The connection between the MAP kinase cascade and light signaling is unknown. This work shows that the MKK3-MPK6 is activated by blue light in a MYC2-dependent manner during Arabidopsis seedling development. MPK6 physically interacts and phosphorylates MYC2, which in turn binds to the promoter of MPK6 and regulates its expression in a feedback mechanism under blue light signaling.
Abstract
Mitogen-activated protein kinase (MAPK) pathways are involved in several signal transduction processes in eukaryotes. Light signal transduction pathways have been extensively studied in plants; however, the connection between MAPK and light signaling pathways is currently unknown. Here, we show that MKK3-MPK6 is activated by blue light in a MYC2-dependent manner. MPK6 physically interacts with and phosphorylates a basic helix-loop-helix transcription factor, MYC2, and is phosphorylated by a MAPK kinase, MKK3. Furthermore, MYC2 binds to the MPK6 promoter and regulates its expression in a feedback regulatory mechanism in blue light signaling. We present mutational and physiological studies that illustrate the function of the MKK3-MPK6-MYC2 module in Arabidopsis thaliana seedling development and provide a revised mechanistic view of photomorphogenesis.
INTRODUCTION
Plants have evolved an intricate molecular network consisting of light sensing photoreceptors, downstream signaling components, and protein degradation machinery to sense the dark-light transitions (Jiao et al., 2007). This network regulates Arabidopsis thaliana seedling growth with two contrasting developmental patterns in the presence and absence of light (Nagy and Schäfer, 2002; Chen and Chory, 2011; Li et al., 2011). Dark-grown seedlings, which have elongated hypocotyls and closed cotyledons with apical hooks, undergo skotomorphogenic growth. Upon exposure to light, plants switch to photomorphogenic growth, during which hypocotyl growth slows down, cotyledons expand, and chlorophyll biosynthesis takes place. Transcriptional regulatory networks play a key role in mediating light signaling through the coordinated activation and repression of a large number of downstream genes (Ma et al., 2001).
The activities of several plant transcription factors that function in light-regulated gene expression are modulated by phosphorylation and dephosphorylation. The phosphorylation of the Arabidopsis G-BOX BINIDING FACTOR1 (GBF1), a bZIP transcription factor by CKII from broccoli (Brassica oleracea), increases its affinity for the G-Box (Klimczak et al., 1992). A well-conserved putative CKII phosphorylation site was also found to be located at the N terminus of another bZIP protein, ELONGATED HYPOCOTYL5 (HY5) (Oyama et al., 1997), indicating that phosphorylation may affect HY5 abundance and activity. Subsequent reports show that HY5 is phosphorylated by a light-regulated kinase in its CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) binding domain, probably by CKII (Hardtke et al., 2000). The phosphorylated and unphosphorylated forms differ in their activity, affinity for target promoters, and degradation. Similarly, the protein abundance of LONG HYPOCOTYL IN FAR-RED1 (HFR1), a basic helix-loop-helix (bHLH) transcription factor, is tightly controlled by phosphorylation and light (Duek et al., 2004; Park et al., 2008). CKII is also known to phosphorylate PHYTOCROME INTERACTING FACTOR1 (PIF1), a negative regulator of photomorphogenesis, which in turn enhances the light-induced degradation of PIF1 to promote photomorphogenesis (Bu et al., 2011).
The mitogen-activated protein kinase (MAPK) cascade is considered to be one of the major signaling systems in eukaryotes (Rodriguez et al., 2010). In plants, the MAPK cascade is involved in various biotic and abiotic stress responses, hormone responses, cell division, and developmental processes (Colcombet and Hirt, 2008; Sinha et al., 2011). MPK3 and MPK6 of group A and MPK4 of group B are the most extensively studied MAPKs in Arabidopsis (Fiil et al., 2009; Andreason and Ellis, 2010). Although the physiological functions of MAPKs in response to external stimuli have been extensively characterized, only a few substrates have been identified. The phosphorylation of ACS6 (1-aminocyclopropane-1-carboxylic acid synthase-6) occurs via three phosphorylation sites by MPK6 and precedes the accumulation of ACS6 and ethylene production (Joo et al., 2008). Dual phosphorylation by the MKK9-MPK3/6 cascade was observed to modulate ETHYLENE INSENSITIVE3 (EIN3) activity during ethylene signaling (Yoo et al., 2008). Additionally, an interaction between the ETHYLENE RESPONSE FACTOR104 (ERF104) transcription factor and MPK6 was detected in vivo, and the release of ERF104 from MPK6 in the nucleus was required for rapid ethylene signaling (Bethke et al., 2009). A large number of putative MAPK substrates were reported using high-throughput protein microarrays (Feilner et al., 2005; Popescu et al., 2009). The identification of phosphorylation sites in the potential substrates of the MAPKs is important for understanding their physiological significance. Phosphorylation of transcription factors by MAPKs can modulate their levels and activities (Tootle and Rebay, 2005). The bHLH transcription factor SPEECHLESS was found to be an in vitro substrate for phosphorylation by the MAPKs (MPK3 and MPK6) (Lampard et al., 2008). ZAT10, a C2H2-type zinc finger transcription factor from Arabidopsis, was identified as a substrate of Arabidopsis MPK3 and MPK6 (MPK3/6) (Nguyen et al., 2012). Recently, it has been reported in rice that a salt-responsive transcription factor, SERF1, binds to the promoter of MAP3K6 and MAPK5 and that SERF1 is also a phosphorylation target of MAPK5 (Schmidt et al., 2013).
Arabidopsis MYC2/ZBF1, a bHLH transcription factor, acts as a negative regulator of blue light-mediated photomorphogenic growth and blue- and far-red-light-regulated gene expression (Yadav et al., 2005). Although MYC2 acts as a negative regulator in cryptochrome-mediated seedling development, it promotes root growth and flowering time and thereby works as both a positive and negative regulator of light signaling (Yadav et al., 2005; Prasad et al., 2012; Gangappa et al., 2013a). Recent studies have also shown that MYC2 regulates SUPPRESOR OF PHYA-105 (SPA1), an associated factor of COP1 ubiquitin ligase in photomorphogenesis (Gangappa et al., 2010; Gangappa and Chattopadhyay, 2010). MYC2 exhibits crosstalk with multiple signaling pathways, such as jasmonic acid (JA), abscisic acid, auxin, ethylene, and gibberellic acid pathways (Kazan and Manners, 2012; Gangappa et al., 2013b). Originally isolated as JIN1/JAI1 (JASMONATE INSENSITIVE1), MYC2/ZBF1 differentially regulates the expression of two groups of JA-induced genes; one group includes genes involved in defense responses against pathogens, which is repressed by MYC2. The second group integrates genes involved in JA-mediated systemic responses to wounding, which is activated by MYC2. Therefore, MYC2 acts as both a positive and negative regulator of JA signaling pathways (Lorenzo et al., 2004; Kazan and Manners, 2011). MYC2 is expressed in the dark and at various wavelengths of light at the seedling stage, and abscisic acid- and JA-mediated induction further increases MYC2 expression (Abe et al., 2003; Anderson et al., 2004; Lorenzo et al., 2004; Yadav et al., 2005). Lower levels of MYC2 accumulation have been reported in the absence of JA (Zhai et al., 2013). A recent report by Chico et al. (2014) shows that MYC2 levels are regulated by the circadian clock, as previously reported by Shin et al. (2012), which shows the involvement of a circadian clock component TIME FOR COFFEE in MYC2 protein regulation. Whereas MYC2 directly represses the expression of PLT1 and PLT2 during jasmonate-mediated regulation of the root stem cell niche, it positively regulates the expression of TPS21 and TPS11 in a gibberellic acid-dependent manner (Chen et al., 2011; Hong et al., 2012). Recently, the interactions between MYC2 and the ethylene-activated transcription factor EIN3 were shown to be involved in hook opening responses; MYC2 represses the DNA binding activity of EIN3 and hence limits the ethylene-induced apical hook curvature responses (Song et al., 2014; Zhang et al., 2014).
The JA-activated MAPK cascade comprising MKK3-MPK6 has been reported to negatively regulate MYC2-mediated JA-dependent gene expression and root growth sensitivity (Takahashi et al., 2007). Although the involvement of MAPK pathways in seedling growth and development has been suggested (Lu et al., 2002; Takahashi et al., 2007), the connection between light signaling pathways and MAPK is currently unknown. In this study, we investigated the possible connection between light and MAPK pathways via MYC2 function. We report that MPK6 is activated by MKK3 in blue light (BL) and that this process is MYC2 dependent. Our study demonstrates that MPK6 physically interacts with and phosphorylates MYC2. We also show that MYC2 interacts with the promoter of MPK6 to regulate its expression. These results collectively highlight the involvement of a feedback regulatory mechanism in the signaling process.
