Expression of Arabidopsis MKK domain deletion variants in three discrete epidermal cell types, coupled with in vitro kinase assays, revealed regions of MKKs that promote signal propagation and regulate protein subcellular localization. Subcellular localization associated with a subset of MKK7/9 functions sheds light onto how signaling specificity is achieved.
Abstract
When multiple mitogen-activated protein kinase (MAPK) components are recruited recurrently to transduce signals of different origins, and often opposing outcomes, mechanisms to enforce signaling specificity are of utmost importance. These mechanisms are largely uncharacterized in plant MAPK signaling networks. The Arabidopsis thaliana stomatal lineage was previously used to show that when rendered constitutively active, four MAPK kinases (MKKs), MKK4/5/7/9, are capable of perturbing stomatal development and that these kinases comprise two pairs, MKK4/5 and MKK7/9, with both overlapping and divergent functions. We characterized the contributions of specific structural domains of these four “stomatal” MKKs to MAPK signaling output and specificity both in vitro and in vivo within the three discrete cell types of the stomatal lineage. These results verify the influence of functional docking (D) domains of MKKs on MAPK signal output and identify novel regulatory functions for previously uncharacterized structures within the N termini of MKK4/5. Beyond this, we present a novel function of the D-domains of MKK7/9 in regulating the subcellular localization of these kinases. These results provide tools to broadly assess the extent to which these and additional motifs within MKKs function to regulate MAPK signal output throughout the plant.
INTRODUCTION
Plants modify their metabolic and developmental programs to respond to the changing environmental conditions they experience during their lifetimes. Mitogen-activated protein kinase (MAPK) cascades comprise one class of intracellular signaling modules used to regulate both developmental processes and environmental stress responses. Canonical MAPK signaling networks are composed of a three-tiered hierarchical system of sequentially acting kinases. In response to an upstream signal, a MAPK kinase kinase (MAPKKK) is activated by phosphorylation and this in turn phosphorylates and activates a MAPK kinase (MKK), which phosphorylates and activates a MAPK. Active MAPKs interact with and phosphorylate a wide range of downstream targets to bring about a specific cellular response (Rodriguez et al., 2010). The Arabidopsis thaliana genome encodes at least 60 MAPKKKs, 10 MKKs and 20 MAPKs, suggesting that discrete combinations of kinases could be used to bring about specific responses. However, both in vitro and in vivo data indicate that multiple MKKs are used to respond to common stimuli and that these MKKs often signal though overlapping downstream MAPKs (Taj et al., 2010). For example, eight out of the 10 Arabidopsis MKKs have been reported to phosphorylate the downstream MAPKs, MPK3 and/or MPK6, and, where characterized, these phosphorylation events bring about both overlapping and discrete cellular responses (Popescu et al., 2009; Rodriguez et al., 2010). This poses the question of how these complex MKK-MAPK relationships are controlled to evoke specific signaling responses.
We know primarily from studies in animals and yeast that MAPK signal fidelity can be generated through posttranslational modifications of the MAPKs, availability of physical association partners, subcellular localization of signaling proteins, and by modulating the amplitude and duration of signals from active kinases (reviewed in Saito, 2010; Wimmer and Baccarini, 2010; Wortzel and Seger, 2011; Ovečka et al., 2014). Each of these mechanisms is dependent upon specific protein-protein interactions. Plants and animals diverged before either became multicellular and their independent evolutionary trajectories (and gene family expansion) have limited the success of sequence homology-based approaches to identify plant MKK interactors. Nonetheless, there is evidence that the protein-protein interaction modes of regulation of MAPK signaling modules, such as inactivation by MAPK phosphatases and control of subcellular localization, are conserved (Takahashi et al., 2010; González Besteiro and Ulm, 2013). Success in finding specificity-providing interactors by in vivo interaction techniques (such as pull-down assays using epitope-tagged kinases) has been similarly elusive. One hypothesis for this is that many of the plant MAPK signaling components are expressed broadly, but their specificity-providing interactors may be expressed only in response to certain stimuli or in restricted cell types and, thus, their abundance would be too low to detect using biochemical methods. Therefore, alternative experimental approaches to studying in vivo specificity in plant MAPK signaling modules are critically needed.
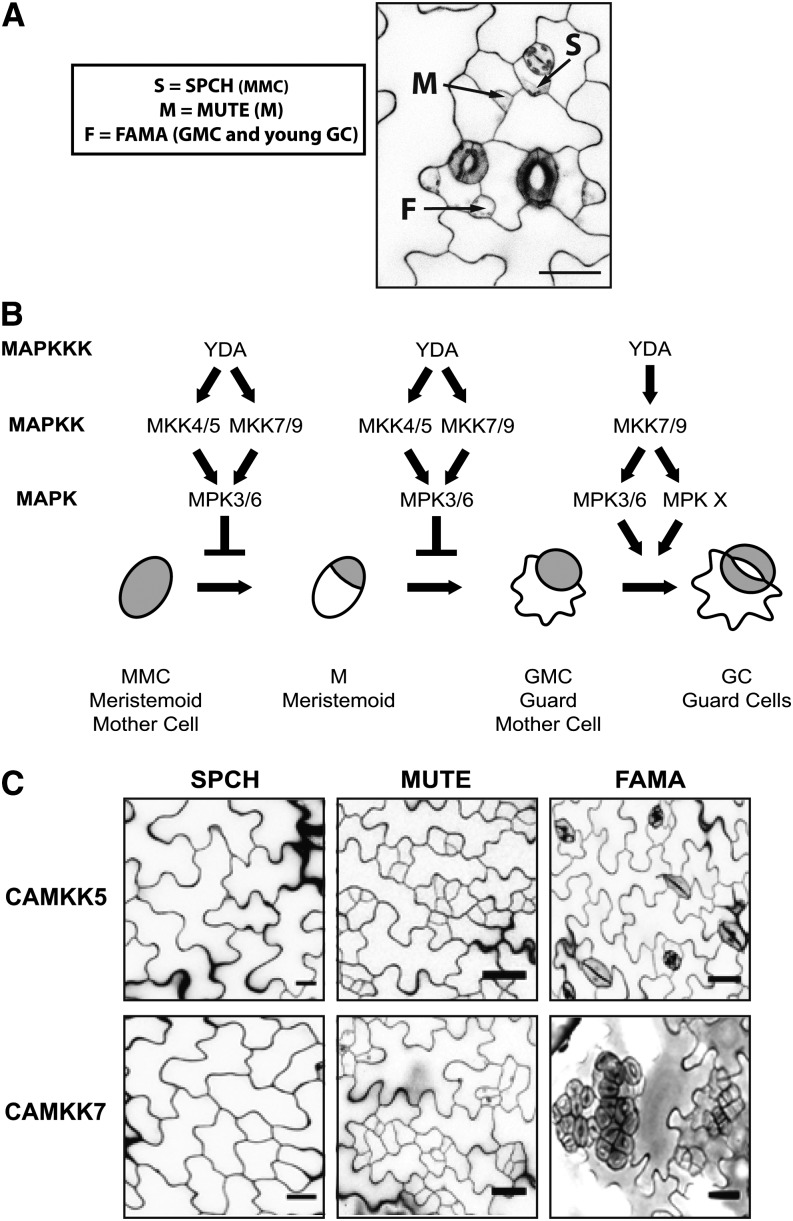
We recently developed a system that allows us to assay the in vivo consequences of MAPK signaling manipulations on developmental events (Lampard et al., 2009). It is based on cell type-specific expression within the epidermal lineage that generates stomata. The formation of stomata involves a stereotyped series of cell division and differentiation steps. Each step is promoted by a discrete bHLH transcription factor, the expression of which is stage specific (Figures 1A and 1B). SPEECHLESS (SPCH) promotes the cell division that leads to entry into the lineage, MUTE regulates the cessation of division and transition of a meristemoid (M) into a guard mother cell (GMC), and FAMA promotes the symmetric cell division of the GMC to form two stomatal guard cells (reviewed in Dong and Bergmann, 2010). Since the expression and activity of each transcription factor is correlated with a specific developmental stage (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007), we will refer to early, mid, and late stages of stomatal development as SPCH, MUTE, and FAMA stages, respectively (Figure 1A).
Figure 1.
Schematic Representation of Stomatal Development and Phenotypes Conferred by MAPK Manipulations.
(A) A 5-DPG abaxial cotyledon depicting the cell types representing the SPCH (S), MUTE (M), and FAMA (F) stages of stomatal development.
(B) The YDA MAPK module that regulates each stage of stomatal development. This module inhibits stomatal initiation or progression when constitutively active variants of YDA or MKK4/5/7/9 are expressed in the SPCH and MUTE stages, respectively. The YDA MAPK module promotes stomatal proliferation and clustering when active during the FAMA stages, but this positive function is specific to YDA and CAMKK7/9.
(C) Confocal images of epidermal phenotypes resulting from manipulations of the YDA MAPK pathway described in (B). In these assays, CAMKK4 and CAMKK5 behave identically, as do CAMKK7 and CAMKK9, and so representative examples from one of each kinase pair are shown.
In all micrographs of the epidermis displayed in figures, cell outlines are visualized with propidium iodide. Bars = 25 μm.
