Abstract
Background
WRKY proteins comprise a large family of transcription factors and play important regulatory roles in plant development and defense response. The WRKY gene family in Salvia miltiorrhiza has not been characterized.
Results
A total of 61 SmWRKYs were cloned from S. miltiorrhiza. Multiple sequence alignment showed that SmWRKYs could be classified into 3 groups and 8 subgroups. Sequence features, the WRKY domain and other motifs of SmWRKYs are largely conserved with Arabidopsis AtWRKYs. Each group of WRKY domains contains characteristic conserved sequences, and group-specific motifs might attribute to functional divergence of WRKYs. A total of 17 pairs of orthologous SmWRKY and AtWRKY genes and 21 pairs of paralogous SmWRKY genes were identified. Maximum likelihood analysis showed that SmWRKYs had undergone strong selective pressure for adaptive evolution. Functional divergence analysis suggested that the SmWRKY subgroup genes and many paralogous SmWRKY gene pairs were divergent in functions. Various critical amino acids contributed to functional divergence among subgroups were detected. Of the 61 SmWRKYs, 22, 13, 4 and 1 were predominantly expressed in roots, stems, leaves, and flowers, respectively. The other 21 were mainly expressed in at least two tissues analyzed. In S. miltiorrhiza roots treated with MeJA, significant changes of gene expression were observed for 49 SmWRKYs, of which 26 were up-regulated, 18 were down-regulated, while the other 5 were either up-regulated or down-regulated at different time-points of treatment. Analysis of published RNA-seq data showed that 42 of the 61 identified SmWRKYs were yeast extract and Ag+-responsive. Through a systematic analysis, SmWRKYs potentially involved in tanshinone biosynthesis were predicted.
Conclusion
These results provide insights into functional conservation and diversification of SmWRKYs and are useful information for further elucidating SmWRKY functions.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1411-x) contains supplementary material, which is available to authorized users.
Background
Salvia miltiorrhiza Bunge (Lamiaceae), known as danshen in Chinese, is one of the most important Traditional Chinese Medicine (TCM) materials. It has been widely used in Chinese medicines treating coronary heart disease, hepatitis, menstrual disorders, menostasis, blood circulation diseases, and other cardiovascular diseases [1]. The main bioactive components of S. miltiorrhiza include the water-soluble (hydrophilic) phenolics, such as rosmarinic acid, salvianolic acid A, salvianolic acid B and lithospermic acid [2], and the lipid-soluble (nonpolar, lipophilic) diterpenoids, known as tanshinones [3]. Enzymes catalyzing the biosynthesis of these bioactive components have been intensively studied recently [4-10]. A large number of genes involved in the biosynthesis of phenolics and terpenoids have been identified through either molecular cloning or transcriptome-wide characterization [3,11-17]. Collectively, S. miltiorrhiza is being developed to be a medicinal model plant [18].
Transcription factors are a class of significant regulators controlling plant growth and development through regulating gene expression at the transcriptional level. They bind to the specific regions, known as cis-elements, in the promoters of genes and then activate or repress the expression of regulated genes in collaboration with other regulatory factors. So far, two large transcription factor gene families, including the plant-specific SQUAMOSA promoter-binding protein-like (SPL) transcription factor gene family and the largest plant transcription factor gene family, termed MYB, have been characterized in S. miltiorrhiza [19,20]. A total of 15 SmSPLs and 110 SmMYBs have been identified from S. miltiorrhiza. SmSPLs are involved in the regulation of developmental timing in S. miltiorrhiza and eight of them are targets of miR156/157 [19]. Similarly, a subset of SmMYBs is regulated by microRNAs, such as miR159, miR319, miR828 and miR858. Many SmMYBs are involved in the biosynthesis of bioactive compounds in S. miltiorrhiza [20].
WRKY is a large transcription factor gene family specific to the green lineage, including green algae and land plants. The first WRKY gene, known as SPF1, was cloned from Ipomoea batatas about twenty year ago [21]. Since then, great progress has been achieved in WRKY gene identification and functional analysis. Plants with WRKYs identified include green alga (Chlamydomonas reinhardtii) [22], moss (Physcomitrella patens) [22], fern (Ceratopteris richardii) [22], pine (Pinus monticola) [23], Arabidopsis [24], tobacco (Nicotiana tabacum) [25-27], rice (Oryza sativa) [28,29], soybean (Glycine max) [30], maize (Zea mays) [31], barley (Hordeum vulgare) [32], grape (Vitis vinifera) [33,34], poplar (Populus trichocarpa) [35], tomato (Solanum lycopersicum) [36], cucumber (Cucumis sativus) [37], coffee (Coffea arabica) [38], and so forth.
Characterization of the identified WRKY genes showed that they were significant regulators involved in plant developmental processes and responsed to biotic and abiotic stresses [39]. The involvement of WRKYs in plant immune response against bacterial, fungal, and viral pathogens has been widely reported [40-51]. Recently, more and more evidence showed the regulatory roles of WRKY in plant response to abiotic stresses. For example, over-expression of three soybean WRKY genes (GmWRKY13, GmWRKY27 and GmWRKY54) in Arabidopsis showed that GmWRKY21-transgenic Arabidopsis plants were tolerant to cold stress, GmWRKY54 conferred salt and drought tolerance, whereas transgenic plants over-expressing GmWRKY13 had increased sensitivity to salt and mannitol stresses and decreased sensitivity to abscisic acid [52]. It suggests the involvement of WRKY genes in multiple abiotic stress-associated signaling pathways and the association of different WRKY members with different abiotic stresses. Moreover, WRKYs associated with same abiotic stress may show different responses. For instance, Arabidopsis WRKY25 and WRKY26 are heat-induced, whereas WRKY33 is heat-repressed [53]. In addition to stress responses, WRKYs also regulate various developmental processes, such as seed dormancy and germination, flowing, fruit maturation, stem elongation, pith secondary cell wall formation, plant senescence, and trichome development [54-58]. It suggests the importance of WRKYs and the complexity of WRKY-associated regulatory networks.
The defining feature of WRKY transcription factors is their DNA-binding domain, known as WRKY domain [39]. It is approximately 60 amino acids in length and includes the conserved amino acid sequence WRKYGQK at the N-terminus and an atypical zinc-finger motif either C2H2 (C–X4–5–C–X22–23–H–X1–H) or C2HC (C–X7–C–X23–H–X1–C) at the C-terminus. The structure of the WRKY domain allows it to specifically interact with W-box and SURE (sugar responsive) cis-elements in the promoter of target genes [59-61]. WRKY can be divided into three groups (Groups I, II and III) based on the number of WRKY domains (two domains in Group I and one in the others) and the pattern of zinc finger motif (C2H2 in Groups I and II and C2HC in Group III) [39,40]. Additionally, Group II WRKY proteins can be further divided into subgroups, including IIa, IIb, IIc, IId and IIe, based on the primary amino acid sequence of the WRKY domain.
Although WRKYs have been identified and characterized in various plant species, no information is available for the WRKY gene family in S. miltiorrhiza. In this study, we cloned and characterized 61 S. miltiorrhiza SmWRKYs.
Results and discussion
Molecular cloning of 61 SmWRKY genes from S. miltiorrhiza
It has been shown that 72 AtWRKY genes exist in the Arabidopsis genome (Additional file 1: Table S1). To identify SmWRKYs, BLAST analysis against the current assembly of the S. miltiorrhiza genome was performed using AtWRKY protein sequences as queries. A total of 61 gene models were predicted for SmWRKYs. The 5’-sequence of many SmWRKYs showed low homology with known plant WRKYs. It might cause errors in computational prediction. To verify the predicted gene models and correct errors of computation, full-length coding sequences (CDSs) of all 61 SmWRKYs were PCR-amplified using the primers listed in Additional file 2: Table S2 and then cloned and sequenced. It resulted in the identification of 61 SmWRKYs, which were named SmWRKY1–SmWRKY61, respectively. The deduced SmWRKY proteins have amino acid numbers from 129 to 706, isoelectric points (pI) from 4.76 to 9.9, and molecular weights (Mw) from 19.9 to 76.2 kDa. All of the 61 cloned CDSs have been submitted to GenBank under the accession numbers shown in Table 1. The number of identified SmWRKYs is comparable with that in Arabidopsis. Comparable gene numbers were also found for the MYB [20], SPL [19], Argonaute (AGO) [62] and RNA-dependent RNA polymerase (RDR) [63] gene families in S. miltiorrhiza and Arabidopsis. It suggests that S. miltiorrhiza and Arabidopsis may have similar number tfmkof gene members for many gene families. Thus, the identified SmWRKYs represent an almost complete set of WRKYs in S. miltiorrhiza, although it may be not a fully complete set.
Table 1.
