Significance
Nonpathogenic Salmonella localize to tumors and can be engineered to secrete anticancer proteins, but tumor-specific expression is essential to prevent systemic toxicity. To reduce unwanted side effects in healthy tissue, we integrated Salmonella with a quorum-sensing (QS) switch that only initiates drug expression in the tightly packed colonies present within tumors. Using an in vitro 3D-tumor-on-a-chip device and in vivo mouse models, we show that QS Salmonella specifically initiates protein expression within cancerous tissue while remaining uninduced in livers. Protein expression was triggered when inducer molecules from enough close neighbors reached a critical concentration. Because of these selective qualities, QS Salmonella are a promising tool for tumor-specific delivery of therapeutic proteins.
Keywords: bacterial anticancer therapy, quorum sensing, Salmonella, cancer, localized drug delivery
Abstract
Salmonella that secrete anticancer proteins have the potential to eliminate tumors, but nonspecific expression causes damage to healthy tissue. We hypothesize that Salmonella, integrated with a density-dependent switch, would only express proteins in tightly packed colonies within tumors. To test this hypothesis, we cloned the lux quorum-sensing (QS) system and a GFP reporter into nonpathogenic Salmonella. Fluorescence and bacterial density were measured in culture and in a tumor-on-a-chip device to determine the critical density necessary to initiate expression. QS Salmonella were injected into 4T1 tumor-bearing mice to quantify GFP expression in vivo using immunofluorescence. At densities below 0.6 × 1010 cfu/g in tumors, less than 3% of QS Salmonella expressed GFP. Above densities of 4.2 × 1010 cfu/g, QS Salmonella had similar expression levels to constitutive controls. GFP expression by QS colonies was dependent upon the distance to neighboring bacteria. No colonies expressed GFP when the average distance to neighbors was greater than 155 µm. Calculations of autoinducer concentrations showed that expression was sigmoidally dependent on density and inversely dependent on average radial distance. Based on bacterial counts from excised tissue, the liver density (0.0079 × 1010 cfu/g) was less than the critical density (0.11 × 1010 cfu/g) necessary to initiate expression. QS Salmonella are a promising tool for cancer treatment that will target drugs to tumors while preventing damage to healthy tissue.
Bacteria that induce expression only in tumors have the potential to solve a critical problem with chemotherapy. Current cancer chemotherapeutic regimens have limited efficacy due to therapeutic resistance and systemic toxicity (1–3), which prevents the use of more aggressive dosage schemes (4). Salmonella are capable of overcoming these limitations because they preferentially accumulate in tumors, actively penetrate tumor tissue, and can be engineered to produce anticancer drugs in situ (5–11). Salmonella that only activate drug expression in tumors and not healthy tissue will reduce toxicity and allow for the use of more aggressive therapeutics. Constitutive, systemic expression of an anticancer drug would be toxic, due to low-level bacterial accumulation in healthy tissue (5). Because Salmonella accumulate almost 10,000-fold higher in tumors than other organs (5, 12), bacteria that sense density would provide a switch to distinguish between healthy and cancerous tissue.
Strict control over protein expression is essential for managing the timing and location of drug production. Precise triggering of expression can boost drug concentration within tumors while minimizing harmful side effects (6). Salmonella can be engineered to induce protein expression in response to molecular triggers, radiation, or hypoxia (6, 11, 13–18). Molecular triggers are limited because small molecules cannot diffuse deep into tissue (19–21). Radiation-inducible promoters are inherently leaky (11), which would lead to unwanted drug expression in healthy tissue. Promoters that respond to hypoxia would have difficulty treating micrometastases less than 2 mm that are typically well oxygenated (22).
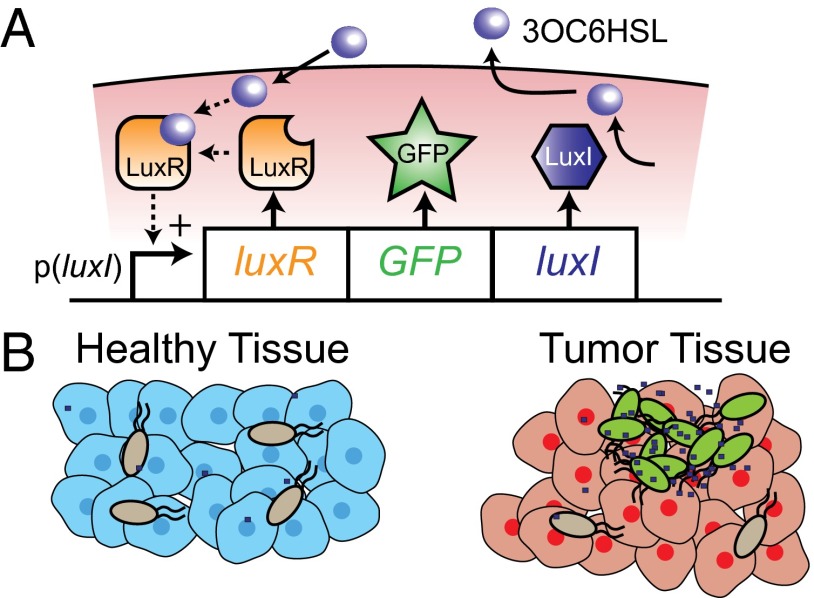
Quorum-sensing (QS) bacteria can change their gene expression based on population density (23). The lux QS system induces expression of bioluminescent genes in marine bacterium Vibrio fischeri. The lux QS system consists of two genes: luxI and luxR (Fig. 1A). Autoinducer synthesis protein LuxI synthesizes the autoinducer N-3(oxohexanoyl)homoserine lactone (3OC6HSL). This autoinducer is specific to V. fischeri and cannot communicate with other species of bacteria (23). Transcriptional regulator protein LuxR activates in the presence of 3OC6HSL and induces transcription by binding to the promoter p(luxI) (24, 25). At low population density, low-level expression of LuxI synthesizes 3OC6HSL, which freely diffuses out of cells. As the population density increases, intracellular 3OC6HSL activates LuxR, creating a positive feedback loop which increases the production of any gene incorporated into the operon (25). The lux QS system has been used in previous research to trigger Escherichia coli invasion into cancer cells (26).
Fig. 1.
