Significance
Across ecology, and particularly within microbial ecology, there is limited understanding of the mechanisms governing the relative influences of stochastic and deterministic processes. Filling this knowledge gap is a major challenge that requires the development of novel conceptual paradigms, experiments, and ecological models. Here we (i) present a conceptual model that couples the stochastic/deterministic balance to primary and secondary ecological succession, thereby integrating previously isolated conceptual domains; (ii) evaluate this model over 105 years of ecosystem development, revealing a systematic shift in the type and strength of ecological selection; and (iii) couple empirical data with a new simulation model to elucidate underlying mechanisms and characterize their scale dependency. The insights and conceptual framework provided here represent a nexus for cross-system integration.
Keywords: community assembly, neutral theory, niche theory, simulation model, evolutionary niche conservatism
Abstract
Ecological succession and the balance between stochastic and deterministic processes are two major themes within microbial ecology, but these conceptual domains have mostly developed independent of each other. Here we provide a framework that integrates shifts in community assembly processes with microbial primary succession to better understand mechanisms governing the stochastic/deterministic balance. Synthesizing previous work, we devised a conceptual model that links ecosystem development to alternative hypotheses related to shifts in ecological assembly processes. Conceptual model hypotheses were tested by coupling spatiotemporal data on soil bacterial communities with environmental conditions in a salt marsh chronosequence spanning 105 years of succession. Analyses within successional stages showed community composition to be initially governed by stochasticity, but as succession proceeded, there was a progressive increase in deterministic selection correlated with increasing sodium concentration. Analyses of community turnover among successional stages—which provide a larger spatiotemporal scale relative to within stage analyses—revealed that changes in the concentration of soil organic matter were the main predictor of the type and relative influence of determinism. Taken together, these results suggest scale-dependency in the mechanisms underlying selection. To better understand mechanisms governing these patterns, we developed an ecological simulation model that revealed how changes in selective environments cause shifts in the stochastic/deterministic balance. Finally, we propose an extended—and experimentally testable—conceptual model integrating ecological assembly processes with primary and secondary succession. This framework provides a priori hypotheses for future experiments, thereby facilitating a systematic approach to understand assembly and succession in microbial communities across ecosystems.
A major goal in microbial community ecology is to understand the processes that underlie observed patterns in species abundances across space and time (1–3). Two types of processes—deterministic and stochastic—influence the assembly of species into communities. Deterministic processes—in which abiotic and biotic factors determine the presence/absence and relative abundances of species—are associated with ecological selection [sensu Vellend (4)]. Stochastic processes include probabilistic dispersal and random changes in species relative abundances (ecological drift) that are not the consequence of environmentally determined fitness (5, 6).
Historically, microbial community assembly has been studied from a deterministic perspective (7, 8), where empirical evidence shows that a variety of environmental factors—such as pH, salinity, and organic carbon—influence community establishment at different scales (9, 10). However, recent studies have provided increasing support for a predominant role of stochasticity in some microbial systems (e.g., ref. 11). As opposed to a dichotomous debate, in which one attempts to reject stochastic processes in favor of deterministic ones (or vice versa), a more comprehensive perspective should integrate both processes and work to understand how and why their relative influences vary across systems, time, and space (3, 6, 12–15).
The study of ecological succession provides an ideal setting for understanding mechanisms that govern community assembly processes through time and space. Although ecological succession in microbial communities has been broadly investigated (16–21), little has been done to formally link this theme with the balance in stochastic/deterministic processes. Only two studies have directly related these conceptual domains, and both have focused on secondary succession (i.e., following disturbance) (2, 20). These studies show that disturbance promotes a time-dependent shift in the stochastic/deterministic balance. A full understanding of linkages among community succession, disturbance, and the assembly processes, however, requires a testable conceptual framework that enables systematic evaluation of the stochastic/deterministic interplay during succession in both pristine and disturbed ecosystems.
Here we set up a framework that integrates the conceptual domains of microbial succession and the balance in stochastic/deterministic ecological processes. We first devised a conceptual model that links environmental heterogeneity to shifts in these assembly processes during microbial primary succession; for this, we purposefully followed the approach used in Ferrenberg et al. (20) to allow a direct linkage between our model and theirs. Alternative hypotheses within the conceptual model were tested by applying an ecological null modeling approach (3) to data from a soil chronosequence spanning 105 years of primary ecosystem succession (22). The analyses revealed scale dependency with respect to how environmental factors govern the interplay between stochastic and deterministic processes. To better understand the mechanisms underlying the observed patterns, we developed an ecological simulation model that revealed how changes in selective environments cause shifts in the processes underlying community assembly. Finally, to facilitate conceptual synthesis and to generate a priori hypotheses for future experiments, we merged our conceptual model—focused on primary succession—with an extended version of the Ferrenberg et al. (20) secondary succession model.
