Abstract
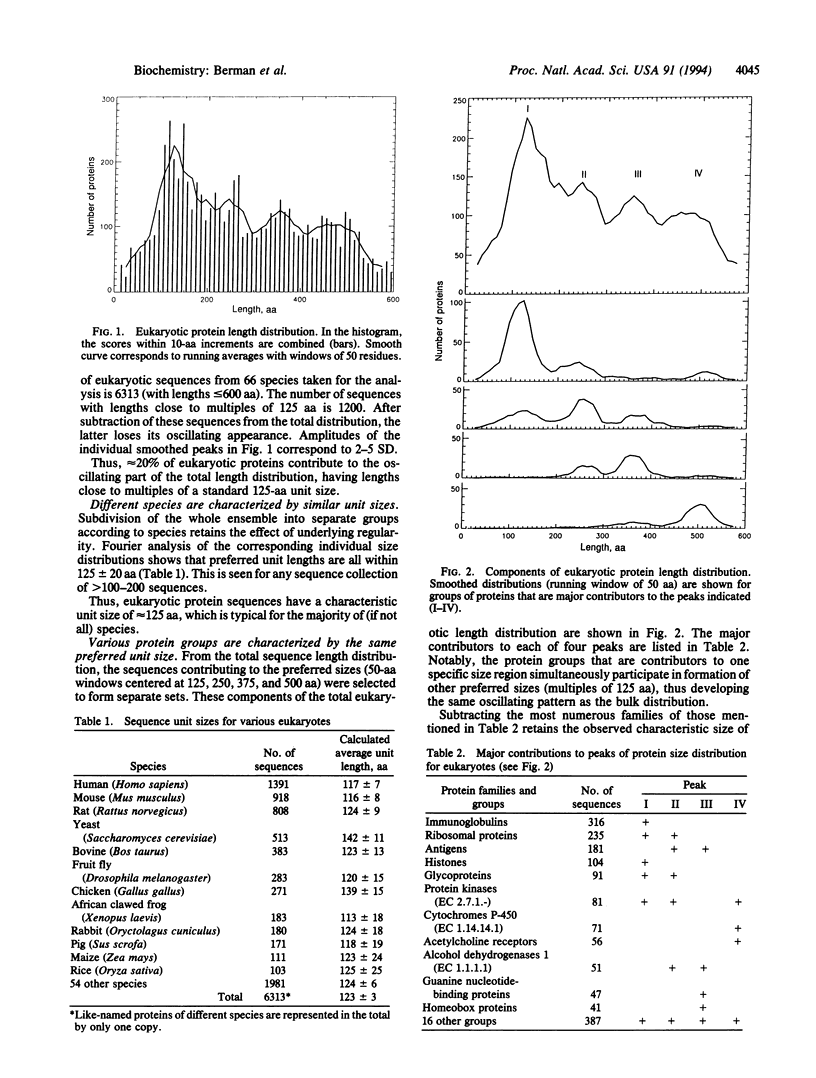
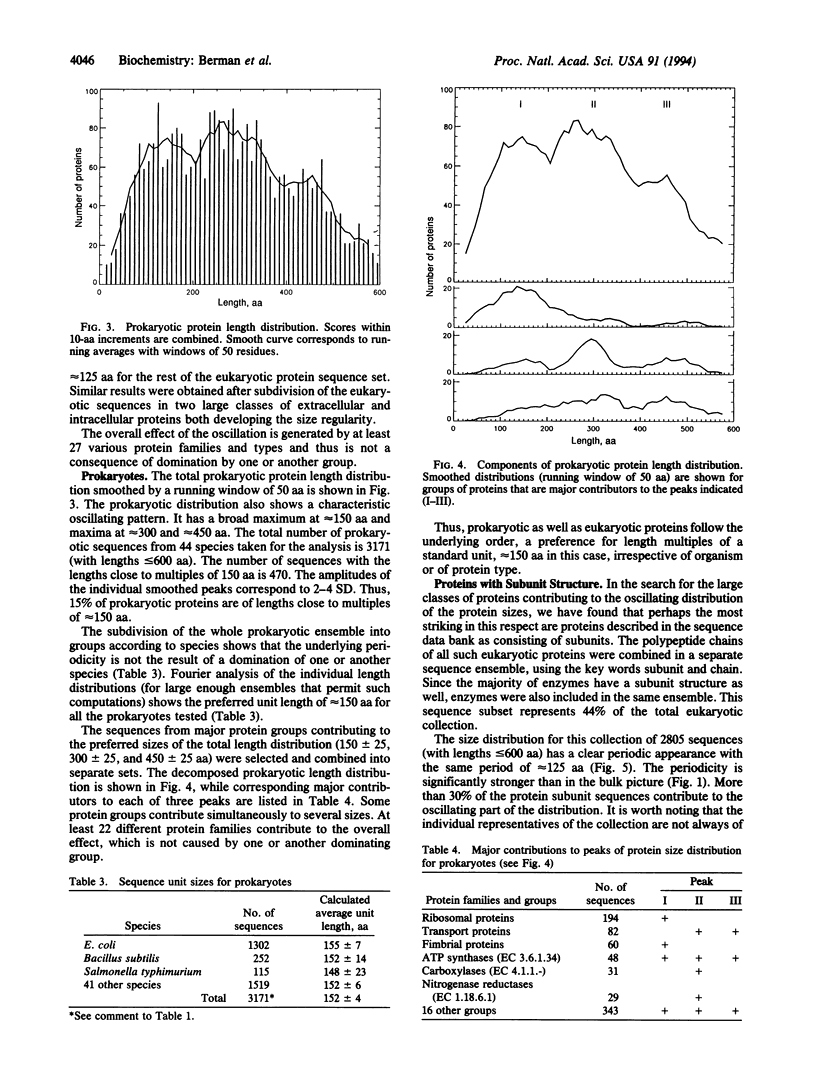
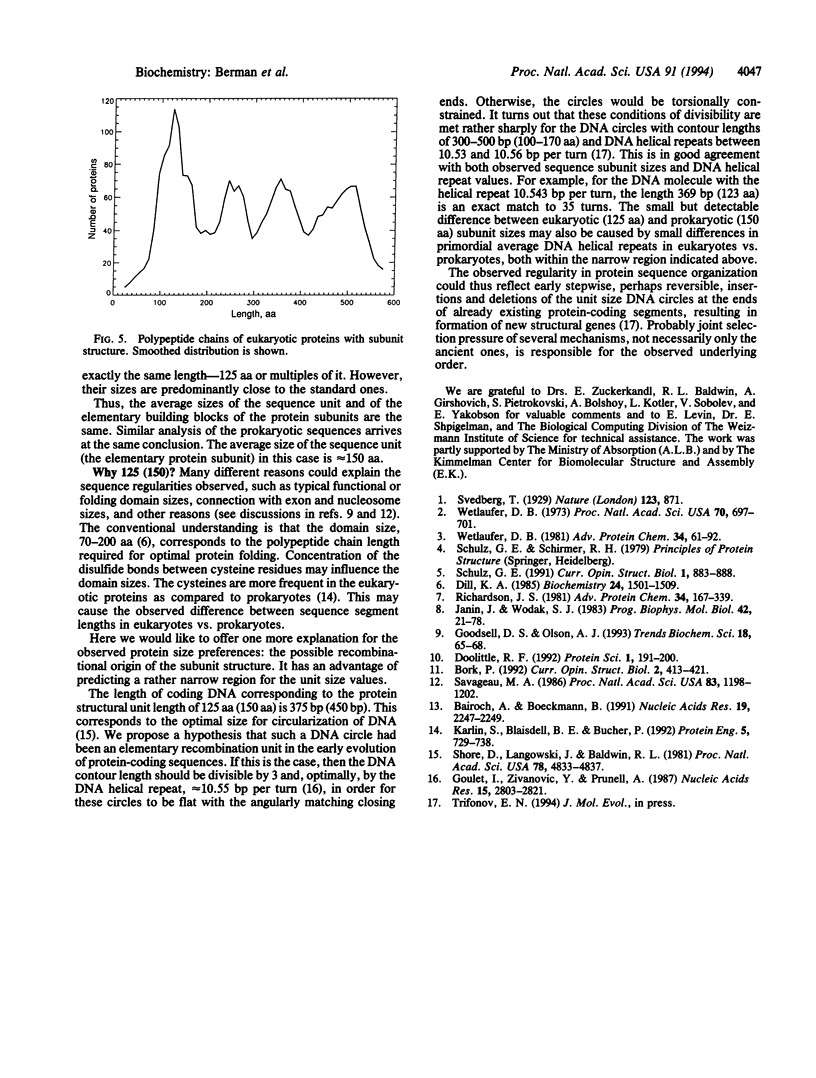
The idea of a possible standard modular structure of proteins has been known since 1929 when it was introduced by Svedberg. It still remains an idea with no quantitative confirmation of universality of such hypothetical organization. From a large collection of nonredundant protein sequences representing > 100 eukaryotic and prokaryotic species, we have obtained the protein sequence length distributions. Mere inspection of these distributions, as well as spectral analysis, shows that 15-30% of proteins, depending on species and sequence types, indeed appear to be made of sequence units with characteristic lengths of approximately 125 aa for eukaryotes and approximately 150 aa for prokaryotes. This underlying order in protein sequence organization is shown to be universal--that is, the weak regularity observed is not caused by a particular dominant species or protein group. Possible mechanisms are discussed that may be responsible for the observed regularity, including a hypothesis about the recombinational nature of such protein sequence organization.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A. Theory for the folding and stability of globular proteins. Biochemistry. 1985 Mar 12;24(6):1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. Stein and Moore Award address. Reconstructing history with amino acid sequences. Protein Sci. 1992 Feb;1(2):191–200. doi: 10.1002/pro.5560010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell D. S., Olson A. J. Soluble proteins: size, shape and function. Trends Biochem Sci. 1993 Mar;18(3):65–68. doi: 10.1016/0968-0004(93)90153-e. [DOI] [PubMed] [Google Scholar]
- Goulet I., Zivanovic Y., Prunell A. Helical repeat of DNA in solution. The V curve method. Nucleic Acids Res. 1987 Apr 10;15(7):2803–2821. doi: 10.1093/nar/15.7.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J., Wodak S. J. Structural domains in proteins and their role in the dynamics of protein function. Prog Biophys Mol Biol. 1983;42(1):21–78. doi: 10.1016/0079-6107(83)90003-2. [DOI] [PubMed] [Google Scholar]
- Karlin S., Blaisdell B. E., Bucher P. Quantile distributions of amino acid usage in protein classes. Protein Eng. 1992 Dec;5(8):729–738. doi: 10.1093/protein/5.8.729. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Savageau M. A. Proteins of Escherichia coli come in sizes that are multiples of 14 kDa: domain concepts and evolutionary implications. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1198–1202. doi: 10.1073/pnas.83.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Langowski J., Baldwin R. L. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetlaufer D. B. Folding of protein fragments. Adv Protein Chem. 1981;34:61–92. doi: 10.1016/s0065-3233(08)60518-5. [DOI] [PubMed] [Google Scholar]
- Wetlaufer D. B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc Natl Acad Sci U S A. 1973 Mar;70(3):697–701. doi: 10.1073/pnas.70.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]