QS bacterial drug delivery. (A) The p(luxI) promoter controls one operon consisting of genes encoding for proteins LuxR, GFP, and LuxI. LuxI produces the communication molecule 3OC6HSL. The p(luxI) promoter responds to LuxR protein bound to 3OC6HSL. As the density of bacteria increases, 3OC6HSL concentration increases within the cell, creating a positive feedback loop that increases transcription of the operon. (B) QS bacteria will only turn on expression in high-density colonies in tumor tissue. Gray and green bacteria represent uninduced and induced bacteria, respectively. Blue dots represent 3OC6HSL.
The spatial distribution of bacteria affects the activation of a QS switch (27). The concentration of a signaling molecule diminishes as it moves away from a cell, which decreases the likelihood of activating the QS switch (28). Clustering of bacterial cells prevents dilution of the signaling molecule and improves QS activation (29). These observations suggest that QS Salmonella would only activate when close to each other in tumor colonies (Fig. 1B).
To create a tumor-sensitive gene expression switch, we integrated the lux QS system and a fluorescence reporter into an attenuated Salmonella cancer vector. We hypothesized that QS Salmonella would (i) induce gene expression in response to high bacterial density, (ii) induce expression as the distance between bacteria decreases, and (iii) only induce expression in tumor tissue. To test this hypothesis, fluorescence and density were measured and compared with constitutive controls to determine the basal level of QS expression. GFP expression was measured in bacterial cultures and an in vitro tumor-on-a-chip device to quantify the density required to trigger expression. QS and constitutive Salmonella were injected into tumor-bearing mice to quantify protein expression in vivo. Bacterial density was measured in tumors and livers. Immunofluorescence was used to quantify the spatial distribution of bacteria and GFP expression within tumors. A mathematical model was created to predict both the density and distribution of bacteria needed to induce protein expression in tissue. QS Salmonella will be an improvement over chemotherapy because it creates a sensitive switch that will only express protein therapeutics in tumors while remaining off in healthy tissue.
Results
Density Dependence of GFP Expression in Vitro.
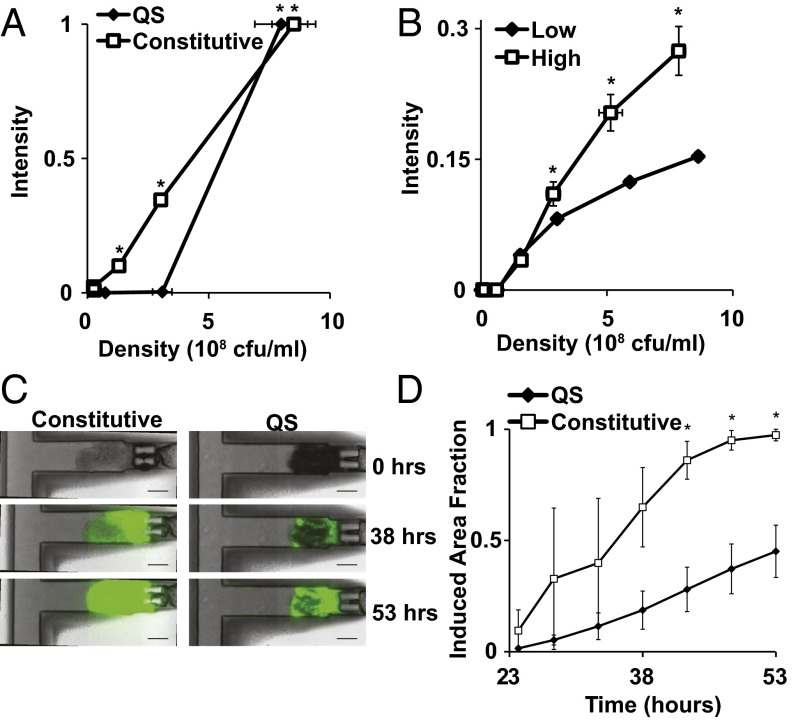
QS Salmonella induced GFP expression both in flasks and in vitro tumor tissue only at high density (Fig. 2). At densities less than 0.5 × 108 cfu/mL, QS Salmonella did not express GFP (Fig. 2A). Above the critical density of 108 cfu/mL, QS Salmonella expressed significant amounts of GFP (Fig. 2A; P < 0.05). Constitutive controls expressed GFP, regardless of density (P < 0.05). Constitutive Salmonella were used as controls because GFP expression was not dependent on an external inducer in these bacteria. GFP expression in constitutive controls was detected at densities as low as 0.25 × 108 cfu/mL (Fig. 2A). The critical density of GFP expression was robust and did not change with culture history (Fig. 2B). For cultures grown to different densities before dilution (0.5 × 108 cfu/mL and 5 × 108 cfu/mL), GFP expression was consistently induced at 108 cfu/mL (Fig. 2B). Cultures grown to higher density before dilution, however, had greater GFP expression with time (Fig. 2B; P < 0.05) because of residual LuxI and LuxR molecules in the bacteria (30).
Fig. 2.
In vitro behavior of QS Salmonella. (A) QS Salmonella only expressed GFP at densities above 1 × 108 cfu/mL (*P < 0.05). Constitutive controls expressed GFP at all densities. (B) Before measurement, QS cultures were grown to either 0.5 × 108 cfu/mL (low density) or 5 × 108 cfu/mL (high density and induced) and then diluted to 0.001 × 108 cfu/mL. High dilution density cultures had greater expression (*P < 0.05). Fluorescence was normalized to constitutive controls at 6 × 108 cfu/mL. (C) 38 h after injection into tumor tissue in a microfluidic tumor-on-a-chip device, QS Salmonella expressed GFP only within distinct bacterial colonies. Constitutive Salmonella expressed GFP throughout the tissue regardless of bacterial concentration. Bacteria took 10 h to colonize tissue and were not present at 0 h. (Scale bar, 100 µm.) (D) Area fraction of tissue with GFP expression was less for tissue treated with QS Salmonella compared with constitutive controls after 40 h (*P < 0.05).