Conceptual Model
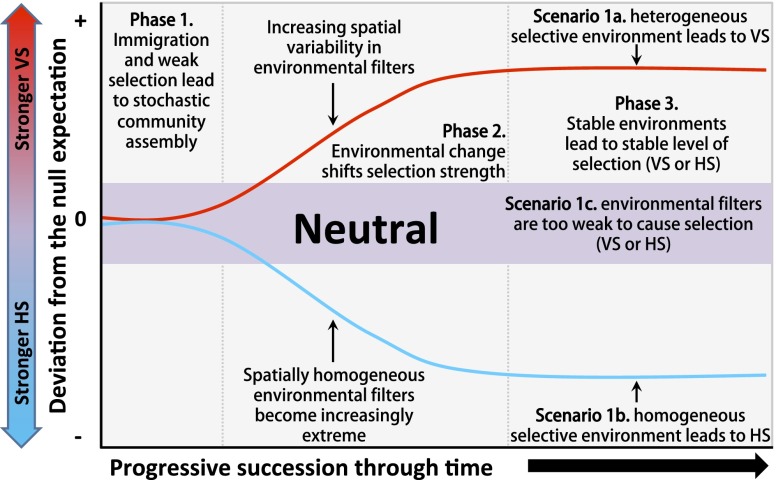
Primary succession in microbial communities has been investigated along receding glacier forelands (20, 23–27), in drinking water biofilms (17, 28–30), and in the rhizosphere (31–34). These studies have collectively revealed a broad suite of patterns associated with shifts in community biomass, diversity, and composition, but none has directly characterized shifts in community assembly processes during succession. However, close examination of the patterns provides indirect insights. We therefore integrate these studies into a conceptual model of predicted trajectories of the stochastic/deterministic balance during microbial primary succession. The model was conceptualized as a time continuum with distinct scenarios playing out across three phases (Fig. 1).
Fig. 1.
Three-phase conceptual model composed of alternative hypotheses related to changes in the strength and type of ecological selection during primary succession. Ecological selection is weak in the center of the vertical axis and is stronger toward both extremes. In phase 1, the initial establishment of microbial communities is expected to be dominated by stochasticity such that turnover in community composition shows little deviation from the null expectation. In phase 2, changes in environmental conditions progressively increase the strength of selection leading to scenarios 1a and 1b; if the environmental factors that change through time do not impose selection, stochastic factors are expected to remain dominant such that the system will remain ecologically neutral [sensu Hubbell (5)] as in scenario 1c. When succession is associated with increasing spatial heterogeneity in selective pressures the variable selection (VS) scenario (1a) is expected; spatial environmental heterogeneity causes turnover in community composition to be greater than the null expectation. In contrast, a spatially homogeneous environment and directional changes—across successional stages—toward increasingly extreme selective conditions lead to the homogeneous selection (HS) scenario (1b); spatial environmental homogeneity causes turnover in community composition to be lower than the null expectation. In phase 3, if succession eventually leads to relatively stable environmental conditions, a relatively stable balance between stochastic/deterministic processes is expected.
Our conceptual model represents a collection of alternative hypotheses focused on the dynamics of the stochastic/deterministic balance during primary succession. We do not attempt to encompass all possible scenarios but focus instead on combining straightforward hypotheses into a testable framework. The model is therefore likely to be supported in some ecosystems and rejected in others; both outcomes are equally informative.
Phase 1: Microbial Community Assembly Is Initially Governed by Stochastic Processes.
It is expected that initial community establishment will be primarily dominated by stochasticity. Jackson (ref. 17, p. 564) observed that during the initial stage of drinking water biofilm formation, communities were “characterized by the colonization of different populations and lack of orderly community structure.” This phenomenon has been confirmed by additional studies (29) and applies to a range of ecosystems. For example, during the early stages of primary succession in a glacier foreland, soil microbial communities are highly diverse and dominated by taxa capable of using many different resources (23, 35). The authors interpreted these findings as evidence for weak competition, which implies weak selection and thus a potentially large influence of stochasticity. It has also been suggested that the sugars released by seedling roots in soil provide a resource-rich environment that reduces competitive pressures, which leads to a dominance of stochasticity during the initial establishment of rhizosphere communities (36–38). More generally, when a broad range of organisms can grow successfully in a given environment, stochasticity is likely to dominate the initial phase of community assembly (6, 14).
Phase 2: Changes in the Local Environment Progressively Increase the Importance of Deterministic Selection.
Following initial microbial community establishment, deterministic selection may become progressively important as organisms affect their environment (e.g., through resource depletion): “following the disordered nature of the early communities, the bacterial assemblage may ‘simplify’ as superior competitors begin to dominate” (ref. 17, p. 563). Thus, as the strength of selection increases, an increasingly large breadth of taxa are excluded. We extend this conceptualization with two hypothesized scenarios that relate to shifts in the level of heterogeneity in the selective environment. In our conceptual model, multiple mechanisms (e.g., abiotic habitat filtering and biotic competition) combine to generate the selective environment.
Under the homogeneous selection scenario (Fig. 1, blue line), the selective environment is spatially homogeneous within each successional stage and does not change significantly during the relatively short time span covered by each stage. The strength of selection does, however, intensify during the longer time span covered by multiple successional stages. This occurs due to directional changes in the mean of one or more selective factors as succession proceeds, which results in specific physiological adaptations being required for positive population growth. For example, soil pH can strongly influence bacterial community composition; thus, a stringent environmental filter is imposed in high- or low-pH conditions (9, 39, 40). The homogeneous selection scenario could therefore emerge if (i) pH was spatially homogeneous within each successional stage and (ii) mean pH became increasingly extreme within later successional stages. The relative influence of homogeneous selection is therefore expected to increase as succession proceeds.