This cell type-specific expression system is unique in that it allows activation of MKK signaling in cells exhibiting important generalizable behaviors (e.g., cell division and fate acquisition) without the induction of cell death or additional deleterious phenotypes commonly associated with broad activation of MKKs (Lampard et al., 2009). This ultimately enables dissection of MKK function in specific cells within the context of the whole plant. Work with this system demonstrated that each stage of stomatal development has the potential to be modulated in specific and predictable patterns by MAPK signaling members (Figure 1C; Lampard et al., 2009). We could confirm that YODA (YDA; MAPKKK) and two known MKKs (MKK4/5; Wang et al., 2007) inhibit entry into the stomatal lineage (SPCH stage; Figure 1C) and demonstrated that activation of these kinases could block stomatal progression at the MUTE stage (Figure 1C). Two additional kinases, MKK7 and MKK9, were found to have overlapping inhibitory functions with MKK4/5 (Figure 1C). Activation of MKK1/2/6 in this system had no impact on stomatal development, highlighting the utility of this system in identifying specific functions of Arabidopsis MKKs that share common downstream signaling components.
During the FAMA stage, we also found unique stomatal-promoting functions of YDA and MKK7/9 (Lampard et al., 2009). These effects were unexpected given the consistent behavior of the so-called “YDA module” (consisting of YDA, MKK4/5, and MPK3/6; Wang et al., 2007) in repressing cell division and differentiation of early stomatal precursors. Importantly, the FAMA stage also separated activities of MKK7/9 from MKK4/5, as activation of the latter kinases had no effect on development (Lampard et al., 2009). Taken together, the stage-specific behaviors of activated MKKs provide a novel system for testing structural contributions of individual domains of MKKs to signaling specificity in vivo.
Here, by exploiting the stomatal system, we complete an analysis of the entire complement of Arabidopsis MKKs and use information derived from their collective behavior to define regions of the proteins that are likely to provide developmental functions. We show that putative docking (D) domains have major roles in signal transduction but exhibit MKK-specific differences in the extent to which they contribute to efficient activation of downstream MPKs. We show that the D-domains of MKK7/9 are important in regulating subcellular localization of the kinases and that this localization is necessary for stomatal promoting activity. Furthermore, we can assign activity to a residue, recently shown to be phosphorylated in a tomato (Solanum lycopersicum) MKK homolog, in regulating divisions. Among domains for which there was no a priori predicted function, we find that N-terminal regions promote MKK4/5 functions in stomatal development, and we disprove the hypothesis that C-terminal extensions in MKK4/5 are cis-acting negative regulators of these kinases within the stomatal lineage. Finally, by testing matched variants of each of the four MKKs in multiple assays, we were also able to test the assumption that the behavior of MKK4 mirrors that of MKK5 and MKK7 mirrors that of MKK9 in the stomatal lineage.
RESULTS
Among the 10 Arabidopsis MKKs, Only MKK4/5/7/9 Affect Stomatal Development
Knowing the contribution of each member of an entire class of signaling proteins to a particular biological event facilitates the search for motifs that promote specificity in signaling modules. Our previous survey of stomatal development included seven of the 10 MKKs encoded in the Arabidopsis genome (Lampard et al., 2009). Of the remaining three MKKs, two (MKK8 and MKK10) are phylogenetically related to MKK7/9, joining them in the Group D subclade (Ichimura et al., 2002). The third, MKK3 (the sole member of Group B), can phosphorylate MPK3 and MPK6 and functions in similar contexts as MKK4/5/7/9 (Ichimura et al., 2002; Dóczi et al., 2007; Takahashi et al., 2007). It is plausible that these MKKs too could impact stomatal development. Therefore, to complete a comprehensive survey of the entire MKK gene family as it relates to stomatal development, we expressed CAMKK3, CAMKK8, and CAMKK10 at the SPCH, MUTE, and FAMA stages and screened >30 T1 plants of each for phenotypic effects. Activation of MKK3/8/10 did not perturb stomatal development at any of the SPCH, MUTE, or FAMA stages (Table 1). Taken together with previous results (Lampard et al., 2009), this suggested that neither similarity within the kinase domain of Group D MKKs (MKK7/8/9/10) nor the ability to phosphorylate common targets (MKK3 versus MKK4/5/7/9) is a good predictor of capacity to affect stomatal development.
Table 1. CAMKK3, CAMKK8, and CAMKK10 Are Not Able to Modulate Stomatal Development.
| Construct | Proportion of T1 Plants with Wild-Type Stomatal Distribution When Kinase Variant Was Expressed in: |
||
|---|---|---|---|
| SPCH | MUTE | FAMA | |
| CAMKK3 | 37/38 | 24/24 | 42/45 |
| CAMKK8 | 30/30 | 33/33 | 29/31 |
| CAMKK10 | 43/44 | 37/41 | 28/28 |
In the few cases where non-wild-type phenotypes were reported, they corresponded to marginal increases in paired stomata, a phenotype that can also occur due to antibiotic selection regimes.
Structural Differences between the MKK4/5 and MKK7/9 Pairs Are Potential Regulators of MKK Signaling
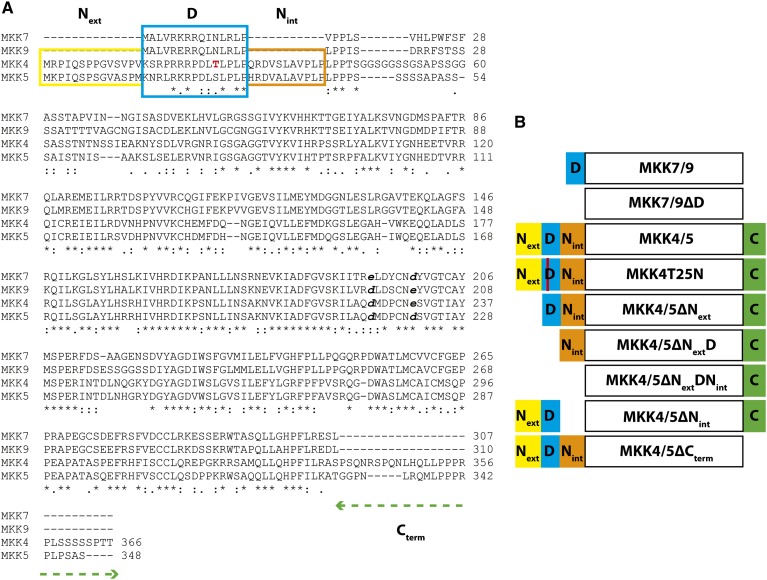
Multiple sequence alignment of the MKK family and representative MKK4/5 putative orthologs identified four regions of greater sequence divergence that might account for the ability of MKK4/5/7/9 to regulate stomatal development or the divergent functions of MKK4/5 and MKK7/9 during the FAMA stage (Figure 2; Supplemental Figure 1 and Supplemental Data Set 1). Three of these differences are N-terminal of the kinase domain, and one such region encodes a predicted D-domain. In other eukaryotes, the D-domains of MKKs, MAPK phosphatases, and substrates of MAPKs enable physical interactions with the complementary common docking domains of specific MAPKs (Grewal et al., 2006; Palacios et al., 2011). The extent to which D-domains contribute to plant MKK-MAPK interactions has not been thoroughly investigated; however, a D-domain is required for the interaction of tomato MKK2 (putatively orthologous to MKK4/5) with its downstream MAPK, SlMPK3 (Oh et al., 2013). The D-domains of MKK7/9 are located near the N terminus of the proteins and are separated from the canonical kinase domain by ∼30 amino acids. In comparison, the D-domains of MKK4/5 are preceded by a conserved 14-amino acid extension (Next) and followed by a 10-amino acid insertion (Nint) before the ∼30-amino acid stretch (Figure 2A). A final structural difference between the kinase pairs is the presence of a C-terminal extension in MKK4/5 (Cterm) that is absent from MKK7/9 (Figure 2A).
Figure 2.
Protein Sequences and Schematic Representation of MKK Domains.
(A) Multiple sequence alignment of Arabidopsis stomatal development-associated MKK proteins. Four variable regions differentiating MKK4/5 from MKK7/9 were examined for their contribution to signaling specificity; Next (yellow), Nint (orange), the D-domain (blue), and the C-terminal extension (green dashed line). Putative 14-3-3 binding site of MKK4 is in red. Full-length protein sequences used in alignment are provided in Supplemental Data Set 1.
(B) Schematic representation of CAMKK variant constructs (domains not to scale) designed to identify the contribution of each region to MKK signaling. The name of each construct is listed within the region depicting the canonical kinase domain of each protein.
