic acid levels and form hypersensitive lesions in the absence of pathogens (Wang et al., 2008). This mutant is defective in one of the three IPCS genes in Arabidopsis and accumulates ceramide, leading to the suggestion that this precursor activates a signaling network that leads to the formation of hypersensitive lesions. Sphingolipid signaling is well characterized in animals, where ceramides are bioactive lipids that can induce apoptosis (Hannun and Obeid, 2008). Similarly, golgi-localized nucleotide sugar transporter1 (gonst1) mutants are defective in GIPC mannosylation and show comparable spontaneous hypersensitive lesions and increased salicylic acid (Mortimer et al., 2013). However, gonst1 mutants did not have increased ceramide levels, indicating that GIPC mannosylation itself, rather than levels of GIPC precursors, is necessary for regulating the hypersensitive response (Mortimer et al., 2013). The mechanisms by which sphingolipids participate in the hypersensitive response are thus very unclear. Future work with iput1 mutants may provide an opportunity to dissect how GIPC glycosylation contributes to changes in various sphingolipid pools and how glycan structures are involved in response to pathogens.
METHODS
Phylogenetic Analysis
Protein sequences were obtained from The Arabidopsis Information Resource (www.arabidopsis.org) or the Sol Genomics Network (http://solgenomics.net/) and were aligned using MUSCLE 3.7 (Edgar, 2004). Phylogenetic trees were built using PhyML 3.0 (Guindon et al., 2010) and viewed using FigTree version 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/). Bootstrap values shown at nodes were calculated using 1000 replicates.
Cloning and Construction of Expression Vectors
Sequences of all primers used in this study can be found in Supplemental Table 1. Arabidopsis thaliana clones were amplified using mixed organ and developmental cDNA libraries as template. The human cDNA hUGTrel7 was obtained from the CCSB Human Orfeome Collection (Thermo Scientific). Entry clones were constructed using Gateway technology (Invitrogen) via BP reaction in pDONR223. The reverse primers contained no stop codon to enable C-terminal fusions. Gene-specific primers were used to amplify cDNA, and PCR products were then reamplified with the Gateway attB-specific primers before the BP recombination reaction. All entry clones were verified by restriction analysis and sequencing. Gateway expression vectors were constructed via LR reaction. The binary vector pEarleyGate101, which contains a 35S promoter and a C-terminal YFP-HA tag, was used for expression in Nicotiana benthamiana (Earley et al., 2006). Yeast expression vectors used were a Gateway-enabled pRS423 with a HIS3 marker (Sikorski and Hieter, 1989) for hUGTrel7 and pDRf1-GW with URA3 or LEU2 markers (Loqué et al., 2007) for UGD2 and IPUT1 cDNA. The sur1Δ knockout yeast strain (MATα Strain YPL057C) was obtained from the Yeast Knockout Collection (Thermo Scientific). For N. benthamiana clones, cDNA from leaves was used as template. The binary vectors pTRV RNA1 and pTRV RNA2 were used for VIGS (Liu et al., 2002). A 400-bp cDNA fragment with sequence similarity to all three IPUT1 homologs was amplified and ligated into the multiple cloning site of pTRV2 using EcoRI and XbaI restriction sites.
Electrospray Ionization Mass Spectrometry
Yeast were grown in appropriate media, pelleted at 2000g, and lipids were extracted three times in 5 volumes of 1-butanol. The extracts were combined, partitioned once against 0.5 volume water to remove salts and proteins, and dried. Dried lipids were resuspended in chloroform/methanol/[4 M ammonium hydroxide plus 1.8 M ammonium acetate] (9:7:2 v/v/v) prior to analysis by mass spectrometry. Samples were analyzed by infusion at 20 μL/min using a Q TRAP LC/MS/MS system (AB SCIEX) equipped with a TurboIonSpray ion source. The QTRAP system was operated in negative ion mode using the enhanced MS (EMS) for MS and enhanced product ion (EPI) for tandem mass spectrometry and a scan rate of 1000 (D/s). The number of scans to sum was set to 2, the scan mode was set to profile, and a dynamic fill time was selected. In EMS mode, the system was set to scan from 800 to 1300 m/z for 0.5 s, whereas for EPI a range of 50 to 1200 m/z was employed and a collision energy of −80 eV was used. The ion spray voltage was set at −4200 V, source temperature (TEM) at 350°C and ion source gas GS1 was 20 and GS2 was set to 7. All data were collected and analyzed using Analyst 1.5.2 (AB SCIEX).