The selective environment may also be spatially heterogeneous, leading to variable selection [sensu Vellend (4)]. In this case, taxa selected for in one place may be selected against in a different place because of spatial variation in the selective environment. We hypothesize a variable selection scenario (Fig. 1, red line) in which spatial environmental heterogeneity increases as succession proceeds. For example, as biofilms develop, new spatially structured ecological niches emerge (e.g., anoxic pockets), and communities are driven toward a 3D architecture where environmental conditions—and thus selective environments—vary (28). In this case, increasing environmental heterogeneity is expected to cause compositional differences across local communities. The relative influence of variable selection is therefore expected to increase as succession proceeds.
Phase 3: Emergence of Stable Environments Leads to Stable Levels of Deterministic Selection.
In the third phase (Fig. 1, phase 3) we hypothesize the relative influences of stochastic and deterministic ecological processes to become relatively stable under both the homogeneous and variable selection scenarios. This assumes that environmental factors that impose selection do not change directionally in their mean or variance. However, some ecosystems may not reach this phase before disturbance.
Neutral Hypothesis.
As an alternative, we hypothesize that some microbial systems are consistently dominated by stochasticity. Such systems might be characterized by abundant resource supply and high levels of organismal dispersal. These characteristics are found, for example, in fluidic systems with constant resource supply (2) such that few microbial taxa are excluded from growing. In this case, species relative abundances will likely be the result of stochastic birth/death events, instead of being environmentally determined (Fig. 1, neutral inset).
Results
Conceptual Model Evaluation.
We evaluated the conceptual model using data on soil bacterial communities and their abiotic environment across 105 years of primary succession in a salt marsh chronosequence (22), which comprised spatial and temporal community turnover within five successional stages (0, 5, 35, 65, and 105 years of soil development). We started by addressing community turnover within each successional stage. We estimated the relative influence of stochastic ecological processes and—when present—the type of deterministic ecological selection (homogeneous or variable). To do so, we combined soil bacterial community data with a previously developed null modeling strategy (3). Second, we used linear regression and statistical model selection to evaluate which abiotic factor was most strongly associated with the relative influence of stochasticity. Below we address the details of these approaches.
First, to infer the relative influences of stochastic and deterministic processes and to differentiate between homogenous and variable selection, we studied community phylogenetic turnover, here defined as the phylogenetic distance separating bacterial operational taxonomic units (OTUs) in one community from OTUs in a second community (41, 42). Using phylogenetic turnover to make ecological inferences requires that phylogenetic distances among taxa approximate differences in the ecological niches they occupy. When this relation is significant, niches are said to have phylogenetic signal (43), and from fundamental evolutionary principles it is expected that microbial niches have phylogenetic signal despite horizontal gene transfer (42). Testing for phylogenetic signal using Mantel correlograms (3, 44, 45) revealed significant positive correlations between differences in OTU environmental optima and OTU phylogenetic distances (P < 0.05) but only across relatively short phylogenetic distances (Figs. S1 and S2). This result is consistent with previous studies on microbial communities in different environments (3, 42, 44, 45) and suggests that within the salt marsh chronosequence, underlying ecological processes can be inferred from analyses of phylogenetic turnover. This assertion was evaluated—and supported—via simulation (SI Materials and Methods, Simulation Model).
To quantify community phylogenetic turnover, we used the abundance weighted β-mean nearest taxon distance (βMNTD). This metric quantifies the phylogenetic distance between assemblages; βMNTD was used because it emphasizes phylogenetic turnover across short phylogenetic distances, where the assumption of phylogenetic signal is strongly supported (Fig. S1). To infer the ecological factor that is primarily responsible for the observed turnover between a given pair of assemblages, we use a null modeling approach that generates an expected level of βMNTD given the dominance of stochastic ecological processes. To quantify the magnitude and direction of deviation between an observed βMNTD value and the null βMNTD distribution, we used the β-nearest taxon index (βNTI). βNTI < −2 or > +2 indicates that βMNTDobs deviates from the mean βMNTDnull by more than two standard deviations; we consider βNTI < −2 or > +2 to indicate significantly less than or greater than expected phylogenetic turnover, respectively, for a given pairwise comparison. The βMNTDnull distribution represents the expected level of phylogenetic turnover given the dominance of stochastic ecological processes such that we further consider a significant deviation (i.e., |βNTI| > 2) to indicate the dominance of deterministic processes and the lack of deviation (i.e., |βNTI| < 2) to indicate the dominance of stochastic processes (3, 42, 46).
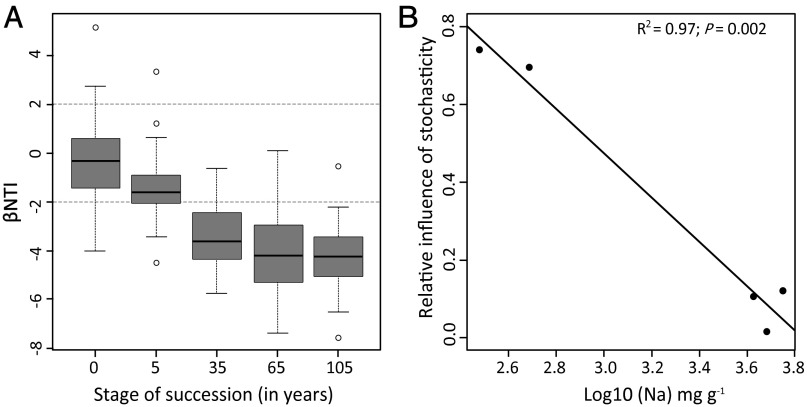
Homogeneous and variable selection should cause less than and greater than expected community turnover, respectively; βNTI < −2 or > +2 is further interpreted as indicating the dominance of homogeneous or variable selection, respectively; this interpretation was evaluated and supported via simulation (SI Materials and Methods, Simulation Model). βNTI values for all possible pairwise comparisons within, but not between, the stages of succession revealed that the βNTI distribution progressively shifted with increasing successional stage, from being primarily consistent with stochastic community assembly (−2 < βNTI < +2) to being consistent with homogeneous selection (βNTI < −2). The means (µ) and standard deviations (σ) were as follows: µ = −0.31 with σ = 1.68, µ = −1.45 with σ = 1.22, µ = −3.46 with σ = 1.23, µ = −4.06 with σ = 1.76, and µ = −4.29 with σ = 1.23 for successional stages 0, 5, 35, 65, and 105 years, respectively (Fig. 2A).