RESULTS
MYC2 Interacts with the Promoter of MPK6 and Regulates Its Expression
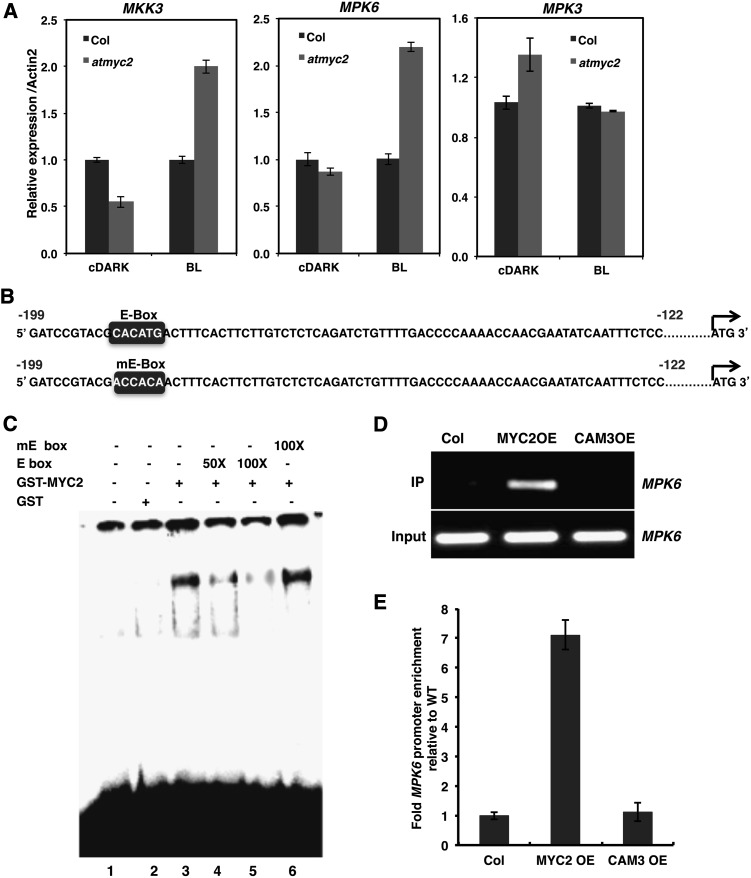
To gain insight into the involvement of MAPKs, especially MKK3 and MPK6, in BL signaling and whether the process is MYC2 dependent, we monitored the transcript levels of MAPKs and MAPKK by quantitative real-time PCR in BL. As shown in Figure 1A, the expression of MPK6 and MKK3, however not MPK3, was increased ∼2-fold in myc2 mutants relative to the wild type in BL. These results indicate that MYC2 negatively regulates the expression of MPK6 and MKK3 in BL.
Figure 1.
MYC2 Directly Binds to the E-Box of MPK6 Promoter.
(A) Real-time PCR analysis of MKK3, MPK6, and MPK3 transcripts in 6-d-old constant dark-grown seedlings transferred to BL (30 μmol m−2 s−1) for 10 min. ACT2, internal control. Error bars indicate sd. n ≥ 3 independent experiments with similar results.
(B) Wild-type (E-box) and mutated (mE-box) E-box containing MPK6 promoter fragments used for the gel shift assays.
(C) Electrophoretic mobility shift (gel shift) assays showing that MYC2 specifically binds to the E box (−181 to −186 bp upstream of ATG). Approximately 500 ng of recombinant protein was added (lanes 3 to 6) to the radioactively labeled 78-bp MPK6 promoter fragment containing the E-box. No protein was added in lane 1, and 500 ng of GST protein was added in lane 2. The 50 and 100 molar excess of wild type E-box containing promoter fragment was added in lanes 4 and 5, and 100 molar excess of mutated E-box containing promoter fragment was added in lane 6 as competitors. The plus and minus signs indicate the presence or absence of competitors, GST-MYC2, or GST.
(D) ChIP assays of the MPK6 promoter from Col, MYC2 overexpresser (MYC2-cMycOE), and Cam3OE transgenic seedlings grown in constant BL (30 μmol m−2 s−1) for 10 d (using antibodies to cMyc). The gel image shows the results of PCR amplification of MPK6 promoter fragment in the immunoprecipitate and input.
(E) Results of real-time qPCR are presented as fold enrichment relative to the wild type (Col). ChIP values were first normalized by the respective input values and then fold enrichment relative to the wild type was calculated. Error bars represent se (n = 3).
To determine whether MYC2 has a direct role in the regulation of MPK6 expression, we performed electrophoretic mobility shift assays (EMSAs). The in silico analysis detected the presence of a (nearest to start site) E-box (CACATG) cis-acting element ∼180 bp upstream of the MPK6 promoter (Figure 1B). We performed EMSA using purified glutathione S-transferase (GST)-MYC2 fusion protein and the 78-bp promoter region (−122 to −199 bp) of MPK6 containing the single E-box. To examine the specificity of this DNA-protein interaction, we used a mutated version of this E-box (mE-box: ACCACA) (Figure 1B). As shown in Figure 1C, GST alone did not show any binding activity (lane 2); however, a low mobility DNA-protein complex was formed with GST-MYC2 (lane 3) and that was competed out by 50 or 100 molar excess of unlabeled MPK6 promoter fragment (lanes 4 and 5) containing this E-box. However, the mE-box fragment was not able to compete with the DNA-protein interaction (lane 6). We also screened for the presence of another E-box in the MPK6 promoter and found an adjacent E-box (E1-box) at the −229 bp position, relative to the ATG start codon. We performed EMSA using purified GST-MYC2 fusion protein and the 57-bp MPK6 promoter region (−199 to −255 bp) containing the E1-box. MYC2 was unable to bind to the E1-box (Supplemental Figures 1A and 1B). These results together suggest that MYC2 binds to the E-box of the MPK6 promoter, which is nearest to the ATG start site.
To further examine the binding of MYC2 to the MPK6 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) experiments. Ten-day-old transgenic Arabidopsis seedlings grown under constant BL (30 μmol m−2 s−1) and overexpressing MYC2 fused to three copies of the c-Myc epitope (MYC2OE) were used in the study. The MYC2-cMyc fusion protein in transgenic plants was immunoprecipitated using the c-Myc antibody. Coimmunoprecipitated genomic DNA with MYC2 was analyzed by RT-PCR and quantitative PCR (qPCR). As shown in Figure 1D, amplification specific to the MPK6 promoter region occurred only in the transgenic MYC2OE plants, but not in the wild type or CAM3-cMyc overexpressor transgenic lines (CAM3OE), which served as the negative control (Abbas et al., 2014). Furthermore, the qPCR analysis revealed that the amount of DNA fragment of the MPK6 promoter coimmunoprecipitated from the transgenic MYC2OE line was ∼7-fold enriched compared with the wild type or CAM3OE line (Figure 1E). Similar results were obtained with dark-to-BL grown seedlings (Supplemental Figures 2B and 2E). Transgenic MYC2OE seedlings grown under constant darkness or red light (60 μmol m−2 s−1) grown did not show amplification specific to the MPK6 promoter region (Supplemental Figures 2A and 2D, and 2C and 2F, respectively). Thus, binding of MYC2 to the MPK6 promoter specifically occurs when the plant is grown in BL. Taken together, these results suggest that MYC2 binds to the promoter of MPK6 and negatively regulates its expression in blue light.
Since MYC2 also negatively regulates the expression of MKK3 in BL, we examined the MKK3 promoter for the presence of the E-box. We detected an E-box −715 bp from the start site (E-box nearest to the start site). ChIP assays were performed to determine the possible binding of MYC2 to the 74-bp MKK3 promoter region (−657 to −730 bp). It was found that MYC2 did not bind to this E-box on the MKK3 promoter. Thus, the regulation of MKK3 may not be through direct binding of MYC2 to the MKK3 promoter (Supplemental Figure 3).
BL-Mediated Activation of MPK6 Is MYC2 Dependent
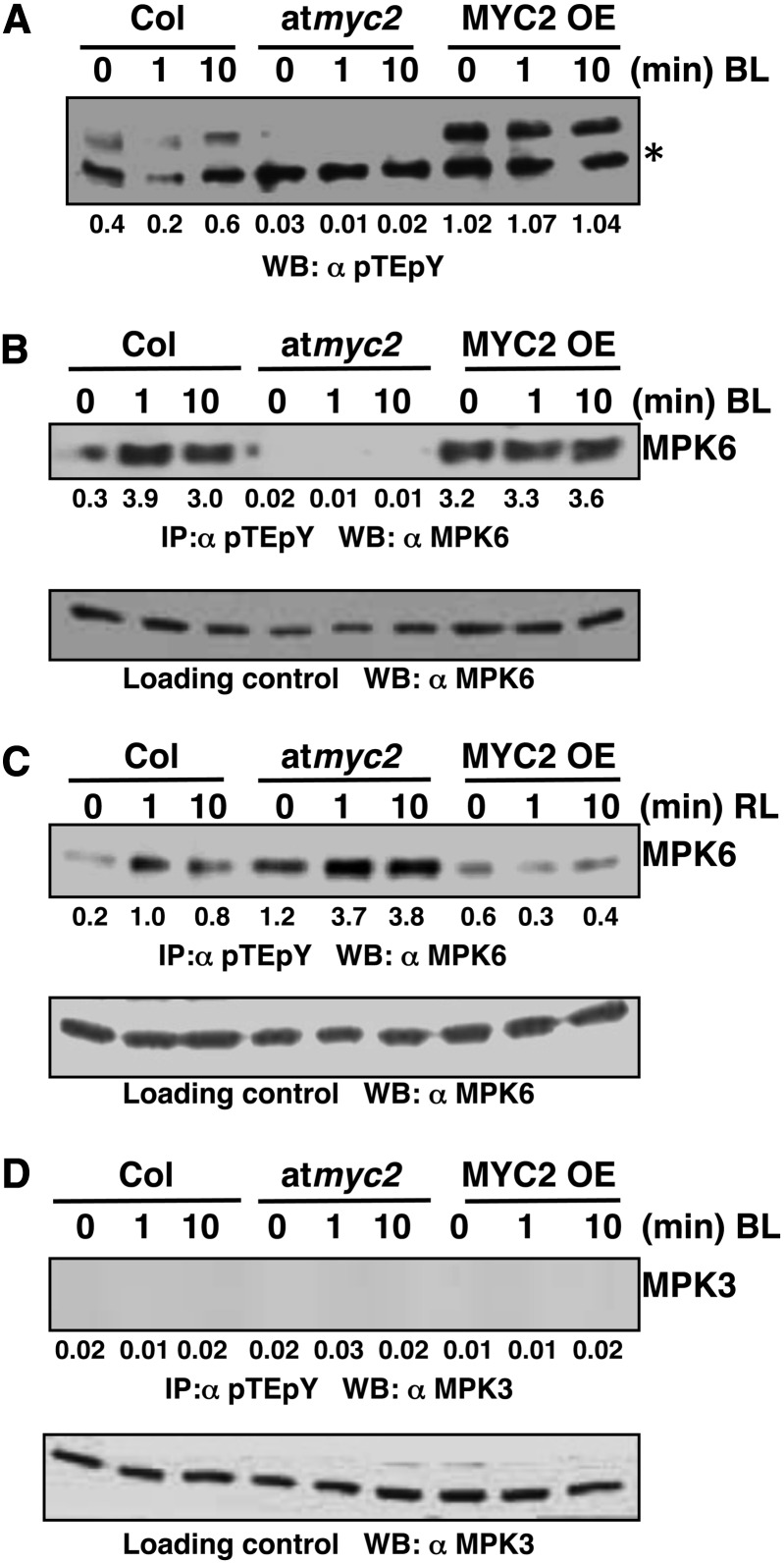
Since MYC2 negatively regulates the transcript accumulation of MPK6 in BL, we asked whether MPK6 was specifically activated in response to BL in a MYC2-dependent manner. To investigate this possibility, we performed an immunoblot analysis of crude protein extracts from Columbia (Col), myc2, and MYC2OE lines grown in darkness for 6 d and then exposed to BL for 1 and 10 min. Proteins were transferred on to the membrane and probed with pTEpY antibody followed by anti-rabbit secondary antibody (Figure 2A). Interestingly, BL activated MAPKs within 10 min in the wild-type plants. However, myc2 lines did not show such activity (Figure 2A). On the other hand, the MYC2OE line had drastically higher activation of MAPKs, even in the control samples (dark-grown seedlings). The overexpression of MYC2, which also binds to and regulates the promoter of MPK6, might account for the high activity in darkness. To substantiate this finding, we performed in-gel kinase assays using Col, myc2, and MYC2OE lines grown in darkness for 6 d and then exposed to BL for 1 and 10 min. The MAPK activity was analyzed using MBP (Myelin Basic Protein), an artificial substrate used to analyze the activity of MAPKs. Interestingly, we found that BL activated MBP phosphorylating protein kinase activity within 10 min in the wild-type plants. However, myc2 lines did not show such activity (Supplemental Figure 4A). On the other hand, the MYC2OE line had drastically higher activity, even in the control samples (dark-grown seedlings). These results suggest that a MAPK is activated in BL and that this activation is likely to be MYC2 dependent.