TLC
Pelleted yeast were extracted in chloroform/methanol/[4 M ammonium hydroxide plus 1.8 M ammonium acetate] (9:7:2 v/v/v) at 40°C and centrifuged to separate the organic and aqueous phases. The aqueous phase was cooled to −20°C and then centrifuged again. The bottom phase was collected, dried, spotted onto silica gel TLC plates, and developed in the same solvent used for extraction.
Plant Material and Transient Expression
Three- to four-week-old N. benthamiana ‘Domin’ plants were infiltrated with Agrobacterium tumefaciens. Agrobacterium strain C58-1 pGV3850 carrying the appropriate vectors was grown to log phase, pelleted at 3500g, and resuspended in infiltration medium consisting of 10 mM MES-KOH, pH 5.6, 10 mM MgCl2, and 200 μM acetosyringone before being infiltrated into the abaxial surfaces of leaves. Agrobacterium carrying the vector with the gene of interest was mixed 1:1 with a strain carrying a plasmid with the p19 gene from Tomato bushy stunt virus (Voinnet et al., 2003). The final optical density at 600 nm (OD600) for each strain was 1.0. As much of each leaf as possible (∼95%) was infiltrated. The expression of genes fused to YFP was verified by monitoring YFP fluorescence with a Leica D4000B epifluorescence microscope and all cells in infiltrated areas were shown to express protein. Entire leaves were harvested 4 d after infiltration.
VIGS
VIGS was performed essentially as described (Burch-Smith et al., 2004). Two-week-old plants were used for VIGS experiments. Agrobacterium strains carrying the IPUT1-TRV RNA2 or GUS-TRV RNA2 vectors were resuspended in infiltration medium as above and mixed 1:1 with Agrobacterium carrying the TRV RNA1 vector for a final OD600 of ∼0.4 for each strain. Cultures were infiltrated into the abaxial surfaces of leaves. Silenced leaves were harvested 10 d after infiltration. Spread of silencing was monitored by infiltrating neighboring plants with a construct designed to silence phytoene desaturase, causing photobleaching.
Microsome Preparation
All microsome preparation steps took place at 4°C. Leaf tissue was ground in buffer containing 50 mM HEPES-KOH, pH 7.0, 400 mM sucrose, 1 mM phenylmethanesulfonyl fluoride, 1% (w/v) polyvinylpolypyrrolidone, and protease inhibitors (Roche Complete protease inhibitor tablets used as instructed by the manufacturer). The homogenate was filtered through two layers of Miracloth (EMD Millipore) and centrifuged at 3000g for 10 min, and the supernatant was then centrifuged at 50,000g for 1 h. The pellet was resuspended in 50 mM HEPES-KOH, pH 7.0, and 400 mM sucrose. Microsomes were used immediately or frozen in liquid nitrogen and stored at −80°C. No significant loss of activity was detected after freezing.