In tissue in a microfluidic device (31), QS Salmonella only expressed GFP in high-density colonies (Fig. 2 C and D). Bacterial accumulation began 10 h after inoculation. By 53 h, tumor tissue containing constitutive bacterial controls expressed GFP throughout the entire tissue (Fig. 2C). Bacteria of both strains colonized the entire tissue. 38 h after bacterial injection, tumor tissue accumulated with QS Salmonella had pockets of GFP expression within distinct colonies. Tissue with sparse colonization contained no GFP expression (Fig. 2C). The area of tissue with GFP expressing bacteria was greater in constitutive controls (97%) than QS Salmonella (45%, Fig. 2D; P < 0.05).
Salmonella Distribution in Tumor-Bearing Mice.
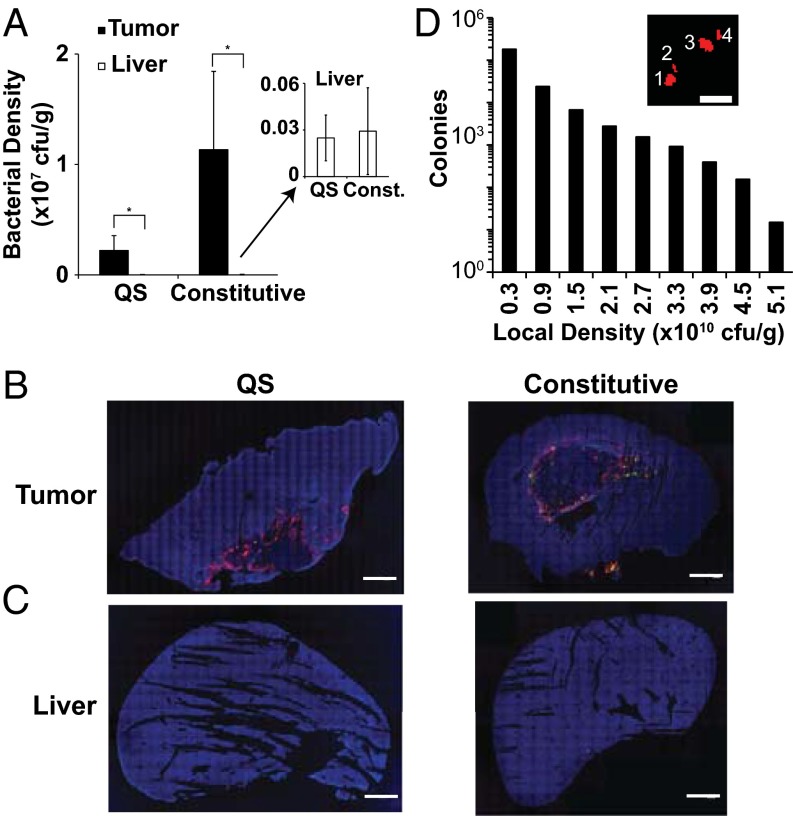
QS Salmonella and constitutive controls preferentially accumulated in tumor tissue compared with healthy tissue (Fig. 3). Bacterial density, based on plating of minced tissue, was 89-fold and 387-fold greater in tumor tissue than liver tissue for QS and constitutive Salmonella, respectively (Fig. 3A; P < 0.05). There was no statistical difference between the QS (n = 5 mice) and constitutive (n = 5) bacterial densities in tumors or livers (Fig. 3A; P > 0.3). GFP was present in all tumors. Expressing colonies are difficult to see in these macroscopic images because of their small size (Fig. S1). Tumor tissue removed at 9 and 24 d after bacterial injection both contained GFP, indicating persistent gene expression over this time range (Fig. 3B). Because of the low density, no Salmonella were observed in liver sections by immunofluorescence (Fig. 3C). In tumors, most colonies formed in regions of low bacterial density. Local density was defined as the number of Salmonella within a 197 µm (rc; 150 pixel) radius around a colony. Colonies were groups of contiguous bacteria distinctly separate from neighbors (Fig. 3D, Inset). Eighty-three percent of QS (n = 84,213 colonies) and constitutive (n = 133,305) Salmonella colonies were at a density of 0.3 × 1010 cfu/g or less (Fig. 3D). Of the QS Salmonella, 0.7% were at densities higher than 3.3 × 1010 cfu/g. The highest density of a QS Salmonella colony was 5.23 × 1010 cfu/g (Fig. 3D).
Fig. 3.
Salmonella distribution within tumor-bearing mice. (A) Bacterial density was higher in tumors (solid bars) than livers (open bars) for mice administered both QS and constitutively expressing Salmonella (*P < 0.05). (Inset) Liver densities were small compared with tumor densities, but bacteria were present in most livers. (B) Salmonella (red) and GFP (green) distribution in 4T1 tumors injected with QS Salmonella or constitutive controls. Sections were counterstained with DAPI (blue). (C) No Salmonella or GFP were observed in any liver section using immunofluoresence. (Scale bar, 5 mm.) (D) Most Salmonella colonies were located in low-density regions. Local density is the number of bacteria within a 197-µm circle surrounding each colony. Colonies were groups of contiguous bacteria distinctly separate from neighbors (Inset). (Scale bar, 25 µm.)
Density-Controlled Protein Expression in QS Colonies in Tumors.
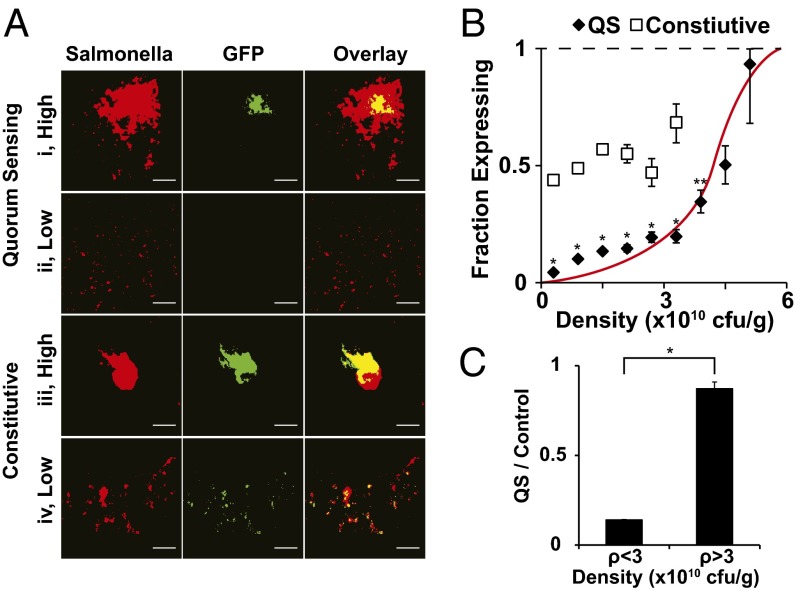
Protein expression by QS Salmonella was dependent on local density (Fig. 4). In high-density regions, QS colonies expressed GFP (Fig. 4 A, i). Local density was high when colonies were large (as in Fig. 4 A, i) or were surrounded by many close neighbors. In low-density regions, small QS colonies did not produce GFP (Fig. 4 A, ii). In comparison, constitutive colonies expressed GFP regardless of size or local density (Fig. 4 A, iii and iv). Both large (Fig. 4 A, iii) and small (Fig. 4 A, iv) constitutive colonies expressed GFP.