Fig. 2.
Patterns of βNTI and stochasticity. (A) Box plots of βNTI distributions across successional stages showing the median (thick black line), the first quartile (lower box bound), the third quartile (upper box bound), the range of data values that deviate from the box no more than 1.5 times the height of the box (vertical dashed lines), and outliers (open circles). Horizontal dashed lines indicate upper and lower significance thresholds at βNTI = +2 and −2, respectively. (B) The estimated relative influence of stochasticity as a function of log-transformed Na concentration per gram of soil. The solid line is the linear regression model, and statistics are provided on the panel.
These results show a time-dependent shift in the relative influence of stochastic and deterministic processes along the chronosequence. As succession proceeds, the relative influence of stochasticity declined, and that of homogeneous selection increased, as in scenario 1b of our conceptual model. We further estimated the relative influence of stochasticity, within each successional stage, as the fraction of βNTI where |βNTI| < 2. These stochasticity estimates were then regressed against each measured abiotic factor or the logarithm of those factors. The best model (based on R2) showed a strong negative relationship between log-transformed sodium (Na) concentration and the relative influence of stochasticity (R2 = 0.97, P = 0.002) (Fig. 2B). This was true despite a concomitant increase (∼1 to 34 g⋅dm−3) in resource supply in the form of soil organic matter content (SOM) (47), contradicting previous work (48) that found stochasticity to become more important with increasing resource availability.
Community Assembly Processes Between Successional Stages.
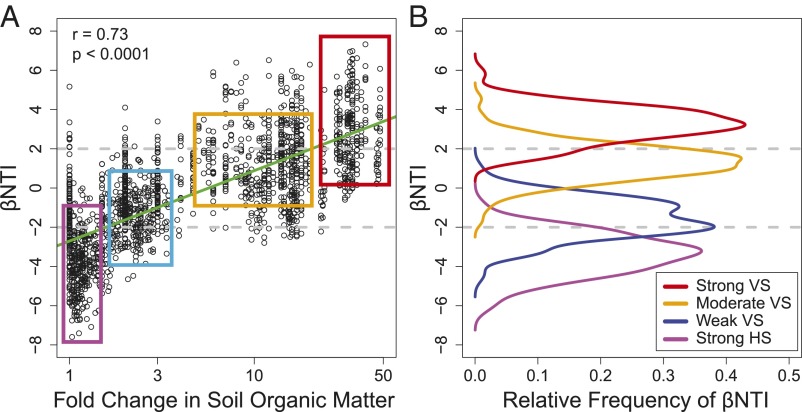
To evaluate potential scale dependency in the mechanisms underlying community assembly, the spatiotemporal scale of analysis was expanded by including between–successional-stage comparisons. For this, βNTI values were determined across all pairwise community comparisons within (∼15 m spatial scale and 6 months temporal scale) and between (up to 8 km spatial scale and 105 years temporal scale) stages of succession. The resulting distance matrix was regressed against distance matrices associated with each measured environmental factor. This analysis showed the larger-scale patterns of βNTI to be most strongly related to changes in log-transformed SOM (r = 0.73), and this relationship was significant via a Mantel test (P < 0.0001). As the change in SOM increased, there was a continuous transition—on average—from βNTI < −2 at the smallest changes in SOM to −2 < βNTI < +2 at intermediate SOM shifts and to βNTI > +2 for the largest shifts in SOM (Fig. 3A). These patterns suggested that increasing shifts in SOM lead to a transition from homogeneous selection (small to no change in SOM), to weak selection and stochasticity (moderate changes in SOM), to variable selection (large changes in SOM).
Fig. 3.
βNTI patterns from empirical comparisons and simulated ecological scenarios. (A) βNTI for all pairwise community comparisons—within and between successional stages—as a function of the change in log-transformed soil organic matter (SOM) concentration between communities, presented as the fold change in SOM. The linear regression model is shown as the green line; statistics are provided on the panel. Horizontal dashed lines indicate the upper (+2) and lower (−2) significance thresholds. Boxes laid over the βNTI data represent our conceptual interpretation of how simulation model outputs (summarized in B) align with the empirical relationship between βNTI and changes in SOM. (B) βNTI distributions obtained from simulated ecological scenarios (see Fig. S3 for a detailed description).
Simulation-Based Evaluation of the Inferences.
