Significance
Altered expression of RNA binding proteins might contribute to cancer development. This study reveals the functional implications and clinical relevance of FXR1, an RNA binding protein, in non-small cell lung cancer (NSCLC). Our results demonstrate that FXR1 promotes tumor progression by regulating two other oncogenes within the same chromosome 3q amplicon. To drive tumor progression, FXR1 forms a new complex with protein kinase C, iota, and posttranscriptionally stabilizes the expression of epithelial cell transforming 2. We show that increased FXR1 expression in NSCLC is a candidate biomarker predictive of poor survival and might represent a novel therapeutic target. In addition, FXR1 expression correlates with poor clinical outcome in multiple human cancers, suggesting broader implications of this RNA binding protein in cancer progression.
Keywords: 3q amplification, FXR1, RNA binding protein, non-small cell lung cancer
Abstract
Aberrant expression of RNA-binding proteins has profound implications for cellular physiology and the pathogenesis of human diseases such as cancer. We previously identified the Fragile X-Related 1 gene (FXR1) as one amplified candidate driver gene at 3q26-29 in lung squamous cell carcinoma (SCC). FXR1 is an autosomal paralog of Fragile X mental retardation 1 and has not been directly linked to human cancers. Here we demonstrate that FXR1 is a key regulator of tumor progression and its overexpression is critical for nonsmall cell lung cancer (NSCLC) cell growth in vitro and in vivo. We identified the mechanisms by which FXR1 executes its regulatory function by forming a novel complex with two other oncogenes, protein kinase C, iota and epithelial cell transforming 2, located in the same amplicon via distinct binding mechanisms. FXR1 expression is a candidate biomarker predictive of poor survival in multiple solid tumors including NSCLCs. Because FXR1 is overexpressed and associated with poor clinical outcomes in multiple cancers, these results have implications for other solid malignancies.
Amplification of the chromosomal region 3q26-29 is the most frequent genomic alteration in primary squamous cell lung cancers (1) and occurs in many other cancers (2). The best studied oncogenes of this amplicon include phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) (3, 4), TP63 (5), sex-determining region Y box 2 (SOX2) (6), epithelial cell transforming 2 (ECT2) (7), and protein kinase C, iota (PRKCI) (8). In an effort to identify oncogenic drivers in lung cancer associated the 3q26-29 amplicon, we previously integrated genomic and gene expression analysis of multiple lung SCC datasets and identified Fragile X-related 1 (FXR1) as a potential new candidate driver gene (9). FXR1 belongs to a small family of RNA-binding proteins that includes Fragile X mental retardation 1 (FMR1) and Fragile X-related 2 (FXR2) (10). Inactivation of FMR1 expression is the cause of the Fragile X syndrome in humans, whereas alterations of FXR1 are yet to be associated with the pathogenesis of human disease. RNA-binding proteins (RBPs) are essential in RNA metabolism, from synthesis to degradation. RBPs coordinate elaborate networks of RNA–protein and protein–protein interactions that link RNA metabolism to signal transduction pathways (11). Aberrant function of RBPs contributes to the progression of many human diseases including cancer. Nevertheless, few RBPs have been identified as oncogenes or tumor suppressors and clinical implications of these cancer related RBPs is largely unknown. FXR1 is highly expressed in vertebrate muscle cells and FXR1 knockout mice die early during embryogenesis, suggesting an essential role for FXR1 in development (12). In this study, we examined whether RNA binding protein FXR1 is a regulator of tumor progression in nonsmall cell lung cancer (NSCLC) and a driver of the 3q amplicon. We tested this hypothesis across a large number of clinical specimens, in gain- and loss-of-function and mechanistic studies in vitro and in vivo. We investigated the translational relevance of our findings in NSCLC tissue microarrays and in datasets of multiple human cancers available in the public domain.
Results
FXR1 Is Coamplified and Coexpressed with ECT2 and PRKCI in SCC of the Lung.
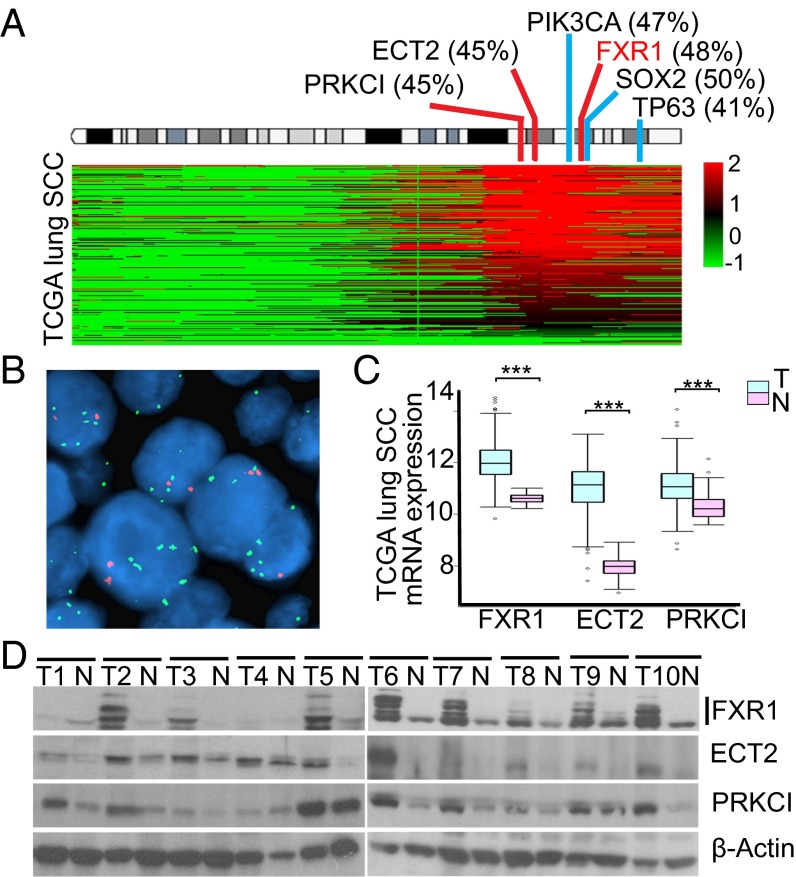