Figure 2.
MPK6 Activation in BL Is MYC2 Dependent.
(A) Immunoblot analysis using pTEpY antibody of 50 μg of total protein from dark-grown Col, myc2, and MYC2 OE lines exposed to BL (30 μmol m−2 s−1) for 0, 1, or 10 min. Upper bands indicate total active MAPKs, and lower bands (asterisk) indicate cross-reacting band. Values below the panel indicate quantification of each band with respect to the loading control (lower cross-reacting band).
(B) Immunoprecipitation assays using 200 μg of total protein from dark-grown Col, myc2, and MYC2OE lines exposed to BL (30 μmol m−2 s−1) for 0, 1, or 10 min. The MPK6 activation was analyzed by immunoprecipitation with pTEpY antibody followed by immunoblot analysis with anti-MPK6. Lower panel shows immunoblot with MPK6 antibody using 20 μg of total protein as the loading control. Values below the panel indicate quantification of each band with respect to MPK6, the loading control.
(C) Immunoprecipitation assay of dark-grown Col, myc2, and MYC2OE lines exposed to RL (60 μmol m−2 s−1) for 0, 1, or 10 min. The specific MPK6 activation was analyzed by immunoprecipitation with pTEpY antibody followed by immunoblot analysis with anti-MPK6. The lower panel shows an immunoblot with MPK6 antibody using 20 μg of total protein as loading control. Values below the panel indicate quantification of each band with respect to MPK6, the loading control.
(D) MPK3 is not activated in BL. Immunoblot probed with anti-MPK3 after immunoprecipitation with pTEpY antibody acts as negative control. Lower panel shows immunoblot analysis with anti-MPK3 using 20 μg of total protein as loading control.
Next, to determine which MAPK was specifically activated in BL and whether the activation was MYC2 dependent, immunoprecipitation (IP) assays were performed using pTEpY antibody followed by immunoblot analysis with MPK6 antibody. The wild type and MYC2OE lines exposed to BL showed a band at 46 kD corresponding to MPK6, whereas this activation was not seen in the myc2 background (Figure 2B). To examine whether this activation was BL specific, we performed similar experiments in which the seedlings were grown in darkness and then exposed to red light (RL). However, no differential activation was observed in Col, myc2, and MYC2OE backgrounds in RL. These results indicate that the specific activation of MPK6 was MYC2 dependent in BL (Figure 2C). To further substantiate this result, we performed similar experiments using MPK3 antibody. However, no signal corresponding to MPK3 was detected (Figure 2D), thereby indicating that MPK6 is specifically activated in BL in a MYC2-dependent manner. We also repeated the same experiment using antiphosphotyrosine antibody (4G10) followed by immunoblot analysis with MPK6 antibody, and a similar trend was observed (Supplemental Figures 4B to 4D).
MKK3 Activates MPK6 in a BL-Dependent Manner
The involvement of MKK3 upstream of MPK6 in JA signaling has already been reported (Takahashi et al., 2007). Since our IP assays indicate that MPK6 is activated in response to BL, we wanted to investigate the involvement of MKK3 in BL signaling. For this, we performed IP assays, in which Col and mkk3 lines were grown in darkness for 6 d and exposed to BL for 1 and 10 min. The immunoprecipitation was performed with pTEpY antibody, followed by immunoblot analysis with MPK6 antibody. Signals corresponding to MPK6 activation were observed in BL-exposed wild-type samples; however, it was not detected either in the dark- or BL-exposed mkk3 background (Figure 3A). These results suggest that MKK3 activates MPK6 in response to BL. To further test whether MKK3-mediated activation of MPK6 is BL specific, we conducted similar experiments where the seedlings were exposed to RL. However, the MPK6 phosphorylation was observed both in wild type and in mkk3 mutant samples (Figure 3B). The experiments were repeated by immunoprecipitating with antiphosphotyrosine antibody (4G10), followed by immunoblot analysis with MPK6 antibody, and similar results were obtained (Supplemental Figures 5A and 5B). These results, taken together, suggest that MKK3-mediated activation of MPK6 is BL specific.
Figure 3.
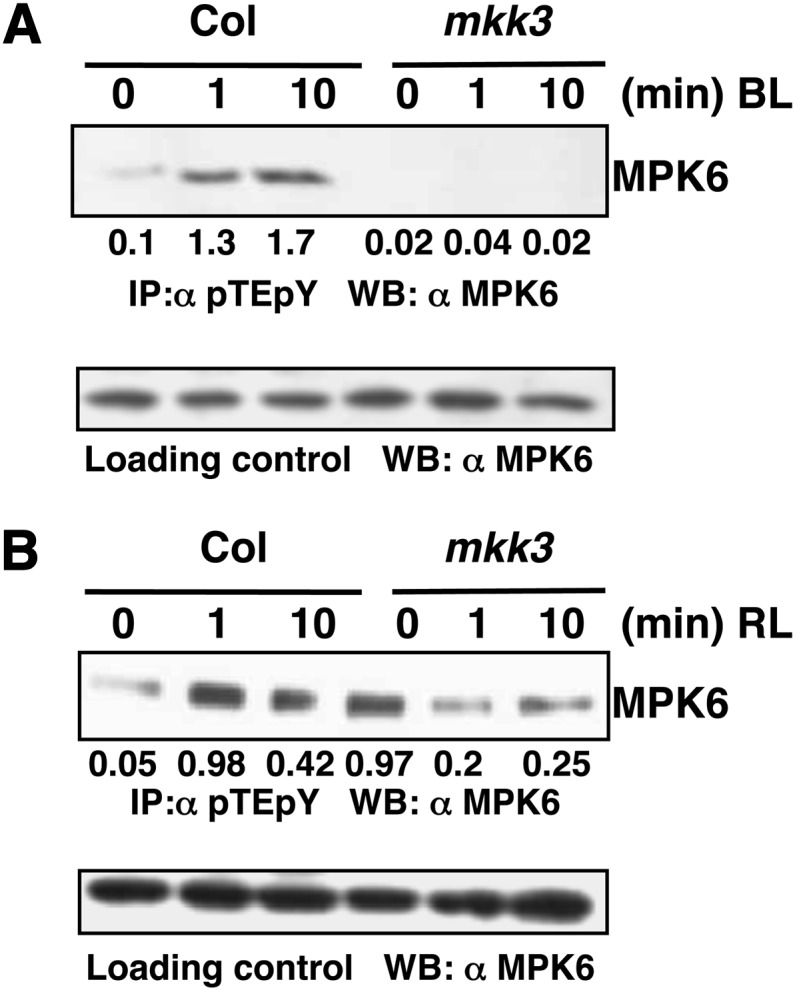
MKK3 Activates MPK6 in BL.
(A) Activation of MPK6 by MKK3 in BL. Immunoprecipitation assays of dark-grown Col, mkk3 mutant lines exposed to BL (30 μmol m−2 s−1) for 0, 1, or 10 min. Experimental details are the same as in Figure 2B. Values below the panel indicate quantification of each band with respect to the MPK6, loading control.
(B) MKK3-dependent activation of MPK6 is not RL specific. Immunoprecipitation assay of dark-grown Col, mkk3 lines exposed to RL (60 μmol m−2 s−1) for 0, 1, or 10 min. Experimental details are the same as in (Figure 2C). Values below the panel indicate quantification of each band with respect to MPK6, the loading control.