We hypothesized that D-domains would mediate functional interactions between the MKKs and MPK3/6 and that these would be essential for MKK4/5/7/9 functions. Because only MKK7/9 are active at the FAMA stage, we hypothesized that MKK4/5 N- and C-terminal extensions serve a negative regulatory role; either actively inhibiting these kinases from participating in a signaling event or passively prohibiting their ability to participate in a signaling complex in which the smaller MKK7/9 proteins fit. To test these hypotheses, we employed a combination of in vitro biochemical assays and in vivo signaling pathway manipulations using CAMKK4/5/7/9 variants lacking each of the differential domains (Next, D-domain, Nint, and Cterm). Because MKK4/5 have three N-terminal segments, we built a panel of deletion variants and one point mutant to identify independent functions of each region (Figure 2B). CAMKK4/5∆Next and CAMKK4/5∆Nint targeted functions distinct from the D-domain, whereas CAMKK4/5∆NextD, CAMKK4/5∆NextDNint, and CAMKK4T25N were used to assess contributions of the D-domain to the MKK4/5 signaling module (Figure 2B). In addition, a pair of deletion constructs, CAMKK4/5∆Cterm, was designed to determine if the C-terminal extensions have regulatory functions on MKK4/5 (Figure 2B). Because the only differential region N-terminal to the MKK7/9 kinase domain is the D-domain, we created MKK7/9∆D variants to test the requirement for the D-domain in these kinases (Figure 2B).
Evaluating the Kinase Activity of MKK Deletion Variants
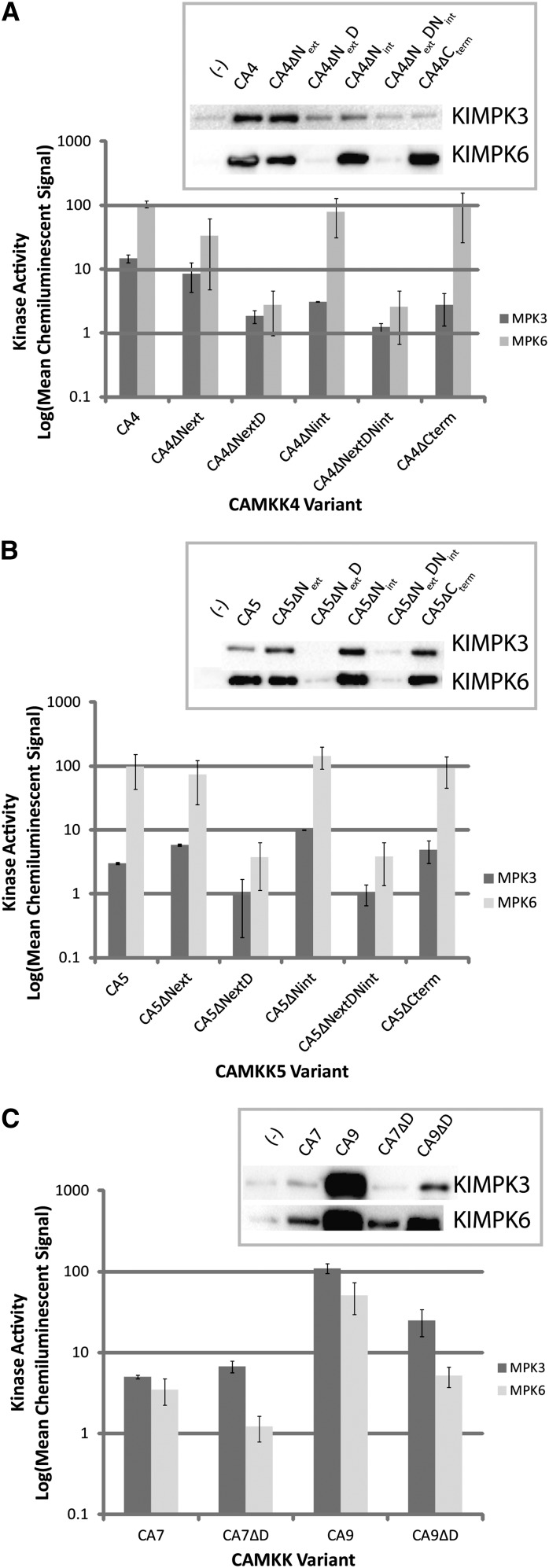
To evaluate whether our variants maintained their in vitro biochemical activity against known targets, each CAMKK4/5/7/9 variant was incubated with KIMPK3 or KIMPK6 and subsequent phosphorylation of the MAPK was detected using anti-phospho-ERK antibody. The relative activity of each CAMKK variant was determined by quantifying chemiluminescent signal intensity (Figure 3).
Figure 3.
In Vitro Kinase Activity of CAMKK Variants against Kinase Inactive MPK3 and KIMPK6.
In all assays, kinase signal is represented in logarithmic scale of the mean chemiluminescent signal from three replicates. Chemiluminescent signal was normalized to the signal from negative control reactions consisting of only KIMPK3 or KIMPK6. Error bars represent se.
(A) Results obtained for CAMKK4 (CA4) variants. Insets are representative immunoblots resulting from in vitro kinase assays using KIMPK3 or KIMPK6 and the specified CAMKK variant.
(B) Results obtained for CAMKK5 (CA5) variants. Insets are representative immunoblots resulting from in vitro kinase assays using KIMPK3 or KIMPK6 and the specified CAMKK variant. CAMKK4/5 D-domain deletion constructs lacked activity toward KIMPK3/6. All other variants retained the ability to phosphorylate KIMPK3/6.
(C) Results obtained for CAMKK7/9 (CA7/9) variants. Insets are representative immunoblots resulting from in vitro kinase assays using KIMPK3 or KIMPK6 and the specified CAMKK variant. D-domains of MKK7/9 are not essential for in vitro kinase activity toward KIMPK3/6.
Based on the behavior of D-domains in other systems, we expected that deletion of the D-domains from MKK4/5/7/9 might prevent physical association of the kinases with their substrates, thus eliminating kinase activity toward their MAPK targets. Indeed, both D-domain deletion variants of CAMKK4/5 (CAMKK4/5∆NextD and CAMKK4/5∆NextDNint) had severely impaired in vitro phosphorylation activity as shown by chemiluminescent signals from MPK3 and MPK6 at or near background levels (Figures 3A and 3B) and in a yeast two-hybrid assay for physical interactions, removal of the D-domain of MKK4 (MKK4ΔNextDNint) resulted in a protein that could no longer interact with MPK6 (Supplemental Figure 2). It was less clear whether the D-domain of CAMKK7/9 was essential for in vitro kinase activity against MPK3/6. Although less efficient than full-length CAMKK9, CAMKK9ΔD did retain some ability to phosphorylate both MPK3 and MPK6 (Figure 3C), implying that in these cases the D-domain is not absolutely essential for kinase activity (Figure 3C). In the case of CAMKK7∆D, there may also be a differential effect on the two substrates: Though not statistically significant, it appeared that CAMKK7∆D was inactive toward MPK6 but the activity toward MPK3 was maintained (Figure 3C). These unanticipated results were consistent with D-domains being important for, but not ultimately required by, MKK7/9 for their phosphorylation of downstream MAPKs.
If our original hypothesis that the additional regions of MKK4/5 function as MKK specificity determinants in vivo is correct, removal of these regions should result in either enhanced or unchanged kinase activity in vitro. Deleting either Next or Nint from CAMKK4/5 had a limited impact on in vitro kinase activity (Figures 3A and 3B). Similarly, the C-terminal extensions of MKK4/5 were not required for kinase activity toward MPK3/6 (Figures 3A and 3B). This is consistent with these regions being in vivo regulatory elements rather than explicitly controlling the kinase-substrate interaction.
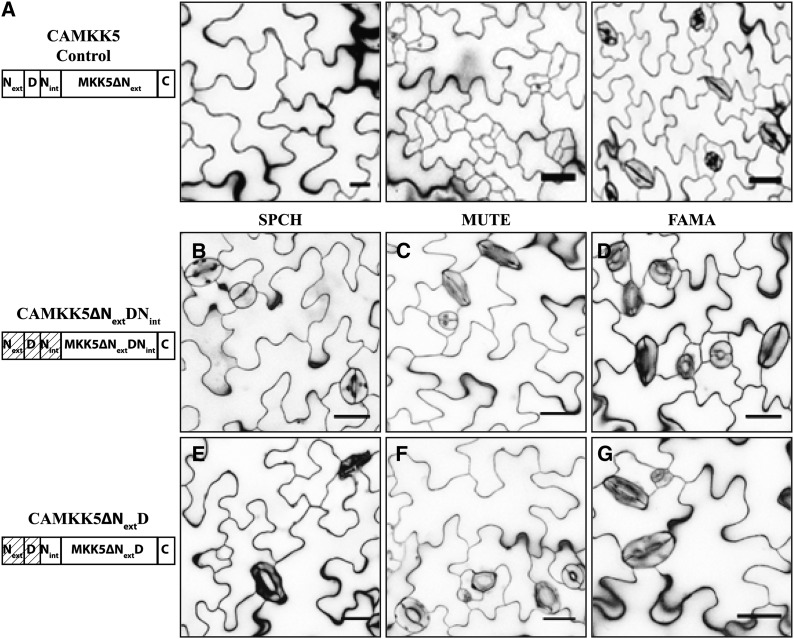
Deletion of the D-Domains Renders Stomatal MKKs Unable to Inhibit Stomatal Development
The in vitro kinase assays suggested that the D-domains of MKK4/5/7/9 are essential for MKK4/5 function and important but not essential for all MKK7/9 functions. To test these predicted functions in plants, we assessed the phenotypic consequences of expressing each CAMKK D-domain deletion variant as a yellow fluorescent protein (YFP) fusion during the SPCH, MUTE, and FAMA stages of stomatal development.