To assess FXR1 copy number (CN) and expression, we performed genome-wide array comparative genomic hybridization (aCGH) and gene expression profiling on twenty-four untreated lung SCCs. High level amplification (log2 ratio > 0.8, 7 of 24) of FXR1 gene was found in 30% of the samples (SI Appendix, Fig. S1A). FXR1 mRNA levels were higher in tumors with CN gain (P < 0.0001; SI Appendix, Fig. S1B). Using The Cancer Genome Atlas (TCGA) lung SCC data, FXR1 amplification was confirmed in 48% of SCCs (Fig. 1A). Significant correlation of CN gain and mRNA expression was observed (R2 = 0.72, P < 0.0001, SI Appendix, Fig. S1C). FXR1 amplification was further verified in 30% (14 of 47) of lung tumors by fluorescence in situ hybridization (FISH) analysis in an independent tissue microarray (TMA) of SCCs (Fig. 1B). To identify potential targets of FXR1, we searched our previously published lung SCC gene expression dataset (13) and found that FXR1 expression was highly correlated with ECT2 and PRKCI (SI Appendix, Table S1). PRKCI is also one of top 20 candidate driver genes we identified using a novel integrative computational analysis (9). ECT2 and PRKCI have been reported to promote lung tumor cell growth through the extracellular signal-regulated kinases (ERK) signaling pathway by forming a complex with PARD6A (14). The overexpression and coexpression of these three genes were validated in two independent gene expression datasets that have both SCCs and matched normal samples including TCGA (Fig. 1C) and GSE31552 (SI Appendix, Fig. S1D and Tables S2 and S3). The coamplification of three genes was observed in TCGA and GSE20393 datasets (SI Appendix, Table S4). In addition, results from ten representative matched normal and lung SCCs samples analyzed by Western blotting confirmed the findings at the protein level (Fig. 1D). In contrast to FXR1, FMR1 and FXR2 genes were neither amplified (SI Appendix, Fig. S1E) nor overexpressed in lung SCCs (SI Appendix, Fig. S1 F and G). Lastly, our findings were confirmed in 147 lung cancer cell lines from the Cancer Cell Line Encyclopedia (15) (SI Appendix, Fig. S1 H and I). Thus, FXR1 is the only FMR1-family member altered in lung SSCs; its amplification and overexpression are associated with those of oncogenes ECT2 and PRKCI.
Fig. 1.
(A) Copy number alteration of FXR1, ECT2, and PRKCI in 343 TCGA lung SCCs (only 182 samples harboring 3q amplicon were shown). “2” is a high-level amplification (log2 ratio > 0.8), “1” indicates a low-level gain (log2 ratio > 0.3), “0” is diploid, “−1” is a single-copy loss (heterozygous deletion). Frequencies of high-level amplification are shown as a percentage of all cases. (B) Representative dual color FISH of the FXR1 gene (3q26.3, green spots) and a chromosome 3 centromeric probe (red spots) in lung SCCs. (C) Lung SCCs (T) express higher FXR1, ECT2, and PRKCI mRNA levels than matched normal tissues (N) in TCGA dataset (n = 50). ***P < 0.0001. (D) Immunoblot analysis of ten primary SCCs and matched normal lung tissue for FXR1, ECT2, PRKCI, and β-actin.
FXR1 Regulates Lung Cancer Cell Growth in Vitro and in Vivo.
To investigate the role of FXR1 in lung cancer progression, we first tested the effect of FXR1 on lung cancer cell growth and survival in cell lines with and without the 3q amplicon (Fig. 2A) by loss of function studies. Using a siRNA approach, we found that FXR1 silencing inhibited cell growth in three FXR1-amplified cell lines but had no significant effect in three nonamplified NSCLC lines (Fig. 2 B and C and SI Appendix, Fig. S2A). FXR1 silencing also induces apoptosis, demonstrated by increased PARP cleavage in H520 cells (Fig. 2D). We further stably transfected H520 cells with three lentiviral FXR1 shRNA constructs knockdown (KD) or a nontarget (NT) shRNA control and demonstrated that FXR1 depletion led to significant inhibition of anchorage-independent colony formation on H520 cells (Fig. 2E and SI Appendix, Fig. S2B). Flow cytometry analysis revealed an increase in the percentage of H520 FXR1-KD cells in S phase, a decrease in G1/G0 and an increase in the sub-G1 population (SI Appendix, Fig. S2C). In addition, FXR1-KD cells exhibited impaired invasion commensurate with FXR1-KD level (SI Appendix, Fig. S2D). To test its tumorigenic properties, we ectopically overexpressed FXR1 into two immortalized human bronchial epithelial cell lines BEAS-2B and HBEC3KT without the 3q amplicon (16) (SI Appendix, Fig. S2E) and observed enhanced anchorage-independent cell growth in soft agar of BEAS-2B (Fig. 2F) but not HBEC3KT overexpressing FXR1.
Fig. 2.