MYC2 Is Phosphorylated by MPK6
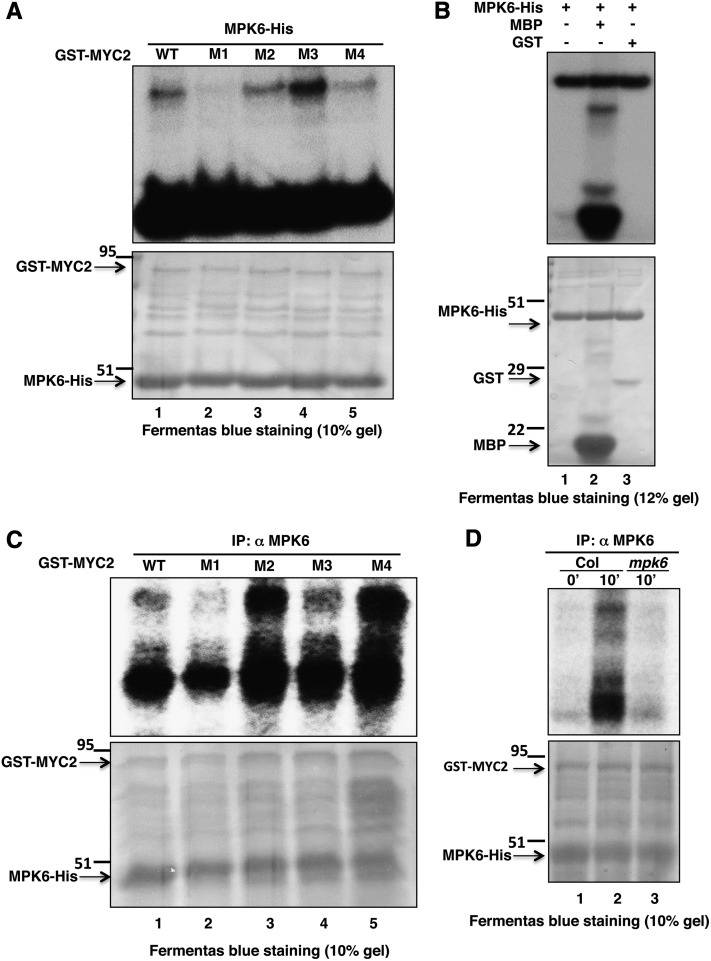
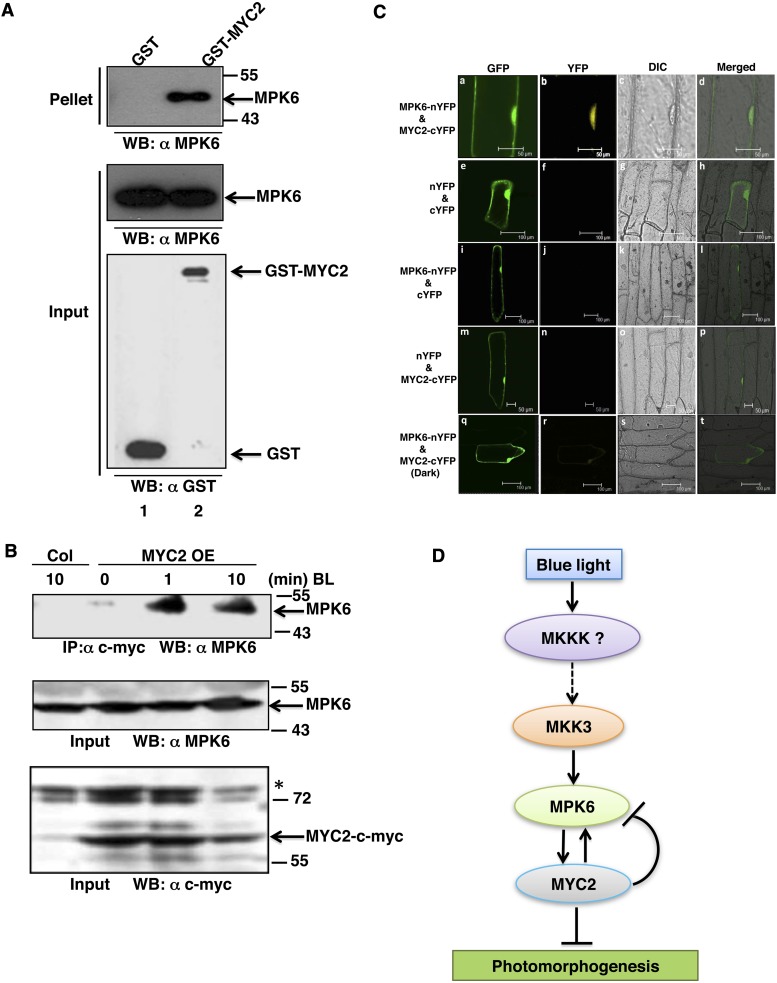
Several earlier reports on light signaling pathways in Arabidopsis suggest that phosphorylation of transcription factors such as GBF1, HY5, and HFR1 influences their functional activity (Klimczak et al., 1992; Hardtke et al., 2000; Duek et al., 2004). In most of these phosphorylation events, casein kinase II has been found to be the kinase involved. Recent studies have suggested that MYC2 phosphorylation at Thr-328 is likely to be functionally coupled with proteolysis and its action to regulate JA-responsive gene transcription (Zhai et al., 2013). However, the kinase involved in the process remains unknown. In this study, since MPK6 phosphorylation and activation were found to be MYC2 dependent, we also wanted to test the phosphorylation of MYC2 by MPK6. To investigate this, an in-solution kinase assay was performed using bacterially expressed MPK6-His and GST-MYC2 protein. The 623-amino acid full-length MYC2/ZBF1 protein was scanned using the Net-Phos K software (Blom et al., 1999) for any putative MPK6 target phosphorylation site. This analysis revealed four putative phosphosites, namely, Ser-123, Thr-287, Thr-328, and Ser-330. These residues were then substituted by Ala individually in each protein as S123A, T287A, T328A, or S330A. The wild type and different site-directed mutant versions of GST-MYC2 protein were used as substrates. GST-MYC2 protein with Ser-123 replaced with Ala (GST-MYC2 M1) did not produce a phosphorylation signal. These results suggest that MPK6 phosphorylates MYC2 at Ser-123 (Figure 4A, lane 2). A control experiment was performed to examine the autophosphorylation of bacterially expressed MPK6 (Figure 4B, lane 1) and to show that it could phosphorylate MBP (Figure 4B, lane 2), while GST served as a negative control (Figure 4B, lane 3).
Figure 4.
MPK6 Phosphorylates MYC2 in BL.
(A) In vitro kinase assay using bacterially expressed MPK6-His and GST-MYC2 (wild-type protein and different site-directed mutagenized versions, i.e., M1-M4 of MYC2 protein, lanes 1 to 5, respectively). GST-MYC2 M1 protein has Ser-123 replaced with Ala, βM2 has Thr-287 replaced with Ala, M3 has Thr-328 replaced with Ala, and M4 has Ser-330 replaced with Ala. Lower panel shows PAGE-blue stained gel showing position of different proteins.
(B) Affinity-purified proteins (MPK6-His and GST) were expressed in Escherichia coli. MPK6-His was incubated alone or in combination with GST or MBP in the kinase reaction mixture to check their phosphorylation by MPK6. The plus and minus signs indicate the presence and absence of proteins, respectively. Aliquots of the samples were separated by SDS-PAGE and subjected to autoradiography. Lower panel shows the PAGE-blue stained gel with the position of different proteins indicated (arrows).
(C) In vitro phosphorylation assay using plant protein. Arabidopsis wild-type (Col) seedlings grown in dark and then exposed to BL (30 μmol m−2 s−1) for 10 min. Immunoprecipitation of crude protein (300 μg) with anti-MPK6 followed by incubation with GST-MYC2 as substrate (wild type or mutated, lanes 1 to 5, respectively) in the kinase reaction mixture to determine phosphorylation activity. PAGE-blue stained gel, with the position of different proteins indicated (arrows).
(D) Immunoprecipitation of MPK6 from Arabidopsis wild-type (Col) seedlings grown in constant darkness (lane 1) and Col and mpk6 seedlings grown in darkness for 6 d and then exposed to BL (30 μmol m−2 s−1) for 10 min (lanes 2 and 3, respectively), followed by incubation with GST-MYC2 as substrate in the kinase reaction mixture to determine phosphorylation activity. PAGE-blue stained gel, with the position of different proteins indicated (arrows).
To substantiate this finding further, plant protein was used for immunoprecipitation of MPK6 using anti-MPK6 antibody followed by in-solution kinase assays using GST-MYC2 and various mutants of GST-MYC2 protein as substrate. In this assay, Arabidopsis wild-type (Col) seedlings were grown in dark for 6 d and then exposed to BL for 10 min. A phosphorylating signal corresponding to the molecular weight of GST-MYC2 was observed, indicating that activated immunoprecipitated MPK6 phosphorylated MYC2 (Figure 4C, lane 1, and lanes 3 to 5). The mutated GST-MYC2 M1 protein with the S123A mutation did not show any phosphorylation signal. These observations suggest that Ser-123 is the site of MYC2 phosphorylation by activated MPK6 in BL. Autophosphorylation of MPK6 was seen in all lanes (Figure 4C, lanes 1 to 5). To further substantiate that the phosphorylation of MYC2 by MPK6 is BL dependent and that specifically MPK6 is targeting MYC2, immunoprecipitation of MPK6 was performed from Col seedlings grown in constant dark and also from the mpk6 mutant exposed to BL (30 μmol m−2 s−1). We found that MPK6 immunoprecipitated from BL-exposed Col seedlings, but not from wild-type or mpk6 seedlings exposed to constant darkness, could phosphorylate MYC2 (Figure 4D, lane 2) in an in vitro phosphorylation assay (Figure 4D, lanes 1 and 3, respectively).