Sequence features of WRKYs in S. miltiorrhiza
| Name | Gene ID | AA Len | pI | Mw (Da) | Group | Conserved motif | Domain pattern | Zinc finger |
|---|---|---|---|---|---|---|---|---|
| SmWRKY1 | KM823124 | 486 | 5.79 | 52075.53 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY2 | KM823125 | 526 | 7.2 | 57916.44 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY3 | KM823126 | 295 | 9.84 | 31785.84 | 2d | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY4 | KM823127 | 292 | 7.05 | 32163.19 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY5 | KM823128 | 283 | 6.6 | 31674.21 | 2e | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY6 | KM823129 | 332 | 9.6 | 36424.23 | 2d | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY7 | KM823130 | 262 | 6.18 | 29689.04 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| SmWRKY8 | KM823131 | 300 | 5.18 | 33675.01 | 2e | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY9 | KM823132 | 269 | 8.91 | 29956.66 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| SmWRKY10 | KM823133 | 343 | 5.37 | 38240.49 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| SmWRKY11 | KM823134 | 349 | 5.34 | 39120.3 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| SmWRKY12 | KM823135 | 211 | 8.11 | 24263.86 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY13 | KM823136 | 435 | 5.7 | 47512.82 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY14 | KM823137 | 243 | 8.19 | 27565.82 | 2c | WRKYGKK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY15 | KM823138 | 549 | 6.39 | 59091.35 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY16 | KM823139 | 321 | 5.02 | 35934.8 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| SmWRKY17 | KM823140 | 157 | 9.46 | 18126.30 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY18 | KM823141 | 333 | 4.76 | 36599.98 | 2e | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY19 | KM823142 | 221 | 9 | 25422.86 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY20 | KM823143 | 328 | 9.56 | 35953 | 2d | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY21 | KM823144 | 309 | 6.18 | 34870.74 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY22 | KM823145 | 407 | 8.72 | 44553.66 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY23 | KM823146 | 341 | 9.61 | 37956.92 | 2d | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY24 | KM823147 | 364 | 7.66 | 40559.08 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY25 | KM823148 | 329 | 5.9 | 36268.73 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY26 | KM823149 | 445 | 6.33 | 49392.18 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY27 | KM823150 | 346 | 9.77 | 37310.40 | 2d | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY28 | KM823151 | 486 | 8.38 | 53222.94 | -- | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY29 | KM823152 | 519 | 6.13 | 63235.29 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY30 | KM823153 | 509 | 6.84 | 55355.55 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY31 | KM823154 | 516 | 8.6 | 55885.41 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY32 | KM823155 | 129 | 9.3 | 15217.26 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY33 | KM823156 | 175 | 9.36 | 19944.38 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY34 | KM823157 | 309 | 8.11 | 34006.08 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| SmWRKY35 | KM823158 | 508 | 6.12 | 55526.63 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY36 | KM823159 | 246 | 6.61 | 27210.05 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY37 | KM823160 | 290 | 6.6 | 32766.16 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY38 | KM823161 | 284 | 9.41 | 30581.63 | 2d | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY39 | KM823162 | 390 | 6.64 | 43510.55 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY40 | KM823163 | 352 | 6.07 | 38849.51 | 2e | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY41 | KM823164 | 587 | 9.12 | 64687.26 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY42 | KM823165 | 706 | 6.02 | 76197.45 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY43 | KM823166 | 179 | 5.39 | 19963.09 | 2c | WRKYGKK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY44 | KM823167 | 265 | 5.73 | 28206.05 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY45 | KM823168 | 336 | 5.95 | 37211.08 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| SmWRKY46 | KM823169 | 306 | 6.41 | 33426.96 | 2e | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY47 | KM823170 | 268 | 5.86 | 30225.72 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY48 | KM823171 | 224 | 5.88 | 25174.62 | 2e | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY49 | KM823172 | 351 | 9.9 | 39451.59 | 2d | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY50 | KM823173 | 291 | 5.45 | 33184.06 | 2e | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY51 | KM823174 | 526 | 7.2 | 57916.44 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY52 | KM823175 | 171 | 6.66 | 19066.92 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY53 | KM823176 | 575 | 6.54 | 62764.4 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY54 | KM823177 | 491 | 7.69 | 53504.9 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY55 | KM823178 | 449 | 9.12 | 49222.93 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY56 | KM823179 | 297 | 5.34 | 33782.19 | 2e | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY57 | KM823180 | 281 | 5.18 | 31675.83 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| SmWRKY58 | KM823181 | 272 | 6.32 | 30091.63 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| SmWRKY59 | KM823182 | 352 | 8.34 | 38285.97 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| SmWRKY60 | KM823183 | 379 | 7.63 | 42242.82 | 1 | 2×[WRKYGQK] | C-X4-C-X22-HXH | C2H2 |
| SmWRKY61 | KM823184 | 168 | 8.96 | 19153.70 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
Classification of the WRKY domains and the WRKY proteins
Transcription factors in a family usually share highly conserved DNA-binding domains. In order to examine the phylogenetic relationships among WRKYs, a neighbor-joining (NJ) phylogenetic tree was constructed for the WRKY domain sequences of AtWRKYs, SmWRKYs, PpWRKYs, GCMa and FLYWCH using MEGA5.0 (Figure 1). According to the classification of AtWRKYs, the WRKY domains were divided into 3 groups (groups 1, 2 and 3). Group 1 WRKY domains come from proteins containing two WRKY domains, one of which is located in the N-terminal (NTWD), while the other one is in the C-terminal (CTWD). The exceptions within this group are the domains from AtWRKY10 and PpWRKY13, each of which possesses a single WRKY domain. Group 1 WRKY domains were further divided into two subgroups, termed group 1 N and group 1C, respectively. Based on their characteristics in the phylogenetic tree, group 2 WRKY domains could be classified into 5 subgroups, including groups 2a, 2b, 2c, 2d and 2e. Multiple sequence alignment of the core WRKY domains, each of which contains approximately 60 residues, showed that 71 of the 74 WRKY domains from 61 SmWRKYs contained the highly conserved sequence WRKYGQK, while the other three (SmWRKY43, SmWRKY52, and SmWRKY61) had WRKYGKK (Figure 2). Of the 61 SmWRKY, 13 are two-WRKY-domain-containing proteins and all of them have the C2H2-type zinc-finger motif (C–X4–C–X22–23–H–X1–H) (Figure 2, Table 2). The other 48 SmWRKY proteins are one-WRKY-domain-containing proteins, 42 of which contain group 2 WRKY domains and have the same type of finger motif (C–X4–5–C23–24–H–X1–H), while the other 6 contain group 3 WRKY domains and have the C2HC zinc finger motif (C–X7–C23–H–X1–C) (Figure 2, Table 2). The distribution of residues in the WRKY domains of S. miltiorrhiza WRKY proteins is quite similar to that of Arabidopsis (Figures 2 and 3), suggesting evolutionary conservation of SmWRKYs and AtWRKYs. Comparing the number of SmWRKYs and AtWRKYs in each group/subgroup showed that the number of SmWRKYs in groups 1 and 2 is similar to that of AtWRKYs in the same group; however the number 6 of SmWRKY members belonging to group 3 is significantly less than the number 14 of AtWRKY members included in the same group. It is consistent with previous results showing group 3 WRKYs to be a newly defined and most dynamic group [22] and suggests the divergence of WRKYs in S. miltiorrhiza and Arabidopsis.
Figure 1.
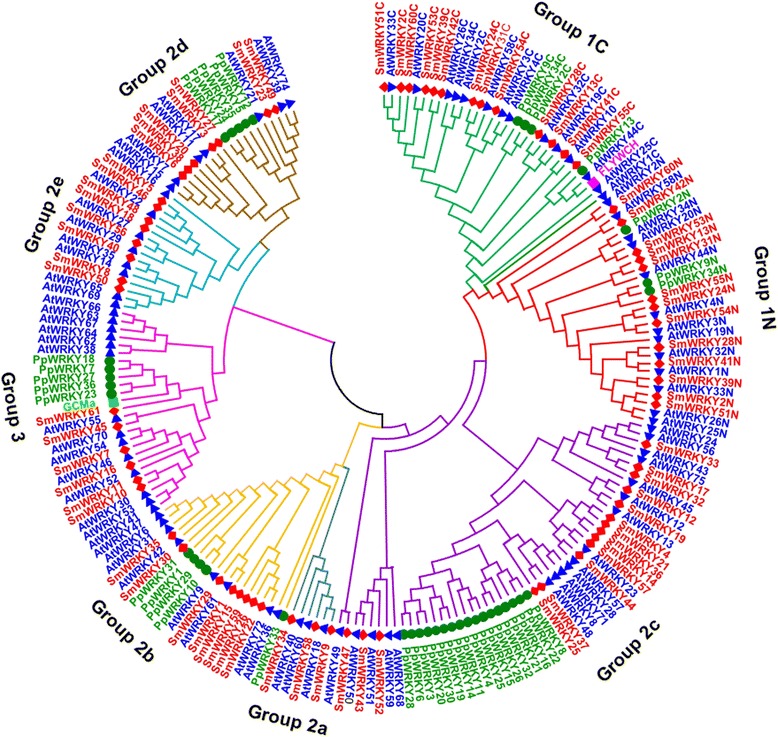
Phylogenetic tree of the WRKY domain from SmWRKYs (red), AtWRKYs (blue). PpWRKYs (dark green), FLYWCH (pink) and GCMa (light green). Groups/subgroups are shown; ‘N’ and ‘C’ indicate the N-terminal and C-terminal WRKY domain of a specific WRKY protein.
Figure 2.
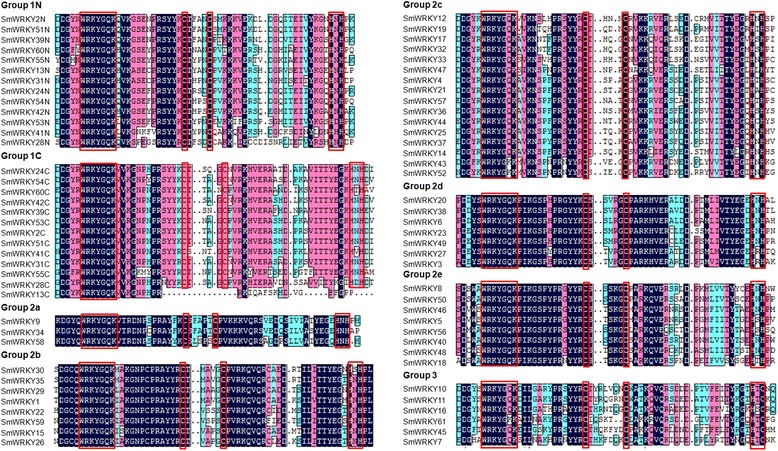
Multiple sequence alignment of the WRKY domain from SmWRKYs. Red box indicates conserved WRKY amino acid signature and zinc-finger motif; Black box indicates conserved amino acids. ‘N’ and ‘C’ indicate the N-terminal and C-terminal WRKY domain of a specific WRKY protein.
Table 2.
Number of WRKY domains from S. miltiorrhiza and Arabidopsis
| Group | Subgroup | Gene number | Consensus | Exception | |||
|---|---|---|---|---|---|---|---|
| AtWRKY | SmWRKY | ||||||
| 1 | 1 N | 27 | 13 | 26 | 13 | C-X4-C-X22-HXH | n = 23, AtWRKY26 |
| 1C | 14 | 13 | C-X4-C-X23-HXH | ||||
| 2 | 2a | 44 | 3 | 42 | 3 | C-X5-C-X23-HNH | |
| 2b | 8 | 8 | C-X5-C-X23-HXH | n = 24, AtWRKY36 | |||
| 2c | 18 | 16 | C-X4-C-X23-HXH | ||||
| 2d | 7 | 7 | C-X5-C-X23-HXH | ||||
| 2e | 8 | 8 | C-X5-C-X23-HXH | ||||
| 3 | 14 | 14 | 6 | 6 | C-X7-C-X23-HXC | n = 22, m = 5, SmWRKY61 | |
| AtWRKY52: HNH for HXC | |||||||
| Total | 85 | 74 | |||||
Figure 3.
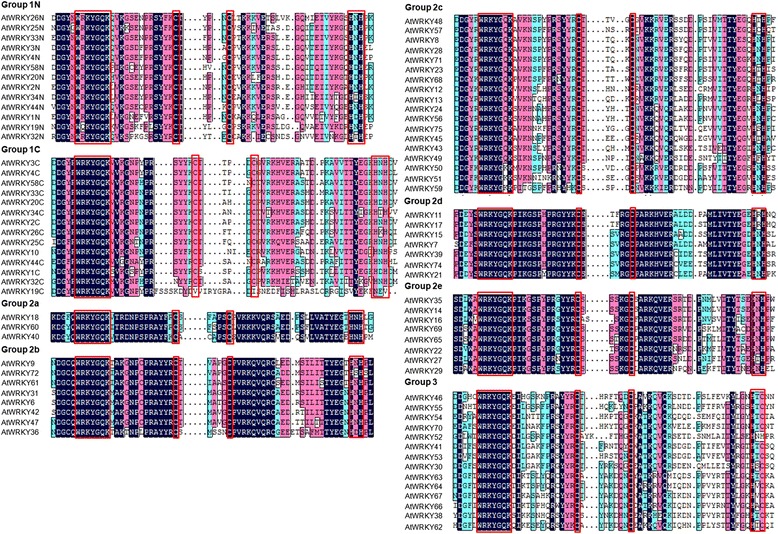
Multiple sequence alignment of the WRKY domain from AtWRKYs. Red box indicates conserved WRKY amino acid signature and zinc-finger motif; Black box indicates conserved amino acids. ‘N’ and ‘C’ indicate the N-terminal and C-terminal WRKY domain of a specific WRKY protein.