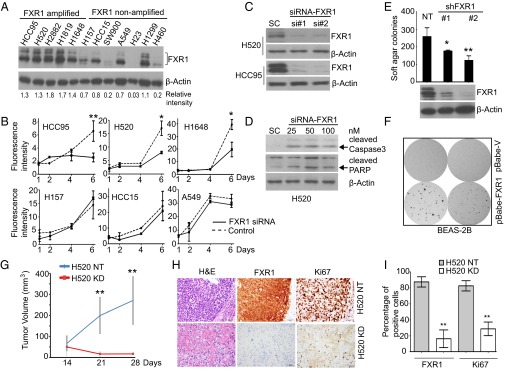
FXR1 regulates lung cancer cell growth in vitro and in vivo. (A) Immunoblot analysis of a panel of NSCLC cell lines for FXR1 and β-actin. Relative FXR1 protein level was normalized to β-actin. (B) Effect of FXR1-specific siRNA on viable cells over time. Cells were measured 1, 2, 4, and 6 d after plating using the Cyquant assay. *P < 0.05; **P < 0.01. (C) Down-regulation of FXR1 protein in H520 and HCC95 cells treated with siRNA against FXR1 after 48 h. SC: scrambled siRNA control. si#1, si#2: two individual FXR1 siRNAs. (D) Activated Caspase 3 and PARP in H520 cells treated with pooled FXR1-siRNA after 72 h. SC: scrambled siRNA control. (E) Effect of FXR1-shRNA on H520 anchorage-independent soft agar colony formation. H520 cells were infected with three lentiviruses containing shRNA targeting FXR1 or a nontarget (NT) sequence. Two stable FXR1 knockdown (KD) cell lines are shown. H520/FXR1 KD cells grow significantly slower than H520/NT cells (*P < 0.05, **P < 0.01). (F) Effect of FXR1 overexpression on BEAS-2B anchorage independent cell growth compared with vector control. pBabe-V, BEAS-2B-vector group; pBabe-FXR1, BEAS-2B cells stably-transfected with pBabe-Flag-FXR1. (G) Effects of FXR1 knockdown on tumorigenicity in nude mice. H520 NT and H520 FXR1 KD cells were injected s.c. into the flanks of nude mice (n = 10). Tumor volume was measured twice a week in all experiments by caliper and calculated by the formula: V = 3.14 (smaller diameter)2(larger diameter)/6. The quantification of tumor volume changes over a 4-wk period is shown; **P < 0.01. (H) Representative H&E staining, immunohistochemical staining of FXR1 and Ki67 in tumors formed by H520 NT and H520 KD in one mouse. Scale bars, 50 μm. (I) Quantification of FXR1 and Ki67 immunohistochemical staining shown by the percentage of positively stained cells compared with total number of cells per field. The P value (Student’s t test) relative to NT is shown. **P < 0.01. Data are mean ± SEM.
To examine the in vivo effect of targeting FXR1 in H520 cells, we injected H520 NT and H520 FXR1-KD cells s.c. into the flanks of athymic nude mice and found H520 FXR1-KD cells exhibited impaired tumor growth compared with control (Fig. 2G and SI Appendix, Fig. S2F). Histological analysis of the xenograft tissues confirmed strong FXR1 protein expression in control tumor tissues but nearly undetectable levels in FXR1-KD xenografts. Ki-67 staining was remarkably reduced in FXR1-KD tumors (Fig. 2 H and I, and SI Appendix, Fig. S2G). The growth inhibition due to loss function of FXR1 in vivo was also observed in another lung cancer cell line H1299 without 3q amplicon but overexpressing FXR1 (SI Appendix, Fig. S2H). Together, these results indicate that FXR1 regulates proliferation and survival of NSCLC cells in vitro and in vivo, a process dependent on FXR1 gene overexpression but not only on genomic amplification.
FXR1 Regulates ECT2 and PRKCI Expression via a FXR1/PRKCI/mRNA Complex in NSCLC Cells.
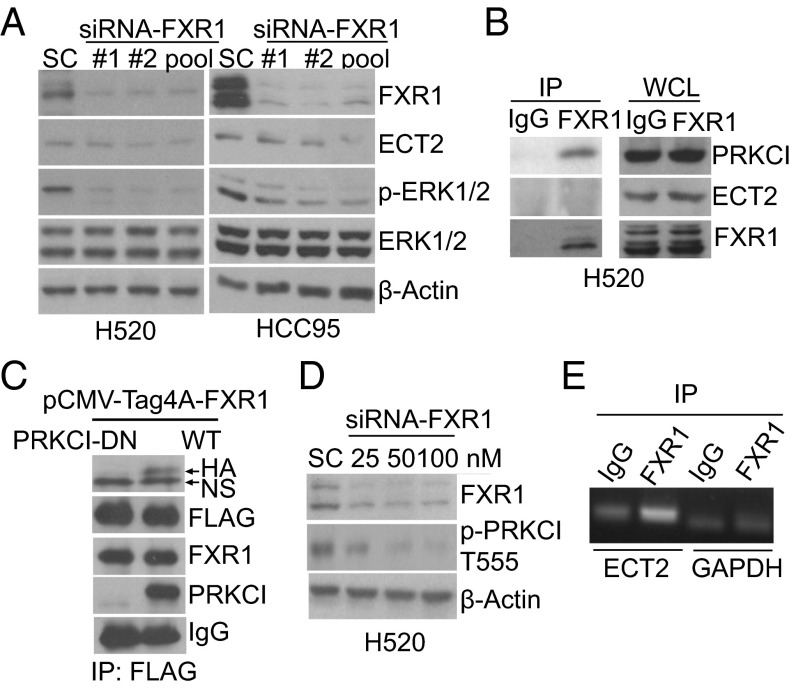
To investigate the mechanisms of FXR1-dependent regulation of lung tumorigenesis, we examined the levels of ECT2 and PRKCI protein expression after knockdown of FXR1 expression in three FXR1-overexpressing NSCLC cell lines. Silencing FXR1 led to reduced expression of ECT2, PRKCI, and phospho-ERK1/2 (Fig. 3A and SI Appendix, Fig. S3A), whereas the converse effect was observed in HBEC3KT cells exogenously overexpressing FXR1 (SI Appendix, Fig. S2E). These findings demonstrate that FXR1 regulates the expression ECT2 and PRKCI.