In both CAMKK4 and CAMKK5, removal of the entire N terminus (CAMKK4/5∆NextDNint) rendered these kinases incapable of inhibiting development during the SPCH and MUTE stages (Figures 4B and 4C show results for CAMKK5ΔNextDNint as representative for this type of variant; CAMKK4 deletion images are in Supplemental Figure 3; quantifications are in Table 2). Expression of these variants at the FAMA stage also had no impact on stomatal development (Figure 4D, Table 2). To ensure that the lack of effect on stomatal development was not due to a failure to express the kinase variant and/or transgene silencing, we verified that each construct was expressed at the correct stage of stomatal development by detecting fluorescence of the kinase variant-YFP fusion protein (Supplemental Figures 3H to 3M). Simultaneous removal of the Next and D-domains from CAMKK4/5 (CAMKK4/5∆NextD) had similar effects to CAMKK4/5∆NextDNint and these variants were also unable to modify any aspect of stomatal development (Figures 4E to 4G, Table 2; Supplemental Figures 3B to 3G). These results are consistent with in vitro results that the D-domain is required for a functional relationship between MKK4/5 and MPK3/6.
Figure 4.
D-Domain Deletion Variants of CAMKK5 Lose the Ability to Inhibit Stomatal Development When Expressed in the SPCH and MUTE Stages.
(A) Protein schematic (left) and images of phenotypes associated with expressing CAMKK5 in SPCH, MUTE, and FAMA stages of stomatal development as indicated.
(B) to (D) The 10-DPG seedlings expressing CAMKK5∆NextDNint in the SPCH (B), MUTE (C), and FAMA (D) stages show wild-type stomatal development patterns.
(E) to (G) The 10-DPG seedlings expressing CAMKK5∆NextD in the SPCH (E), MUTE (F), and FAMA (G) stages show wild-type stomatal development patterns.
A D-domain is required for stomatal inhibition by CAMKK5 and also does not prevent CAMKK5 from promoting stomatal clusters. Hashed boxes in schematic representations of kinase variants indicate the region(s) that were removed. All images are of 10-DPG abaxial cotyledons, and bars = 25 μm.
Table 2. Summary of in Vitro and in Vivo Behaviors of CAMKK Variants.
| Kinase Variant | In Vitro Activity on: (Fold over Background) |
Number of Independent T1 Plants Showing Altered Stomatal Development When Expressed in Stage: |
Visualize YFP in: |
|||||
|---|---|---|---|---|---|---|---|---|
| MPK3 | MPK6 | SPCH (Inhibit) | MUTE (Inhibit) | FAMA (Clusters) | SPCH | MUTE | FAMA | |
| MKK4 (Lampard et al., 2009) | 14.7 | 103.6 | Arrest | Arrest | Wild type | − | − | + |
| MKK4ΔNext | 8.4 | 33.6 | 2/42 | 1/31 | 0/25 | − | − | − |
| 4/12 T2s had paired stomata | 8/13 T2s had paired stomata | |||||||
| MKK4ΔNint | 3.1 | 79.5 | 2/48 | 1/67 | 0/30 | ++ | ++ | ++ |
| 4/10 T2s had paired stomata | 6/12 T2s had paired stomata | |||||||
| MKK4ΔNextD | 1.8 | 2.7 | 0/30 | 0/28 | 0/30 | ++ | ++ | ++ |
| MKK4ΔNextDNint | 1.2 | 2.6 | 0/27 | 0/22 | 0/26 | ++ | ++ | ++ |
| MKK4ΔCterm | 2.7 | 91.7 | 6/6 | 4/4 | 0/18 | − | − | ++ |
| MKK4T25N | 9.0 | 63.8 | 8/10 | N/A | 6/8a | ++ | N/A | ++ |
| MKK5 (Lampard et al., 2009) | 3.0 | 97.9 | Arrest | Arrest | Wild type | − | − | ++ |
| MKK5ΔNext | 5.8 | 74.1 | 1/32 | 2/45 | 0/28 | + | + | + |
| 3/7 T2s had paired stomata | 6/15 T2s had paired stomata | |||||||
| MKK5ΔNint | 10.0 | 144.5 | 3/62 | 1/55 | 0/28 | ++ | ++ | ++ |
| 9/12 T2s had paired stomata | 5/12 T2s had paired stomata | |||||||
| MKK5ΔNextD | 0.9 | 3.7 | 0/34 | 0/21 | 0/27 | ++ | ++ | ++ |
| MKK5ΔNextDNint | 1.0 | 3.8 | 0/36 | 0/24 | 0/31 | ++ | ++ | ++ |
| MKK5ΔCterm | 4.9 | 93.0 | 5/5 | 8/8 | 0/20 | − | − | ++ |
| MKK7 (Lampard et al., 2009) | 5.0 | 3.5 | Arrest | Arrest | Clusters | − | − | ++ |
| MKK7ΔD | 110.2 | 51.2 | 2/69 | 4/55 | 24/69b | + | + | + |
| MKK9 (Lampard et al., 2009) | 6.8 | 1.2 | Arrest | Arrest | Clusters | − | − | − |
| MKK9ΔD | 24.8 | 5.2 | 2/45 | 2/39 | 17/58b | ++ | ++ | ++ |
Some data in YFP visualization columns were taken from those presented by Lampard et al. (2009). Fluorescence scale is −, negative; +, weak; ++, strong. N/A, not applicable; this construct was not generated.
These plants showed paired stomata.
Stomatal clusters qualitatively smaller than those induced by full-length CAMKK7/9. Wild-type MKK variant phenotypes are as described (Lampard et al., 2009).
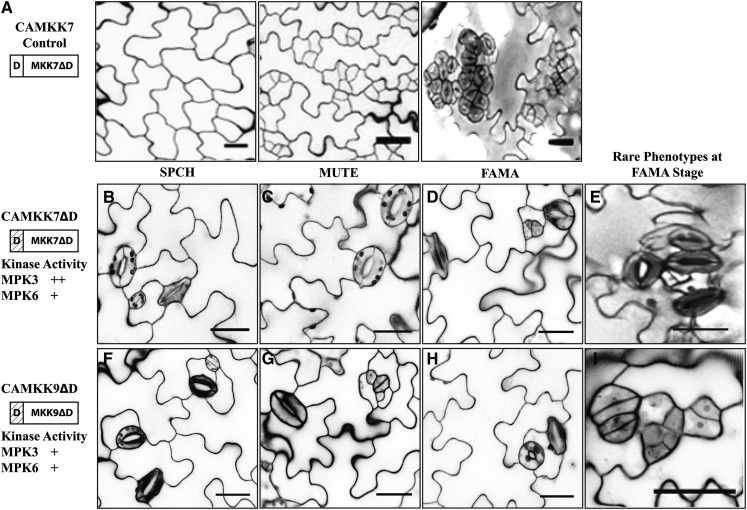
As with CAMKK4/5 variants lacking D-domains, CAMKK7/9ΔD variants were generally unable to inhibit stomatal development in the SPCH and MUTE stages (Figures 5B, 5C, 5F, and 5G, Table 2; Supplemental Figures 4B, 4C, 4E, and 4F). However, these variants retained greater activity during the FAMA stage. While the majority of plants expressing CAMKK7/9ΔD at the FAMA stage appeared wild-type (Figures 5D and 5G, Table 2), some plants produced small clusters of stomata (Figures 5E and 5I). In this less frequent class of plants, the extent of stomatal clustering was less severe, with each cluster containing fewer stomata than plants expressing full-length CAMKK7/9 (Figures 5A, 5E, and 5I). The phenotypes observed following expression of CAMKK7/9ΔD at the FAMA stage, combined with the in vitro kinase activities of CAMKK7/9ΔD, suggest that, unlike MKK4/5, stomatal proliferation is not entirely dependent on D-domain-based interactions with downstream MAPKs.
Figure 5.
The D-Domains of MKK7/9 Are Not Absolutely Necessary for CAMKK7/9 Functions in Stomatal Development.
(A) Protein schematic (left) and images of phenotypes associated with expressing CAMKK7 in the SPCH, MUTE, and FAMA stages of stomatal development as indicated.
(B) and (C) The cotyledon epidermis of plants expressing CAMKK7ΔD in the SPCH (B) and MUTE (C) stages generally have wild-type stomatal distribution.
(D) and (E) The cotyledon epidermis of plants expressing CAMKK7ΔD in the FAMA stage. The majority of these plants have wild-type stomatal development but occasionally have limited clusters of stomata.
(F) to (I) Cotyledon epidermis of plants expressing CAMKK9ΔD plants in the SPCH (F), MUTE (G), and FAMA ([H] and [I]) stages. CAMKK9ΔD has impacts on stomatal development in the same fashion as CAMKK7ΔD.
The relative activity of each kinase variant in in vitro kinase assays is depicted as follows (+, less active than full length; ++, comparable activity to the full-length CAMKK). Hashed boxes in schematic representations of kinase variants indicate the region(s) that were removed. All images are of 10-DPG abaxial cotyledons, and bars = 25 μm.