In order to investigate whether the phylogenies are different between the WRKY domains and the corresponding WRKY proteins, we constructed an NJ tree based on the full-length amino acid sequences of SmWRKYs, AtWRKYs, PpWRKYs, GCMa and FLYWCH (Figure 4). The results showed that the phylogenetic tree of WRKY proteins was quite similar to the tree of WRKY domains with little difference observed (Figures 1 and 4). For instance, AtWRKY1, AtWRKY32 and SmWRKY28 having two WRKY domains and AtWRKY10 belonging to group 1 WRKY domains form separated clades outside group 1. AtWRKY19 and FLYWCH with the WRKY domain belonging to group 1, AtWRKY16 with the WRKY domain belonging to group 2e, and AtWRKY52 and GCMa with the WRKY domain belonging to group 3 form separated clades outside groups 1, 2 and 3. These results indicate the difference between the WRKY domain and the sequence outside the WRKY domain. Based on the NJ tree constructed with full-length amino acid sequences, we identified 17 pairs of orthologous WRKY genes in S. miltiorrhiza and Arabidopsis (Table 3). It suggests that many SmWRKYs and AtWRKYs are evolutionarily conserved.
Figure 4.
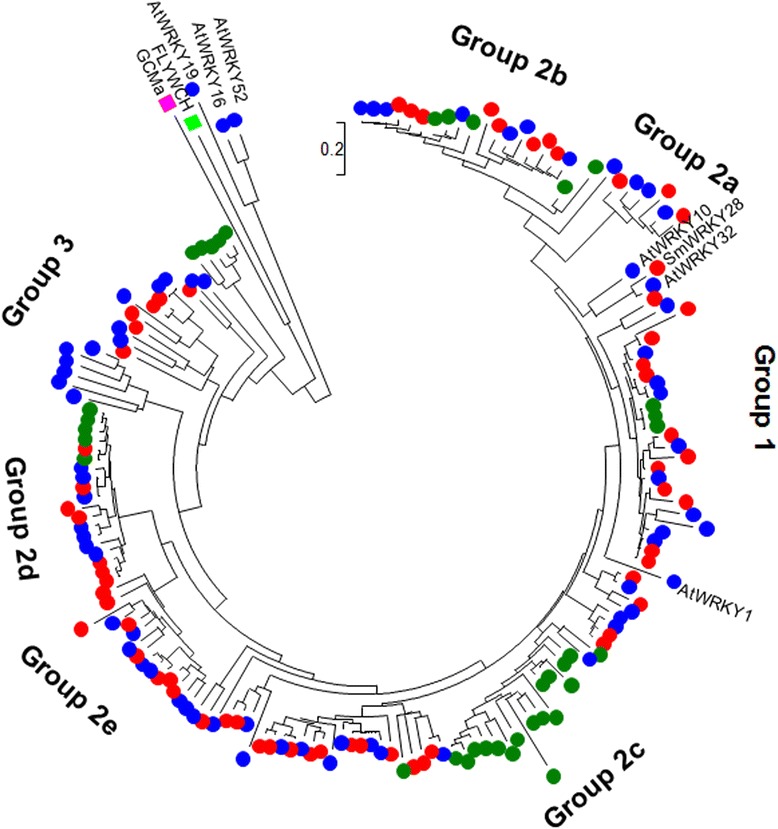
Phylogenetic analysis of SmWRKYs (red), AtWRKYs (blue). PpWRKYs (dark green), FLYWCH (light green) and GCMa (pink) proteins. The unrooted phylogenetic tree was constructed using the neighbor-joining method. The names of WRKY proteins not included in a group are shown.
Table 3.
WRKY orthologs in S. miltiorrhiza and Arabidopsis
| SmWRKYs | Putative Arabidopsis orthologs | Phylogenetic group in the NJ tree |
|---|---|---|
| SmWRKY55 | AtWRKY44 | Group 1 |
| SmWRKY54 | AtWRKY3/AtWRKY4 | Group 1 |
| SmWRKY53 | AtWRKY20 | Group 1 |
| SmWRKY51/SmWRKY2 | AtWRKY33 | Group 1 |
| SmWRKY28 | AtWRKY32 | Out of group 1 |
| SmWRKY34/SmWRKY58 | AtWRKY40 | Group 2a |
| SmWRKY59 | AtWRKY72 | Group 2b |
| SmWRKY57 | AtWRKY23 | Group 2c |
| SmWRKY12 | AtWRKY12 | Group 2c |
| SmWRKY19 | AtWRKY13 | Group 2c |
| SmWRKY47 | AtWRKY49 | Group 2c |
| SmWRKY49 | AtWRKY39 | Group 2d |
| SmWRKY56 | AtWRKY29 | Group 2e |
| SmWRKY40 | AtWRKY35/AtWRKY14 | Group 2e |
| SmWRKY8 | AtWRKY27 | Group 2e |
| SmWRKY45 | AtWRKY55 | Group 3 |
| SmWRKY16 | AtWRKY30 | Group 3 |
Analysis of conserved motifs
In addition to the WRKY domain, other conserved motifs could be important for the diversified functions of WRKY proteins from S. miltiorrhiza and Arabidopsis [64,65]. Using the MEME program, we identified a total of 20 conserved motifs in WRKYs from S. miltiorrhiza and Arabidopsis (Figure 5). The length of motifs varies from 8 to 150 amino acids and the number of motifs in each WRKY varies between 2 and 11. The majority of the identified motifs were found in more than one subgroup of WRKYs. Many AtWRKYs in a subgroup contain the same motif(s) as their SmWRKYs orthologues in the subgroup. It suggests the conservation of motifs in S. miltiorrhiza and Arabidoopsis WRKYs belonging to a subgroup.
Figure 5.
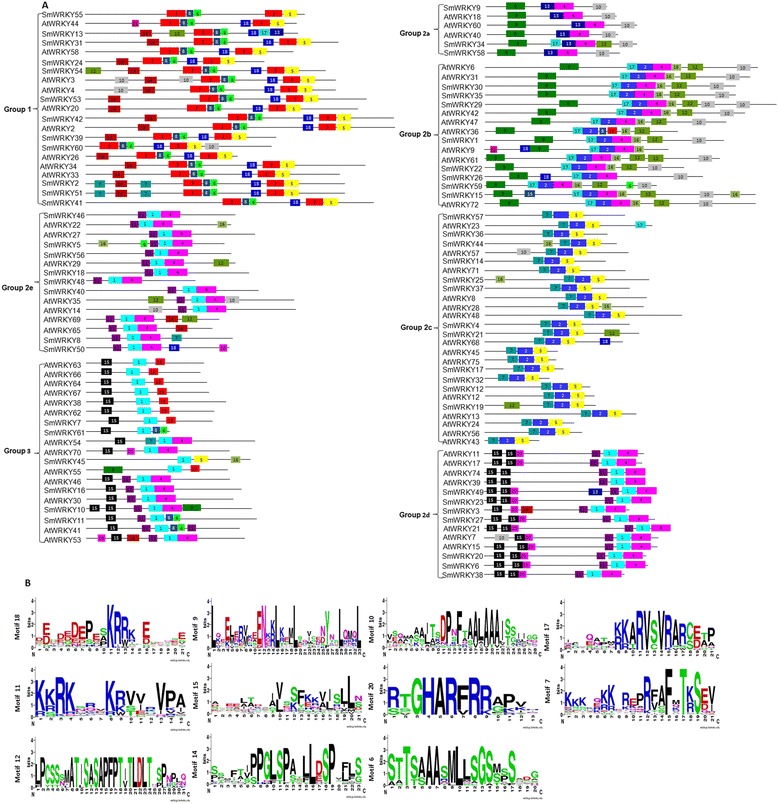
Architecture of conserved protein motifs in SmWRKYs and AtWRKYs. A: Architecture of conserved protein motifs in SmWRKYs and AtWRKYs from different groups (or subgroups). Motifs represented with boxes are predicted using MEME. The number in boxes (1–20) represents motif 1–motif 20, respectively. Box size indicates the length of motifs; B: Sequence logo of eleven conserved motifs, including motif 7, motif 9–motif 12, motif 14–motif 18 and motif 20. The logos were created on the WebLogo server (http://weblogo.berkeley.edu/logo.cgi). Bits represent the conservation of sequence at a position.
Among the 20 conserved motifs, 9 motifs, including motifs 1, 2, 3, 4, 5, 6, 8, 13, and 19, are located in the WRKY domain, while the other 11 are located outside the domain. Most WRKY proteins in a group have similar motif compositions (Figure 5). For instance, motif 18 exists in almost all of group 1 WRKY proteins except for SmWRKY55. Motif 14 exists in 16 of the 21 group 1 S. miltiorrhiza and Arabidopsis WRKY proteins. Motifs 9 and 10 commonly exist in groups 2a and 2b, two close subgroups in the phylogenetic tree. Motifs 12, 16 and 17 exist in most WRKY proteins of group 2b but only in a few members of 2a. Motif 7 exists in all of the WRKY proteins belonging to group 2c. Motif 11 is shared by proteins belonging to groups 2d and 2e. Additionally, motif 15, known as the Ca2+-dependent CaM-binding domain (CaMBD) [66], commonly exists in most WRKY proteins belonging to groups 2d and 3. Group 2d WRKY proteins usually contain two motif 15 s, while the majority of group 3 proteins contain only one. Motif 20, known as the HARF motif [40,67], exists in 5 of 7 AtWRKYs and all 7 SmWRKYs in group 2d. The results indicate functional similarities of WRKY proteins belonging to a group. Group-specific motifs may attribute to functional divergence of WRKYs.
Selective constraints on SmWRKY genes
In order to preliminarily examine the evolutionary mechanism of WRKYs, we test the hypothesis of positive selection acting on SmWRKY genes using site models and branch-site models in PAML [68] developed by Nielsen and Yang [69] and Yang et al. [70]. Codon substitution models [71] M0, M1a, M2a, M3, M7 and M8 were applied to the alignments and these models assume variation in ω among sites. The parameter estimates, log-likelihood and the LRT tests for these models are shown in Table 4. To examine how dN/dS ratios differed among codon positions, we compared models M0 and M3. The log likelihood of M0 for SmWRKY sequences was l = −5119.032, with an estimate of ω = 0.038. The low ω value suggests a strong action of purifying selection in the evolution of SmWRKY analyzed. M3 (discrete) assumes a general discrete distribution with three site classes (p0, p1, p2). The log likelihood of M3 was l = −4864.540, with an estimate of ω0 = 0.00052, ω1 = 0.02439, ω2 = 0.08755 (Table 4). Consistent with M0, the data from M3 also suggest that all codons are under purifying selection. Additionally, the value of twice the log likelihood difference (2ΔlnL) between M3 and M0 is 508.98. It is strongly statistically significant (p < 0.01) and suggests the overall level of selective constraints fluctuated.
Table 4.