Fig. 3.
FXR1 regulates ECT2 and PRKCI expression via distinct mechanisms. (A) Down-regulation of ECT2 and phospho-ERK1/2 in FXR1 knockdown NSCLC cells. (B) Immunoprecipitation (IP) of FXR1 reveals that FXR1 binds to PRKCI protein but not to ECT2 in H520 cells. (C) HEK293T cells were cotransfected with Flag-tagged pCMV-Tag4A-FXR1 and either HA-tagged pHACE-PRKCI-WT or pHACE-PRKCI-DN for 48h. Cell lysates were then subjected to IP with anti-HA, anti-Flag, anti-FXR1 or anti-PRKCI. The IP experiments reveal that FXR1 binds to active PRKCI but not the PRKCI DN mutant. NS, nonspecific band. (D) Reduced phosphorylated PRKCI T555 is observed in FXR1 knockdown H520 cells. (E) IP using either anti-FXR1 or normal IgG in H520 under conditions that preserve the association of RNA-binding proteins with target mRNAs. IP was followed by RT-PCR analysis to detect endogenous ECT2 mRNA; PCR products were resolved by electrophoresis in 1.5% agarose gels stained with ethidium bromide.
To gain insights into the mechanisms by which FXR1 regulates ECT2 and PRKCI, we used a series of reciprocal immunoprecipitation (IP) experiments and found that FXR1 physically binds to PRKCI protein but not to ECT2 or PRAD6A protein in NSCLC cells (Fig. 3B and SI Appendix, Fig. S3 B–F). The binding of FXR1 to PRKCI was not observed in immortalized normal lung epithelial cells (SI Appendix, Fig. S3G). We further found that FXR1 is phosphorylated and binds to phosphorylated PRKCI in NSCLC cells (SI Appendix, Fig. S3 H–L). When a Flag-tagged full length of FXR1 and a hemagglutinin-tagged (HA) constitutively active wild type (WT) of PRKCI (17) or a HA-tagged dominant negative (DN) mutant of PRKCI were cotransfected into HEK293T cells, we found that FXR1 binds only to active WT-PRKCI but not PRKCI DN mutant (Fig. 3C and SI Appendix, Fig. S3M). These results indicate that FXR1 directly binds to active PRKCI and this binding might be a common event in NSCLC cells. In addition, we discovered that the knockdown of FXR1 leads to the down-regulation of phosphorylated PRKCI in H520 cells (Fig. 3D). In contrast, the inhibition of PKC activity with chelerythrine resulted in a time- and dose-dependent decrease in phosphorylated PRKCI/T555, FXR1 and ECT2 expression (SI Appendix, Fig. S3 N and O). Altogether, these results suggest that FXR1 is phosphorylated by PRKCI and that there is a reciprocal regulation between PRKCI and FXR1 in lung cancer cells.
Because FXR1 regulates ECT2 expression without binding to the ECT2 protein, we tested whether FXR1 binds to ECT2 mRNA. We recovered endogenous ECT2 mRNA transcripts by RT-PCR in H520 cell lysates immunoprecipitated with anti-FXR1 (Fig. 3E and SI Appendix, Fig. S3P). To further determine the role of FXR1 in ECT2 gene expression, we performed actinomycin D chase experiments (18) to measure ECT2 mRNA half-life in H520-FXR1-KD cells and found that the half-life of ECT2 mRNA in H520-FXR1-KD cells was much shorter than those observed in control cells (P < 0.01, SI Appendix, Fig. S3Q). These results demonstrate that FXR1 regulates ECT2 expression by binding to and stabilizing ECT2 mRNA in H520 cells.
FXR1 Overexpression Predicts Overall Survival in NSCLC.