IPC Glucuronosyltransferase Assay
Assay reactions included 50 mM HEPES-KOH, pH 7.0, 5 mM MnCl2, 1 mM DTT, 400 mM sucrose, 100 μg N. benthamiana microsomal proteins, and 4.4 μM UDP-[14C]GlcA (1480 Bq; MP Biomedicals) in a total volume of 50 μL. Reactions were mixed and incubated at 21°C for 1 h, and the reaction was then stopped by the addition of 300 μL 1-butanol. The butanolic phase was partitioned against 300 μL water, and then an additional 300 μL 1-butanol was added and the partition was repeated. The butanolic phases were combined, dried, spotted onto silica gel TLC plates (Sigma-Aldrich), and developed in chloroform/methanol/[4 M ammonium hydroxide plus 1.8 M ammonium acetate] (9:7:2 v/v/v). Plates were exposed to phosphor screens for 2 to 5 d before imaging with a Typhoon 8600 Variable Mode phosphor imager. Band intensities were quantified using ImageJ software following instructions given in the user guide (http://imagej.nih.gov/ij/index.html) (Schneider et al., 2012). Mild alkaline hydrolysis was performed by incubating samples in methanolic 0.1 M potassium hydroxide for 1 h at 21°C, neutralizing with acetic acid, and drying the samples. After this treatment, samples were resuspended in butanol and partitioned against water to remove salts.
Detection of Proteins by Immunoblot
Yeast proteins were extracted from yeast as described (Kushnirov, 2000). N. benthamiana microsomal proteins were washed twice with 80% acetone to remove residual membranes before analysis. Proteins were resolved by SDS-PAGE on 7% to 15% gradient gels and blotted onto nitrocellulose membranes (GE Healthcare). Yeast protein blots were probed with a 1:10,000 dilution of an antibody raised in rabbit against the attB2 Gateway site (Invitrogen) (Eudes et al., 2011) followed by a 1:20,000 dilution of goat anti-rabbit IgG conjugated to horseradish peroxidase (Sigma-Aldrich). N. benthamiana protein blots were probed with a 1:1000 dilution of rat monoclonal anti-HA antibody (Sigma-Aldrich) followed by a 1:15,000 dilution of goat anti-rat IgG conjugated to horseradish peroxidase (Sigma-Aldrich). Chemiluminescence was detected using ECL Plus detection reagent (GE Healthcare). Blots were imaged using a ChemiDoc-It 600 Imaging System (UVP).
Expression Analysis
For quantitative PCR, RNA was extracted from Arabidopsis or N. benthamiana tissue using an RNeasy Plant Mini Kit (Qiagen). One microgram of RNA was used as template in a reverse transcriptase reaction using an iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was done with Absolute SYBR Green ROX Mix (ABgene) on a StepOnePlus Real-time PCR system (Applied Biosystems) according to conditions described earlier (Czechowski et al., 2005) using StepOne 2.0 software (Applied Biosystems). Primers for Arabidopsis quantitative PCR reference genes were taken from (Valdivia et al., 2013). Quantitative PCR data were quantified using the comparative Ct method (Schmittgen and Livak, 2008) using geometric averaging of the three reference genes as the common reference (Vandesompele et al., 2002). For pollen expression analysis, RNA was extracted from pollen at different stages of development as described (Honys and Twell, 2004). Samples of 750 ng of total RNA for pollen stages were reverse transcribed in a 20 μL reaction using Superscript II RNase H reverse transcriptase (Invitrogen) and an oligo(dT) primer as per the manufacturer’s instructions. For PCR amplification, 1 μL of a 103 diluted cDNA was used in a 25-μL reaction using Biotaq DNA polymerase (Bioline) and 12.5 pmol of each primer. PCR conditions were as follows: 96°C for 1 min, 30 cycles of 96°C for 30 s, 55°C for 30 s, and 72°C for 40 s followed by 5 min at 72°C. Histone H3 (At4g40040) was used as a control as described (Brownfield et al., 2009).