Fig. 4.
Density dependence of GFP expression by QS Salmonella. (A) Colonies of QS and constitutive Salmonella (red) in 4T1 tumors and associated GFP (green) for areas of low and high density. Yellow indicates areas of colocalization between Salmonella and GFP. (Scale bar, 100 µm.) (B) GFP expression dependence on Salmonella density. The relationship between expression and density was linear for constitutive controls (n = 133,305 colonies) and sigmoidal (red line) for QS Salmonella (n = 84,213 colonies). At all densities less than 4.2 × 1010 cfu/g, the fraction of GFP-expressing QS colonies was less than the constitutive control colonies at the same density (*P < 0.05). Below 4.2 × 1010 cfu/g, the expressing fraction of QS colonies was less than control colonies at the lowest density, 0.3 × 1010 cfu/g (* and **P < 0.05). (C) The ratio of QS to control expressing fractions was significantly greater at densities above 3.0 × 1010 cfu/g compared with the densities below this threshold (*P < 0.05). Above 3.0 × 1010 cfu/g the QS:control ratio was close to 1.
The relationship between the fraction of GFP-expressing QS colonies and local density was sigmoidal (Fig. 4B). In comparison, the relationship for constitutive controls was linear and constant across all densities. At low densities, the fraction of induced QS colonies was low. Below a density of 0.6 × 1010 cfu/g, the induced fraction was 10-fold lower than controls (Fig. 4B; P < 0.05). At higher densities, the induced fraction was close to the maximum value of 1. Above 4.8 × 1010 cfu/g, 93% of QS colonies were induced. The difference between colonies at low (<0.6 × 1010 cfu/g) and high (>4.8 × 1010 cfu/g) density was 21-fold (P < 0.05). The fractions of expressing QS colonies at densities less than 4.2 × 1010 cfu/g were all less than the fraction of expressing control colonies at the lowest density of 0.3 × 1010 cfu/g (P < 0.05). Below a threshold density of 3.0 × 1010 cfu/g, the fraction of induced QS colonies was seven times less than constitutive controls (Fig. 4C; P < 0.05). Above this threshold, the fraction of induced QS colonies increased sixfold (P < 0.05) and was equivalent to constitutive controls (Fig. 4B; P < 0.05).
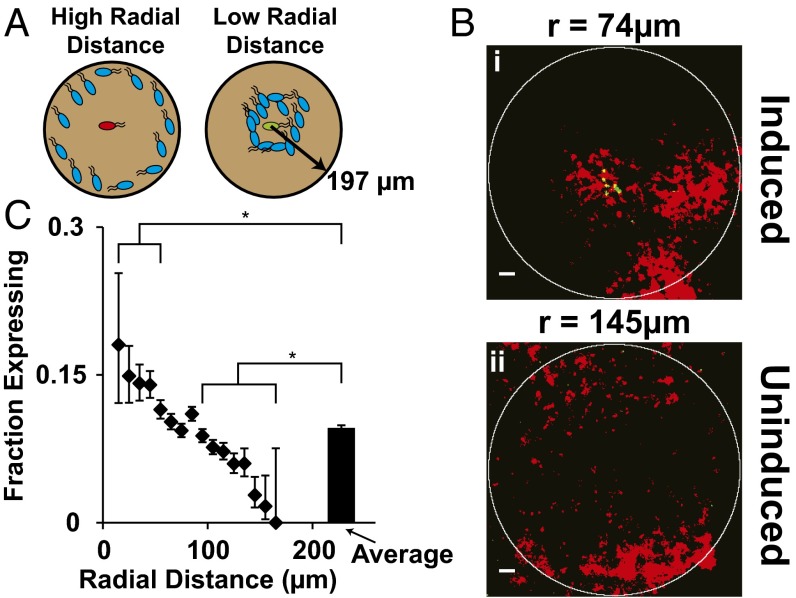
Proximity Between Colonies Controlled Expression.
The percentage of QS colonies expressing GFP was greater for colonies closely surrounded by neighbors (Fig. 5). The key descriptor of spatial distribution was average radial distance, which was defined as the location-weighted average of distances between a colony and all neighboring bacteria within 197 µm (Fig. 5A). Colonies with equal densities but different average radii had different GFP-expression patterns (Fig. 5B). A colony with close neighbors, at an average radial distance of 74 µm, expressed GFP (Fig. 5 B, i). In comparison, a colony at the same density, but with distant neighbors (at a radius of 145 µm, or 71 µm farther away) was not induced (Fig. 5 B, ii). The average radius to neighboring bacteria affected GFP expression (Fig. 5C) in colonies in regions with density greater than 0.11 × 1010 cfu/g. In this range, the percentage of colonies expressing GFP was linearly and inversely dependent on average radius. At low densities, below 0.11 × 1010 cfu/g, induction was sparse (Fig. 4B) and not correlated with radius. The average expression fraction for all moderate and high-density colonies was 0.09 (Fig. 5C). The fractions of induced colonies with close (3 < r < 58 µm) and distant (87 < r < 166 µm) neighbors were significantly greater (P < 0.05) and less (P < 0.05) than the average, respectively. The lowest and highest radii measured were 3 and 166 µm. No colonies with neighbors farther away than 155 µm expressed GFP.
Fig. 5.