In the simulation model we assembled local communities under four ecological scenarios (Fig. 3B and Fig. S3). The four scenarios were based on conceptual interpretations of the observed βNTI patterns in the empirical data. That is, ecological rules governing simulated community assembly were developed from the βNTI-based inferences. The simulated scenarios are referred to with names similar to our conceptual model, but they are distinct from these in that succession (or dynamic changes) in the selective environment is not simulated. In the homogeneous selection scenario, local communities were assembled within a single (arbitrary) environment. The other three scenarios invoked variable selection by assembling communities across two environments, and the strength of variable selection was manipulated by changing how different the two environments were from each other (Fig. S3).
Distributions of βNTI from the simulation model had the following means (µ) and standard deviations (σ): µ = −3.39 with σ = 1.04 from the strong homogeneous selection scenario (Fig. S3, purple panel), µ = −1.58 with σ = 1.00 from the weak variable selection scenario (Fig. S3, blue panel), µ = 1.29 with σ = 0.94 from the moderate variable selection scenario (Fig. S3, orange panel), and µ = 3.23 with σ = 0.89 from the strong variable selection scenario (Fig. S3, red panel).
The βNTI distributions arising from the simulation corresponded closely to the shifts in βNTI associated with changes in SOM (Fig. 3 and Fig. S3) and support our inference that a continuous transition from βNTI < −2 to βNTI > +2 indicates that with progressive shifts in SOM, there is a transition from homogeneous selection, to stochasticity (i.e., weak selection), to variable selection. In addition, the simulations support our within-stage inferences of a transition from stochasticity (|βNTI| < 2 in stages 0 and 5 years) to strong homogeneous selection (βNTI < −2 in stages 35, 65, and 105 years) as succession proceeds. The simulation outcomes further indicate that robust ecological inferences can be made via βNTI-based analyses when there is phylogenetic signal across relatively short phylogenetic distances, as is the case in both the empirical and simulated regional species pools (Figs. S1 and S2).
Discussion
Consistency Between the Conceptual Model and Empirical Data.
In line with our conceptual model (Fig. 1), we observed a greater influence of stochasticity in the initial successional stages of the salt marsh chronosequence (stages 0 and 5 years). This result is consistent with previous indirect evidence from studies of microbial primary succession across diverse systems (17, 23, 29, 35–38). Initial stages of the chronosequence studied here showed diverse bacterial communities (47), in accordance with observations from glacier forelands (23, 35) but in contrast to those from biofilms (17, 29) and plant rhizospheres (36, 37). These high levels of stochasticity and diversity may result from the initial physical structure of the salt marsh, which is formed through sand accumulation and sedimentation and is subjected to regular overflows by seawater (22). As a consequence, these stages are characterized by bare sand and relatively nutrient poor soils that are regularly saturated with seawater but that rapidly dry following seawater retreat (22, 47). This frequent tidal regimen may promote a dynamic environment with many opportunities for successful immigration (dispersal followed by establishment). Thus, random dispersal through both aerial and seawater vectors may be an important ecological factor at these sites. In addition, the physicochemical conditions (e.g., pH and Na concentration) are not extreme in the initial stages (47) such that a strong environmental filter is not present. We suggest that edaphic properties during early succession lead to weak selection and high immigration rates such that random ecological drift governs spatiotemporal shifts in species abundances.
Deterministic selection was found to become increasingly strong and homogeneous toward later successional stages, which aligns with the homogeneous selection scenario in our conceptual model. Further analyses suggested that the progressive accumulation of Na was related to the decrease in stochasticity and the concomitant increase of homogeneous selection. In our system, initial stages had relatively low levels of Na (∼1.8–2.4% by weight) compared with intermediate and late successional stages (∼13.8–14.4%) (47). Sodium concentrations above 13% impose a stringent filter on microbial communities, reflecting the need for physiological adaptations such as the biosynthesis of compatible solutes and the ‘salt-in’ strategy (49, 50).
Although our analyses encompassed 105 years of succession, we could not discern whether the relationship between stochasticity and Na was truly linear or if there was a threshold at which a small increase in Na concentration led to a large decrease in stochasticity. Future work that more finely partitions the Na concentration gradient could be used to identify such thresholds. We also note that there is covariation among physicochemical variables in our system (47) such that caution is warranted in terms of concluding that Na concentration is the physicochemical variable that results in strong homogeneous selection. On the other hand, available literature provides evidences that support our interpretation (9, 10, 51–53).
Contrary to macroecological theory in which the relative influence of stochasticity is thought to increase with resource supply (48, 54), we observed a negative resource supply–stochasticity relationship. This points to an interaction between resource supply and other physicochemical conditions that impose selection. More specifically, we suggest that increased resource supply can increase stochasticity under physicochemical conditions that do not impose strong selection; when physicochemical conditions are extreme and require specific physiological adaptations, changes in resource supply should have little influence over levels of stochasticity. From a macroecological perspective, high levels of diversity may therefore be maintained in regions characterized by high resource supply and relatively benign abiotic conditions, consistent with Chase’s (48) assertion that high productivity enhances tropical biodiversity through elevated levels of stochasticity.
Evidence for Scale Dependency in the Mechanisms Underlying Ecological Selection.