Tests for positive selection among codons of WRKY genes using site models
| Model | lnL | Estimates of parameter 1 | 2ΔlnL | Positive selection sites 2 | |
|---|---|---|---|---|---|
| Frequency | dN/dS | ||||
| M0(one-ratio) | −5119.032 | p = 1.000 | ω = 0.03756 | 508.984 (M3 vs. M0)** | Not allowed |
| M3(discrete) | −4864.540 | p0 = 0.30493 | ω0 = 0.00052 | None | |
| p1 = 0.32215 | ω1 = 0.02439 | ||||
| p2 = 0.37292 | ω2 = 0.08755 | ||||
| M1a(nearly neutral) | −5119.251 | p0 = 0.93054 | ω0 = 0.04576 | 0 (M2a vs. M1a) | Not allowed |
| p1 = 0.06946 | ω1 = 1.00000 | ||||
| M2a(positive selection) | −5119.251 | p0 = 0.93052 | ω0 = 0.04576 | None | |
| p1 = 0.03470 | ω1 = 1.00000 | ||||
| p2 = 0.03478 | ω2 = 1.00000 | ||||
| M7(beta) | −4857.207 | p = 0.39586 | 0.002 (M8 vs. M7) | Not allowed | |
| q = 9.86708 | |||||
| M8(beta & ω) | −4857.206 | p0 = 0.99999 | ω = 1.35981 | None | |
| p = 0.39661 | |||||
| p1 = 0.00001 | |||||
| q = 9.86579 | |||||
Note: *p < 0.05 and **p < 0.01 (x 2 test).
1ω was estimated under model M0,M3,M7, and M8; p and q are the parameters of the beta distribution.
2The number of amino acid sites estimated to have undergone positive selection.
To test whether positive selection promoted divergence between genes, the codon substitution models that allow positive selection (M2a and M8) and that hypothesize nearly neutral selection (M1a and M7) were compared (M2a vs. M1a and M8 vs. M7; Table 4). The log likelihood of M1a and M2a for SmWRKY sequences was l = −5119.251. However, no site was positively selected at a level of 95%. M7 and M8 fitted the sequences better than M0, M3, M1a and M2a with values of l = −4857.207 and −4857.206, respectively (Table 4). In both cases, no significant evidence of positive selection was found.
Branch-site models aim to detect positive selection affecting a few sites along particular lineages and allow ω ratios to vary among sites and lineages simultaneously [68]. It seems that the branch-site models are most suitable for describing evolutionary processes of the WRKY gene family. Therefore, we analyzed positively selected amino acid sites of SmWRKYs using the improved branch-site model [72]. The branches being tested for positive selection were used as the foreground, while all other branches on the tree were used as the background. The Bayes empirical Bayes (BEB) method was used to calculate the posterior probabilities. A codon is probably from the site class of positive selection if LRT suggested the presence of codons under positive selection on the foreground branch [73]. The parameter estimates for lineages under positive selection are given in Table 5. A total of 19 residues were found to be under positive selection (p > 90%). It includes 6 in group 2c and 10 in group 2d. The other three residues were found in group 2b, group 2e and group 3. No residues in group 1 and group 2a were found to be under positive selection. The results suggest that different WRKY groups may have different evolutionary rates. Groups 2c and 2d could be confronted with strong positive Darwinian selection, since many highly significant positive sites were detected at the 0.01 significance level (Table 5). The evolution in the other groups seems to be more conservative.
Table 5.
Parameters estimation and likelihood ratio tests for the branch-site models
| Branch-site model | |||||
|---|---|---|---|---|---|
| Foreground branches | Estimates of parameter | Positive delection sites(BEB) | |||
| Site class 0 | Site class 1 | Site class 2a | Site class 2b | ||
| P0 = 0.10272 | P1 = 0.36090 | P2a = 0.11884 | P2b = 0.41754 | ||
| Group 1 | ω0(b) = 0.05880 | ω1(b) = 1.00000 | ω2a(b) = 0.05880 | ω2b(b) = 1.00000 | None |
| ω0(f) = 0.05880 | ω1(f) = 1.00000 | ω2a(f) = 1.00000 | ω2b(f) = 1.00000 | ||
| P0 = 0.45646 | P1 = 0.17985 | P2a = 0.26089 | P2b = 0.10279 | ||
| Group 2a | ω0(b) = 0.05165 | ω1(b) = 1.00000 | ω2a(b) = 0.05165 | ω2b(b) = 1.00000 | None |
| ω0(f) = 0.05165 | ω1(f) = 1.00000 | ω2a(f) = 1.00000 | ω2b(f) = 1.00000 | ||
| P0 = 0.42991 | P1 = 0.34434 | P2a = 0.12535 | P2b = 0.10040 | ||
| Group 2b | ω0(b) = 0.11089 | ω1(b) = 1.00000 | ω2a(b) = 0.11089 | ω2b(b) = 1.00000 | 359 G* |
| ω0(f) = 0.11089 | ω1(f) = 1.00000 | ω2a(f) = 999.00000 | ω2b(f) = 999.00000 | ||
| P0 = 0.20480 | P1 = 0.56991 | P2a = 0.05956 | P2b = 0.16573 | 171 K**, 181 Q**, | |
| Group 2c | ω0(b) = 0.06509 | ω1(b) = 1.00000 | ω2a(b) = 0.06509 | ω2b(b) = 1.00000 | 192S**, 210 A**, |
| ω0(f) = 0.06509 | ω1(f) = 1.00000 | ω2a(f) = 5.55679 | ω2b(f) = 5.55679 | 237 S**, 243 I** | |
| P0 = 0.55167 | P1 = 0.13832 | P2a = 0.24786 | P2b = 0.06215 | 25 N**, 26 I**, 34 C**, 79 S**, | |
| Group 2d | ω0(b) = 0.08684 | ω1(b) = 1.00000 | ω2a(b) = 0.08684 | ω2b(b) = 1.00000 | 148 T**, 208 G**, 214 D**, |
| ω0(f) = 0.08684 | ω1(f) = 1.00000 | ω2a(f) = 214.85997 | ω2b(f) = 214.85997 | 269 H**, 363 C**, 395 N** | |
| P0 = 0.27330 | P1 = 0.60581 | P2a = 0.03758 | P2b = 0.08331 | ||
| Group 2e | ω0(b) = 0.08686 | ω1(b) = 1.00000 | ω2a(b) = 0.08686 | ω2b(b) = 1.00000 | 196 E* |
| ω0(f) = 0.08686 | ω1(f) = 1.00000 | ω2a(f) = 44.60691 | ω2b(f) = 44.60691 | ||
| P0 = 0.42970 | P1 = 0.32076 | P2a = 0.14288 | P2b = 0.10666 | ||
| Group 3 | ω0(b) = 0.07702 | ω1(b) = 1.00000 | ω2a(b) = 0.07702 | ω2b(b) = 1.00000 | 107 F* |
| ω0(f) = 0.07702 | ω1(f) = 1.00000 | ω2a(f) = 999.00000 | ω2b(f) = 999.00000 | ||
Note: *p < 0.05 and **p < 0.01 (x2 test).
Site class: The sites in the sequence evolve according to the same process, the transition probability matrix is calculated only once for all sites for each branch.
b: Background ω.
f: Foreground ω.
Positive delection sites: The number of amino acid sites estimated to have undergone positive selection.
BEB: Bayes Empirical Bayes.
Functional divergence analysis (FDA) of SmWRKY proteins
Using DIVERGE 2.0 that evaluates shifted evolutionary rate and altered amino acid property after gene duplication [74,75], we carried out posterior analysis for estimation of Type-I and Type-II functional divergence between SmWRKY clusters. The estimation was based on the WRKY protein neighbor-joining tree consisting of three major groups (group 1, group 2a–e, and group 3) (Figure 4). Comparison among SmWRKY subgroups showed that all of the coefficients for the type I functional divergence (θI) were greater than zero (Additional file 3: Table S3). The θI values of eight group pairs, including 1/2e, 1/3, 2a + b/2d, 2a + b/2e, 2a + b/3, 2c/2e, 2c/3 and 2d/3, were ranged from 0.219 to 0.772 and were statistically significant (P < 0.01) (Additional file 3: Table S3). It indicates that significant site-specific changes may have happened at certain amino acid sites between these group pairs, leading to a subgroup-specific functional evolution after their diversification.
Type II functional divergence (θII) values in six group pairs, including 1/2c, 1/2d, 2a + b/2d, 2c/2d and 2d/3, were also greater than zero and ranged from 0.017 to 0.234 (Additional file 4: Table S4). It indicates a radical shift in amino acid properties. In order to extensively reduce positive false, Qk > 0.8 and 1.0 were empirically used as cutoff in the identification of the Type-I and Type-II functional divergence-related residues between gene groups, respectively (Figures 6 and 7). Detailed analysis showed that the number and the distribution of predicted residues for functional divergence in group pairs were different (Additional file 3: Table S3 and Additional file 4: Table S4) and the residues predominantly existed in the WRKY domain. It suggests that these residues probably play important roles in functional divergence of WRKYs during the evolutionary process.
Figure 6.
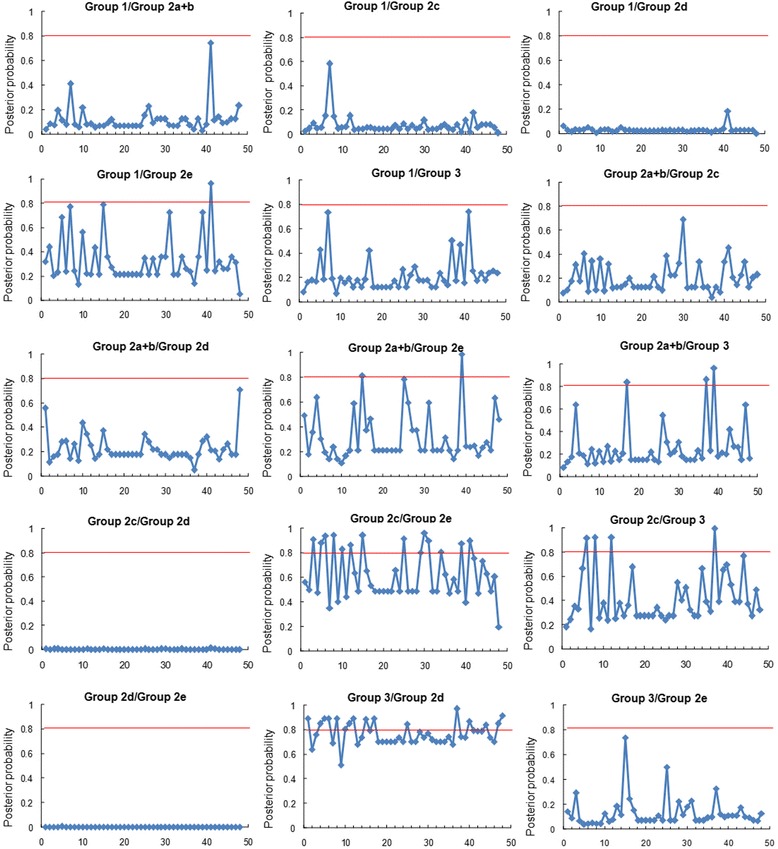
Site-specific prediction for type-I functional divergence between groups of SmWRKYs. The X-axis represents locations of sites. The Y-axis represents the probability of each group. The red line indicates cutoff = 0.80.