A 14-3-3 Binding Site in the D-Domain of MKK4 Contributes to MKK4 Function
The differential requirement for D-domains between MKK7/9 and MKK4/5 prompted us to look more closely at this domain. MKK4/5, but not MKK7/9, contain a S/T within the D-domain. In SIMKK2, the putative tomato MKK4/5 ortholog, phosphorylation at this site is required for interaction with a 14-3-3 protein, TOMATO FOURTEEN-THREE-THREE SEVEN (Sl-TFT7; Figure 2; Oh et al., 2013). Therefore, to test the relevance of this site for in vivo function, we modified Arabidopsis CAMKK4 to contain the MKK7/9 residue (asparagine [N]; CAMKK4T25N) and asked if this chimera would now acquire stomatal clustering activity during the FAMA stage. We verified that CAMKK4T25N retained in vitro kinase activity against KIMPK3/6 (Table 2) and that it inhibited stomatal development when expressed in the SPCH stage (Supplemental Figure 6A; Table 2). Interestingly, plants expressing CAMKK4T25N during the FAMA stage showed an increase in the production of stomatal pairs (Supplemental Figures 6B to 6D; Table 2). This phenotype could be interpreted as a weak clustering phenotype, consistent with the residue providing some specificity between MKK classes.
N-Terminal Regions Outside of the D-Domains of MKK4/5 Promote MKK4/5 Signaling
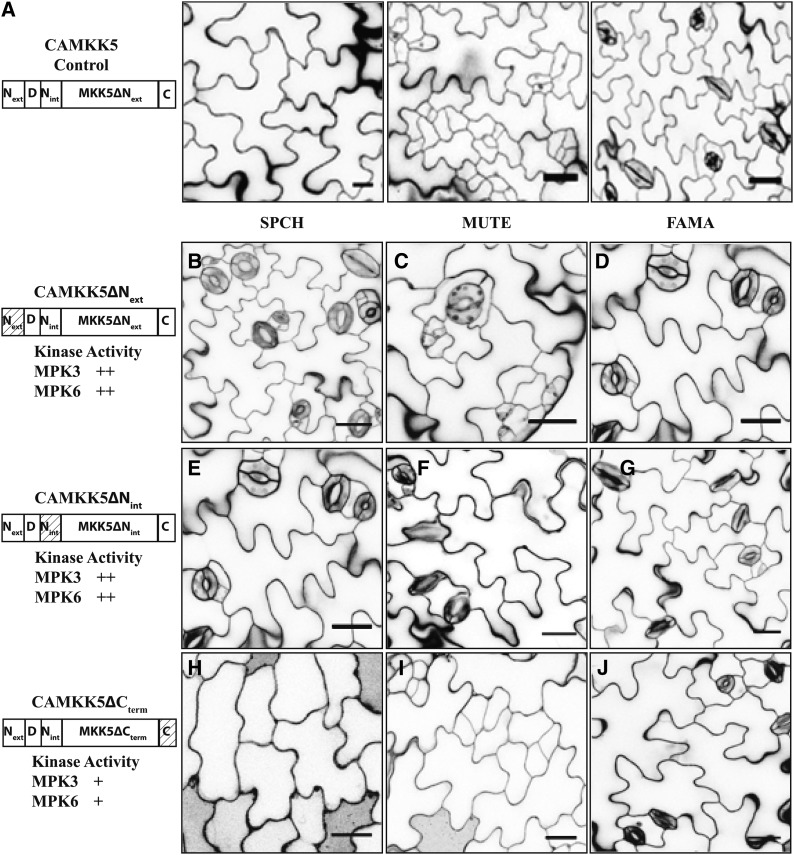
Both CAMKK4/5ΔNext and CAMKK4/5ΔNint retained kinase activity toward MPK3/6 in vitro. Therefore, it was plausible that Nint and Next, as features not found in MKK7/9, differentiate between kinase group activities at the FAMA stage. If so, removal of these regions would be expected to promote stomatal clustering. Expression of CAMKK4/5∆Next at the FAMA stage, however, had no capacity to induce stomatal clustering (Figure 6D, Table 2; Supplemental Figure 5). Unexpectedly, CAMKK4/5∆Next were also largely unable to inhibit stomatal development during the SPCH or MUTE stages (Figures 6B and 6C, Table 2; Supplemental Figure 5), even though their kinase activity was indistinguishable from full-length CAMKK4/5 in vitro (Figures 3A and 3B). These data indicate that Next plays a positive role in CAMKK4/5 activity in vivo but is not a feature distinguishing MKK4/5 from MKK7/9 in the FAMA stage.
Figure 6.
N-Terminal Extensions in CAMKK4/5 Promote Stomatal Inhibition in the SPCH and MUTE Stages.
(A) Protein schematic (left) and images of phenotypes associated with expressing CAMKK5 in the SPCH, MUTE, and FAMA stages of stomatal development as indicated.
(B) to (D) The 10-DPG seedlings expressing CAMKK5∆Next in the SPCH (B), MUTE (C), and FAMA (D) stages show wild-type stomatal development patterns.
(E) to (G) The 10-DPG seedlings expressing CAMKK5∆Nint in the SPCH (E), MUTE (F), and FAMA (G) stages show wild-type stomatal development patterns. Thus, both Next and Nint have a positive function in inhibiting stomatal development but do not prevent CAMKK4/5 from inducing stomatal clustering in the FAMA stage.
(H) to (J) The 10-DPG seedlings expressing CAMKK5∆Cterm in the SPCH (H), MUTE (I), and FAMA (J) stages show the same stomatal development patterns as controls (A).
The relative activity of each kinase variant in in vitro kinase assays is depicted as follows (+, less active than full length; ++, comparable activity to the full-length CAMKK). Hashed boxes in schematic representations of kinase variants indicate the region(s) that were removed. All images are of 10-DPG abaxial cotyledons, and bars = 25 μm.
We next tested the function of CAMKK4/5ΔNint deletion variants in each stomatal lineage cell type. No plants expressing CAMKK4/5ΔNint during the FAMA stage showed stomatal clustering (Figure 6G, Table 2; Supplemental Figure 5). Similar to CAMKK4/5ΔNext, the majority of plants expressing CAMKK4/5∆Nint in the SPCH or MUTE stages displayed wild-type stomatal development, while a small proportion of T1 plants had arrested stomatal development (Figures 6E and 6F, Table 2; Supplemental Figure 5). Therefore, as with the Next region, Nint enhances the function of CAMKK4/5 in the SPCH and MUTE stages but neither Next or Nint is responsible for preventing MKK4/5 from inducing stomatal clustering.
The C-Terminal Extensions on MKK4/5 Are Not Required for Control of Stomatal Development
Since MKK7/9 lack a C-terminal extension and MKK4/5∆Cterm variants maintained full kinase activity, we did not expect the elimination of the C-terminal extension from CAMKK4/5 to lessen the inhibition of stomatal development at the SPCH or MUTE stages. Indeed, stomatal development was arrested in plants expressing CAMKK4/5∆Cterm at these stages (Figures 6H and 6I, Table 2; Supplemental Figures 7B, 7C, 7E, and 7F). We then tested the possibility that the region prevents CAMKK4/5 from inducing stomatal clustering at the FAMA stage; however, FAMApro:CAMKK4/5∆Cterm variants do not alter stomatal development (Figure 6J, Table 2; Supplemental Figures 7D and 7G). Thus, the C-terminal region may be dispensable for signal transduction during stomatal development.
MKK7/9 D-Domains Impact Subcellular Localization and Stomatal Clustering Ability
Because all MKK variants were YFP tagged for expression in planta, we uncovered another, unanticipated, mode of regulation related to subcellular localization. We found full-length CAMKK7 in punctate cytoplasmic structures (Figure 7A; Lampard et al., 2009) that colocalize with the mitochondrial marker MitoTracker-Orange (Figures 7B to 7D). Moreover, time-lapse imaging of CAMKK7 (Supplemental Movie 1 and Supplemental Figure 8) reveals that its dynamics mirror those of mitochondria (Supplemental Movie 2 and Supplemental Figure 8) in the stomatal lineage. Variants such as CAMKK7ΔD that lose stomatal cluster-promoting activity, however, are not localized to MitoTracker-positive puncta (Figures 7G and 7H). To further test whether activity correlates with subcellular localization, we examined the localization of other MKKs. Previous microscopy capabilities were insufficient for detection of CAMKK9 (Lampard et al., 2009); however, with the use of ultrasensitive hybrid detectors, we could see CAMKK9-YFP in GMCs and guard cells (Supplemental Figure 9A). CAMKK9 is diffuse in the cytoplasm but, like CAMKK7, is also in punctate structures resembling mitochondria. As with CAMKK7ΔD, the punctate localization is specifically lost in the inactive CAMKK9ΔD (Supplemental Figures 9D to 9F), providing another link between stomatal clustering activity and specific subcellular localization. CAMKK4/5, which are incapable of inducing stomatal clustering in the FAMA stage, also do not localize to mitochondria (Supplemental Figures 9B and 9C).
Figure 7.
Colocalization of Active MKKs with Mitochondria and Correlation of Subcellular Localization with Activity in Stomatal Assays.
(A) Confocal image of a mature guard cell expressing CAMKK7-YFP (green), which localizes to punctate structures.
(B) and (C) Confocal image of mature guard cells expressing WTMKK7-YFP (B) or a catalytically inactive (KI)-CAMKK7 (C). These MKK7 variants also localize to punctate structures.