MYC2 Genetically Interacts with MPK6 and MKK3 in BL-Mediated Photomorphogenic Growth
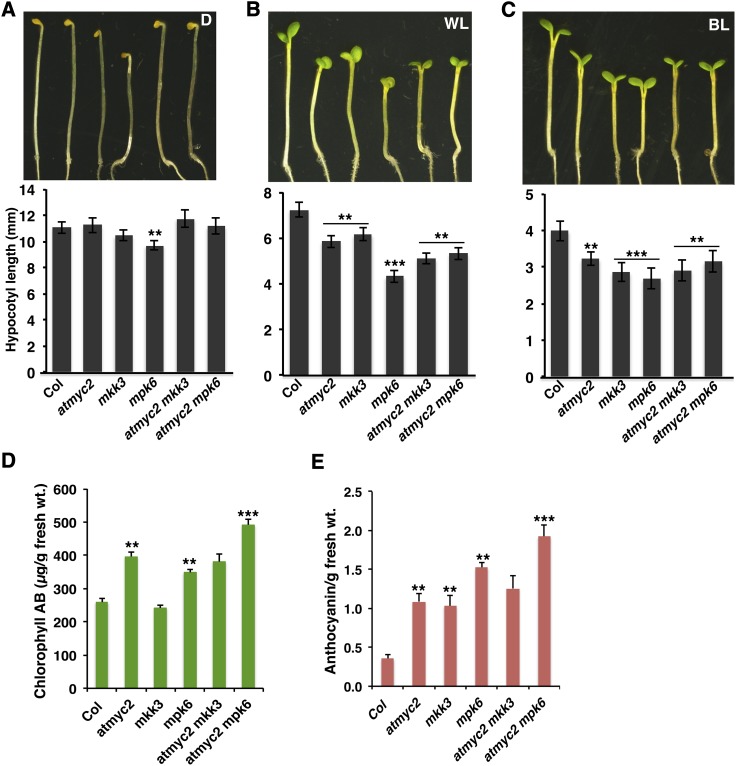
MYC2 encodes a bHLH transcription factor that plays a predominant role in BL-mediated photomorphogenic growth (Yadav et al., 2005; Gangappa et al., 2010a; Prasad et al., 2012). Since MKK3-MPK6 is found to be molecularly connected to MYC2 in a BL-specific manner, we wanted to determine the physiological significance of such an interaction. We generated two sets of double mutants, namely, myc2 mpk6 and myc2 mkk3 through genetic crosses and examined the morphology of the mutant seedlings in darkness and under different light conditions. Hypocotyl measurement of 6-d-old dark-grown seedlings revealed that mkk3 and mpk6 single mutants had weak and strong photomorphogenic phenotypes, respectively, in comparison with the wild type (Col), whereas myc2 mutants displayed a hypocotyl length similar to that of the wild type. Analysis of double mutants revealed that additional mutation of myc2 suppressed the weak and strong phenotype of mkk3 and mpk6, respectively, in darkness (Figure 5A). Examination of hypocotyl length of 6-d-old seedlings in white light (WL) revealed that all the three single mutants, mkk3, mpk6, and myc2, had significantly shorter hypocotyls than did the wild type. The myc2 mkk3 double mutants displayed significantly shorter hypocotyls than did their respective single mutants, suggesting that the function of MYC2 and MKK3 is additive for the suppression of WL-mediated inhibition of hypocotyl elongation. However, myc2 mpk6 double mutants displayed similar hypocotyl length to myc2 single mutants, suggesting that MYC2 functions downstream of MPK6 to regulate hypocotyl growth (Figure 5B). Under BL, all three single mutants, myc2, mkk3, and mpk6, displayed shorter hypocotyls than the corresponding wild type (Figure 5C). The myc2 mkk3 and myc2 mpk6 double mutants displayed similar hypocotyl length to myc2 single mutants in BL, suggesting that MYC2 functions downstream of MKK3 and MPK6 to regulate BL-mediated hypocotyl growth (Figure 5C).
Figure 5.
Genetic Interactions of myc2 with mpk6 and mkk3 in Darkness and White and Blue Light.
(A) to (C) Phenotype of 6-d-old wild-type and different mutant seedlings grown in constant darkness, WL (10 μmol m−2 s−1), and BL (30 μmol m−2 s−1), respectively. Quantification of hypocotyl length of seedlings shown below the photographs. Error bars represent sd (n ≥ 27). Asterisks indicate genotypes that differ significantly (Student’s t test, **P ≤ 0.01 and ***P ≤ 0.001) from the wild type (Col).
(D) and (E) Accumulation of chlorophyll (D) and anthocyanin (E) in 6-d-old seedlings grown in BL (30 µmol m−2 s−1). Error bars represent se (n = 3). Asterisks indicate genotypes that differ significantly (Student’s t test, **P ≤ 0.01 and ***P ≤ 0.001) from the wild type (Col).
[See online article for color version of this figure.]
We next examined the morphology of 6-d-old seedlings grown in RL. The mpk6 mutants displayed significantly shorter hypocotyls than the wild type; however, the hypocotyls of the myc2 and mkk3 mutants did not differ from those of the wild type (Supplemental Figure 6A). The myc2 mpk6 double mutants displayed similar hypocotyl lengths as the wild type in RL, suggesting that the additional mutation in MYC2 suppresses the mpk6 phenotypes in RL (Supplemental Figure 6A). Measurement of hypocotyl length in far-red light showed that mpk6 and mkk3 mutants had significantly shorter hypocotyls than the wild type (Supplemental Figure 6B). The myc2 mpk6 and myc2 mkk3 double mutants displayed similar hypocotyl lengths as mpk6 and mkk3, respectively, suggesting that the additional mutation in MYC2 does not affect the mpk6 and mkk3 phenotypes in far-red light (Supplemental Figure 6B).
The MKK3-MPK6-MYC2-Mediated Regulation of Light-Inducible Genes and HY5, GBF1, and SPA1 Expression
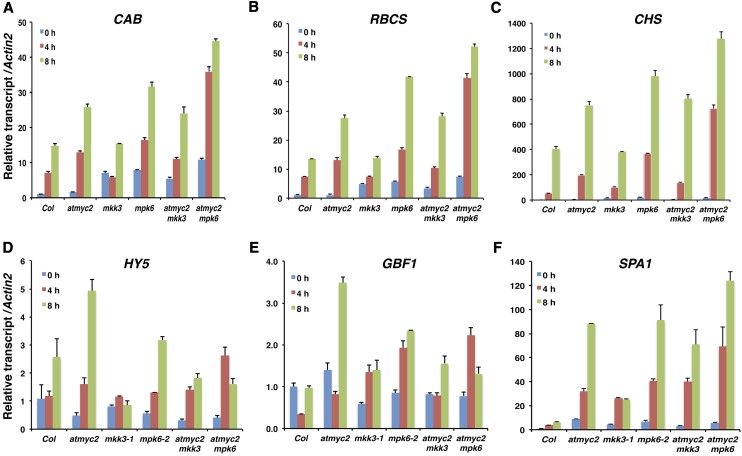
MYC2 represses BL-mediated expression of light-inducible genes (Yadav et al., 2005). We monitored the expression of light-inducible genes such as CAB1, RBCS-1A, and CHS in various single and double mutants. For these experiments, dark-grown seedlings were transferred to BL for 4 and 8 h, and the transcript levels were monitored by quantitative real-time PCR. MYC2 and MPK6 function in an additive manner to regulate the expression of CAB, RBCS, and CHS in BL. Similar results were obtained when chlorophyll a/b content or anthocyanin content was analyzed in these mutants under BL (Figures 5D and 5E). As shown in Figures 6A to 6C, myc2 and mpk6 mutants accumulated higher amounts of CAB1, RBCS-1A, and CHS transcripts than did the wild type. However, the transcript levels of these gens remained similar in wild-type and mkk3 mutant backgrounds. The level of CAB1, RBCS-1A, and CHS transcripts drastically increased in myc2 mpk6 double mutants.
Figure 6.
MKK3-MPK6-MYC2-Mediated Regulation of Genes Involved in Light Signaling.
(A) to (C) Expression of light-inducible genes in various mutant backgrounds. Real-time PCR analysis of CAB1, RBCS-1A, and CHS transcript levels, respectively, in 6-d-old seedlings grown under constant darkness and then transferred to BL (30 μmol m−2 s−1) for 0, 4, and 8 h. Actin2 was used as a control. Error bars represent sd. n = 3 independent experiments, with similar results.
(D) to (F) Expression of regulatory genes in light signaling pathways. Experimental details are the same as in (A) to (C).
To determine whether the MKK3-MPK6-MYC2 pathway regulates the expression of the regulatory genes of light signaling pathways, we examined the transcript levels of HY5 (a key positive regulator at various wavelengths of light), GBF1 (a bZIP transcription factor that acts as a negative regulator of blue light), and SPA1 (a negative regulator, known to work in concert with MYC2) (Chattopadhyay et al., 1998; Mallappa et al., 2006, 2008; Gangappa et al., 2010a) in various single and double mutants of myc2, mkk3, and mpk6. HY5 expression was strongly increased in myc2 mutants; however, it was suppressed in the mkk3 mutant background after an 8-h exposure to BL. Mutation of MPK6 did not affect HY5 expression. The level of expression of HY5 in myc2 mpk6 and myc2 mkk3 double mutants was found to be similar to that in the wild-type background, suggesting an antagonistic relationship between MYC2 with MPK6 and MKK3 in the regulation of HY5 expression (Figure 6D). GBF1 expression was elevated in myc2 and mpk6 mutants at 8 h. There was no significant difference in the expression of GBF1 in the mkk3 mutant background compared with the wild type. The level of GBF1 expression in the myc2 mpk6 and myc2 mkk3 double mutants remained similar to that in the wild-type background, thereby suggesting an antagonistic relationship between MYC2 with MPK6 and MKK3 in the regulation of GBF1 expression (Figure 6E). The expression of SPA1 was drastically increased in the myc2 and mpk6 mutants. Moreover, SPA1 expression was increased significantly in mkk3 mutants. The myc2 mkk3 double mutants showed similar levels of SPA1 as the myc2 mutant background. However, the level of expression was further increased in myc2 mpk6 double mutants compared with the single mutants (Figure 6F). Therefore, MYC2 antagonistically regulates MKK3 and MPK6 in the regulation of HY5 and GBF1 expression, whereas MYC2 acts additively with MPK6 in the regulation of SPA1.