Figure 7.
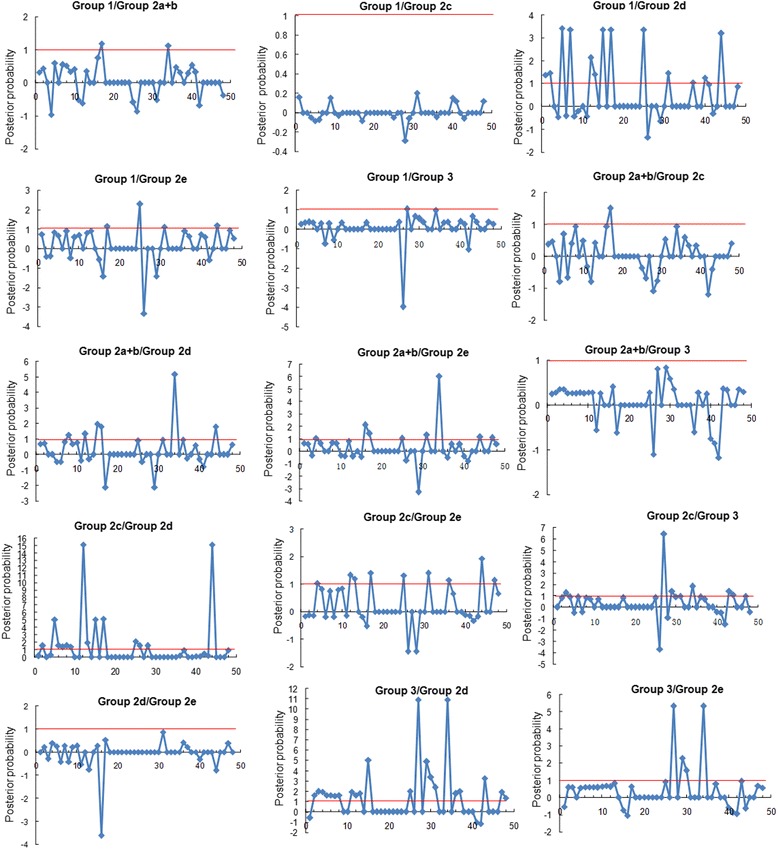
Site-specific profile for predicting critical amino acid residues responsible for the type-II functional divergence between groups of SmWRKYs. The X-axis represents locations of sites. The Y-axis represents the probability of each group. The red line indicates cutoff = 1.0.
Tissue-specific expression of SmWRKYs
It has been shown that a number of WRKY proteins are involved in plant developmental processes [54,76,77]. In order to preliminarily understand the roles of WRKYs in S. miltiorrhiza development, we analyzed the expression of SmWRKYs in roots, stems, leaves and flowers of S. miltiorrhiza plants. All of the 61 SmWRKYs identified were expressed in at least a tissue analyzed and exhibited differential expression patterns (Figure 8). Of the 61 SmWRKYs, 22 (36.1%) showed predominant expression in roots, 13 (21.3%) in stems, 4 (6.6%) in leaves and 1 (1.6%) in flowers. The other 21 (34.4%) were mainly expressed in at least two tissues analyzed, indicating these genes are likely to play a more ubiquitous role in S. miltiorrhiza. Furthermore, some SmWRKYs in a group shared similar expression patterns, while the others were not. For example, SmWRKY2, SmWRKY24, SmWRKY39, SmWRKY54 and SmWRKY55, belonging to group 1, were predominantly expressed in roots, while the other group 1 members, such as SmWRKY42, SmWRKY13 and SmWRKY60, were mainly expressed in stems, leaves and flowers, respectively (Figure 8). It suggests that SmWRKYs belonging to a group do not necessarily indicate their functions in the same tissues. However, it has been shown that the tissues-specific expression patterns appear to be consistent with their role in the tissues. For example, VvWRKY01, belonging to group 2c, is involved in the regulation of lignin biosynthesis [78]. Over-expression of VvWRKY01 in tobacco resulted in the alteration of expression patterns of genes involved in lignin biosynthesis pathway [78]. Similarly, SmWRKY12, SmWRKY19 and SmWRKY47 in group 2c were predominantly expressed in stems (Figure 8). It indicates the putative roles of these SmWRKYs in the regulation of lignin biosynthesis in S. miltiorrhiza. AtWRKY6, a member of group 2b, and AtWRKY53 and AtWRKY70, two AtWRKYs in group 3, are an important regulator in senescent leaves [55,76,79-81]. Of them, AtWRKY6 acts in the upstream of SIRK during leaf senescence [55]. SmWRKY59 belonging to the same WRKY group of AtWRKY6 and SmWRKY7 included in group 3 could be regulators of leaf senescence in S. miltiorrhiza, since both of them showed predominant expression in leaves (Figure 8). It has been shown that AtWRKY44 included in group 1 is involved in trichome differentiation [54]. Several SmWRKYs in group 1, such as SmWRKY2, SmWRKY39, SmWRKY24, SmWRKY54 and SmWRKY55, were highly expressed in roots with abundant root hairs, and SmWRKY13 belonging to the same group was highly expressed in leaves with abundant of trichomes (Figure 8). It implies that these SmWRKY may be associated with trichome development in S. miltiorrhiza.
Figure 8.
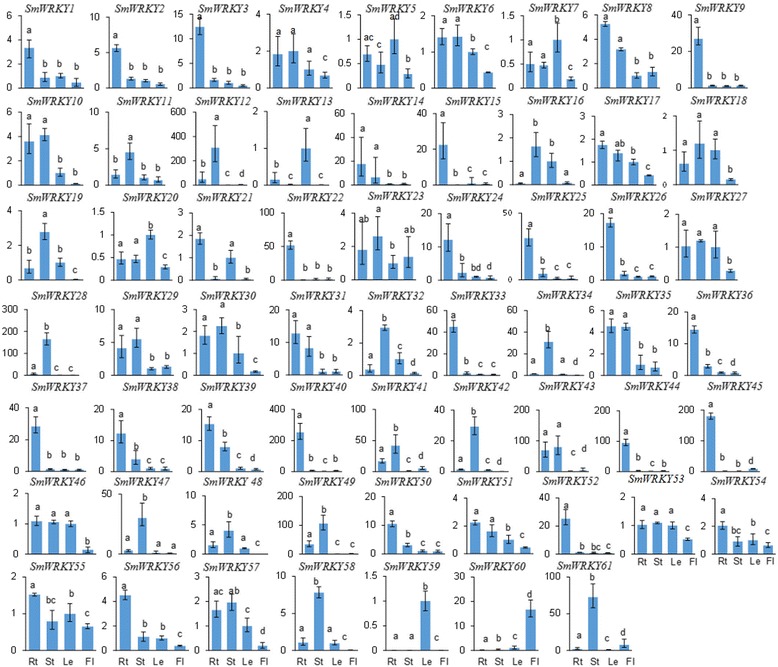
Expression patterns of SmWRKY genes in roots (Rt), stems (St), leaves (Le) and flowers (Fl) of 2-year-old, field-grown S. miltiorrhiza Bunge (line 993). The expression level of SmWRKYs was analyzed by the quantitative RT-PCR method. Y-axis indicates relative expression levels. X-axis indicates different tissues. SmUBQ10 was used as the reference gene. Transcript levels in leaves were arbitrarily set to 1 and the levels in other tissues were given relative to this. Error bars represent standard deviations of mean value from three biological and three technical replicates. ANOVA (analysis of variance) was calculated using SPSS. P < 0.05 was considered statistically significant.
Methyl jasmonate (MeJA)-responsive SmWRKYs
MeJA is a key signaling molecule involved in plant response to stress and in regulating secondary metabolite production in many plant species, including S. miltiorrhiza [3,14]. More than 50% (39 of 74) of AtWRKYs were MeJA-responsive in Arabidopsis [82]. More than 1/3 of CrWRKY were regulated by MeJA in hairy roots of Catharanthus roseus [83]. Additionally, CaWRKY27 in Capsicum annuum [84], GbWRKY1 in Gossypium barbadense [85] and GhWRKY3 in Gossypium hirsutum [86] were also regulated by MeJA. In order to test whether WRKYs were responsive to MeJA treatment in S. miltiorrhiza, the expression level of SmWRKYs in roots of plantlets treated with MeJA was analyzed using the quantitative RT-PCR method. MeJA treatment showed a wide variety of SmWRKY gene expression profiles (Figure 9). Significant expression level changes were observed for 49 SmWRKYs, of which 26 were up-regulated, 18 were down-regulated, while the other 5, including SmWRKY1, SmWRKY15, SmWRKY17, SmWRKY20 and SmWRKY24, were either up-regulated or down-regulated at different time-points of treatment (Figure 9). It suggests that about 80% of the SmWRKYs analyzed are MeJA-responsive. Examination of the number of SmWRKYs with significant expression level changed at different time-points of treatment showed that the expression of 28, 43, 25 and 23 SmWRKYs was changed after MeJA treatment for 12, 24, 36, and 48 hours, respectively (Figure 9). It suggests that the majority of SmWRKYs have altered expression levels at the time-point of 24 h-treatment. The number of up-regulated SmWRKYs was 13, 26, 15, and 13 at the time-point of 12, 24, 36, and 48 hours, respectively, while that of down-regulated was 15, 17, 10, and 10, respectively. Additionally, 8 SmWRKYs, including SmWRKY9, SmWRKY13, SmWRKY14, SmWRKY25, SmWRKY32, SmWRKY38, SmWRKY44 and SmWRKY52, were significantly up-regulated at all time-points of MeJA treatment, while 7, including SmWRKY7, SmWRKY33, SmWRKY47, SmWRKY49, SmWRKY53, SmWRKY54 and SmWRKY58, were down-regulated (Figure 9). It suggests that the number of up-regulated SmWRKYs is slightly more than down-regulated.
Figure 9.
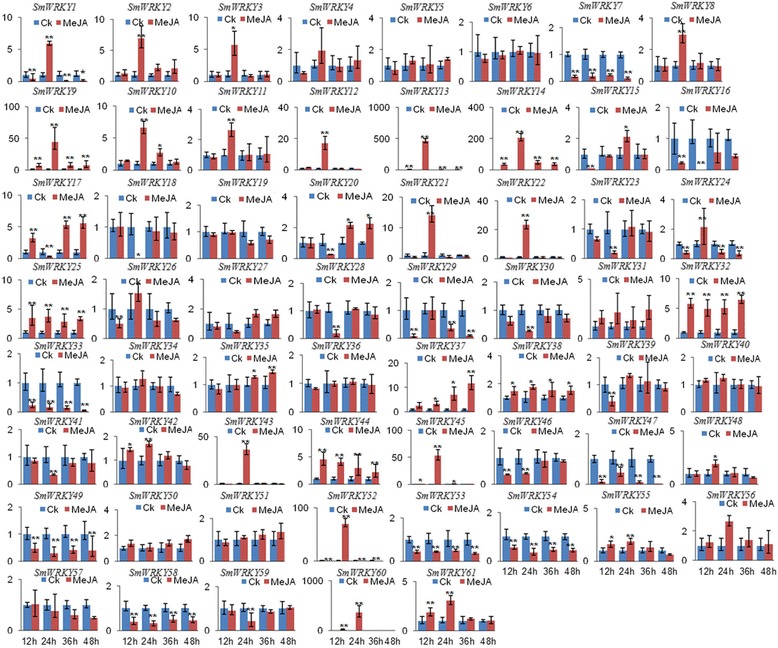
Quantitative RT-PCR analysis of SmWRKY gene expression in S. miltiorrhiza roots treated with MeJA. Fold changes of SmWRKYs in roots of S. miltiorrhiza plantlets treated with MeJA for 12, 24, 36 and 48 h are shown. SmUBQ10 was used as the reference gene. The level of transcripts in roots treated with carrier solution (CK) was arbitrarily set to 1 and the levels in roots treated with MeJA were given relative to this. Mean values and SDs were obtained from three biological and three technical replicates. Y-axis indicates relative expression levels. X-axis indicates different time-points of MeJA treatment. ANOVA (analysis of variance) was calculated using SPSS. P < 0.05 (*) and P < 0.01(**) were considered statistically significant and extremely significant, respectively.