(D) Confocal image of a mature guard cell expressing CAMKK7∆D. This MKK7 variant localizes to larger structures not consistent with mitochondria.
(E) and (F) Confocal images of a pair of guard cells expressing CAMKK7-YFP that have been treated with MitoTracker Orange. The punctuate structures described in (A) and shown in (E) stain positively with MitoTracker Orange (F).
(G) Overlay of MitoTracker Orange and YFP signals described in (E) and (F). Arrow indicates enlargement of the region measured (yellow) to show colocalization of fluorescent signal. Merging of signals suggests CAMKK7 colocalizes with mitochondria. Bar = 15 μm.
(H) Graph depicting fluorescence signals measured along yellow line (G) for YFP (E), MitoTracker (F), and overlaid signals (G). The pattern of fluorescence intensities suggests that CAMKK7-YFP and MitoTracker Orange signals colocalize. Note that CAMKK7∆D in (D) colocalizes with large cytosolic structures and not mitochondria-like structures. The CAMKK7∆D construct fails to induce CAMKK7-like stomatal clusters. All images were collected at 400× magnification and are of 5-DPG abaxial cotyledons.
A recent report described localization of MPKs to punctate structures in response to activation (Ovečka et al., 2014). To test whether mitochondrial localization of CAMKK7 is a property of their constitutive activation or, alternatively, of their kinase activity, we monitored the localization of nonmodified MKK7-YFP and kinase-inactive (KI), yet constitutively active, variants (KI-CAMKK7-YFP) that fail to impact stomatal development at any stage. These two variants showed subcellular localization patterns similar to that of CAMKK7 (Figures 7E and 7F), though neither induced stomatal clustering. These results indicate that stomatal clustering requires both correct MKK subcellular localization and kinase activity.
DISCUSSION
Previously, we uncovered roles for four MKKs, MKK4/5/7/9, in controlling various aspects of stomatal development (Lampard et al., 2009), yet many questions regarding the generation of MAPK signaling fidelity remained. For example, it was unknown what enables these four MKKs, but not other MKKs that share common substrates, to specifically function within each stomatal lineage cell type. Furthermore, the means by which FAMA-staged cells are responsive to MKK7/9 activity but not MKK4/5 activity remained uncharacterized. Beyond their functions in controlling stomatal development, these MKKs function broadly in other signaling pathways and in other cell types (Rodriguez et al., 2010). Therefore, answering these questions could provide general insights into how MAPK signaling networks are regulated in planta. We exploited the power of the Arabidopsis stomatal lineage to study how MAPK signal output is influenced by discrete structural domains of MAPK signaling components: By activating MAPK signaling using CAMKK deletion variants in three discrete cell types, we identified protein regions and residues involved in the regulation of MKK4/5 activity for which there were no a priori predicted functions. More importantly, we established that MKK4/5 and MKK7/9 have different requirements for their D-domains in planta and that D-domains of plant MKKs are involved in controlling signaling output through at least two mechanisms: (1) as a result of posttranslational modification and (2) through influencing subcellular localization of MAPK signaling components. The high degree of sequence conservation of MKK gene family members among plants suggests that these mechanisms are conserved across species.
MKK4/5 and MKK7/9 differ primarily by the presence of N- and C-terminal extensions on MKK4/5. Removing these extensions from CAMKK4/5 had little impact on kinase activity in vitro, yet when tested in vivo, alterations to the N terminus had significant effects on signaling. Both N-terminal extensions (Next and Nint) of MKK4/5 enhanced the ability of CAMKK4/5 to inhibit stomatal development in the SPCH and MUTE stages. This could indicate that Next and Nint function to provide a particular three-dimensional structure to facilitate binding of the MKK via the D-domain to downstream MAPKs, thus enabling signal propagation. However, this does not account for the retention of in vitro catalytic activity on MPK3/6 by CAMKK4/5∆Next/int variants. Alternatively, both Next and Nint may have specific structural roles independent from the D-domain. For example, these domains may be required to mediate interactions with signaling partners such as putative scaffold protein(s) or additional regulatory proteins in vivo. If this were the case, stochastic alterations in CAMKK4/5∆Next/int localization or interactions could occasionally lead to normal signaling but also could inactivate signaling by sequestering downstream MAPKs in nonproductive complexes. Such effects were indeed observed, as plants expressing CAMKK4/5∆Next or CAMKK4/5∆Nint during the SPCH and MUTE stages occasionally showed paired and triplet stomata (Table 2), a phenotype comparable to that of a dominant-negative YDA variant expressed during the SPCH stage (Lampard et al., 2009; Lampard, 2009). The infrequent ability of CAMKK4/5∆Next and CAMKK4/5∆Nint to inhibit stomatal development could result from the transgenes being expressed at sufficient levels to negate the necessity of a scaffold protein and/or since MKKs are known to function as dimers (Ohren et al., 2004), CAMKK4/5∆Next/int variants may have been recruited to the proper signaling complex as a dimer with endogenous MKK4/5, where they were able to induce inhibition of stomatal development.
Thus far, we find no role for the C-terminal extensions in modulating CAMKK4/5 activity during stomatal development. These extensions, however, are conserved among plant MKK4/5 homologs, and a predicted function of these, based on comparison with mammalian MKKs, is that they could mediate activation by upstream activating MAPKKKs (Takekawa et al., 2005). Since our use of CAMKK4/5 variants abrogates the necessity for interaction with and phosphorylation by any upstream MAPKKK(s), clarification of the functional roles of MKK4/5 C-terminal extensions will require further studies examining the effects of both expressing CAMKK4/5∆Cterm variants in additional cell types and assaying the functions of WT-MKK4/5∆Cterm variants within the stomatal lineage.
The CAMKK4/5 D-domain deletion variants used in this study concurrently removed one or both of Next/int, which should result in a nonfunctional kinase that also fails to interact with a signaling complex. Therefore, when expressed in plants, the expected phenotype would be a failure to inhibit stomatal development without seeing any dominant negative-like phenotypes (paired or triplet stomata). Our observations that all CAMKK4/5 D-domain deletion variants lacked biochemical activities in vitro, failed to interact with MPK6 via yeast two-hybrid analysis and had no biological activities in vivo are therefore consistent with MKK D-domains mediating the physical and subsequent interactions with downstream MAPKs and with MKK4/5 interacting with a putative scaffold protein via Next/int (Grewal et al., 2006; Saito, 2010). Future studies examining protein complex purification from isolated SPCH and/or MUTE stage stomatal lineage cells may ultimately reveal the nature of this YODA-MAPK module scaffold protein.
Removal of the D-domain from MKK7/9 diminished, but did not eliminate, functionality in vivo, suggesting that unlike MKK4/5, the D-domains of MKK7/9 are not absolutely required for catalytic activity toward KIMPK3/6. Intriguingly, these data are consistent with a scaffold protein influencing inhibition of stomatal development via MKK7/9: In yeast, elimination of the D-domain from the MKK, Ste7, only decreases the pheromone response and complete abolishment requires the additional deletion of the Ste5 scaffold protein (Bardwell et al., 2001). Since MKK7/9∆D variants retain in vitro catalytic activity toward MPK3/6, it is plausible that inhibition of stomatal development by CAMKK7/9 is influenced by a yet to be identified Ste5-like scaffold protein.
Interestingly, the extent to which removal of the D-domain from MKK7/9 impacted in vivo function was cell type specific: While inhibition of stomatal development was severely compromised in the SPCH and MUTE stages, roughly one-third of plants expressing CAMKK7/9∆D during the FAMA stage of stomatal development showed limited stomatal clustering. This could indicate that a MAPK(s) other than MPK3/6 is involved in promoting stomatal clustering at the FAMA stage and this activity is not dependent upon the D-domain. Such a scenario is plausible, as MKK7 and MKK9 have been reported to interact with Group D MAPKs: MKK7 with MPK15 and MKK9 with both MPK17 and MPK19 (Lee et al., 2008). These interactions are presumably D-domain independent as Group D MAPKs lack common docking domains that are required for interaction with D-domains (Ichimura et al., 2002). The yeast MAPK phosphatase Msg5 illustrates this paradigm; Msg5 interacts with and can dephosphorylate each of the MAPKs, Fus3, Kss1, and Slt2 (Palacios et al., 2011). The interactions with Fus3 and Kss1 require the D-domain of Msg5 but the interaction with Slt2 is D-domain independent (Palacios et al., 2011).