MYC2 Functions Downstream to MKK3 and MPK6 in JA-Mediated Root Growth Inhibition
Whereas myc2 mutants are less sensitive, mpk6 and mkk3 mutants are hypersensitive to JA-mediated root growth inhibition (Yadav et al., 2005; Takahashi et al., 2007). We examined the JA-mediated root growth response in myc2 mkk3 and myc2 mpk6 double mutants after 10, 12, 14, and 16 d in constant BL (Supplemental Figure 7A) and constant WL (Supplemental Figure 7B). The root phenotype of myc2 mpk6 resembled that of myc2. The root length of myc2 mkk3 was also found to be comparable to that of myc2; however, no difference in root length was observed among different mutants in the absence of JA application. These results suggest that myc2 is epistatic to mkk3 and mpk6 in JA-mediated root growth inhibition in BL and WL.
To determine how the expression of JA-regulated genes is affected in myc2 mkk3 and myc2 mpk6 double mutants, we used qPCR analysis to examine the expression of VSP2 (Vegetative Storage Protein-2) and b-CHI (b-Chitinase), which are involved in JA signaling pathways (Boter et al., 2004; Lorenzo et al., 2004). Whereas the JA-mediated induction of b-CHI was drastically reduced in mkk3 and mpk6 mutants, the expression of b-CHI was induced by JA treatment in myc2 single mutants. The level of b-CHI expression in myc2 mkk3 and myc2 mpk6 double mutants was similar to that in myc2 mutants (Supplemental Figure 8A). Our qPCR analysis suggested that JA treatment induced VSP2 expression in the wild type and mkk3 and mpk6 single mutants, but not in myc2. In myc2 mkk3 and myc2 mpk6 double mutants, the expression of VSP2 was found to be similar to that in myc2 (Supplemental Figure 8B). These results together with the root growth studies suggest that MYC2 functions downstream of MKK3 and MPK6 in JA-mediated root growth inhibition.
MYC2 Physically Interacts with MPK6
The phosphorylation of MYC2/ZBF1 by MPK6 indicates direct interactions between these two proteins. To examine the physical interactions between MYC2 and MPK6, we performed in vitro pull-down assays. GST or GST-MYC2 bound to glutathione beads was incubated individually with equimolar amounts of MPK6-His protein. The beads were subsequently washed and immunoblot analysis was performed using anti-MPK6 antibody. MPK6 was retained by GST-MYC2 protein, but not in the column containing GST alone (Figure 7A). These results indicate direct protein-protein interaction between MYC2 and MPK6 in vitro. To further investigate the interaction between MYC2 and MPK6, we performed coimmunoprecipitation assays using total plant protein extract. The total plant protein extract prepared from the MYC2-cMyc overexpresser transgenic seedlings exposed to 1 or 10 min of BL was immunoprecipitated using protein-A agarose beads coupled to c-Myc antibody. Immunoblot analysis was performed using anti-MPK6 antibody. MYC2OE seedlings treated with BL for 1 and 10 min exhibited bands corresponding to MPK6, suggesting that MYC2 interacts with MPK6 in planta (Figure 7B).
Figure 7.
MYC2 Physically Interacts with MPK6 in BL.
(A) In vitro binding of MYC2 and MPK6. GST or GST-MYC2 bound to GST beads was incubated with equimolar amounts of MPK6-His protein. SDS-PAGE was performed followed by immunoblot analysis with anti-MPK6 antibody. Supernatant (20 μL) was loaded and immunoblot was performed with MPK6 antibody and GST antibody to show the amounts of bait and prey present, respectively.
(B) In vivo binding of MYC2 to MPK6. Coimmunoprecipitation of dark-grown Col (10 min) and MYC2OE lines (0, 1, and 10 min) exposed to BL (30 μmol m−2 s−1). Total protein was incubated with anti-cMyc antibody coupled to protein A agarose beads. SDS-PAGE followed by immunoblot analysis with anti-MPK6. Total protein (50 μg) was loaded and probed with anti-MPK6 and anti-cMyc antibody to show bait and prey amounts, respectively.
(C) MYC2 physically interacts with MPK6 in live onion epidermal cells. (a) GFP channel fluorescence produced by GFP of the pCAMBIA-1302 vector, serving as a control for transformation. (b) YFP channel image produced by reconstruction of YFP when the MPK6-YFP n-ter and MYC2-YFP c-ter constructs were cotransfected into onion epidermal cells. (c) and (d) Bright-field image and merged image of (a) and (b), respectively. The lower panels serve as controls: (e) to (h) show cells cotransfected with empty BiFC vectors. (i) to (l) show cells cotransfected with the MPK6-YFP n-ter construct and empty YFP c-ter vector. (m) to (p) show cells cotransfected with the empty YFP n-ter vector and MYC2 YFP c-ter construct. (q) to (t) show cells cotransfected with the MPK6-YFP n-ter and MYC2-YFP c-ter constructs in constant dark, respectively.
(D) MAPK signaling cascade in blue light. The model shows the involvement of the MKK3-MPK6 module upstream of MYC2 in response to blue light. Furthermore, MYC2 regulates the expression of MPK6 in a negative feedback loop mechanism.
To further substantiate the physical interactions between MPK6 and MYC2, we performed bimolecular fluorescence complementation (BiFC) experiments. Full-length MPK6 coding sequence (CDS) was fused to the N terminus of YFP in the pUC-SPYNE vector, and MYC2 full-length CDS was fused to the C terminus of YFP in the pUC-SPYCE vector. These constructs were cobombarded into onion epidermal cells along with the pCAMBIA 1302 vector, which has a green fluorescent protein (GFP) tag and was used as a positive control for transformation (Figure 7C, a). The interaction between MPK6 and MYC2 produced strong YFP fluorescence in the nucleus (Figure 7C, b). The bright-field image and image merged with fluorescence confirm the nuclei positions (Figure 7C, c and d). Taken together, these results demonstrate that MPK6 physically interacts with MYC2 in vivo. Cotransfection of empty BiFC vectors (Figure 7C, e to h), cotransfection of the MPK6-YFP n-ter construct and empty YFP c-ter vector (Figure 7C, i to l), cotransfection of the empty YFP n-ter vector and MYC2 YFP c-ter construct (Figure 7C, m to p), and cotransfection of the MPK6-YFP n-ter and MYC2-YFP c-ter constructs (constant dark) (Figure 7C, q to t), respectively, served as controls.
DISCUSSION
MYC2 is a bHLH transcription factor, which acts as a negative regulator of BL-mediated photomorphogenic growth in BL (Yadav et al., 2005; Gangappa et al., 2010). This study provides evidence that MYC2 works in concert with the MKK3-MPK6 module in a feedback loop mechanism to regulate BL-mediated photomorphogenic growth and gene expression. While the MKK3-MPK6 module phosphorylates MYC2, MYC2 specifically interacts with the MPK6 promoter and regulates its expression. Furthermore, this study demonstrates that the activation of MPK6 in BL is MYC2 dependent.
The JA-inducible expression of MYC2/JIN1 and VSP2 genes has been shown to be increased in either mkk3 or mpk6 mutants, but reduced in either MKK3 or MPK6 overexpresser transgenic lines. Based on these gene expression and JA-mediated root growth inhibition studies, the MKK3-MPK6 cascade has been shown to negatively regulate the JA-mediated responses of MYC2 (Takahashi et al., 2007). Since MYC2 acts as a point of crosstalk in multiple signaling pathways, including light and JA responses, we investigated the possible involvement of MAPK signaling components and MYC2 in response to light. Our quantitative real-time PCR studies suggest that MYC2 negatively regulates the transcript accumulation of both MPK6 and MKK3. Furthermore, the in-gel kinase assays demonstrate that MAPKs are activated in response to BL and this activation is MYC2 dependent. The immunoblot analyses with pTEpY antibody and the IP kinase assays specifically demonstrate that the activation of MPK6 in BL is MYC2 dependent. Thus, on one hand MYC2 negatively regulates the transcript accumulation of MKK3 and MPK6; on the other hand, it positively affects the activation of MPK6 in BL. The IP experiments involving mkk3 mutant show that BL induces the activation of MKK3, which further activates downstream MPK6. Once activated, MPK6 phosphorylates MYC2, and at the same time MYC2 is involved in the regulation of MPK6 in BL. In jasmonate signal-induced plant immunity, MYC2 is phosphorylated at the T328 motif for its proteolysis (Zhai et al., 2013). It is evident from our study that MPK6 phosphorylates MYC2. We found that out of four putative MAPK phosphorylation motifs present in MYC2, S123 is specifically phosphorylated by MPK6. This observation indicates that MPK6 is not involved in phosphorylation-coupled proteolysis of MYC2; rather, it activates MYC2 by phosphorylating it at S123. Therefore, a feedback loop mechanism operates in which MYC2 regulates the expression of MPK6 in BL, and the activated MPK6 phosphorylates MYC2. Interdependent and independent networks of phosphorylation have been demonstrated in eukaryotes. For example, a complex interdependent phosphorylation system has been shown to function in ChK2 activity in cellular DNA damage repair (Guo et al., 2010).