Yeast extract and Ag+-responsive SmWRKYs
In order to further investigate the roles of SmWRKYs in S. miltiorrhiza, transcriptome-wide analysis of SmWRKY expression in response to yeast extract and Ag+ treatment was performed. RNA-seq data of S. miltiorrhiza hairy roots treated with or without yeast extract (100 μg/ml) and Ag+ (30 μM) were downloaded from GenBank under the accession number SRR924662 [12]. RNA-seq reads from non-treated (0 hpi) and treated for 12 h (12 hpi), 24 h (24 hpi) and 36 h (36 hpi) were mapped to SmWRKYs using the SOAP2 software [87]. The log-2-transformed RPKM (RNA-seq reads mapped to a SmWRKY per total million reads from a treatment per kilobases of the SmWRKY length) value of SmWRKYs varied between −3.04 and 8.38 (Additional file 5: Table S5). Using a cutoff of RPKM value >2.0, a total of 49 SmWRKYs were found to be expressed in hairy roots. Fisher’s exact test showed that 42 of the 49 SmWRKYs were differentially expressed (Additional file 5: Table S5). It includes 17 significantly up-regulated, 19 significantly down-regulated and 6 significantly up- or down-regulated at different time-points, suggesting the majority of the identified SmWRKYs were responsive to yeast extract and Ag+ treatment.
SmWRKY candidates potentially involved in tanshinone biosynthesis
Terpenoids are plant secondary metabolites with significant physiological and ecological functions and great economic values, and a class of terpenoids, known as tanshinones, is the main bioactive compounds in S. miltiorrhiza. Increasing evidence demonstrates the importance of WRKY genes in the biosynthesis of secondary metabolites, such as terpenoid indole alkaloid in Catharanthus roseus [83]. Additionally, it has been shown that Gossypium arboretum GaWRKY1, which belongs to group 2a, participates in sesquiterpene biosynthesis in cotton by controlling (+)-δ-cadinene synthase (CAD1) activity [88]. PqWRKY1, a member of group 2d, responds to MeJA treatment and is a positive regulator of osmotic stress response and triterpene ginsenoside biosynthesis in Panax quinquefolius [89]. AaWRKY1 and CrWRKY, belonging to group 3, are the other two terpenoid biosynthesis-related WRKYs. Artemisia annua AaWRKY1 was highly expressed in glandular secretory trichomes (GSTs), where the sesquiterpene artemisinin was synthesized [90]. AaWRKY1 might be strongly induced by MeJA and could bind to the W-box in the promoter of ADS gene encoding amorpha-4, 11-diene synthase, a key enzyme in the artemisinin biosynthesis pathway [90]. CrWRKY1 was preferentially expressed in roots of C. roseus and also induced by MeJA [91]. It controlled terpenoid indole alkaloid biosynthesis through positive regulation of DXS and SLS genes involved in the terpenoid pathway and AS and TDC genes involved in the indole pathway [91]. Thus, SmWRKYs included in group 2a, 2d and 3 probably have an evolutionarily conserved role in regulating terpenoid biosynthesis in S. miltiorrhiza.
Terpenoid tanshinones have been mainly produced and accumulated in roots of field-grown S. miltiorrhiza during the fast growing period from June to September [92-94]. The process of tanshinone production may be stimulated by MeJA, yeast extract and Ag+ [10,12,95]. Among the 61 identified SmWRKYs, sixteen showed similar responses to the MeJA treatment and the yeast extract and Ag+ treatment (Figure 9, Additional file 5: Table S5). SmWRKY2, SmWRKY3, SmWRKY9, SmWRKY11, SmWRKY25, SmWRKY32, SmWRKY37, SmWRKY43 and SmWRKY52 were up-regulated by both the MeJA treatment and the yeast extract and Ag+ treatment, while SmWRKY23, SmWRKY26, SmWRKY33, SmWRKY41, SmWRKY47, SmWRKY53 and SmWRKY59 were down-regulated (Figure 9, Additional file 5: Table S5). Among the sixteen SmWRKYs, eight, including six up-regulated (SmWRKY2, SmWRKY3, SmWRKY9, SmWRKY25, SmWRKY37 and SmWRKY52) and two down-regulated (SmWRKY26 and SmWRKY33), were predominantly expressed in roots of field-grown S. miltiorrhiza in August (Figure 8), suggesting their specific roles in roots. SmWRKY2, SmWRKY3, SmWRKY9 and SmWRKY26 are members of groups 1, 2d, 2a and 2b, respectively, while SmWRKY25, SmWRKY33, SmWRKY37 and SmWRKY52 are members of group 2c (Figures 1 and 4). Thus, SmWRKY3 and SmWRKY9 are two SmWRKYs (1) with similar responses to the MeJA treatment and the yeast extract and Ag+ treatment, (2) having root-specific expression, and (3) belonging to group 2a, 2d or 3 with members probably playing an evolutionarily conserved role in regulating terpenoid biosynthesis. It implicates that SmWRKY3 and SmWRKY9 are very likely to be activators in tanshinone production. Notably, we may not exclude the possibility that some other SmWRKYs are also involved in tanshinone biosynthesis based on the data currently available. Further analysis of transgenic S. miltiorrhiza plants with over-expressed or silenced SmWRKYs may help to elucidate their function.
Divergence of paralogous SmWRKY genes
Gene duplication is an important event for gene family expansion and functional diversity during evolution [65,96,97]. A total of 42 (68.85% of 61) SmWRKY genes appear to be duplicated (Additional file 5: Table S5). In order to preliminarily reveal the mechanism of functional diversity (nonfunctionalization, subfunctionalization and neofunctionalization [98]) of these genes after duplication, the synonymous (Ks) and non-synonymous (Ka) substitution rate were calculated using the CDS of paralogous SmWRKY genes (Additional file 6: Table S6). The Ka/Ks ratios for all of the 21 paralogous SmWRKY gene pairs were less than one with 5 pairs even close to zero. It suggests that the WRKY genes from S. miltiorrhiza have experienced strong purifying selection pressure. Some closely related gene pairs displayed different expression patterns, indicating functional divergences occurred. For example, SmWRKY13 was expressed dominantly in leaves, whereas the other member of the SmWRKY13/SmWRKY31 gene pair, SmWRKY31, was expressed mainly in roots and stems (Figure 8). In addition, SmWRKY13, but not SmWRKY31, was significantly up-regulated by MeJA (Figure 9). Expression patterns of other paralogous genes, such as SmWRKY23/49, SmWRKY30/35, SmWRKY41/55, SmWRKY42/53, and SmWRKY43/SmWRKY52, were also different (Figures 8 and 9). It indicates that many SmWRKY gene pairs are divergent under the purifying pressure [99].
Conclusions
In this study, we cloned a total of 61 SmWRKY genes. The cloned genes and the deduced proteins were characterized through a comprehensive approach, including phylogenetic tree construction, WRKY domain characterization, conserved motif identification, selective constraint analysis, functional divergence analysis, and expression profiling. We showed that many SmWRKYs and AtWRKYs were evolutionarily conserved. The WRKY domains could be divided into 3 groups (1, 2 and 3) and 8 subgroups (1 N, 1C, 2a, 2b, 2c, 2d, 2e and 3). Each group of WRKY domains contains characteristic conserved sequences. Additionally, sequence outside the WRKY domain might contribute to the difference between the phylogenetic tree constructed with the WRKY domains and that constructed with the whole WRKY proteins. A total of 20 conserved motifs were identified, of which group-specific motifs might attribute to functional divergence of WRKYs. We identified 17 pairs of orthologous SmWRKY and AtWRKY genes and 21 pairs of paralogous SmWRKY genes. Selective constraint analysis and functional divergence analysis showed that the SmWRKY subgroup genes have experienced strong positive selection and diverged in function. Gene expression profiles suggest that the majority of 61 SmWRKY genes are tissue-specific and MeJA- and yeast extract and Ag+-responsive. These results provide insights into functional conservation and diversification of SmWRKYs and are useful for further investigating SmWRKY functions in S. miltiorrhiza development and defense response.
Methods
Database search and sequence annotation
The nucleotide sequences and amino acids of 72 AtWRKY genes were obtained from the Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org/). S. miltiorrhiza WRKY (SmWRKY) genes were predicted by tBLASTn [100] search of AtWRKY [101] homologs against the current S. miltiorrhiza genome assembly, which covers about 92% of the entire genome and 96% of the protein-coding genes [18]. An e-value cut-off of 1e-10 was applied to the homologue recognition. The retrieved sequences were used for gene model prediction on the GENSCAN web server (http://genes.mit.edu/GENSCAN.html). Full-length CDSs of SmWRKYs were amplified by reverse transcription-PCR using the primers listed in Additional file 2: Table S2. PCR products were gel-purified, cloned, and then sequenced. The theoretical isoelectric point (pI) and molecular weight (Mw) were predicted using the Compute pI/Mw tool on the ExPASy server (http://web.expasy.org/compute_pi/) [102].
Multiple sequence alignment, phylogenetic analysis and motif detection
PpWRKY genes were obtained from Physcomitrella patens v3.1 (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ppatens_er). Human FLYWCH CRAa (EAW85450) and GCMa (BAA13651) were obtained from NCBI (http://www.ncbi.nlm.nih.gov/protein/). Multiple sequence alignment of the WRKY domain from 61 S. miltiorrhiza SmWRKYs and 72 Arabidopsis AtWRKYs was performed using CLUSTALW with BOXSHADE (http://bioweb.pasteur.fr) [103]. Phylogenetic trees were constructed using MEGA 5.0 with the neighbor-joining method [104]. Bootstrap test was replicated 1000 times. Motifs were detected using MEME 5.0 [105].
Plant materials and MeJA treatment
Roots, stems, leaves and flowers from 2-year-old, field-grown S. miltiorrhiza Bunge (line 993) plants were collected in August, 2012 and stored in liquid nitrogen until use. Plantlets cultivated in vitro were grown at 25°C with a photoperiod of 16 h light and 8 h dark for six weeks and treated with 200 μM methyl jasmonate (MeJA) for 12, 24, 36 and 48 h as described previously [3,63]. Plantlets treated with carrier solution were used as controls. Roots of plantlets with or without MeJA treatment were collected and stored in liquid nitrogen until use. Three independent biological replicates were carried out for each experiment.