Irrespective of which kinases are downstream of MKK7/9 during the FAMA stage, our findings suggest that the divergence of function between the MKK4/5 and MKK7/9 pairs at this stage is specifically related to regulation of subcellular localization of MKKs. Stomatal clustering was correlated with CAMKK7/9 displaying mitochondrial colocalization, while all MKK variants incapable of promoting stomatal clustering (CAMKK7/9∆D and all CAMKK4/5 variants) showed alternative subcellular localization patterns in all stomatal lineage cell types. Recent findings indicate that in plants, similar to other eukaryotes, relocalization of MAPK components upon activation is needed for proper signal output (Ovečka et al., 2014). Here, we show that mitochondrial localization and kinase activity are required for MKK7/9 to promote stomatal clustering. Multiple associations between MAPK signaling and mitochondrial localization have been reported in mammals, and these links involve signaling of MAPK components from both within and on the outer surface of these organelles (Wortzel and Seger, 2011). Intriguingly, human MEK1/2 (MKK) and ERK1/2 display translocation to mitochondria in response to treatment with H2O2, and differing levels of H2O2 are known to both promote and inhibit the cell cycle via these MAPKs (Galli et al., 2008). The method of translocation is unknown as neither MEK1/2 nor ERK1/2 contain mitochondrial transit peptides. Since MKK7/9 activity is associated with uncontrolled cell divisions (similar to tumor formation), it is tempting to speculate that MKK7/9 are involved in controlling the cell cycle via MPK3/6 following a similar mitochondrial translocation mechanism. In support of this, both MPK3 and MPK6 are involved in redox signaling (Lee and Ellis, 2007), and MPK3 has been specifically linked with controlling the response to H2O2 in stomata (Gudesblat et al., 2007).
If MPK3 and MPK6 are the MPKs controlling stomatal clustering in the FAMA stage, localization of MKK7/9 and downstream MAPKs to the mitochondria via a scaffold protein could account for both how CAMKK7/9∆D variants retain a limited ability to drive the formation of stomatal clusters and why CAMKK4/5 fail to induce stomatal clustering. Here, the putative scaffold protein would exclude MKK4/5 while selectively maintaining a pool of MKK7/9 at mitochondria via interactions with the D-domain. The retention of an ability to induce limited stomatal clusters by CAMKK7/9∆D variants could then be the result of being recruited to the scaffold as a dimer with endogenous MKK7/9. Our observation of a range of phenotypes in plants expressing CAMKK7/9∆D, including wild-type stomatal patterning, paired stomata, and small clusters, supports this. Further studies characterizing the nature of MKK7/9 and downstream component targeting to mitochondria and precise analysis of whether signals originate from within or on the surface of the mitochondria will allow for more thorough refinement of this model.
Our studies indicating possible dual roles for D-domains of MKKs in controlling interactions with downstream MAPKs and protein localization/interactions contribute to the growing evidence that the D-domains of plant MKKs are multifunctional (Oh and Martin, 2011; Oh et al., 2013). In tomato, Sl-MKK2 contains a D-domain that is essential for interaction with both the downstream MAPK, SlMPK3, and the 14-3-3 protein TFT7 (Oh and Martin, 2011; Oh et al., 2013). The biological relevance of this overlapping function has yet to be determined, but TFT7 has been proposed to serve as a scaffold protein between Sl-MKK2 and the upstream MAPKKK, Sl-MAPKKKα (Oh and Martin, 2011), while a functional D-domain is required for the Sl-MKK2 to induce programmed cell death via Sl-MPK3 (Oh et al., 2013). MKK4/5 are orthologous to Sl-MKK2 and contain a conserved 14-3-3 binding site within the D-domains. Given the putative scaffolding role of TFT7 and the report that human 14-3-3ζ prevents mitochondrial translocation of the cell death-related protein BCL2-ASSOCIATED AGONIST OF CELL DEATH (Lim et al., 2013), we sought to identify whether this 14-3-3 binding site plays a role in preventing MKK4/5 from inducing stomatal clustering. Initial observation of plants expressing a CAMKK4T25N variant containing an altered D-domain that (1) abolished the 14-3-3 binding site and (2) made it more similar to the D-domains of MKK7/9 suggested that CAMKK4T25N mimicked MKK7/9 during the FAMA stage, as plants expressing this variant had an epidermis containing paired and tripled stomata. However, further examination suggests these developmental abnormalities arise via a different type of failure to control cell division/differentiation than that mediated by CAMKK7/9. CAMKK4T25N did not colocalize with mitochondria, and pairs of stomata resulting from CAMKK4T25N expression during the FAMA stage appeared developmentally distinct from those induced by CAMKK7/9 (Supplemental Figure 5). In the former, pairs at 7 d postgermination (DPG) often contain one large and one smaller stomata, whereas in the latter, stomata within pairs or clusters are of similar size leading to the possibility that studying these variants within the stomatal lineage has uncovered novel roles of MKK4/5 in regulating cell division and/or differentiation.
In this study, CAMKK4 and CAMKK5 variants were examined in parallel. These two kinases have often been considered interchangeable, though specific data supporting this assumption in vivo were lacking (Asai et al., 2002; Wang et al., 2007). Here, we provide evidence that, at least in one developmental context, MKK4 and MKK5 mediate the same activities, providing some of the strongest evidence that assumptions about functional redundancy are not unreasonable. Intriguingly, despite a dearth of reports showing overlapping functions for MKK7 and MKK9, these kinases appeared to function interchangeably in the context of controlling stomatal development. Though MKK7 and MKK9 share mitochondrial localization, the subtle differences in their subcellular localization patterns should be examined in the context of additional aspects of MKK7/9 signaling.
Several points of evidence from this study suggest that structural regions outside of the canonical kinase domains of MKKs mediate signal output and specificity through controlling interactions with regulatory scaffold proteins. Furthermore, in the case of MKK7/9, these domains dictate discrete subcellular localization patterns that are associated with specific signaling outcomes in specialized cell types. These results likely have implications beyond MAPK control of stomatal development on multiple levels. First, the molecular tools generated here can be coupled with additional cell type-specific promoters to characterize (1) how broadly these structures function to control MAPK signal output in different cell types and (2) additional structures of MKKs involved in controlling signal specificity. Beyond this, associating a specific signaling outcome (stomatal clustering) to a specific cell type (guard mother cell) and subcellular localization (mitochondria) can lead to more directed approaches to finally answer questions regarding the existence of MAPK scaffolding proteins in plants.
METHODS
Construction of CAMKK Variants
cDNA corresponding to MKK8 in pCR8 (Lee et al., 2008) was used as a starting template. Elimination of stop codons and conversion of activation sequences to render the kinases constitutively active was completed as previously described (Lampard et al., 2009) using oligonucleotides listed in Supplemental Table 1. cDNA encoding CAMKK3 was provided by Brian Ellis, and cDNA encoding CAMKK10 was kindly supplied by Sorina Popescu and previously described by Popescu et al. (2009).
Deletion variants for CA-MKK4/5/7/9 were created using cDNA clones in pENTR-D-TOPO (Life Technologies) as previously described (Lampard et al., 2009). Deletion constructs were created using Accuprime Pfx (Life Technologies) and oligonucleotides listed in Supplemental Table 1. Briefly, deletion variants were amplified by PCR and the resultant amplicons were cloned into pENTR-D-TOPO. For all N-terminal variants with the exception of the ∆Nint variants, new start codons were added to the 5′ ends of forward oligonucleotides (Supplemental Table 1). ∆Nint variants were constructed using fusion PCR using overlapping oligonucleotides lacking the deleted sequences (Supplemental Table 1).
Creation and Phenotypic Analysis of Transgenic Arabidopsis thaliana Plants
For expression of CAMKK variants in the stomatal lineage, SPCH, MUTE, or FAMA promoters were ligated into the NotI site within the pENTR-D-TOPO plasmid immediately upstream of the cDNA sequences (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007). Each promoter-CAMKK construct was then recombined into pHGY (Kubo et al., 2005). For CAMKK4/5∆Nint variants, sequences recombined along with either the SPCH, MUTE, or FAMA promoter sequences contained within pDONR_P4_P1R (Life Technologies) into R4pGWB540 (Nakagawa et al., 2008) for Agrobacterium tumefaciens-mediated transformation. Columbia-0 was the ecotype used as the wild type for all transgenic plants.
Plants were selected on antibiotic and analyzed as T1s (except as noted). Confocal images of seedlings at 5 and 10 DPG were collected using a Leica SP5 confocal microscope equipped with HyD detectors with excitation/emission spectra of 514/520 to 540 for YFP, 565/575 to 600 for MitoTracker Orange, and 565/580 to 610 for propidium iodide counterstaining. Images were processed in ImageJ (NIH). For mitochondrial staining, seedlings were immersed in 200 nM MitoTracker Orange CMTMRos (Life Technologies) and placed in a vacuum infiltrator for 5 min followed by incubation at room temperature for 25 min.
Recombinant Protein Expression and Purification
N-terminal glutathione S-transferase (GST)-tagged versions of CAMKK variants were made in pDEST15 via LR recombination and introduced into Escherichia coli BL21 codon plus DE3 RIL cells (Agilent Technologies). Recombinant proteins were expressed as follows: a 5-mL overnight culture of transformed cells in Luria-Bertani broth was diluted 1:50 in 250 mL prewarmed (37°C) Luria-Bertani broth. The resultant cell suspension was grown while shaking at 250 rpm at 37°C until an OD600 of 0.5 to 1.0 was reached. Heterologous gene expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (0.5 mM final concentration) followed by continued growth while shaking at 250 rpm at 37°C for four hours. E. coli cells were harvested by centrifugation at 4000g for 10 min and stored at −80°C until recombinant protein purification.