Dependence on spatial distribution. (A) A QS Salmonella colony (Center) was more likely to express GFP if the average radial distance to its neighbors was shorter. Red, green, and blue bacteria represent uninduced, induced, and neighboring colonies, respectively. Density and radial distance was measured within circles of radius 197 µm (150 pixels) around colonies. (B) GFP expression was different for two colonies at the same density (1.2 × 1010 cfu/g) with different spatial distributions. A colony with an average radial distance to its neighbors of 74 µm expressed GFP (B, i), but a colony with an average radial distance of 145 µm did not (B, ii). (Scale bars, 100 µm.) (C) The fraction of colonies expressing GFP was greater for colonies with close compared with far neighbors, and with densities greater than 0.11 × 1010 cfu/g (n = 50,145 colonies). The expressing fraction was greater than the average for colonies with average radial distances less than 58 µm (*P < 0.05) and less than the average for distances greater than 87 µm (*P < 0.05). At an average distance of 155 µm, the fraction expressing was zero.
Production and Diffusion of 3OC6HSL in Tumor Tissue.
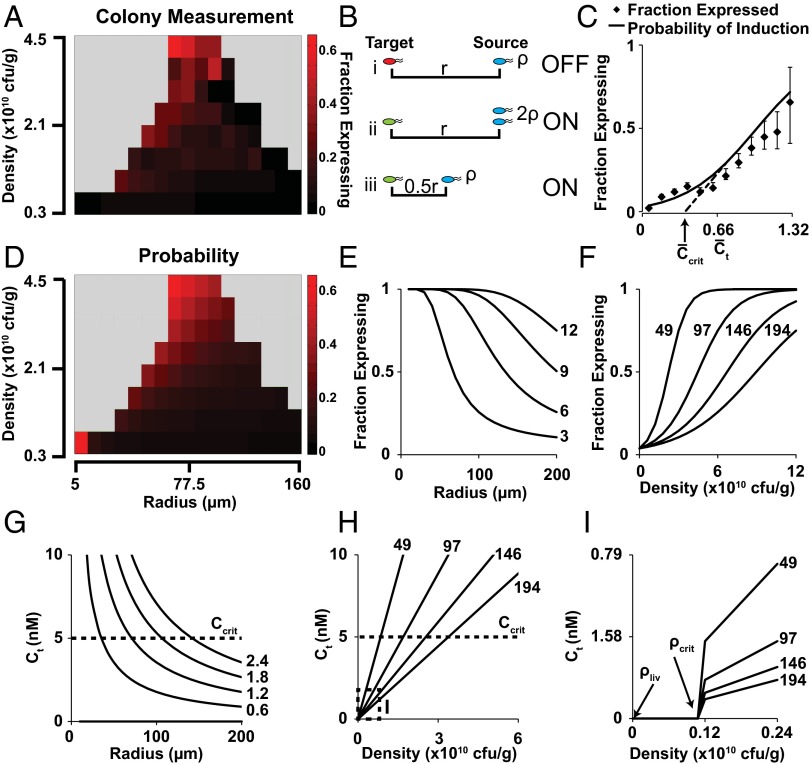
The density and spatial distribution of bacteria in tumors predicted protein expression by individual colonies (Fig. 6). These two dependencies showed how QS controlled expression. At both higher density and shorter average distance between bacteria, the fraction of induced colonies was greater (Fig. 6A). For QS bacteria, protein expression was induced by 3OC6HSL. In tumors, two mechanisms controlled the concentration of 3OC6HSL: production by surrounding bacteria and diffusion through interstitial tissue (Fig. 6B). A target colony surrounded by few distant colonies (Fig. 6 B, i) would have had a low local 3OC6HSL concentration (Fig. 6B). A colony with twice the number of source colonies (Fig. 6 B, ii) would have had double the 3OC6HSL concentration. Similarly, a colony that was closer to source colonies (Fig. 6 B, iii) would have had a higher 3OC6HSL concentration.
Fig. 6.
Calculated 3OC6HSL concentration predicts GFP expression. (A) The fraction of colonies (n = 84,213) that expressed GFP was greater (more red) at high densities (Top) and short radial distances (Left). (B) Compared with an uninduced colony (B, i, red), a target colony was more likely to express GFP (green) if (B, ii) it was surrounded by more source colonies (blue) or (B, iii) the distance to source colonies was shorter. (C) The fraction of expressing colonies and the predicted probability of induction (α), were lower for colonies with lower calculated concentrations of 3OC6HSL (P < 1 × 10−15). The critical 3OC6HSL concentration (Ccrit) was = 0.38. (D) The predicted fraction of expressing colonies, α, was higher at high density and low average radial distance, matching colony measurements in tumor sections (A). (E and F) Predicted α-fractions were lower at longer average radial distances (E) and higher at higher densities (F). Numbers to the right are density (×1010 cfu/g) for E and G, and radius (µm) for F, H, and I. (G and H) The predicted 3OC6HSL concentration at target colonies () was lower at higher average radial distances (G), and higher at higher densities (H). (I) Expanded range of H. At low densities below ρcrit = 0.11 × 1010 cfu/g, the predicted 3OC6HSL concentration was zero. At the maximum possible bacterial density in liver (ρliv = 0.00792 × 1010 cfu/g), no 3OC6HSL was produced, regardless of spatial distribution.
To quantify these mechanisms, the 3OC6HSL concentration around source and target colonies was modeled as a coupled production–diffusion system.
| [1] |
Around a source colony, the 3OC6HSL concentration (Cs) was dependent on the production rate (m) and an effective diffusion coefficient (Deff) in heterogeneous tumor tissue (SI Materials and Methods). Above a critical density (), 3OC6HSL was produced at the maximum rate (mmax). Production decreased at low density with sensitivity (σ). At steady state (see SI Materials and Methods for derivation), the normalized source-colony concentration () was inversely related to normalized radial distance () by dimensionless production–diffusion (Q).
| [2] |
The reference concentration (Cq) was the concentration at which 50% of QS colonies were induced. The concentration at each target colony () was equal to the contribution of 3OC6HSL from the total number of source colonies () within radius rc (Fig. 6 B, ii). Each target colony was at an average radial distance () from all surrounding source colonies. The probability that a target colony was induced (α) was dependent on the 3OC6HSL concentration () and the minimum probability (β; Fig. 6C).
| [3] |
At increasing 3OC6HSL concentrations, the probability of GFP expression (α) and the fraction of induced colonies both increased (Fig. 6C). Dimensionless production–diffusion, Q, was 1.34 (P < 1 × 10−15), indicating that the system was moderately diffusion limited (Table 1). The normalized critical induction concentration () was 0.38 (Fig. 6C). Based on previously measured values of Ccrit (30, 32) and Deff (12, 32–34), mmax was 53,000 molecules·s−1 per bacterium.