The mpk6 single mutants had shorter hypocotyls than did wild-type seedlings in BL, WL, and RL as well as in dark conditions. Both the mkk3 and mpk6 single mutants showed shorter hypocotyls in BL and genetically interact with myc2. These results suggest the involvement of MKK3 and MPK6 in photomorphogenic growth in a pathway mediated by MYC2. Analysis of expression of light-inducible genes suggests that MKK3 function is dependent of MYC2, whereas MPK6 is likely to function independently of MYC2 to regulate CAB, RBCS, and CHS expression either in dark- or BL-grown seedlings. Thus, MPK6-mediated regulation of hypocotyl growth depends on MYC2; however, MPK6-mediated expression of light-inducible genes is independent of MYC2. The VSP2 and b-CHI gene expression studies also support the previously reported observation that MYC2 functions downstream of the MKK3-MPK6 module in JA signaling pathways (Takahashi et al., 2007).
The coimmunoprecipitation and BiFC assays demonstrate the physical interactions between MYC2 and MPK6. The DNA-protein interaction studies by gel shift assays show that MYC2 directly binds to the E-box at the −199 bp position, but not to the E-box present further upstream (−230 bp) of the MPK6 promoter. This binding activity is further substantiated by the ChIP assays. Moreover, the ChIP experiments with constant BL and dark-to-BL grown samples demonstrate that the process of MYC2 binding to the MPK6 promoter is BL specific. The protein-protein interaction studies suggest that MYC2 and MPK6 physically interact with each other. Thus, on one hand, MPK6 regulates the activity of MYC2 by phosphorylation, whereas on the other hand, MYC2 binds to the promoter of MPK6 and regulates its transcription. The IP assays show that MKK3 activates MPK6 in BL, and further that the activation of MPK6 in BL is MYC2 dependent. Collectively, it is tempting to speculate that in the absence of downstream substrate (MYC2), MKK3 does not phosphorylate MPK6. Once MYC2 is phosphorylated by MPK6, the phosphorylated MYC2 may bind to the MPK6 promoter and negatively regulate its expression (Figure 7D). A similar phenomenon has been reported in rice (Oryza sativa), where a salt-responsive transcription factor, ERF1, binds to the promoter of MAP3K6 and MAPK5, while MAPK5 in turn phosphorylates and activates ERF1 (Schmidt et al., 2013). Recent studies have shown that the proteasomal activity is important for MYC2 function. In other words, MYC2 proteolysis is tightly linked to its transcriptional activity (Zhai et al., 2013). Therefore, a feedback loop regulation operates between MYC2 and MPK6 in BL. Taken together, this study demonstrates that MYC2 acts as a point of crosstalk between MAPK and light signaling pathways.
METHODS
Plant Material and Growth Conditions
Seeds were surface-sterilized (2% sodium hypochlorite and 0.02% Triton X-100 solution), plated on the Murashige and Skoog (MS) medium, and kept in dark at 4°C for stratification. The seedlings were grown for 6 d in darkness and then exposed to BL (30 μmol m−2 s−1) or RL (60 μmol m−2 s−1) for the indicated periods of time. To generate myc2 mpk6 and myc2 mkk3 double mutants, mpk6-2 and mkk3-1 mutant seeds were obtained from ABRC and made homozygous. myc2-3 mutant plants were crossed individually with mpk6-2 and mkk3-1 mutants. To determine the genotype at the MYC2 locus, seeds were plated on MS (kanamycin, 25 μg/μL) and the seedlings that had kanamycin resistance were further tested for MPK6 or MKK3 mutation by genomic PCR using gene-specific and T-DNA-specific primers (Supplemental Table 1). To study the root growth response to methyl jasmonate, different genotypes (Col and single and double mutants) were plated on MS without (control) or with 20 μM of methyl jasmonate (Sigma-Aldrich). Plates were photographed and root length was measured on 10, 12, 14, and 16 d after germination.
Gene Expression Analysis
Wild-type and different mutant seedlings were grown in the dark for 6 d and then given light or JA treatment for specific time periods. The total RNA was extracted using the RNeasy plant mini kit (Qiagen). cDNA was synthesized from 1 µg of total RNA using RT-AMV reverse transcriptase (Titan One Tube RT-PCR System; Roche Applied Science). Real-time PCR analyses were performed using the Thermal Cycler Applied Biosystem Step One and Light Cycler Fast start DNA Masterplus SYBR Green 1 systems (Roche Applied Science). The fold expression of different genes was determined using gene-specific primers. The common 2−∆∆CT algorithm was used to analyze the relative changes in gene expression. Actin2, a housekeeping gene that is assumed to be uniformly expressed in all samples, was used as the internal control. ∆Ct value is calculated by normalizing samples Ct to Actin2 Ct values. This value for different samples was then normalized to Ct value of the experimental control (such as wild-type Col dark-grown samples as in Figure 1A), and the ∆∆Ct value was obtained. Fold expression was calculated by the formula, 2−∆∆CT, which was plotted on the graph.
Electrophoretic Mobility Shift (Gel Shift) Assay
GST-MYC2 and GST were induced and overexpressed in Escherichia coli and affinity purified following the manufacturer’s protocol (Amersham Biosciences). The MPK6 and MKK3 promoter fragment containing the E-boxes was amplified, cloned in pBluescript SK+ digested with XbaI and HindIII, purified, and 3′ end-labeled with [α-32P]dCTP for use as a probe for the DNA binding assays. Approximately 25 ng of labeled DNA fragment was incubated with purified GST-MYC2 or GST alone (100 ng) in the presence of 1 µg of poly dI-dC and 1× binding buffer (75 mM HEPES, pH 7.5, 350 mM KCl, 2.5 mM EDTA, 10% glycerol, 5 mM DTT, and 10 mM MgCl2) in a reaction volume of 30 μL for 30 min at room temperature. Competition assays were performed using 50 and 100 molar excess of cold E-box and 100 molar excess of mutated E-box containing MPK6 promoter fragments. The reactions were fractionated on a native polyacrylamide gel (10% acrylamide, 0.5× TBE, and 2.5% glycerol), dried, and autoradiographed.
ChIP Assays
The ChIP assays were performed as described by Gangappa et al. (2010a) with some modifications. Transgenic (MYC2-cMyc and CAM3-cMyc) and nontransgenic wild-type lines were used for the experiment. Dark-grown samples were grown in continuous dark conditions for 10 d, and BL- and RL-grown samples were grown in continuous BL (30 μmol m−2 s−1) and continuous RL (60 μmol m−2 s−1), respectively, for 10 d. Dark-to-blue light or dark-adapted samples were grown for 4 d in continuous BL (to obtain more tissue as dark-grown seedlings yield very little tissue), followed by 6 d in darkness, and then exposed for 10 min to BL (30 μmol m−2 s−1). The immunoprecipitated products were subjected to real-time PCR analysis for examining the relative enrichment of the promoter fragment using Light Cycler Fast-Start DNA Masterplus SYBR Green1 (Roche) using primers that were used to amplify the promoter fragment in gel shift assay. The c-Myc polyclonal antibodies (Sigma-Aldrich) were used for the immunoprecipitation experiments.
In-Gel Kinase Assay and IP Assay
For in-gel and IP assay, Arabidopsis thaliana seedlings (control and treated) were homogenized in MAPK extraction buffer (50 mM HEPES-KOH, pH 7.6, 5 mM EDTA, 5 mM EGTA, 10 mM DTT, 10 mM Na3VO4, 10 mM NaF, 50 mM β-glycerophosphate, 10% glycerol, 7.5% PVPP, 1 mM phenylmethylsulfonyl fluoride, and 1× plant protease inhibitor). The protocol followed was as described in Rao et al. (2011). Anti- pTEpY from Cell Signaling Technology was used for immunoprecipitation. Rabbit polyclonal anti-MPK6 and anti-MPK3 antibodies were obtained from Sigma-Aldrich.
Phosphorylation Assays of MYC2
To establish the phosphorylation status of MYC2 by MPK6, an in-solution kinase assay was performed using GST-MYC2 and MPK6-His fusion proteins expressed in bacteria and MPK6 protein immunoprecipitated from Arabidopsis plants. Arabidopsis wild type (Col) plants were homogenized in MAPK extraction buffer. Total protein extract (300 μg) was incubated overnight with anti-MPK6 antibody at 4°C, followed by incubation with protein-A sepharose beads for 2 h the next day. After several washes, beads with immunoprecipitated MPK6 were subjected to an in-solution kinase assay. The beads were incubated with GST (control) and GST-MYC2 (in the ratio of 1:3) as substrate, in a reaction mix that contained radiolabeled [γ-32P]ATP for 30 min at 28°C. The beads were then denatured and run on 12% SDS-PAGE. The gel was stained using PAGE-blue protein staining solution (Fermentas) to show the position of different proteins in the gel. For the mutated GST-MYC2 protein, different primers were used for site-directed mutagenesis by inverse PCR. The pGEX4T-2 clone harboring the wild-type MYC2 insert was used as a template for inverse PCR. The PCR product was digested with DpnI restriction enzyme. The digested product was used for E. coli BL21 transformation and protein expression. The mutated proteins were purified and further used for in vitro kinase assay.
In Vitro Binding Assays
DNA fragment encoding full-length MYC2 was cloned into pGEX-4T2 vector to yield a fusion with the GST protein. GST-MYC2 was overexpressed and purified from bacteria using Glutathione sepharose 4B beads (Amersham Biosciences). The MPK6 CDS was cloned into the pET-20b (+) vector with a 6× His tag at the C-terminal end of the protein. MPK6-His protein was overexpressed and purified from bacteria using Ni-NTA agarose beads (Qiagen). For the in vitro binding experiment, GST and GST-MYC2 prey proteins were bound to glutathione beads by incubation in phosphate buffered saline (pH 7.4) for 3 h at 4°C. Excess unbound protein was washed off and MPK6-His bait protein was added in an equimolar ratio to all reactions and incubated overnight in 300 μL binding buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM PMSF, and 1× protease inhibitors cocktail [Sigma-Aldrich]) at 4°C. The next day, glutathione beads were collected by brief centrifugation and (supernatant was collected separately and saved for further analysis) washed thrice with 1 mL of wash buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 10% glycerol, and 0.075% Tween 20). The pellet was resuspended in 1× SDS loading buffer, boiled for 10 min, and analyzed by SDA-PAGE for protein binding. Both pellet and supernatant (5%) were analyzed by probing with anti-MPK6 antibody.