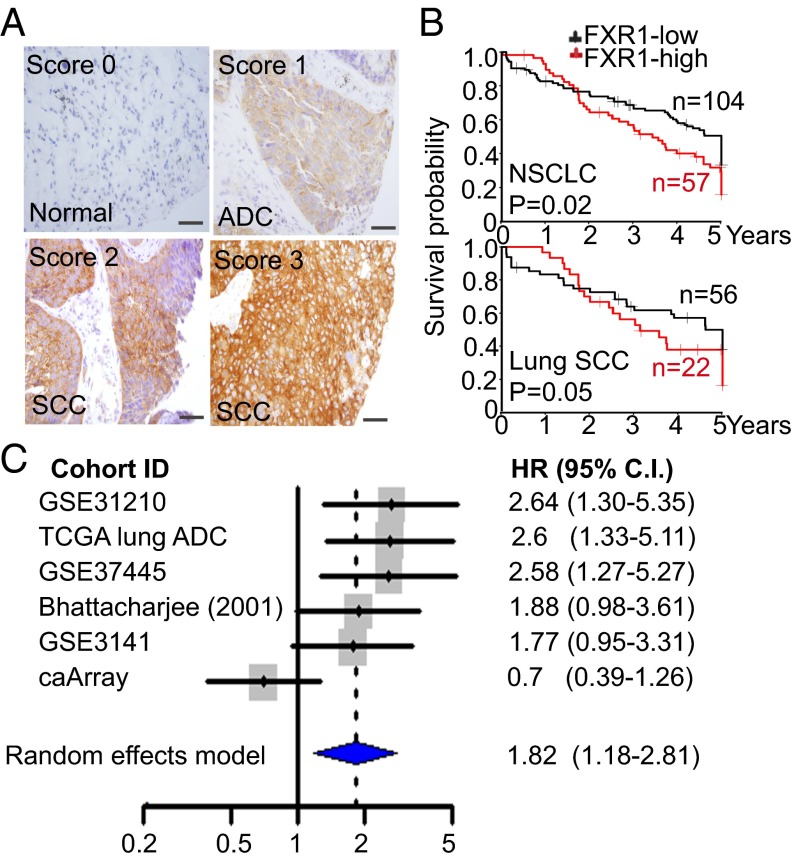
To test the translational potential of FXR1 as a biomarker, we studied the association of FXR1 expression with clinical outcomes by immunohistochemistry in 292 NSCLCs (SI Appendix, Table S5). FXR1 protein was localized to the cytoplasm in lung cancer cells (Fig. 4A). FXR1 expression was significantly and positively correlated with squamous histology, disease stage and smoking history (SI Appendix, Fig. S4). Elevated FXR1 expression was associated with shorter overall survival (OS) independent of age, sex, stage, and smoking history (P = 0.0024, HR, 1.28, 95% CI, 1.09–1.49, SI Appendix, Table S6). Multivariate analysis further indicates that FXR1 expression is a better predictor for OS than other clinical variables tested in patients with stage I NSCLC treated surgically as primary treatment modality (HR, 1.31, 95% CI, 1.05–1.63, P = 0.018), particularly in lung SCC subtype (HR, 1.44, 95% CI, 1.03–2.01, P = 0.032, Fig. 4B, SI Appendix, Tables S7 and S8). To evaluate the clinical relevance of FXR1 expression in lung adenocarcinoma (ADC), we analyzed FXR1 mRNA expression in six independent cohorts consisting of 697 stage I lung ADC patients (SI Appendix, Table S9). A meta-analysis indicated higher FXR1 mRNA is associated with shorter OS in lung ADC stage I patients (Fig. 4C). Together, these results suggest that FXR1 expression is a novel prognostic biomarker in NSCLC.
Fig. 4.
FXR1 overexpression is associated with poor overall survival in NSCLC. (A) Representative immunohistochemical staining of FXR1 protein expression in sections of formalin-fixed paraffin-embedded normal lung tissue, lung ADC, and SCCs. (B) Kaplan–Meier plots of overall survival of stage I NSCLC patients (n = 161) or SCC (n = 78) stratified by FXR1 protein expression. The log-rank P values are shown. (C) FXR1 mRNA level is associated with poor overall survival in stage I lung ADC patients indicated by a meta-analysis in six independent cohorts (SI Appendix, Table S9). Patients who received adjuvant chemotherapy were excluded.
FXR1 Overexpression Correlates with Poor Clinical Outcomes in Multiple Cancers.
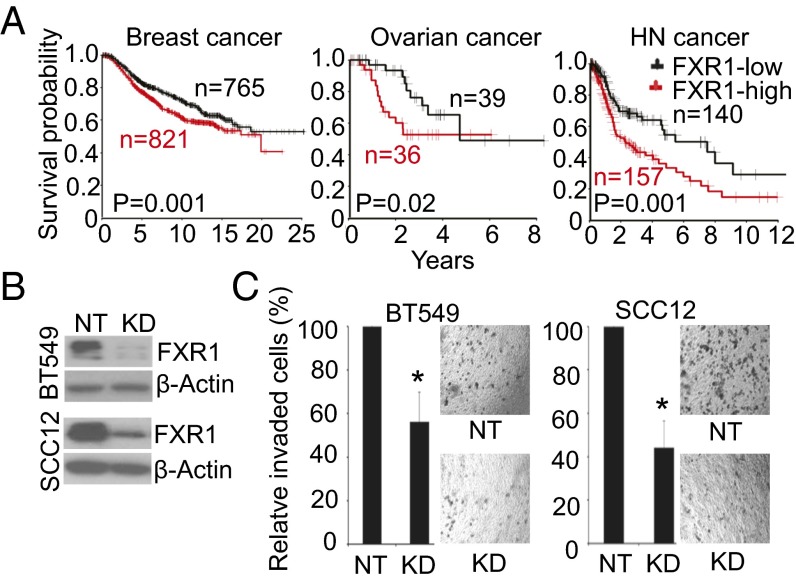
To determine whether FXR1 may be involved in other solid tumors, we interrogated its gene copy number alteration and gene expression in 18 types of human cancers in the TCGA database. We found FXR1 amplification in 19–48% of SCCs of different origin including lung, cervix, and head and neck squamous carcinoma (HNSC) (SI Appendix, Fig. S5A and Table S10). Elevated FXR1 mRNA levels were observed in 10 types of cancers including breast, ovarian and HNSC cancers compared with control tissues (SI Appendix, Table S10 and Fig. S5B). In breast cancer, FXR1 mRNA expression was tested as a prognostic biomarker using multivariate Cox proportional hazards analysis in eight independent breast cancer cohorts including GSE3494 (n = 250), GSE4922 (n = 249), GSE2034 (n = 286), GSE2603 (n = 121), GSE11121 (n = 200), TCGA (n = 483), TRANSBIG consortium (n = 198), and METABRIC consortium (n = 1,992, Fig. 5A) (SI Appendix, Tables S11–S18). The association of FXR1 mRNA with poor outcome was significantly in all these cohorts (SI Appendix, Tables S19–S26). In ovarian cancer, FXR1 mRNA level was significantly associated with worse relapse free survival (RFS) in early stage of ovarian cancer patients in combined GSE9891 and TCGA cohorts (HR, 5.38, 95% CI 1.95–14.82, P = 0.001, Fig. 5A and SI Appendix, Tables S27 and S28). In HNSC, multivariate Cox analysis indicates that FXR1 mRNA level was independently associated with poor overall survival (TCGA, HR, 1.56, P = 0.037, Fig. 5A) and metastasis-free survival (E-TABM-302, HR, 2.25, P = 0.01) (SI Appendix, Tables S29–S32).