Table 1.
Autoinducer transport parameters
| Name | Parameter | Value |
| Dimensionless production–diffusion | Q | 1.34 |
| Critical density | ρcrit | 0.11 × 1010 cfu/g |
| Density sensitivity | σ | 6.84 × 103 cfu/g |
| Minimum probability | β | −3.2 |
The predicted fraction of induced colonies was greater at high density and low radius (Fig. 6D). This dependence was caused by the proportional and inverse relationships of 3OC6HSL concentration to and , respectively (Eq. 2). At small radii, the predicted fraction of induced colonies was close to 1, regardless of density (Fig. 6E). Similarly, at low density, the predicted fraction of induced colonies was close to zero, regardless of radius (Fig. 6F). At high average radii between colonies, a greater density was required to produce a greater than and induce expression (Fig. 6G). Inversely, at low radii, 3OC6HSL concentration was less dependent on density (Fig. 6H). Below the critical density (ρcrit = 0.11 × 1010 cfu/g), protein production (m, Eq. 1) and the 3OC6HSL concentration were both zero (Fig. 6I). The dependence of production (m) on density was almost binary because the sensitivity (σ = 6.84 × 103 cfu/g) was nearly six orders of magnitude smaller than ρcrit (Table 1).
Based on the number of bacteria within the liver (1.51 × 104 cfu/g; Fig. 3A), ∼47 bacteria were located within each liver section. The absence of visible colonies in the immunofluorescent liver images (Fig. 3D) indicates that bacteria were sparsely distributed as individuals within the tissue. In the extreme case that these bacteria were all located within a single colony, the density would be 7.92 × 107 cfu/g. Because this density (ρliv) was less than ρcrit (Fig. 6I), 3OC6HSL production was independent of radius and spatial distribution (Fig. 6I). At this maximum possible liver density, production, 3OC6HSL concentration, and protein expression would have all been zero.
Discussion
Administering Salmonella with the ability to change gene expression in a density-dependent manner will initiate protein expression within tumors and has the potential to reduce systemic toxicity. We have shown that Salmonella integrated with a QS trigger turn on protein expression in tightly packed high-density colonies within tumors, while remaining off in low-density colonies. A mathematical model of 3OC6HSL concentration in tumor tissue was used to determine the mechanisms of QS protein expression. The model predicted that QS Salmonella will not trigger protein expression in healthy tissue. When Salmonella were administered with a constitutive trigger, protein expression was observed in low-density colonies and in individual Salmonella with no surrounding neighbors. A bacterial cancer therapy with a QS triggering system will prevent therapeutic protein release in healthy tissue and maximize therapeutic effect in tumors.
The density of QS Salmonella in livers and the critical density needed to trigger the QS system render the possibility of gene expression unlikely in healthy tissue. Mathematical modeling predicts that QS Salmonella would remain off (Fig. 6I) at the density measured in liver tissue (Fig. 3A). Constitutive controls, on the other hand, expressed GFP at the lowest possible detectable density in tumor tissue (Fig. 4B), indicating that the constitutive Salmonella would express GFP everywhere, including the liver. Constitutive expression of toxic proteins in livers or other healthy organs, even at low rates, could have detrimental effects on the host. QS Salmonella can overcome these therapeutic limitations by specifically triggering drug expression within tumors without causing unintended side effects. It is also unlikely that Salmonella would grow in off-target tissues to densities that would induce expression. Experiments with cynomolgus monkeys have shown that, after initial accumulation following injection, Salmonella are eliminated from most organs by 30 d (35).
QS Salmonella have important advantages over other proposed mechanisms of bacterial drug delivery. Integrating Salmonella with a robust QS triggering system enables the use of aggressive therapeutic proteins, such as Staphylococcus aureus α-hemolysin (SAH). SAH kills cells quickly and is effective against therapeutically resistant tissue (36, 37). Systemic delivery of ubiquitously toxic molecules is not a viable treatment strategy because all tissues would be damaged. When controlled by the QS system, SAH is only released from high-density colonies, where it kills cancer cells and tumor tissue (Fig. S2). At bacterial densities considerably higher than the density in livers, SAH is not produced and no toxicity is observed (Fig. S2).
With QS, no external inducer is needed to initiate expression after colonization. Previous strategies with external inducers have been problematic. Inducers must overcome both clearance from the body and diffusion barriers into tissue. Without the need for an external inducer, a QS system is not reliant on the delivery of a small molecule to maintain therapeutic expression levels. Persistent gene expression was observed in tumor tissue as late as 24 d after injection (Fig. 3C). From a clinical standpoint, this enables continual drug production and an increased therapeutic effect over a longer period.
The sensitivity of this density-dependent switch suggests that QS Salmonella will turn on in undetected metastatic legions. The QS system turns on at a critical density of 0.11 × 1010 cfu/g in tumor tissue (Fig. 6H). In previous work, Salmonella were shown to accumulate in liver metastases, at a density of 5.28 × 1010 cfu/g (7). Assuming a uniform distribution, all colonies at this density would have 3OC6HSL concentrations over Ccrit (Fig. 6G). In comparison, Salmonella were at a density of 0.06 × 1010 cfu/g in the surrounding hepatic parenchyma (7), almost half the density required for the QS system to activate (Fig. 6H).
Increasing the number of colonies within tumor tissue has the potential to increase drug production, due to the important roles diffusion and bacterial spatial distribution play in triggering the QS system. QS Salmonella turn on protein expression at densities of 108 cfu/mL in flasks (Fig. 2A), but mixing ensures that 3OC6HSL is well distributed and not affected by diffusion. In tumor tissue, however, the QS switch turned on at densities of 11 × 108 cfu/mL, almost 10-fold higher than in flasks (Fig. 6H), assuming a tissue density of 1 g/mL. The increase in bacterial density was caused by the distance necessary for 3OC6HSL to diffuse through tissue once it was produced. Below this critical density, there were not enough individuals producing 3OC6HSL to induce expression, no matter how tightly packed they were (Fig. 6I). In addition, 3OC6HSL must overcome loss through hyperpermeable blood vessels, dissipation by lymphatic flows, and the heterogeneous environment of tumor tissue.