Coimmunoprecipitation Assays
Total protein from wild-type and MYC2-overexpresser (with c-Myc tag) lines was extracted from seedlings grown in constant darkness for 6 d or exposed to BL (30 μmol m−2 s−1) for required time points, in a buffer containing 50 mM Tris-Cl (pH 7.5), 100 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitors cocktail (Sigma-Aldrich). Cell debris was removed by centrifugation at 10,000g for 10 min. Total protein concentration was estimated by the Bradford assay (Sigma-Aldrich). One milligram of protein from both the wild type and MYC2OE was incubated overnight with anti-c-Myc antibodies at 4°C. The next day, beads and supernatant were collected separately by brief centrifugation and beads were washed three times with 1 mL of wash buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 10% glycerol, and 0.075% Tween 20). The pellet was resuspended in 1× SDS loading buffer, boiled for 10 min, and analyzed by SDS-PAGE for protein binding. Both pellet and supernatant (5%) were analyzed by probing with anti-MPK6 antibody. Immunoblot analysis of plant protein with anti-c-Myc antibody was performed to show the prey protein.
BiFC Experiment
For BiFC experiments, full-length CDS of MPK6 was cloned in the pUC-SPYNE vector at BamHI and SalI sites to obtain MPK6-YFP n-ter fusion protein. Similarly, the full-length CDS of MYC2 was cloned in pUC-SPYCE at SpeI and ClaI sites to obtain MYC2-YFP c-ter fusion protein. For transient expression, particle bombardment was performed as described by Singh et al. (2012). After bombardment, the plates were exposed to BL (30 μmol m−2 s−1) for 24 h before confocal imaging. A confocal scanning microscope (Leica TCS SP2 AOBS system) with YFP filters was used for imaging. The fluorescent excitations using an argon laser source at 514 nm while emissions of 527 nm was used. The dark treatment plates were kept in constant darkness for 48 h before confocal imaging. The GFP fluorescence of the pCAMBIA-1302 vector was used as a marker for transformed cells.
Chlorophyll and Anthocyanin Estimation
Chlorophyll and anthocyanin levels were measured following protocols as described by Holm et al. (2002).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MYC2 (At1g32640), MPK6 (At2g43790), MKK3 (At5g40440), CAB1 (At1g29930), RBCS1A (At1g67090), CHS (At5g13930), bCHI (At3g12500), VSP2 (At5g24770), HY5 (At5g11260), GBF1 (At4g36730), SPA1 (At2g46340), and Actin2 (At3g18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. MYC2 Binds to the E-Box but Not the E1-Box of MPK6 Promoter.
Supplemental Figure 2. MYC2 Binds to the E-Box of MPK6 Promoter in Blue Light.
Supplemental Figure 3. MYC2 Does Not Bind to the E-box of MKK3 Promoter.
Supplemental Figure 4. Activation of MPK6 in Blue Light Is MYC2 Dependent.
Supplemental Figure 5. MKK3 Activates MPK6 in Blue Light.
Supplemental Figure 6. Genetic Interactions of myc2 with mpk6 and mkk3 in Red Light and Far-Red Light.
Supplemental Figure 7. JA Responses in myc2 mkk3 and myc2 mpk6 Mutants.
Supplemental Figure 8. Expression of JA Marker Genes in Various Mutant Backgrounds.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
This work was supported by a research grant of the Department of Science and Technology, Government of India, to A.K.S. and S.C. V.S. is a recipient of a fellowship from the Council of Scientific and Industrial Research, Government of India, while B.R. is a recipient of fellowship from Department of Biotechnology, Government of India.
AUTHOR CONTRIBUTIONS
V.S., A.K.S., and S.C. designed the research. V.S. and B.R. carried out the experiments. V.S., A.K.S., and S.C. analyzed the data and wrote the manuscript.
Glossary
- bHLH
basic helix-loop-helix
- MAPK
mitogen-activated protein kinase
- JA
jasmonic acid
- BL
blue light
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
- EMSA
electrophoretic mobility shift assay
- Col
Columbia
- IP
immunoprecipitation
- RL
red light
- WL
white light
- BiFC
bimolecular fluorescence complementation
- CDS
coding sequence
- MS
Murashige and Skoog
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abbas N., Maurya J.P., Senapati D., Gangappa S.N., Chattopadhyay S. (2014). Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26: 1036–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreason E., Ellis B. (2010). Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15: 106–113. [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294: 1351–1362. [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Zhu L., Huq E. (2011). Multiple kinases promote light-induced degradation of PIF1. Plant Signal. Behav. 6: 1119–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Ang L.H., Puente P., Deng X.W., Wei N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21: 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico J.M., Fernández-Barbero G., Chini A., Fernández-Calvo P., Díez-Díaz M., Solano R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26: 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413: 217–226. [DOI] [PubMed] [Google Scholar]
- Duek P.D., Elmer M.V., van Oosten V.R., Fankhauser C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14: 2296–2301. [DOI] [PubMed] [Google Scholar]
- Feilner T., et al. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4: 1558–1568. [DOI] [PubMed] [Google Scholar]
- Fiil B.K., Petersen K., Petersen M., Mundy J. (2009). Gene regulation by MAP kinase cascades. Curr. Opin. Plant Biol. 12: 615–621. [DOI] [PubMed] [Google Scholar]
- Gangappa S.N., Chattopadhyay S. (2010). MYC2, a bHLH transcription factor, modulates the adult phenotype of SPA1. Plant Signal. Behav. 5: 1650–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Prasad V.B.R., Chattopadhyay S. (2010). Functional interconnection of MYC2 and SPA1 in the photomorphogenic seedling development of Arabidopsis. Plant Physiol. 154: 1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Maurya J.P., Yadav V., Chattopadhyay S. (2013a). The regulation of the Z- and G-box containing promoters by light signaling components, SPA1 and MYC2, in Arabidopsis. PLoS ONE 8: e62194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Srivastava A.K., Maurya J.P., Ram H., Chattopadhyay S. (2013b). Z-box binding transcription factors (ZBFs): a new class of transcription factors in Arabidopsis seedling development. Mol. Plant 6: 1758–1768. [DOI] [PubMed] [Google Scholar]
- Guo X., Ward M.D., Tiedebohl J.B., Oden Y.M., Nyalwidhe J.O., Semmes O.J. (2010). Interdependent phosphorylation within the kinase domain T-loop regulates CHK2 activity. J. Biol. Chem. 285: 33348–33357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54: 129–140. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2011). The interplay between light and jasmonate signalling during defence and development. J. Exp. Bot. 62: 4087–4100. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17: 22–31. [DOI] [PubMed] [Google Scholar]
- Klimczak L.J., Schindler U., Cashmore A.R. (1992). DNA binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell 4: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.A., Ho T.H., Ho S.L., Yu S.M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14: 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Li J., Qu L., Hager J., Chen Z., Zhao H., Deng X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallappa C., Singh A., Ram H., Chattopadhyay S. (2008). GBF1, a transcription factor of blue light signaling in Arabidopsis, is degraded in the dark by a proteasome-mediated pathway independent of COP1 and SPA1. J. Biol. Chem. 283: 35772–35782. [DOI] [PubMed] [Google Scholar]
- Mallappa C., Yadav V., Negi P., Chattopadhyay S. (2006). A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J. Biol. Chem. 281: 22190–22199. [DOI] [PubMed] [Google Scholar]
- Nagy F., Schäfer E. (2002). Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 53: 329–355. [DOI] [PubMed] [Google Scholar]
- Nguyen X.C., Kim S.H., Lee K., Kim K.E., Liu X.M., Han H.J., Hoang M.H.T., Lee S.W., Hong J.C., Moon Y.H., Chung W.S. (2012). Identification of a C2H2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Rep. 31: 737–745. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Ding L., Dai M., Lin R., Wang H. (2008). Multisite phosphorylation of Arabidopsis HFR1 by casein kinase II and a plausible role in regulating its degradation rate. J. Biol. Chem. 283: 23264–23273. [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B.R., Kumar S.V., Nandi A., Chattopadhyay S. (2012). Functional interconnections of HY1 with MYC2 and HY5 in Arabidopsis seedling development. BMC Plant Biol. 12: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao K.P., Vani G., Kumar K., Wankhede D.P., Misra M., Gupta M., Sinha A.K. (2011). Arsenic stress activates MAP kinase in rice roots and leaves. Arch. Biochem. Biophys. 506: 73–82. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Schmidt R., Mieulet D., Hubberten H.M., Obata T., Hoefgen R., Fernie A.R., Fisahn J., San Segundo B., Guiderdoni E., Schippers J.H.M., Mueller-Roeber B. (2013). Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25: 2115–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Heidrich K., Sanchez-Villarreal A., Parker J.E., Davis S.J. (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24: 2470–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Ram H., Abbas N., Chattopadhyay S. (2012). Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J. Biol. Chem. 287: 25995–26009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A.K., Jaggi M., Raghuram B., Tuteja N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6: 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Huang H., Gao H., Wang J., Wu D., Liu X., Yang S., Zhai Q., Li C., Qi T., Xie D. (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K. (2007). The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle T.L., Rebay I. (2005). Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. BioEssays 27: 285–298. [DOI] [PubMed] [Google Scholar]
- Yadav V., Mallappa C., Gangappa S.N., Bhatia S., Chattopadhyay S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Yan L., Tan D., Chen R., Sun J., Gao L., Dong M.Q., Wang Y., Li C. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9: e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhu Z., An F., Hao D., Li P., Song J., Yi C., Guo H. (2014). Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26: 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.