RNA extraction and quantitative real-time reverse transcription-PCR (qRT-PCR)
Total RNA was extracted from plant tissues using the Quick RNA Isolation Kit (Huayueyang, China). RNA integrity was analyzed on a 1.2% agarose gel. RNA quantity was determined using a NanoDrop 2000C Spectrophotometer (Thermo Scientific, USA). cDNA synthesis was carried out using Superscript III Reverse Transcriptase (TaKaRa, China). qRT-PCRs were performed using the SYBR premix Ex Taq™ kit (TaKaRa, China) and carried out in triplicate for each tissue sample. Gene-specific primers (Additional file 7: Table S7) were designed using Primer Premier 5.0. The length of amplicons is between 80 bp and 250 bp. SmUBQ10 was selected as a reference gene as described previously [3]. Three independent biological replicates were performed. Statistical analysis was carried out as described [20]. Briefly, standardization of gene expression data was performed from three biological replicates as described [106]. 2-ΔΔCq was used to achieve results for relative quantification. For statistical analysis, ANOVA (analysis of variance) was calculated using SPSS (Version 19.0, IBM, USA).
Analysis of SmWRKY expression in response to yeast extract and Ag+ treatment
RNA-seq data for S. miltiorrhiza hairy roots treated with yeast extract (100 μg/ml) and Ag+ (30 μM) were downloaded from GenBank under the accession number SRR924662 [12]. RNA-seq reads from non-treated (0 hpi) and treated for 12 h (12 hpi), 24 h (24 hpi) and 36 h (36 hpi) were mapped to SmWRKYs using the SOAP2 software [87] and analyzed as described previously [107]. The parameter v cutoff of 3 and parameter r cutoff of 2 were applied. SmWRKYs with the RPKM value greater than 2 were analyzed for differential expression using Fisher’s exact test. P < 0.05 was considered as differentially expressed.
Ka and Ks calculation
Paralogous SmWRKY genes were inferred from phylogenetic analysis. Non-synonymous (Ka) and synonymous (Ks) substitution of each paralogous gene pair were also determined by PAL2NAL program (http://www.bork.embl.de/pal2nal/) [108], which is based on the codon model program in PAML [68].
Tests of positive selection
To determine whether the WRKY gene family exhibited evidence of positive selection under the site model and branch-site model [71], we applied the codeml program in PAML v4.8 to test the hypothesis of positive selection. An unrooted phylogenetic tree of SmWRKYs was reconstructed using the maximum likelihood method. In the site model, M0 (one ratio), M1a (neutral), M2a (selection), M3 (discrete), M7 (beta) and M8 (beta + ω > 1) were applied to the alignments, and we detected variation in the ω parameter among sites using the LRT for M1a vs. M2 a, M0 vs. M3 and M7 vs. M8. Branch-site model [72] was used to compare the non-synonymous/synonymous substitution rate ratio (Ka/Ks) between clades or sequences. The ratio of nonsynonymous-to-synonymous for each branch under model A was calculated. Posterior probabilities (Qks) were calculated using the BEB method [68].
Estimation of functional divergence
The software DIVERGE2 was used to detect the functional divergence among members of SmWRKY subgroups [74]. The method is based on maximum likelihood procedures to estimate significant changes in the site-specific shift. The coefficients of Type-I and Type-II functional divergence (θI and θII) between two clusters were calculated. The coefficients of Type-I and Type-II functional divergence (θI and θII) greater than 0 indicates that site specific altered selective constraints or a radical shift of amino acid physiochemical property occurred after gene duplication and/or speciation [74]. Large posterior probability (Qk) indicates a high possibility that the functional constraint (or the evolutionary rate) and/or the radical change in the amino acid property of a site is different between two clusters [74].
Availability of supporting data
SmWRKY sequences supporting the results of this article are available in GenBank under accession numbers KM823124–KM823184. The other supporting data are included within the article and its additional files.
Acknowledgements
We thank Prof. Shilin Chen and the sequencing group in our institute for kindly providing the S. miltiorrhiza genome sequence. We appreciate Prof. Xian’en Li for providing S. miltiorrhiza plants. This work was supported by grants from the Natural Science Foundation of China (grant no. 31370327), the Beijing Natural Science Foundation (grant nos. 5112026 and 5152021), the Major Scientific and Technological Special Project for Significant New Drugs Creation (grant no. 2012ZX09301002-001-031), the Program for Changjiang Scholars and Innovative Research Team in University (grant no. IRT1150), and the Research Fund for the Doctoral Program of Higher Education of China (grant no. 20111106110033).
Abbreviations
- AS
Anthranilate synthase
- BEB
Bayes empirical bayes
- CAD1
(+)-δ-cadinene synthase
- CDS
Coding sequence
- Ka
Non-synonymous
- Ks
Synonymous
- DXS
1-deoxy-D-xylulose 5-phosphate synthase
- GST
Glandular secretory trichome
- LRT
Likelihood ratio test
- MeJA
Methyl jasmonate
- Mw
Molecular weight
- pI
Isoelectric points
- SLS
Secologanin synthase
- TCM
Traditional Chinese medicine
- TDC
Tryptophan decarboxylase
Additional files
Sequence features of WRKYs in A. thaliana. Some sequence features of WRKYs in A. thaliana are shown.
Primers used for cloning of SmWRKY CDSs. Complete set of primers used for amplification of SmWRKY CDSs.
Maximum likelihood estimation of the coefficient of Type-I functional divergence (θ) from pairwise comparisons between WRKY groups. The coefficient of Type-I between WRKY groups is shown.
Estimation of the coefficient of Type-II functional divergence (θ) from pairwise comparisons between WRKY groups. The coefficient of Type-II between WRKY groups is shown.
Log-2 transformed RPKM value for SmWRKYs in hairy roots treated with or without yeast extract and Ag+. The transformed RPKM value and differential expression of SmWRKYs are shown.
Ka/Ks and divergence analysis of paralogous SmWRKY genes. Ka, Ks and Ka/Ks are shown.
Primers used for qRT-PCR analysis of SmWRKY genes. Complete set of primers used for qRT-PCR of SmWRKY genes.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CL contributed to RNA extraction, coding sequence (CDS) cloning, qRT-PCR and bioinformatics analysis, and participated in writing the manuscript. DL analyzed the RNA-seq data. FS prepared the plantlets treated with methyl jasmonate (MeJA). SL designed the experiment, participant in bioinformatics analysis, and wrote the manuscript. All authors have read and approved the version of manuscript.
Contributor Information
Caili Li, Email: licaili390@163.com.
Dongqiao Li, Email: butterfly_qiao@163.com.
Fenjuan Shao, Email: sfjstar@126.com.
Shanfa Lu, Email: sflu@implad.ac.cn.
References
- 1.Wu WY, Wang YP. Pharmacological actions and therapeutic applications of Salvia miltiorrhiza depside salt and its active components. Acta Pharmacol Sin. 2011;33:1119–30. doi: 10.1038/aps.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang XH, Morris-Natschke SL, Lee KH. Developments in the chemistry and biology of the bioactive constituents of Tanshen. Med Res Rev. 2007;27:133–48. doi: 10.1002/med.20077. [DOI] [PubMed] [Google Scholar]
- 3.Ma Y, Yuan L, Wu B, Li X, Chen S, Lu S. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J Exp Bot. 2012;63:2809–23. doi: 10.1093/jxb/err466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao X, Shi M, Cui L, Xu C, Zhang Y, Kai G. Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol Appl Biochem. 2014. doi:10.1002/bab.1236. [DOI] [PubMed]
- 5.Zhang Y, Yan YP, Wu YC, Hua WP, Chen C, Ge Q, et al. Pathway engineering for phenolic acid accumulations in Salvia miltiorrhiza by combinational genetic manipulation. Metab Eng. 2014;21:71–80. doi: 10.1016/j.ymben.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Shi M, Luo X, Ju G, Yu X, Hao X, Huang Q, et al. Increased accumulation of the cardio-cerebrovascular disease treatment drug tanshinone in Salvia miltiorrhiza hairy roots by the enzymes 3-hydroxy-3-methylglutaryl CoA reductase and 1-deoxy-D-xylulose 5-phosphate reductoisomerase. Funct Integr Genomics. 2014;14:603–15. doi: 10.1007/s10142-014-0385-0. [DOI] [PubMed] [Google Scholar]
- 7.Di P, Zhang L, Chen J, Tan H, Xiao Y, Dong X, et al. 13C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem Biol. 2013;8:1537–48. doi: 10.1021/cb3006962. [DOI] [PubMed] [Google Scholar]
- 8.Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L, Wang Y, et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci U S A. 2013;110:12108–13. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao G, Shi R, Tao R, Fang Q, Jiang X, Ji H, et al. Cloning, molecular characterization and functional analysis of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase (HDR) gene for diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza Bge. f. alba. Plant Physiol Biochem. 2013;70:21–32. doi: 10.1016/j.plaphy.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, et al. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett. 2009;11:5170–3. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo H, Zhu Y, Song J, Xu L, Sun C, Zhang X, et al. Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol Plant. 2014;152:241–55. doi: 10.1111/ppl.12193. [DOI] [PubMed] [Google Scholar]
- 12.Gao W, Sun HX, Xiao H, Cui G, Hillwig ML, Jackson A, et al. Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza. BMC Genomics. 2014;15:73. doi: 10.1186/1471-2164-15-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Ding G, Lin H, Cheng H, Kong Y, Wei Y, et al. Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS ONE. 2013;8:e80464. doi: 10.1371/journal.pone.0080464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou X, Shao F, Ma Y, Lu S. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: genome-wide characterization, molecular cloning and expression analysis. Mol Biol Rep. 2013;40:4301–10. doi: 10.1007/s11033-013-2517-3. [DOI] [PubMed] [Google Scholar]
- 15.Wenping H, Yuan Z, Jie S, Lijun Z, Zhezhi W. De novo transcriptome sequencing in Salvia miltiorrhiza to identify genes involved in the biosynthesis of active ingredients. Genomics. 2011;98:272–9. doi: 10.1016/j.ygeno.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Yan Y, Wang Z, Tian W, Dong Z, Spencer DF. Generation and analysis of expressed sequence tags from the medicinal plant Salvia miltiorrhiza. Sci China Life Sci. 2010;53:273–85. doi: 10.1007/s11427-010-0005-8. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Sun C, Luo HM, Li XW, Niu YY, Chen SL. Transcriptome characterization for Salvia miltiorrhiza using 454 GS FLX. Yao Xue Xue Bao. 2010;45:524–9. [PubMed] [Google Scholar]
- 18.Song JY, Luo HM, Li CF, Sun C, Xu J, Chen SL. Salvia miltiorrhiza as medicinal model plant. Yao Xue Xue Bao. 2013;48:1099–106. [PubMed] [Google Scholar]
- 19.Zhang L, Wu B, Zhao D, Li C, Shao F, Lu S. Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. J Integr Plant Biol. 2014;56:38–50. doi: 10.1111/jipb.12111. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Lu S. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genomics. 2014;15:277. doi: 10.1186/1471-2164-15-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishiguro SN, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol Gen Genet. 1994;244:563–71. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 2005;5:1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JJ, Ekramoddoullah AKM. Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome. 2009;52:77–88. doi: 10.1139/G08-106. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Wang M, Zhang X, Hao B, Kaushik SK, Pan Y. WRKY gene family evolution in Arabidopsis thaliana. Genetica. 2011;139:973–83. doi: 10.1007/s10709-011-9599-4. [DOI] [PubMed] [Google Scholar]
- 25.Hara K, Yagi M, Kusano T, Sano H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet. 2000;263:30–7. doi: 10.1007/pl00008673. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Yang P, Fan B, Chen Z. An oligo selection procedure for identification of sequence- specific DNA-binding activities associated with the plant defence response. Plant J. 1998;16:515–22. doi: 10.1046/j.1365-313x.1998.00311.x. [DOI] [PubMed] [Google Scholar]