Fig. 5.
FXR1 is associated with poor outcome in multiple human cancers. (A) FXR1 overexpression is associated with worse disease specific free survival in METABRIC breast cancer cohort, worse relapse-free survival in early stage of ovarian cancer (combined GSE9891 and TCGA) and worse overall survival in TCGA HNSC cohort. The log-rank P values are shown. The datasets were described in detail in SI Appendix, Tables S17, S27, and S29. (B) Down-regulation of FXR1 protein in BT549 and SCC12 cells treated with shRNA against FXR1. NT: nontarget shRNA control. KD: shRNA FXR1 knockdown. (C) Effect of FXR1-shRNA on BT549 and SCC12 cell invasion. Significant differences between NT and FXR1 KD cells are indicated; *P < 0.05.
Finally, to demonstrate that FXR1 overexpression was a biological plausible biomarker of tumor progression in these other tumor types, we examined its expression in seven breast cancer cell lines, five HNSC lines and two ovarian cancer cell lines (SI Appendix, Fig. S5C). shRNA knockdown of FXR1 expression in three cell lines (Fig. 5B and SI Appendix, Fig. S5D), each of different histological subtype and overexpressing FXR1 protein (without genomic amplification) significantly impaired cell invasion of BT549 (breast cancer line) and SCC12 (HNSC line) in vitro (Fig. 5C). The growth inhibition of A2780 (ovarian cancer line) and SCC12 upon FXR1 depletion were also confirmed in mouse xenograft models (SI Appendix, Fig. S5 E–G). Together, these results suggest that FXR1 is a newly identified candidate prognostic marker and may play critical role in other human cancers including breast, ovarian and head and neck cancers.
Discussion
Here we described, to our knowledge for the first time, the functional and clinical significance of FXR1 overexpression in NSCLC and other solid tumors. We and others have previously identified FXR1 gene is located at the peak of 3q26-29 amplicon in squamous cell carcinoma of the lung (9, 19). FXR1 promotes lung epithelial cell growth and its overexpression is critical for lung cancer cell proliferation, survival and invasion. We discovered that FXR1 engages with two novel binding partners (PRKCI and ECT2), genes located in the same 3q amplicon, thereby providing a mechanism of action and opening new potential therapeutic avenues in 3q amplified lung tumors. Loss of FXR1 substantially reduced the size of multiple types of tumors in mouse xenograft models. FXR1 is highly expressed in many human cancers and its overexpression is associated with poor outcomes in NSCLC, breast, ovarian cancers and HNSC (Table 1), suggesting a role across multiple tumor types. In contrast, the expression of FMR1 and FXR2 is significantly lower than FXR1 in normal or squamous carcinoma cells (SI Appendix, Fig. S6). Only FXR1 expression correlates with smoking history (SI Appendix, Fig. S7) and predicts poor prognosis in lung adenocarcinoma and HNSC (SI Appendix, Tables S33 and S34), further suggesting a unique role of FXR1 in the pathogenesis of airway epithelial cell cancers (20).
Table 1.
FXR1 expression is associated with poor outcomes in multiple human cancers
| Cancer type | Outcome | Sample size | HR* | P | Biomarker | Platform |
| NSCLC stage I | Overall survival | 161 | 1.31 | 0.018 | Protein | IHC |
| Lung SCC stage I | Overall survival | 78 | 1.44 | 0.032 | Protein | IHC |
| Lung ADC stage I† | Overall survival | 697 | 1.82 | 0.007 | mRNA | Microarray and RNA-seq |
| Breast cancer (TCGA) | Overall survival | 483 | 2.75 | 0.03 | mRNA | RNA-seq |
| Breast cancer (METABRIC) | Disease-specific free survival | 1,992 | 1.43 | 0.03 | mRNA | Microarray |
| Ovarian cancer (stage I+II)‡ | Relapse-free survival | 81 | 5.38 | 0.001 | mRNA | Microarray |
| Head and neck SCC (TCGA) | Overall survival | 300 | 1.54 | 0.04 | mRNA | RNA-seq |
| Head and neck SCC§ | Metastasis-free survival | 81 | 2.25 | 0.01 | mRNA | Microarray |
| Total | 3,795 |
Hazard ratio (HR) was calculated using multivariate Cox proportional-hazards analysis.
HR was derived from a meta-analysis of six independent lung ADC cohorts (SI Appendix, Table S9).
HR was derived from a combined two independent cohorts (GSE9881 and TCGA).
HR was derived from E-TABM-302 cohort.