Sphingolipidomics
N. benthamiana leaf tissue was lyophilized and powderized using a mortar and pestle. Sphingolipids were extracted from 30 mg of the powderized tissue using isopropanol/hexane/water (55:20:25 v/v/v) followed by 33% methylamine treatment as previously described (Markham and Jaworski, 2007) and resuspended in tetrahydrofuran/methanol/water (2:1:2 v/v/v) containing 0.1% formic acid. Sphingolipids were analyzed using a Shimadzu Prominence UPLC coupled with a QTRAP4000 mass spectrometer (AB SCIEX) as described (Kimberlin et al., 2013). Instrument potentials and chromatography conditions for the initial detection of N-acetyl-sugar-containing GIPCs were as for hexose(R1:OH)-glucuronic acid-inositol-phosphoceramides (GlcOH-GlcA-IPCs) described previously (Kimberlin et al., 2013). Multiple reaction monitoring (MRM) Q1/Q3 transitions to detect N-acetyl-sugar-containing GIPCs (GlcNAc-GlcA-IPCS) were calculated by adding 41 mass units to the Q1 ion of previously described GlcOH-GlcA-IPC MRM Q1/Q3 transitions (Markham and Jaworski, 2007). GlcN-GlcA-IPC MRM Q1/Q3 transitions were calculated by subtracting one mass unit from the Q1 ion of GlcOH-GlcA-IPC species and are distinguished from GlcOH-GlcA-IPCs by longer elution times. The retention time and M+H mass of the IPCs were first confirmed by precursor ion scanning for the t18:1/h24:0 backbone (precursors of m/z 664.6) combined with chromatographic separation of GIPCs as described (Kimberlin et al., 2013). Based on this information, MRM Q1/Q3 transitions for IPC detection were calculated based on known structures and fragmentation patterns and are given in Supplemental Table 2.
Arabidopsis Mutant Isolation and Transmission
To determine transmission efficiency (TE), seeds (iput1-2) were sown on solid medium (0.8% [w/v] agar) containing half-strength Murashige and Skoog (Murashige and Skoog, 1962) salts including vitamins (Sigma-Aldrich) and sucrose (1% (w/v) with Sulfadiazine (4-amino-N-2-pyrimidinylbenzene-sulfonamide; Sigma-Aldrich) at 5 mg/L. Seeds (iput1-1) were sown in soil (John Innes No. 1) in a growth room (20°C, 100 μmol s−1 m−2, 16-h-light/8-h-dark, 60% humidity). Three-week-old plants were sprayed with BASTA at a concentration of 75 mg/L. TE through male and female gametophytes was determined by crossing iput1/+ heterozygotes with wild-type plants and scoring resistance of progeny to the appropriate selection as described (Howden et al., 1998). TE was calculated as (number of resistant seedlings/number of susceptible seedlings) × 100.
Microscopy Analyses
Analysis of nuclear DNA in mature pollen with DAPI was performed as previously described (Park et al., 1998). Fluorescein diacetate staining to monitor pollen viability was performed by incubating freshly isolated pollen in 0.3 M mannitol solution containing 2 μg mL fluorescein diacetate as described (Goubet et al., 2003).
Pollen Germination
Pollen were collected and germinated as described (Boavida and McCormick, 2007). Liquid germination media consisted of 0.01% boric acid, 5 mM CaCl2, 5 mM KCl, 1 mM MgSO4, and 10% sucrose at pH 7.5. Pollen was allowed to germinate at 22°C for 6 to 8 h, and then images were taken using a Leica D4000B epifluorescence microscope coupled with a Leica DC500 camera.
Accession Numbers
Sequence data from this article can be found at The Arabidopsis Information Resource under accession number At5g18480 (IPUT1). The accession numbers of the Arabidopsis mutants used in this study are iput1-1 (SAIL.532.E01-N822575) and iput1-2 (GABI-KAT 856G03).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Production of GlcA-IPC in sur1Δ Yeast Expressing UGD2, hUGTrel7, and IPUT1.
Supplemental Figure 2. Homologs of GUX, PGSIP, and IPUT1 Proteins in N. benthamiana.
Supplemental Figure 3. Diagram of Insertions in iput1-1 and iput1-2 Alleles.
Supplemental Figure 4. Tube Growth Rate of Pollen from iput1-1/+ and iput1-2/+ Arabidopsis Plants.
Supplemental Figure 5. Expression of IPUT1 in Different Arabidopsis Tissues.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental Table 2. Parameters for MRM Detection of IPCs in Positive Ion Mode.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny Presented in Figure 1.