Administration of QS Salmonella with exogenous lipid A (12) and enhanced motility (12, 38) would increase bacterial density and improve distribution within tumors. Combined, these effects could increase density 253-fold and increase overall drug production. Mathematical modeling predicted that as the density of bacteria increased, bacteria did not have to be as tightly packed together to induce the QS switch (Fig. 6G). Therefore, coupling enhanced-motility QS Salmonella with lipid A administration could have an exponential effect on drug production.
Conclusion
Salmonella integrated with a QS triggering system creates a drug delivery vehicle that improves upon existing therapies. QS Salmonella only initiate protein production within tumors and not in healthy tissue. These bacteria maintain continuous therapeutic production due to persistent expression. No external inducer is required to initiate drug production. The QS switch is not dependent on cell surface markers that are unique to specific tumor types. Because of these targeting abilities, QS Salmonella are a promising tool to deliver therapeutic proteins and treat cancerous tissue and metastases.
Materials and Methods
Detailed methods are found in SI Materials and Methods.
In Vitro Bacterial Density and GFP Expression Analysis.
A robust density switch was created in Salmonella by transforming a QS architecture, in which all genes are under control of the p(luxI) bidirectional promoter (Fig. S3), into VNP200010 (msbB−, purI−, xyl−, asd−) a nonpathogenic Salmonella strain (Fig. 1A). To measure density dependence of GFP expression, Salmonella were grown from single colonies in flasks and optical density and fluorescence were measured hourly. A microfluidic tumor-on-a-chip device containing LS174T colon carcinoma cells was used to measure bacterial protein expression in tissue. Salmonella were administered to devices for 1 h and then switched to bacteria-free medium. Transmitted and fluorescence images were acquired for 30 h, starting 23 h after bacterial inoculation (Olympus), and were analyzed using ImageJ (NIH Research Services Branch).
In Vivo Salmonella Administration and Analysis.
QS and control Salmonella were injected via the tail vein into mice with 500-mm3 s.c. 4T1 mammary tumors. Mice were killed when tumors reached 2,000 mm3. Excised tumors and livers were cut in half. One half was plated on LB-agar plates and colonies were counted after 24 h. The other half was embedded in paraffin, sectioned, and probed with antibodies against GFP and Salmonella. Fluorescence images were thresholded and colonies were identified. Local density was determined by counting the number of bacteria in a 150-pixel (197-µm) circle around each colony. Average radial distances of neighboring bacteria were determined by counting Salmonella within 5-pixel-wide annuli and weighting by the annulus area. Colonies were considered to be induced if a GFP pixel was within 25 pixels. All animal procedures were approved by Baystate Medical Center Institutional Animal Care and Use Committee, and were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (39).
Mathematical Modeling of 3OC6HSL Diffusion.
A mathematical model was used to predict the concentration of 3OC6HSL in tumor tissue, which has an analytical solution (SI Materials and Methods). Parameters ρcrit, σ, β, and Q were determined by binomial regression of the logistic probability function (Eq. 3) and the predicted 3OC6HSL concentrations (Eq. 2) for each colony. The value of Ccrit was determined by linearly extrapolating from the concentration of maximum slope in Eq. 3 to the concentration at which α = 0.
Statistical Analysis.
For results obtained in bacterial cultures, microfluidic devices, and tissue plating experiments, errors are reported as SEMs. Hypotheses were tested using Student’s t test with a significance level of P < 0.05. For results obtained by colony analysis in tumor sections, errors are reported as 95% Clopper–Pearson binomial confidence intervals, with individual colonies as biological replicates. Hypotheses were tested using Fisher’s exact test with a significance level of P < 0.05.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from the NIH (Grants R01CA120825 and R01CA188382).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP294373, KP294374, and KP294375).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414558112/-/DCSupplemental.
References
- 1.Jain RK. The next frontier of molecular medicine: Delivery of therapeutics. Nat Med. 1998;4(6):655–657. doi: 10.1038/nm0698-655. [DOI] [PubMed] [Google Scholar]
- 2.Brown JM, Giaccia AJ. The unique physiology of solid tumors: Opportunities (and problems) for cancer therapy. Cancer Res. 1998;58(7):1408–1416. [PubMed] [Google Scholar]
- 3.Tannock IF, Lee CM, Tunggal JK, Cowan DSM, Egorin MJ. Limited penetration of anticancer drugs through tumor tissue: A potential cause of resistance of solid tumors to chemotherapy. Clin Cancer Res. 2002;8(3):878–884. [PubMed] [Google Scholar]
- 4.Sakhrani NM, Padh H. Organelle targeting: Third level of drug targeting. Drug Des Devel Ther. 2013;7:585–599. doi: 10.2147/DDDT.S45614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes NS, Munn LL, Fukumura D, Jain RK. Sparse initial entrapment of systemically injected Salmonella typhimurium leads to heterogeneous accumulation within tumors. Cancer Res. 2003;63(17):5188–5193. [PubMed] [Google Scholar]
- 6.Ganai S, Arenas RB, Forbes NS. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br J Cancer. 2009;101(10):1683–1691. doi: 10.1038/sj.bjc.6605403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganai S, Arenas RB, Sauer JP, Bentley B, Forbes NS. In tumors Salmonella migrate away from vasculature toward the transition zone and induce apoptosis. Cancer Gene Ther. 2011;18(7):457–466. doi: 10.1038/cgt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasinskas RW, Forbes NS. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 2007;67(7):3201–3209. doi: 10.1158/0008-5472.CAN-06-2618. [DOI] [PubMed] [Google Scholar]
- 9.Kasinskas RW, Forbes NS. Salmonella typhimurium specifically chemotax and proliferate in heterogeneous tumor tissue in vitro. Biotechnol Bioeng. 2006;94(4):710–721. doi: 10.1002/bit.20883. [DOI] [PubMed] [Google Scholar]