FMR members regulate mRNA translation through mRNA-protein or microRNA–protein complexes (21, 22). To date, the FXR1-associated complex has not been identified in human cancer cells. In this report we provide mechanistic studies suggesting a novel role of FXR1 in the progression of NSCLC. FXR1 regulates ERK signaling pathway through interaction with PRKCI and ECT2, two known oncogenes within same 3q26-29 amplicon (7, 23). PRKCI has been reported to be activated by PI3 Kinase, PDK1, RAS and SRC either alone or in association with the PAR complex, although the signaling mechanisms involved appear to differ in different tumor types (24). Our results demonstrate that PRKCI is a new binding partner of FXR1 in lung cancer cells. The detailed binding sites of FXR1 to PRKCI and the effect of FXR1-PRKCI interaction on FXR1-bound transcripts in lung cancer cells remain to be elucidated. ECT2, a proposed oncogene in NSCLC (7), is the other newly discovered binding partner of FXR1. ECT2 is phosphorylated by PRKCI and associated with PAR complex to drive transformed lung cancer cell growth and invasion via activation of ERK signaling cascade (14, 25). Here, we provide compelling evidence for ECT2 as a novel mRNA target of FXR1 in lung cancer. The change in the expression of FXR1 involved in SCC exemplifies how a derangement in the mRNA stabilization process may be associated with tumor progression. Together, our findings unravel a new important mechanistic insight into how FXR1 participates in transformation by forming a FXR1/PRKCI/mRNA complex. Further profiling of FXR1 RNA protein complexes to identify potential binding proteins or mRNAs in cancer versus normal cells may provide new insights into the role of FXR1 in the regulation of specific signaling pathways in human cancer.
The importance of FXR1 in this study is suggested by its cooperation with other driver genes in the 3q amplicon. We report the results of functional studies in five cell lines overexpressing FXR1 with or without gene amplification and confirmed the critical driver properties provided by overexpression of FXR1. Our results demonstrate that FXR1 is a critical member of this large amplicon (176 genes included between 3q26-29) and contributes to the pathogenesis of lung cancer. The single gene driver theory has been challenged by recent studies (26, 27), supporting that in fact multiple oncogenes work in concert to drive the cancer, sometime as a driver, sometime a passenger gene. Genomic amplifications happen in different size (Mb) and intensity (copy number between 4 and 25). The mechanisms underlying these differences are poorly understood. Chromothripsis is a possible mechanism to explain these genomic amplifications (28). Other multistep evolutionary process starting from single chromosome ancestral amplicons could explain the process (29). Therefore, in large amplicon like this one (∼32 Mb), the pressure may be such that it is not one gene but a set of genes that can drive the genomic alteration and that many neighboring genes are indeed coamplified as bystanders of the process. However, some of the bystanders functionally are critical to tumor progression. These genes, drivers and passengers, might be functionally associated and their cooperation might be a key mechanism to promote cancer progression. More evidence that multiple oncogenic drivers in 3q amplicon cooperate to promote tumorigenesis is indeed emerging. Justilien et al. first reported a mechanism by which PRKCI forms a complex with ECT2-PAR6A to drive transformed growth through activation of a RAC1-PAK-MEK1-ERK1,2 signaling axis in NSCLC (14, 25). Recently, the same group reported a functional link of PRKCI and SOX2 to activate hedgehog signaling in lung SCC (30). Hagerstrand et al. reported SEC62 and SKIL as new cancer drivers in 3q amplicon and co-overexpression of SEC62 and SKIL induces transformation independently of PIK3CA or SOX2 (31). Interestingly, PIK3CA, ECT2, and SKIL were among 34 3q genes identified as potential mRNA targets of FXR1 in human embryonic kidney 293 cells (32), further implying the regulatory mechanisms driven by RNA binding protein FXR1 other than coamplification may play a role in cancer. Here we are first to describe a new oncogenic role of an RNA binding protein FXR1 linking to PRKCI and ECT2 in NSCLC. We also demonstrate that although genomic amplification is a key mechanism of FXR1 gene overexpression, it is the gene expression level that drives tumor progression in vitro and in vivo.
In conclusion, our results present direct evidence that the RNA binding protein FXR1 is a novel candidate oncogene interacting with two other oncogenes ECT2 and PRKCI located on same 3q26-29 amplicon in lung cancer. Future investigations are warranted to explore whether FXR1 may have utility as a biomarker predictive poor outcomes of human cancers and may represent a novel therapeutic target.
Materials and Methods
Biospecimens.
Lung cancer tissues were collected from surgical specimens through the Specialized Program of Research Excellence (SPORE) in lung at Vanderbilt University Medical Center and the Department of Veterans Affairs (VA) Medical Center in Nashville, Tennessee. All samples were reviewed by a pathologist (R.E). 292 lung cancer tissues contained in tumor tissue microarrays were used for the evaluation of FXR1 protein expression using immunohistochemistry. Clinical characteristics of these patients are described in SI Appendix, Table S5. All primary tumors were fresh-frozen, with efforts made to use samples with tumor content >70%. Studies using human biospecimens were approved by the Vanderbilt University Internal Review Board and the Nashville VA Internal Review Board.
Gene Copy Number and Expression Data Analysis.
Array CGH and mRNA microarray expression data obtained (n = 24) in our laboratory have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under the following accession numbers: GSE40048 and GSE40074. aCGH data were analyzed using Agilent DNA Analytics Software (version 4.0) with ADM-2 algorithm. The average log2 ratio of 0.8 was defined as the cutoff for amplification or 0.3 for low level gain. mRNA expression data were normalized and analyzed by Agilent GeneSpring11. The analysis on other types of human cancer cohorts publically available including 18 TCGA datasets was described in SI Appendix.
Statistical Analysis.