Supplemental Data Set 2. Alignments Used to Generate the Phylogeny Presented in Supplemental Figure 2.
Supplemental Data Set 3. Summary of Sphingolipidomic Data on IPUT1-Overexpressing and IPUT1-Silencing N. benthamiana Plants.
Supplementary Material
Acknowledgments
We thank Sherry Chan for technical assistance and Tsan-Yiu Chiu for providing vectors. This work was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through Contract DE-AC02-05CH11231 between Lawrence Berkeley National Lab and the U.S. Department of Energy and by National Science Foundation Graduate Research Fellowship Program Grant DGE 1106400 (E.A.R.) and National Science Foundation Grant MCB 115850 (E.B.C.).
AUTHOR CONTRIBUTIONS
E.A.R., B.E., G.P.M., S.S., K.M.C., H.K., D.T., C.J.P., P.D.A., P.D., J.L.H., and H.V.S. designed research. E.A.R., B.E., G.P.M., R.E.C., S.S., K.M.C., E.B.C., and H.K. performed research. E.A.R., B.E., G.P.M., R.E.C., E.B.C., and H.V.S. analyzed data. E.A.R. and H.V.S. wrote the article.
Glossary
- GIPC
glycosyl inositol phosphorylceramide
- LCB
long-chain sphingobase
- IPC
inositol phosphorylceramide
- IPCS
inositol phosphorylceramide synthase
- TLC
thin-layer chromatography
- VIGS
virus-induced gene silencing
- GUS
β-glucuronidase
- DAPI
4′,6-diamidino-2-phenylindole
- MRM
multiple reaction monitoring
- TE
transmission efficiency
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Beeler T.J., Fu D., Rivera J., Monaghan E., Gable K., Dunn T.M. (1997). SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37 ° C, is required for mannosylation of inositolphosphorylceramide. Mol. Gen. Genet. 255: 570–579. [DOI] [PubMed] [Google Scholar]
- Boavida L.C., McCormick S. (2007). Technical advance: temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582. [DOI] [PubMed] [Google Scholar]
- Borner G.H.H., Sherrier D.J., Weimar T., Michaelson L.V., Hawkins N.D., Macaskill A., Napier J.A., Beale M.H., Lilley K.S., Dupree P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137: 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley P.E., Li Y.O., Murphy S.M., Sumner C.M., Lynch D.V. (2003). Complex sphingolipid synthesis in plants: characterization of inositolphosphorylceramide synthase activity in bean microsomes. Arch. Biochem. Biophys. 417: 219–226. [DOI] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Borg M., Sidorova A., Mori T., Twell D. (2009). A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 5: e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Anderson J.C., Martin G.B., Dinesh-Kumar S.P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39: 734–746. [DOI] [PubMed] [Google Scholar]
- Buré C., Cacas J.-L., Wang F., Gaudin K., Domergue F., Mongrand S., Schmitter J.-M. (2011). Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 25: 3131–3145. [DOI] [PubMed] [Google Scholar]
- Cacas J.-L., Buré C., Furt F., Maalouf J.-P., Badoc A., Cluzet S., Schmitter J.-M., Antajan E., Mongrand S. (2013). Biochemical survey of the polar head of plant glycosylinositolphosphoceramides unravels broad diversity. Phytochemistry 96: 191–200. [DOI] [PubMed] [Google Scholar]
- Carter H.E., Strobach D.R., Hawthorne J.N. (1969). Biochemistry of the sphingolipids. 18. Complete structure of tetrasaccharide phytoglycolipid. Biochemistry 8: 383–388. [DOI] [PubMed] [Google Scholar]
- Chao D.-Y., et al. (2011). Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell 23: 1061–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M., Berbezy P., Vyas D., Coates S., Barsby T. (2005). Reduced expression of a protein homologous to glycogenin leads to reduction of starch content in Arabidopsis leaves. Plant Sci. 168: 501–509. [Google Scholar]
- Chen M., Markham J.E., Cahoon E.B. (2012). Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 69: 769–781. [DOI] [PubMed] [Google Scholar]
- Chen M., Han G., Dietrich C.R., Dunn T.M., Cahoon E.B. (2006). The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18: 3576–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59: 547–572. [DOI] [PubMed] [Google Scholar]