- 10.Loeffler M, Le’Negrate G, Krajewska M, Reed JC. IL-18-producing Salmonella inhibit tumor growth. Cancer Gene Ther. 2008;15(12):787–794. doi: 10.1038/cgt.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuyts S, et al. The use of radiation-induced bacterial promoters in anaerobic conditions: A means to control gene expression in clostridium-mediated therapy for cancer. Radiat Res. 2001;155(5):716–723. doi: 10.1667/0033-7587(2001)155[0716:tuorib]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Zhang M, Swofford CA, Forbes NS. Lipid A controls the robustness of intratumoral accumulation of attenuated Salmonella in mice. Int J Cancer. 2014;135(3):647–657. doi: 10.1002/ijc.28700. [DOI] [PubMed] [Google Scholar]
- 13.Ryan RM, et al. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther. 2009;16(3):329–339. doi: 10.1038/gt.2008.188. [DOI] [PubMed] [Google Scholar]
- 14.Mengesha A, et al. Development of a flexible and potent hypoxia-inducible promoter for tumor-targeted gene expression in attenuated Salmonella. Cancer Biol Ther. 2006;5(9):1120–1128. doi: 10.4161/cbt.5.9.2951. [DOI] [PubMed] [Google Scholar]
- 15.Arrach N, Zhao M, Porwollik S, Hoffman RM, McClelland M. Salmonella promoters preferentially activated inside tumors. Cancer Res. 2008;68(12):4827–4832. doi: 10.1158/0008-5472.CAN-08-0552. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman RM, Hayashi K, Zhao M. Therapeutic targeting of tumors with imageable GFP expressing Salmonella typhimurium auxotrophic mutants. Proc SPIE. 2008;6868:68680K. [Google Scholar]
- 17.Theys J, et al. Clostridium as a tumor-specific delivery system of therapeutic proteins. Cancer Detect Prev. 2001;25(6):548–557. [PubMed] [Google Scholar]
- 18.Nuyts S, et al. Radio-responsive recA promoter significantly increases TNFalpha production in recombinant clostridia after 2 Gy irradiation. Gene Ther. 2001;8(15):1197–1201. doi: 10.1038/sj.gt.3301499. [DOI] [PubMed] [Google Scholar]
- 19.Seri K, et al. L-arabinose selectively inhibits intestinal sucrase in an uncompetitive manner and suppresses glycemic response after sucrose ingestion in animals. Metabolism. 1996;45(11):1368–1374. doi: 10.1016/s0026-0495(96)90117-1. [DOI] [PubMed] [Google Scholar]
- 20.Foley JE, Cushman SW, Salans LB. Glucose transport in isolated rat adipocytes with measurements of L-arabinose uptake. Am J Physiol. 1978;234(2):E112–E119. doi: 10.1152/ajpendo.1978.234.2.E112. [DOI] [PubMed] [Google Scholar]
- 21.Loessner H, et al. Remote control of tumour-targeted Salmonella enterica serovar Typhimurium by the use of L-arabinose as inducer of bacterial gene expression in vivo. Cell Microbiol. 2007;9(6):1529–1537. doi: 10.1111/j.1462-5822.2007.00890.x. [DOI] [PubMed] [Google Scholar]
- 22.Simonsen TG, Gaustad J-V, Rofstad EK. Development of hypoxia in a preclinical model of tumor micrometastases. Int J Radiat Oncol Biol Phys. 2010;76(3):879–888. doi: 10.1016/j.ijrobp.2009.09.045. [DOI] [PubMed] [Google Scholar]
- 23.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 24.Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: Acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 25.Sitnikov DM, Schineller JB, Baldwin TO. Transcriptional regulation of bioluminesence genes from Vibrio fischeri. Mol Microbiol. 1995;17(5):801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 26.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355(4):619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 27.West SA, Winzer K, Gardner A, Diggle SP. Quorum sensing and the confusion about diffusion. Trends Microbiol. 2012;20(12):586–594. doi: 10.1016/j.tim.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10(8):365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 29.Hense BA, et al. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol. 2007;5(3):230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 30.Haseltine EL, Arnold FH. Implications of rewiring bacterial quorum sensing. Appl Environ Microbiol. 2008;74(2):437–445. doi: 10.1128/AEM.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toley BJ, Ganz DE, Walsh CL, Forbes NS. Microfluidic device for recreating a tumor microenvironment in vitro. J Vis Exp. 2011;(57):e2425. doi: 10.3791/2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trovato A, et al. Quorum vs. diffusion sensing: A quantitative analysis of the relevance of absorbing or reflecting boundaries. FEMS Microbiol Lett. 2014;352(2):198–203. doi: 10.1111/1574-6968.12394. [DOI] [PubMed] [Google Scholar]
- 33.Thurber GM, Weissleder R. A systems approach for tumor pharmacokinetics. PLoS ONE. 2011;6(9):e24696. doi: 10.1371/journal.pone.0024696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart PS. A review of experimental measurements of effective diffusive permeabilities and effective diffusion coefficients in biofilms. Biotechnol Bioeng. 1998;59(3):261–272. doi: 10.1002/(sici)1097-0290(19980805)59:3<261::aid-bit1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Clairmont C, et al. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis. 2000;181(6):1996–2002. doi: 10.1086/315497. [DOI] [PubMed] [Google Scholar]
- 36.Swofford CA, St Jean AT, Panteli JT, Brentzel ZJ, Forbes NS. Identification of Staphylococcus aureus α-hemolysin as a protein drug that is secreted by anticancer bacteria and rapidly kills cancer cells. Biotechnol Bioeng. 2014;111(6):1233–1245. doi: 10.1002/bit.25184. [DOI] [PubMed] [Google Scholar]
- 37.St Jean AT, Swofford CA, Panteli JT, Brentzel ZJ, Forbes NS. Bacterial delivery of Staphylococcus aureus α-hemolysin causes regression and necrosis in murine tumors. Mol Ther. 2014;22(7):1266–1274. doi: 10.1038/mt.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toley BJ, Forbes NS. Motility is critical for effective distribution and accumulation of bacteria in tumor tissue. Integr Biol (Camb) 2012;4(2):165–176. doi: 10.1039/c2ib00091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. National Research Council (2011) Guide for the Care and Use of Laboratory Animals (The National Academies Press, Washington, DC) 8th Ed. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.