The relevance of FXR1 protein or mRNA expression to the clinical parameters of cancer patients were compared using Student’s t test, Wilcoxon two-sample test, or Kruskal–Wallis test. Pearson correlation was used for gene expression correlation analysis. The relationship of clinical outcomes such as overall survival, relapse free survival or distant recurrence free survival to FXR1 expression was assessed with the Cox proportional hazards regression and the Kaplan-Meier method. All statistics analysis was done using R package (www.R-project.org). Differences were considered statistically significant if the P value was <0.05.
Supplementary Material
Acknowledgments
We thank Drs. Kendal Broadie, Christine M. Lovly, Joseph M. Amann, David P. Carbone, Bapsi Chakravarthy, and Jennifer M. Giltnane for helpful discussion of the results. We thank Drs. Jennifer A. Pietenpol and Dineo Khabele for providing breast, head and neck, and ovarian cancer cell lines. We are also grateful for the datasets publically available made by scientific community including the TCGA Research Network (cancergenome.nih.gov/). This research was supported by National Cancer Institute Grant RO1 CA102353, Department of Defense CDMRP LC090615P3, and Vanderbilt SPORE in lung cancer CA 090949 (to P.P.M.). This work was partially supported by Vanderbilt Clinical and Translational Science Award UL1TR000445 from the National Center for Advancing Translational Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.D.M. is a guest editor invited by the Editorial Board.
Data Deposition: Microarrays have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession nos. GSE40048 and GSE40074).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421975112/-/DCSupplemental.
References
- 1.Qian J, Massion PP. Role of chromosome 3q amplification in lung cancer. J Thoratic Oncol. 2008;3(3):212–215. doi: 10.1097/JTO.0b013e3181663544. [DOI] [PubMed] [Google Scholar]
- 2.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massion PP, et al. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: Implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62(13):3636–3640. [PubMed] [Google Scholar]
- 4.Massion PP, et al. Early involvement of the phosphatidylinositol 3-kinase/Akt pathway in lung cancer progression. Am J Respir Crit Care Med. 2004;170(10):1088–1094. doi: 10.1164/rccm.200404-487OC. [DOI] [PubMed] [Google Scholar]
- 5.Massion PP, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63(21):7113–7121. [PubMed] [Google Scholar]
- 6.Bass AJ, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41(11):1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul. 2010;50(1):190–200. doi: 10.1016/j.advenzreg.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regala RP, et al. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65(19):8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, et al. Integrative genomics analysis identifies candidate drivers at 3q26-29 amplicon in squamous cell carcinoma of the lung. Clin Cancer Res. 2013;19(20):5580–5590. doi: 10.1158/1078-0432.CCR-13-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14(21):5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24(8):416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Mientjes EJ, et al. Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum Mol Genet. 2004;13(13):1291–1302. doi: 10.1093/hmg/ddh150. [DOI] [PubMed] [Google Scholar]
- 13.Yamagata N, et al. Analysis of RNA and protein expression patterns in human lung cancer using cDNA microarrays and MALDI-MS. Proceedings of the 92nd annual meeting of the American Association for Cancer Research. 2001;42:610. [Google Scholar]
- 14.Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene. 2009;28(41):3597–3607. doi: 10.1038/onc.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez RD, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64(24):9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 17.Soh JW, Weinstein IB. Roles of specific isoforms of protein kinase C in the transcriptional control of cyclin D1 and related genes. J Biol Chem. 2003;278(36):34709–34716. doi: 10.1074/jbc.M302016200. [DOI] [PubMed] [Google Scholar]
- 18.Leclerc GJ, Leclerc GM, Barredo JC. Real-time RT-PCR analysis of mRNA decay: half-life of Beta-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int. 2002;2(1):1. doi: 10.1186/1475-2867-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comtesse N, et al. Frequent overexpression of the genes FXR1, CLAPM1 and EIF4G located on amplicon 3q26-27 in squamous cell carcinoma of the lung. Int J Cancer. 2007;120(12):2538–2544. doi: 10.1002/ijc.22585. [DOI] [PubMed] [Google Scholar]
- 20.Spira A, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13(3):361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 21.Mortensen RD, Serra M, Steitz JA, Vasudevan S. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs) Proc Natl Acad Sci USA. 2011;108(20):8281–8286. doi: 10.1073/pnas.1105401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128(6):1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields AP, Regala RP. Protein kinase C iota: Human oncogene, prognostic marker and therapeutic target. Pharmacol Res. 2007;55(6):487–497. doi: 10.1016/j.phrs.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moscat J, Diaz-Meco MT, Wooten MW. Of the atypical PKCs, Par-4 and p62: recent understandings of the biology and pathology of a PB1-dominated complex. Cell Death Differ. 2009;16(11):1426–1437. doi: 10.1038/cdd.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Justilien V, Jameison L, Der CJ, Rossman KL, Fields AP. Oncogenic activity of Ect2 is regulated through protein kinase C iota-mediated phosphorylation. J Biol Chem. 2011;286(10):8149–8157. doi: 10.1074/jbc.M110.196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendall J, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104(42):16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rui L, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang CZ, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27(23):2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.L’Abbate A, et al. Genomic organization and evolution of double minutes/homogeneously staining regions with MYC amplification in human cancer. Nucleic Acids Res. 2014;42(14):9131–9145. doi: 10.1093/nar/gku590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Justilien V, et al. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell. 2014;25(2):139–151. doi: 10.1016/j.ccr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagerstrand D, et al. Systematic interrogation of 3q26 identifies TLOC1 and SKIL as cancer drivers. Cancer Dis. 2013;3(9):1044–1057. doi: 10.1158/2159-8290.CD-12-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492(7429):382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.