- Coskun Ü., Grzybek M., Drechsel D., Simons K. (2011). Regulation of human EGF receptor by lipids. Proc. Natl. Acad. Sci. USA 108: 9044–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursol S., Le Stunff H., Lynch D.V., Gilroy S., Assmann S.M., Spiegel S. (2005). Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol. 137: 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C.R., Han G., Chen M., Berg R.H., Dunn T.M., Cahoon E.B. (2008). Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 54: 284–298. [DOI] [PubMed] [Google Scholar]
- Dunkley T.P.J., Watson R., Griffin J.L., Dupree P., Lilley K.S. (2004). Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell. Proteomics 3: 1128–1134. [DOI] [PubMed] [Google Scholar]
- Dunn T.M., Haak D., Monaghan E., Beeler T.J. (1998). Synthesis of monohydroxylated inositolphosphorylceramide (IPC-C) in Saccharomyces cerevisiae requires Scs7p, a protein with both a cytochrome b5-like domain and a hydroxylase/desaturase domain. Yeast 14: 311–321. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A., Baidoo E.E., Yang F., Burd H., Hadi M.Z., Collins F.W., Keasling J.D., Loqué D. (2011). Production of tranilast [N-(3′,4′-dimethoxycinnamoyl)-anthranilic acid] and its analogs in yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 89: 989–1000. [DOI] [PubMed] [Google Scholar]
- Goubet F., Misrahi A., Park S.K., Zhang Z., Twell D., Dupree P. (2003). AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 131: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X.L., Wenk M.R. (2006). Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast 23: 465–477. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hannun Y.A., Obeid L.M. (2008). Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- Hillig I., Leipelt M., Ott C., Zähringer U., Warnecke D., Heinz E. (2003). Formation of glucosylceramide and sterol glucoside by a UDP-glucose-dependent glucosylceramide synthase from cotton expressed in Pichia pastoris. FEBS Lett. 553: 365–369. [DOI] [PubMed] [Google Scholar]
- Honys D., Twell D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul K., Lester R.L. (1975). Characterization of inositol-containing phosphosphingolipids from tobacco leaves: isolation and identification of two novel, major lipids: N-acetylglucosamidoglucuronidoinositol phosphorylceramide and glucosamidoglucuronidoinositol phosphorylceramide. Plant Physiol. 55: 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul K., Lester R.L. (1978). Isolation of six novel phosphoinositol-containing sphingolipids from tobacco leaves. Biochemistry 17: 3569–3575. [DOI] [PubMed] [Google Scholar]
- Kimberlin A.N., Majumder S., Han G., Chen M., Cahoon R.E., Stone J.M., Dunn T.M., Cahoon E.B. (2013). Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell 25: 4627–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghammer M., Tenhaken R. (2007). Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J. Exp. Bot. 58: 3609–3621. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16: 857–860. [DOI] [PubMed] [Google Scholar]
- Lalanne E., Honys D., Johnson A., Borner G.H.H., Lilley K.S., Dupree P., Grossniklaus U., Twell D. (2004). SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J.T., Luo S., Lu H., Greenberg J.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17: 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li R.-L., Zhang L., Wang Q.-L., Niehaus K., Baluška F., Šamaj J., Lin J.-X. (2009). Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J. 60: 303–313. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30: 415–429. [DOI] [PubMed] [Google Scholar]
- Loqué D., Lalonde S., Looger L.L., von Wiren N., Frommer W. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446: 195–198. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Jaworski J.G. (2007). Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21: 1304–1314. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Li J., Cahoon E.B., Jaworski J.G. (2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281: 22684–22694. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Lynch D.V., Napier J.A., Dunn T.M., Cahoon E.B. (2013). Plant sphingolipids: function follows form. Curr. Opin. Plant Biol. 16: 350–357. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Molino D., Gissot L., Bellec Y., Hématy K., Marion J., Belcram K., Palauqui J.-C., Satiat-Jeunemaître B., Faure J.D. (2011). Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23: 2362–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer J.C., Miles G.P., Brown D.M., Zhang Z., Segura M.P., Weimar T., Yu X., Seffen K.A., Stephens E., Turner S.R., Dupree P. (2010). Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc. Natl. Acad. Sci. USA 107: 17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina J.G., Okada Y., Wansadhipathi-Kannangara N.K., Pratt S., Shams-Eldin H., Schwarz R.T., Steel P.G., Fawcett T., Denny P.W. (2010). Functional analyses of differentially expressed isoforms of the Arabidopsis inositol phosphorylceramide synthase. Plant Mol. Biol. 73: 399–407. [DOI] [PubMed] [Google Scholar]
- Mortimer J.C., et al. (2013). Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 25: 1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka M., Kawakita M., Ishida N. (2001). Molecular characterization of human UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, a novel nucleotide sugar transporter with dual substrate specificity. FEBS Lett. 495: 87–93. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Obeid L.M., Okamoto Y., Mao C. (2002). Yeast sphingolipids: metabolism and biology. Biochim. Biophys. Acta 1585: 163–171. [DOI] [PubMed] [Google Scholar]
- Oikawa A., Joshi H.J., Rennie E.A., Ebert B., Manisseri C., Heazlewood J.L., Scheller H.V. (2010). An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS ONE 5: e15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.K., Howden R., Twell D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799. [DOI] [PubMed] [Google Scholar]
- Parsons H.T., et al. (2012). Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 159: 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotto S., Donovan N., Drobak B.K., Brewin N.J. (1995). Differential expression of a glycosyl inositol phospholipid antigen on the peribacteroid membrane during pea nodule development. Mol. Plant Microbe Interact. 8: 560–568. [Google Scholar]
- Pinto W.J., Srinivasan B., Shepherd S., Schmidt A., Dickson R.C., Lester R.L. (1992). Sphingolipid long-chain-base auxotrophs of Saccharomyces cerevisiae: genetics, physiology, and a method for their selection. J. Bacteriol. 174: 2565–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie E.A., Hansen S.F., Baidoo E.E.K., Hadi M.Z., Keasling J.D., Scheller H.V. (2012). Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol. 159: 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Toomre D. (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1: 31–39. [DOI] [PubMed] [Google Scholar]
- Sperling P., Heinz E. (2003). Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim. Biophys. Acta 1632: 1–15. [DOI] [PubMed] [Google Scholar]
- Sperling P., Franke S., Lüthje S., Heinz E. (2005). Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 43: 1031–1038. [DOI] [PubMed] [Google Scholar]
- Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29: 417–426. [DOI] [PubMed] [Google Scholar]
- Ternes P., Feussner K., Werner S., Lerche J., Iven T., Heilmann I., Riezman H., Feussner I. (2011). Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol. 192: 841–854. [DOI] [PubMed] [Google Scholar]
- Tsegaye Y., Richardson C.G., Bravo J.E., Mulcahy B.J., Lynch D.V., Markham J.E., Jaworski J.G., Chen M., Cahoon E.B., Dunn T.M. (2007). Arabidopsis mutants lacking long chain base phosphate lyase are fumonisin-sensitive and accumulate trihydroxy-18:1 long chain base phosphate. J. Biol. Chem. 282: 28195–28206. [DOI] [PubMed] [Google Scholar]
- Valdivia E.R., Herrera M.T., Gianzo C., Fidalgo J., Revilla G., Zarra I., Sampedro J. (2013). Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J. Exp. Bot. 64: 1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: H0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wang W., et al. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20: 3163–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zäuner, S., Ternes, P., and Warnecke, D. (2010). Biosynthesis of sphingolipids in plants (and some of their functions). In Sphingolipids as Signaling and Regulatory Molecules, C. Chalfant and M. Poeta, eds (New York: Springer), pp. 249–263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.