- 27.Yoda H, Ogawa M, Yamaguchi Y, Koizumi N, Kusano T, Sano H. Identification of early-responsive genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol Genet Genomics. 2002;267:154–61. doi: 10.1007/s00438-002-0651-z. [DOI] [PubMed] [Google Scholar]
- 28.Ryu HS, Han M, Lee SK, Cho JI, Ryoo N, Heu S, et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006;25:836–47. doi: 10.1007/s00299-006-0138-1. [DOI] [PubMed] [Google Scholar]
- 29.Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005;12:9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- 30.Yin G, Xu H, Xiao S, Qin Y, Li Y, Yan Y, et al. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013;13:148. doi: 10.1186/1471-2229-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei KF, Chen J, Chen YF, Wu LJ, Xie DX. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012;19:153–64. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu UH, Harter K, Jansson C, et al. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics. 2008;9:194. doi: 10.1186/1471-2164-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Vannozzi A, Wang G, Liang Y, Tornielli BG, Zenoni S, et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic Res. 2014;16:1. doi: 10.1038/hortres.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo C, Guo R, Xu X, Gao M, Li X, Song J, et al. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot. 2014;65:1513–28. doi: 10.1093/jxb/eru007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, et al. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012;31:1199–217. doi: 10.1007/s00299-012-1241-0. [DOI] [PubMed] [Google Scholar]
- 36.Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol Genet Genomics. 2012;287:495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- 37.Ling J, Jiang WJ, Zhang Y, Yu HJ, Mao ZC, Gu XF, et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics. 2011;12:471. doi: 10.1186/1471-2164-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramiro D, Jalloul A, Petitot AS, Grossi De Sác MF, Maluf MP, Fernandez D. Identification of coffee WRKY transcription factor genes and expression profiling in resistance responses to pathogens. Tree Genet Genomes. 2010;6:767–81. [Google Scholar]
- 39.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 41.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51:21–37. doi: 10.1023/a:1020780022549. [DOI] [PubMed] [Google Scholar]
- 42.Xu X, Chen C, Fan B, Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18:1310–26. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Brader G, Kariola T, Palva T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006;46:477–91. doi: 10.1111/j.1365-313X.2006.02712.x. [DOI] [PubMed] [Google Scholar]
- 44.Oh SK, Yi SY, Yu SH, Moon JS, Park JM, Choi D. CaWRKY2, a chili pepper transcription factor, is rapidly induced by incompatible plant pathogens. Mol Cells. 2006;22:58–64. [PubMed] [Google Scholar]
- 45.Zheng Z, Mosher SL, Fan B, Klessig DF, Chen Z. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 2007;7:2. doi: 10.1186/1471-2229-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Z, Qamar SA, Chen Z, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 47.Beyer K, Binder A, Boller T, Colling M. Identification of potato genes induced during colonization by Phytophthora infestans. Mol Plant Pathol. 2001;2:125–34. doi: 10.1046/j.1364-3703.2001.00059.x. [DOI] [PubMed] [Google Scholar]
- 48.Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant-Microbe Interact. 2003;16:295–305. doi: 10.1094/MPMI.2003.16.4.295. [DOI] [PubMed] [Google Scholar]
- 49.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10:366–71. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 50.Knoth C, Ringler J, Dangl JL, Eulgem T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant-Microbe Interact. 2007;20:120–8. doi: 10.1094/MPMI-20-2-0120. [DOI] [PubMed] [Google Scholar]
- 51.Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–55. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 53.Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thalianaWRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–52. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 54.Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–75. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–49. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot. 2012;63:2667–79. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu F, Huaxia Y, Lu W, Wu C, Cao X, Guo X. GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 2012;12:144. doi: 10.1186/1471-2229-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Y, Hu R, Wang H, Cao Y, He G, Fu C, et al. MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plant Sci. 2013;212:1–9. doi: 10.1016/j.plantsci.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 59.Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brand LH, Kirchler T, Hummel S, Chaban C, Wanke D. DPIELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods. 2010;6:25. doi: 10.1186/1746-4811-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun C, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–92. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao F, Lu S. Genome-wide identification, molecular cloning, expression profiling and posttranscriptional regulation analysis of the Argonaute gene family in Salvia miltiorrhiza, an emerging model medicinal plant. BMC Genomics. 2013;14:512. doi: 10.1186/1471-2164-14-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shao F, Lu S. Identification, molecular cloning and expression analysis of five RNA-dependent RNA polymerase genes in Salvia miltiorrhiza. PloS One. 2014;9:e95117. doi: 10.1371/journal.pone.0095117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lynch M, O’Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 66.Park CY, Lee JH, Yoo JH, Moon BC, Choi MS, Kang YH, et al. WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 2005;579:1545–50. doi: 10.1016/j.febslet.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 67.Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005;137:176–89. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 69.Nielsen R, Yang Z. Likelihood models for detecting positively amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–36. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J Mol Evol. 2000;51:423–32. doi: 10.1007/s002390010105. [DOI] [PubMed] [Google Scholar]
- 71.Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–49. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–9. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 73.Yang Z, Wong WSW, Nielsen R. Bayes empirical Bayes inferences of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–18. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 74.Gu X. Statistical methods for testing functional divergence after gene duplication. Mol Biol Evol. 1999;16:1664–74. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- 75.Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol. 2006;23:1937–45. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- 76.Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol. 2004;55:853–67. doi: 10.1007/s11103-004-2142-6. [DOI] [PubMed] [Google Scholar]
- 77.Luo M, Dennis ES, Berger F, Peacock WP, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:17531–6. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guillaumie S, Mzid R, Méchin V, Léon C, Hichri I, Destrac-Irvine A, et al. The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol Biol. 2010;72:215–34. doi: 10.1007/s11103-009-9563-1. [DOI] [PubMed] [Google Scholar]
- 79.Robatzek S, Somssich IE. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence- related processes. Plant J. 2001;28:123–33. doi: 10.1046/j.1365-313x.2001.01131.x. [DOI] [PubMed] [Google Scholar]
- 80.Ulker B, Shahid Mukhtar M, Somssich IE. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta. 2007;226:125–37. doi: 10.1007/s00425-006-0474-y. [DOI] [PubMed] [Google Scholar]
- 81.Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G, Humbeck K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 2009;58:333–46. doi: 10.1111/j.1365-313X.2008.03782.x. [DOI] [PubMed] [Google Scholar]
- 82.Schluttenhofer C, Pattanail S, Patra B, Yuan L. Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genomics. 2014;15:502. doi: 10.1186/1471-2164-15-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Z, Wang X, Xue J, Meng L, Li R. Identification and expression analysis of WRKY transcription factors in medicinal plant Catharanthus roseus. Chin J Biotech. 2013;29:785–802. [PubMed] [Google Scholar]
- 84.Dong F, Wang Y, She J, Lei Y, Liu Z, Eulgem T, et al. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol Plant. 2014;150:397–411. doi: 10.1111/ppl.12093. [DOI] [PubMed] [Google Scholar]
- 85.Shu-Ling Z, Xing-Fen W, Yan Z, Jian-Feng L, Li-Zhu W, Dong-Mei Z, et al. GbWRKY1, a novel cotton (Gossypium barbadense) WRKY gene isolated from a bacteriophage full-length cDNA library, is induced by infection with Verticillium dahliae. Indian J Biochem Biophys. 2012;49:405–13. [PubMed] [Google Scholar]
- 86.Guo R, Yu F, Gao Z, An H, Cao X, Guo X. GhWRKY3, a novel cotton (Gossypium hirsutum L.) WRKY gene, is involved in diverse stress responses. Mol Biol Rep. 2011;38:49–58. doi: 10.1007/s11033-010-0076-4. [DOI] [PubMed] [Google Scholar]
- 87.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 88.Xu YH, Wang JW, Wang S, Wang JY, Chen XY. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol. 2004;135:507–15. doi: 10.1104/pp.104.038612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun Y, Niu Y, Xu J, Li Y, Luo H, Zhu Y, et al. Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tiss Org Cult. 2013;114:269–77. [Google Scholar]
- 90.Ma D, Pu G, Lei C, Ma L, Wang H, Guo Y, et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009;50:2146–61. doi: 10.1093/pcp/pcp149. [DOI] [PubMed] [Google Scholar]
- 91.Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Sing SK, Yuan L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011;157:2081–93. doi: 10.1104/pp.111.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ni X, Su J. Active constituents of above-ground portion and root of Salvia miltiorrhiza. Chinese Pharm J. 1995;30:336–8. [Google Scholar]
- 93.Xu C, Shu Z, Wang Y, Miao F, Zhou L. The accumulation rule of the main medicinal components in different organs of Salvia miltiorrhiza Bunge. and Salvia miltiorrhiza Bunge. F. alba. Lishizhen Med Materia Medica Res. 2010;21:2129–32. [Google Scholar]
- 94.Liu F, Cui LJ, He G, Yang ZM. Dynamic changes in several effective components in different vegetative organs of Salvia miltiorrhiza Bge cultivars in different seasons. Plant Sci J. 2011;29:93–8. [Google Scholar]
- 95.Ge XC, Wu JY. Tanshinone production and isoprenoid pathways in Salvia miltiorrhiza hairy roots induced by Ag+ and yeast elicitor. Plant Sci. 2005;168:487–91. [Google Scholar]
- 96.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–37. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 97.He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–64. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, et al. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23:469–78. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- 99.Wang YP, Wang XY, Tang HB, Tan X, Ficklin SP, Feltus FA, et al. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS One. 2011;6:e28150. doi: 10.1371/journal.pone.0028150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Song Y, Gao J. Genome-wide analysis of WRKY gene family in Arabidopsis lyrata and comparison with Arabidopsis thaliana and Populus trichocarpa. Chin Sci Bull. 2014;8:1–12. [Google Scholar]
- 102.Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–39. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- 103.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evo. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–73. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Willems E, Leyns L, Vandesompele J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal Biochem. 2008;379:127–9. doi: 10.1016/j.ab.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 107.Li D, Shao F, Lu S. Identification and characterization of mRNA-like noncoding RNAs in Salvia miltiorrhiza. Planta. 2015. doi:10.1007/s00425-015-2246-z. [DOI] [PubMed]
- 108.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–12. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]