Our analyses revealed that selection was primarily imposed by Na at a relatively small scale (i.e., within successional stages; spatial scale up to 15 m, temporal scale up to 6 months) but that it was imposed by SOM at a larger scale (i.e., among successional stages; spatial scale up to 8 km, temporal scale up to 105 years). We hypothesize that this scale dependence arises because (i) there are locations within the larger-scale domain where Na concentrations are relatively low and thus do not impose ecological selection and (ii) changes in SOM across the larger-scale domain result in different selective environments across successional stages, but the strength of selection imposed by SOM does not vary substantially across successional stages. From this perspective, the strength of selection imposed by Na or SOM is hypothesized to be dependent (Na) or independent (SOM) of their respective concentrations. Application of the simulation model provided an initial evaluation of this hypothesis and showed that progressive shifts in an environmental variable that imposes a consistent strength of selection are expected to cause βNTI patterns that closely align with our among-stage βNTI observations (Fig. 3). Although these simulation results are not definitive, they corroborate the above conceptual inferences and provide a point of departure for future experimental research.
Although the larger-scale analyses point to an important influence of SOM, we cannot distinguish among potential mechanisms that may be mediated by shifts in SOM concentration and/or composition. We note, however, that underlying physiological mechanisms are likely related to the composition of SOM, which is a heterogeneous mixture of organic compounds derived from marine and terrestrial sources and that are found at different states of degradation/lability (55). Spatiotemporal shifts in SOM composition—that covary with SOM concentration—may therefore cause shifts in the selective environment and, in turn, promote turnover in soil bacterial community composition among successional stages (56). Manipulative experiments are required to characterize the degree to which selection is imposed by SOM concentration versus composition.
Conclusions and the Path Forward: Integrating Primary and Secondary Succession with Ecological Processes that Govern Microbial Community Assembly.
In this study we aimed to synthetize and evaluate concepts related to shifts in the strength and type of ecological processes governing microbial community assembly during primary succession. In this effort, we set up a framework that allows the balance between stochastic/deterministic processes and the mechanisms mediating their relative influences to be systematically quantified and understood. Empirical and simulation analyses revealed that the relative influences of underlying mechanisms are scale-dependent. This is a critical result that resonates with previous work (57) and that will strongly influence the conceptual foundation of studies linking ecological processes to community assembly. We showed that microbial communities do not follow macroecological assumptions because the mechanisms driving stochasticity at a small scale were not related to resource supply.
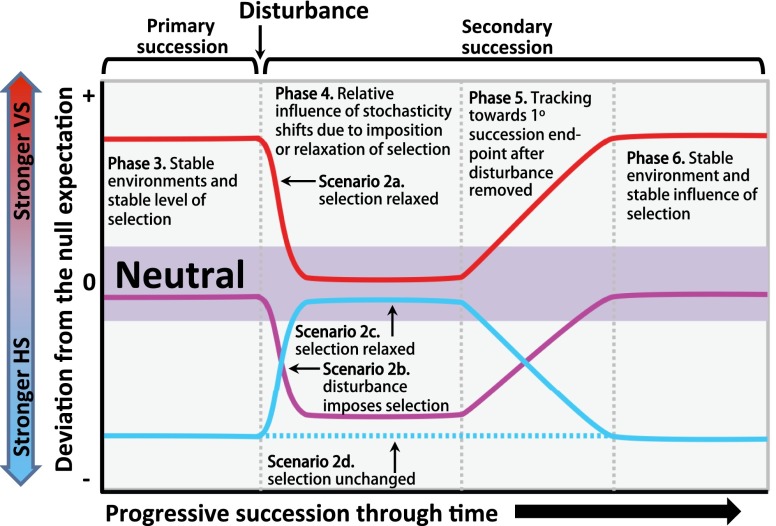
We further hypothesize that the position of a community along the stochastic/deterministic continuum during primary succession will influence how community assembly processes operate during secondary succession (i.e., following disturbance). We used this hypothesis to couple our primary succession conceptual model with an extended version of the secondary succession model provided by Ferrenberg et al. (20) (Fig. 4). As summarized in Fig. 4, this broader conceptual model provides an experimentally testable framework composed of a priori hypotheses that represent a theoretical basis for future experiments aimed at linking microbial community assembly with successional dynamics across pristine and disturbed systems. A systematic approach to understanding community assembly and succession in microbiomes across ecosystems is thereby enabled. Relevant new studies designed to test the broader conceptual model will significantly advance fundamental knowledge of ecological systems, within and beyond microbial ecology.
Fig. 4.
Hypothesized conceptual model linking primary and secondary succession to the stochastic/deterministic balance. Phase 3 is the final phase of primary succession and is consistent with phase 3 in Fig. 1; vertical axis is as in Fig. 1. Following a disturbance event the relative influence of stochastic factors can shift in ways that are dependent on both the outcome of primary succession and the type of disturbance. In scenario 2a (red line), primary succession has resulted in strong variable selection. A significant shift away from strong variable selection—following disturbance—is expected if the environment is homogenized; the system may become dominated by strong homogeneous selection (not displayed) or become neutral if the resulting environmental condition does or does not, respectively, impose strong selection. In scenario 2b (purple line), a strong influence of stochastic factors has been maintained throughout primary succession, and the disturbance itself imposes a strong and spatially homogeneous selective pressure similar to patterns observed in soil microbial communities following fire (20). In scenarios 2c and 2d (blue lines), primary succession has resulted in strong homogeneous selection as in our field system. In scenario 2c, disturbance removes the strong selective pressure that developed over the course of primary succession; in our field system this could occur if shifts in topography lead to an increased frequency of tidal inundation at an older part of the chronosequence, thereby causing a decline in Na concentration. In scenario 2d, disturbance does not impact the primary selective pressure such that strong homogeneous selection is expected to be maintained; in our field system this could occur if SOM was artificially added to the later successional stages—high Na concentrations in late primary successional stages impose a dominant selective pressure that would not be alleviated by the addition of SOM.