GST-tagged MAPK proteins were purified batch-style using glutathione-Sepharose 4B beads (GE Healthcare) according to the manufacturer's protocols. Recombinant proteins were stored at −80°C until further use. To ensure equal loading of kinase variants, 500 ng of each GST-tagged variant was resolved on SDS-PAGE gels and subsequently transferred to polyvinylidene fluoride membranes. Membranes were probed with anti-GST antibody (kindly provided by Or Gozani, Department of Biology, Stanford University) and signal intensity was analyzed using a Bio-Rad ChemiDoc XRS+ system and ImageLab Software (Bio-Rad). The volume of each kinase variant used in the in vitro kinase assays was adjusted to ensure equal loading.
In Vitro Kinase Assays
In vitro kinase assays to assess the ability of CAMKK variants to phosphorylate either kinase inactive (KI) MPK3 or MPK6 were performed as follows: 250 ng of recombinant CAMKK variant MAPK and 500 ng of KIMPK were diluted to a total volume of 20 μL in 10× kinase buffer (250 mM Tris HCl, pH 7.5, 10 mM EGTA, 10 mM DTT, 50 mM MgCl2, 2 mM ATP, 50 mM β-glycerophosphate, 1 mM sodium orthovanadate, and 10× Protease inhibitor) and water. Reactions were incubated at 30°C for 30 min and were then stopped by the addition of 6× Laemmli buffer followed by incubation at 95°C for 10 min. Samples were resolved on SDS-PAGE gels, and separated proteins were transferred to polyvinylidene fluoride membranes. Blots were probed using anti-Phospho-p44/42 MAPK antibody (Erk1/2; Thr-202/Tyr-204; Cell Signaling Technology) that specifically recognizes the phosphorylated forms of MPK3 and MPK6. Band intensity was detected and analyzed using ImageLab (Bio-Rad). Each reaction was performed in triplicate, and mean signal intensity was normalized to the average signal of MPK3 or MPK6 incubated in kinase buffer in the absence of any MKK variant.
Yeast Two-Hybrid Assays
MKK4, CAMKK4, and MKK4ΔNextDNint each were cloned into pENTR according to the manufacturer's protocols. Each MKK4 variant was subsequently inserted into pDEST22 via LR recombination. MPK6 was transferred from pENTR to pDEST32 also via LR recombination. The yeast strain MaV203 was cotransformed with a single MKK4-pDEST32 variant and MPK6-pDEST22. Cotransformed yeast cells were selected for by growth on minimal media plates lacking Trp and Leu (SC-Leu-Trp). The level of interaction between MKK4 variants and MPK6 was scored using minimal media plates lacking Leu, Trp, and Ura (SC-Leu-Trp-Ura) with positive interactions showing growth on these plates.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes studied in this work are: MKK4 (At1g51660), MKK5 (At3g21220), MKK7 (At1g18350), MKK9 (At1g73500), MPK3 (At3g45640), and MPK6 (At2g43790).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Analysis of MKKs.
Supplemental Figure 2. Validation of Expression of D-Domain Deletion Variants of CAMKK4/5.
Supplemental Figure 3. Validation of Expression of D-Domain Deletion Variants of CAMKK7/9.
Supplemental Figure 4. Validation of Expression of N-Terminal Deletion Variants of CAMKK4/5.
Supplemental Figure 5. Evaluation of the Role of T25 in Regulating CAMKK4 Function within the Stomatal Lineage.
Supplemental Figure 6. The C-Terminal Extensions of CAMKK4/5 Are Not Associated with the Control of Stomatal Development.
Supplemental Figure 7. CAMKK Variants That Are Associated with Stomatal Clustering Are Localized to Punctate Structures.
Supplemental Figure 8. Deletion of the D-Domain from MKK4 Prevents Interaction with MPK6.
Supplemental Figure 9. Movement of CAMKK7 in Mature Guard Cells Resembles Movement of Mitochondria.
Supplemental Table 1. Oligonucleotides Used in This Study.
Supplemental Data Set 1. Protein Sequences of MKKs Used in Alignments.
Supplemental Movie 1. Movement of CAMKK7 in Guard Cells: Cotyledon of 7 DPG Plant Expressing FAMAproCAMKK7-YFP.
Supplemental Movie 2. Movement of MitoTracker Orange-Labeled Mitochondria in Guard Cells.
Supplementary Material
Acknowledgments
We thank Sorina Popescu (Boyce Thompson Institute) for the gift of the MKK10 clones. This work was supported in part by DOE-FG02-06ER15810. D.C.B. is a Gordon and Bettye Moore Foundation investigator of the Howard Hughes Medical Institute.
AUTHOR CONTRIBUTIONS
G.R.L., D.L.W., and D.C.B. designed the research, analyzed data, and wrote the article. G.R.L. and D.L.W. performed research.
Glossary
- MAPK
mitogen-activated protein kinase
- MAPKKK
MAPK kinase kinase
- MKK
MAPK kinase
- GMC
guard mother cell
- DPG
7 days postgermination
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Bardwell A.J., Flatauer L.J., Matsukuma K., Thorner J., Bardwell L. (2001). A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276: 10374–10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóczi R., Brader G., Pettkó-Szandtner A., Rajh I., Djamei A., Pitzschke A., Teige M., Hirt H. (2007). The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Bergmann D.C. (2010). Stomatal patterning and development. Curr. Top. Dev. Biol. 91: 267–297. [DOI] [PubMed] [Google Scholar]
- Galli S., Antico Arciuch V.G., Poderoso C., Converso D.P., Zhou Q., Bal de Kier Joffé E., Cadenas E., Boczkowski J., Carreras M.C., Poderoso J.J. (2008). Tumor cell phenotype is sustained by selective MAPK oxidation in mitochondria. PLoS ONE 3: e2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Besteiro M.A., Ulm R. (2013). Phosphorylation and stabilization of Arabidopsis MAP kinase phosphatase 1 in response to UV-B stress. J. Biol. Chem. 288: 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S., Molina D.M., Bardwell L. (2006). Mitogen-activated protein kinase (MAPK)-docking sites in MAPK kinases function as tethers that are crucial for MAPK regulation in vivo. Cell. Signal. 18: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat G.E., Iusem N.D., Morris P.C. (2007). Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol. 173: 713–721. [DOI] [PubMed] [Google Scholar]
- Ichimura K., Shinozaki K., Tena G., Sheen J., Henry Y., Champion A., Kreis M., Zhang S., Hirt H., MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7: 301–308. [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R. (2009). The missing link?: Arabidopsis SPCH is a MAPK specificity factor that controls entry into the stomatal lineage. Plant Signal. Behav. 4: 425–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., Lukowitz W., Ellis B.E., Bergmann D.C. (2009). Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21: 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Ellis B.E. (2007). Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J. Biol. Chem. 282: 25020–25029. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Huh K.W., Bhargava A., Ellis B.E. (2008). Comprehensive analysis of protein-protein interactions between Arabidopsis MAPKs and MAPK kinases helps define potential MAPK signalling modules. Plant Signal. Behav. 3: 1037–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G.E., Piske M., Johnson J.D. (2013). 14-3-3 proteins are essential signalling hubs for beta cell survival. Diabetologia 56: 825–837. [DOI] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nakamura S., Tanaka K., Kawamukai M., Suzuki T., Nakamura K., Kimura T., Ishiguro S. (2008). Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci. Biotechnol. Biochem. 72: 624–629. [DOI] [PubMed] [Google Scholar]
- Oh C.S., Martin G.B. (2011). Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. J. Biol. Chem. 286: 14129–14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C.S., Hwang J., Choi M.S., Kang B.C., Martin G.B. (2013). Two leucines in the N-terminal MAPK-docking site of tomato SlMKK2 are critical for interaction with a downstream MAPK to elicit programmed cell death associated with plant immunity. FEBS Lett. 587: 1460–1465. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohren J.F., et al. (2004). Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol. 11: 1192–1197. [DOI] [PubMed] [Google Scholar]
- Ovečka M., et al. (2014). Salt-induced subcellular kinase relocation and seedling susceptibility caused by overexpression of Medicago SIMKK in Arabidopsis. J. Exp. Bot. 65: 2335–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios L., Dickinson R.J., Sacristán-Reviriego A., Didmon M.P., Marín M.J., Martín H., Keyse S.M., Molina M. (2011). Distinct docking mechanisms mediate interactions between the Msg5 phosphatase and mating or cell integrity mitogen-activated protein kinases (MAPKs) in Saccharomyces cerevisiae. J. Biol. Chem. 286: 42037–42050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505. [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Saito H. (2010). Regulation of cross-talk in yeast MAPK signaling pathways. Curr. Opin. Microbiol. 13: 677–683. [DOI] [PubMed] [Google Scholar]
- Taj G., Agarwal P., Grant M., Kumar A. (2010). MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signal. Behav. 5: 1370–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K. (2007). The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Soyano T., Kosetsu K., Sasabe M., Machida Y. (2010). HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51: 1766–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa M., Tatebayashi K., Saito H. (2005). Conserved docking site is essential for activation of mammalian MAP kinase kinases by specific MAP kinase kinase kinases. Mol. Cell 18: 295–306. [DOI] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer R., Baccarini M. (2010). Partner exchange: protein-protein interactions in the Raf pathway. Trends Biochem. Sci. 35: 660–668. [DOI] [PubMed] [Google Scholar]
- Wortzel I., Seger R. (2011). The ERK cascade: Distinct functions within various subcellular organelles. Genes Cancer 2: 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.