Materials and Methods
Details for all methods are provided in SI Materials and Methods. Briefly, soil samples were collected in triplicated plots at five stages of soil development in a salt marsh chronosequence, estimated as 0, 5, 35, 65, and 105 years of soil development. Total soil DNA was extracted using a MoBio PowerSoil DNA extraction kit (MoBio Laboratories), and communities were profiled targeting the V4–V6 region of the bacterial 16S rRNA using a Roche GS-FLX 454 automated pyrosequencer running the Titanium chemistry. Sequence data were analyzed in QIIME (Quantitative Insights Into Microbial Ecology) (58). Samples were individually subjected to measurements of soil physical structure (clay:silt:sand %) and chemical content of total organic matter (OM), nitrate (N-NO3−), ammonium (N-NH4+), sulfate (S-SO4), sodium (Na), and pH.
To test for phylogenetic signal (in both empirical and simulation data analyses), we used phylogenetic Mantel correlograms, as described elsewhere (3, 44, 45). To characterize the turnover in phylogenetic community composition, we quantified the β-mean nearest taxon distance (βMNTD), calculated as follows:
where is the relative abundance of OTU i in community k, nk is the number of OTUs in k, and is the minimum phylogenetic distance between OTU i in community k and all OTUs j in community . βMNTD was calculated using the R function comdistnt (abundance.weighted = TRUE; package picante) (SI Materials and Methods).
To quantify the magnitude and direction of deviation between an observed βMNTD value and the null βMNTD distribution, we used the β-nearest taxon index (βNTI), calculated as follows:
where βMNTDobs is observed βMNTD, βMNTDnull are null values of βMNTD, and sd indicates the standard deviation of the βMNTDnull distribution. We quantified βNTI for all pairwise comparisons, using a separate null model for each comparison.
Detailed information on the development of the simulation model, including the regional species pool evolution and local community assembly, is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Allan Konopka, Jim Fredrickson, Steve Lindemann, and Stephanie Jurburg for critical reading of the manuscript. We also thank the Nederlandse Vereniging voor Natuurmonumenten for granting us access to the salt marsh. This research was supported by the Netherlands Organisation for Scientific Research (NWO) and the Soil Biotechnology Foundation. A portion of the research described in this paper was conducted under the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory (PNNL), a multiprogram national laboratory operated by Battelle for the US Department of Energy. J.C.S. is grateful for the support of the Linus Pauling Distinguished Postdoctoral Fellowship program at PNNL. A portion of the research was performed using Institutional Computing at PNNL.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414261112/-/DCSupplemental.
References
- 1.Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2010;4(3):337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- 2.Zhou J, et al. Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci USA. 2014;111(9):E836–E845. doi: 10.1073/pnas.1324044111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stegen JC, et al. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013;7(11):2069–2079. doi: 10.1038/ismej.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vellend M. Conceptual synthesis in community ecology. Q Rev Biol. 2010;85(2):183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 5.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton Univ Press; Princeton, NJ: 2001. [DOI] [PubMed] [Google Scholar]
- 6.Chase JM, Myers JA. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc Lond B Biol Sci. 2011;366(1576):2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baath E. Adaptation of soil bacterial communities to prevailing pH in different soils. FEMS Microbiol Ecol. 1996;19(4):227–237. [Google Scholar]
- 8.Torsvik V, Øvreås L, Thingstad TF. Prokaryotic diversity—Magnitude, dynamics, and controlling factors. Science. 2002;296(5570):1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 9.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103(3):626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104(27):11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caruso T, et al. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 2011;5(9):1406–1413. doi: 10.1038/ismej.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chisholm RA, Pacala SW. Theory predicts a rapid transition from niche-structured to neutral biodiversity patterns across a speciation-rate gradient. Theor Ecol. 2011;4(2):195–200. [Google Scholar]
- 13.Langenheder S, Székely AJ. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 2011;5(7):1086–1094. doi: 10.1038/ismej.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chase JM. Drought mediates the importance of stochastic community assembly. Proc Natl Acad Sci USA. 2007;104(44):17430–17434. doi: 10.1073/pnas.0704350104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers JA, Harms KE. Seed arrival and ecological filters interact to assemble high-diversity plant communities. Ecology. 2011;92(3):676–686. doi: 10.1890/10-1001.1. [DOI] [PubMed] [Google Scholar]
- 16.Fierer N, Nemergut D, Knight R, Craine JM. Changes through time: Integrating microorganisms into the study of succession. Res Microbiol. 2010;161(8):635–642. doi: 10.1016/j.resmic.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Jackson CR, Churchill PF, Roden EE. Successional changes in bacterial assemblage structure during epilithic biofilm development. Ecology. 2001;82(2):555–566. [Google Scholar]
- 18.Tscherko D, Rustemeier J, Richter A, Wanek W, Kandeler E. Functional diversity of the soil microflora in primary succession across two glacier forelands in the Central Alps. Eur J Soil Sci. 2003;54(4):685–696. [Google Scholar]
- 19.Schütte UM, et al. Bacterial succession in a glacier foreland of the High Arctic. ISME J. 2009;3(11):1258–1268. doi: 10.1038/ismej.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrenberg S, et al. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 2013;7(6):1102–1111. doi: 10.1038/ismej.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bengtsson MM, Sjøtun K, Lanzén A, Ovreås L. Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J. 2012;6(12):2188–2198. doi: 10.1038/ismej.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olff H, De Leeuw J, Bakker JP, Platerink RJ, van Wijnen HJ. Vegetation succession and herbivory in a salt marsh: Changes induced by sea level rise and silt deposition along an elevational gradient. J Ecol. 1997;85(6):799–814. [Google Scholar]
- 23.Sigler WV, Zeyer J. Microbial diversity and activity along the forefields of two receding glaciers. Microb Ecol. 2002;43(4):397–407. doi: 10.1007/s00248-001-0045-5. [DOI] [PubMed] [Google Scholar]
- 24.Nicol GW, Tscherko D, Embley TM, Prosser JI. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ Microbiol. 2005;7(3):337–347. doi: 10.1111/j.1462-2920.2005.00698.x. [DOI] [PubMed] [Google Scholar]
- 25.Deiglmayr K, Philippot L, Tscherko D, Kandeler E. Microbial succession of nitrate-reducing bacteria in the rhizosphere of Poa alpina across a glacier foreland in the Central Alps. Environ Microbiol. 2006;8(9):1600–1612. doi: 10.1111/j.1462-2920.2006.01051.x. [DOI] [PubMed] [Google Scholar]
- 26.Nemergut DR, et al. Microbial community succession in an unvegetated, recently deglaciated soil. Microb Ecol. 2007;53(1):110–122. doi: 10.1007/s00248-006-9144-7. [DOI] [PubMed] [Google Scholar]
- 27.Zumsteg A, et al. Bacterial, archaeal and fungal succession in the forefield of a receding glacier. Microb Ecol. 2012;63(3):552–564. doi: 10.1007/s00248-011-9991-8. [DOI] [PubMed] [Google Scholar]
- 28.Jackson CR. Changes in community properties during microbial succession. Oikos. 2003;101(2):444–448. [Google Scholar]
- 29.Martiny AC, Jørgensen TM, Albrechtsen H-J, Arvin E, Molin S. Long-term succession of structure and diversity of a biofilm formed in a model drinking water distribution system. Appl Environ Microbiol. 2003;69(11):6899–6907. doi: 10.1128/AEM.69.11.6899-6907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Revetta RP, et al. Establishment and early succession of bacterial communities in monochloramine-treated drinking water biofilms. FEMS Microbiol Ecol. 2013;86(3):404–414. doi: 10.1111/1574-6941.12170. [DOI] [PubMed] [Google Scholar]
- 31.Mahaffee WF, Kloepper JW. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field grown cucumber (Cucumis sativus L.) Microb Ecol. 1997;34(3):210–223. doi: 10.1007/s002489900050. [DOI] [PubMed] [Google Scholar]
- 32.Wieland G, Neumann R, Backhaus H. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl Environ Microbiol. 2001;67(12):5849–5854. doi: 10.1128/AEM.67.12.5849-5854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marschner P, Neumann G, Kania A, Weiskopf L, Lieberei R. Spatial and temporal dynamics of the microbial community structure in the rhizosphere of cluster roots of white lupin (Lupinus albus L.) Plant Soil. 2002;246(2):167–174. [Google Scholar]
- 34.Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8(4):790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sigler WV, Crivii S, Zeyer J. Bacterial succession in glacial forefield soils characterized by community structure, activity and opportunistic growth dynamics. Microb Ecol. 2002;44(4):306–316. doi: 10.1007/s00248-002-2025-9. [DOI] [PubMed] [Google Scholar]
- 36.Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem. 2013;288(7):4502–4512. doi: 10.1074/jbc.M112.433300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaparro JM, et al. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE. 2013;8(2):e55731. doi: 10.1371/journal.pone.0055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inceoğlu Ö, Al-Soud WA, Salles JF, Semenov AV, van Elsas JD. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS ONE. 2011;6(8):e23321. doi: 10.1371/journal.pone.0023321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75(15):5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousk J, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4(10):1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 41.Graham CH, Fine PV. Phylogenetic beta diversity: Linking ecological and evolutionary processes across space in time. Ecol Lett. 2008;11(12):1265–1277. doi: 10.1111/j.1461-0248.2008.01256.x. [DOI] [PubMed] [Google Scholar]
- 42.Stegen JC, Lin X, Konopka AE, Fredrickson JK. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012;6(9):1653–1664. doi: 10.1038/ismej.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol Lett. 2008;11(10):995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, et al. Phylogenetic beta diversity in bacterial assemblages across ecosystems: Deterministic versus stochastic processes. ISME J. 2013;7(7):1310–1321. doi: 10.1038/ismej.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindemann SR, et al. The epsomitic phototrophic microbial mat of Hot Lake, Washington: Community structural responses to seasonal cycling. Front Microbiol. 2013;4:323. doi: 10.3389/fmicb.2013.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardy OJ. Testing the spatial phylogenetic structure of local communities: Statistical performances of different null models and test statistics on a locally neutral community. J Ecol. 2008;96(5):914–926. [Google Scholar]
- 47.Dini-Andreote F, et al. Dynamics of bacterial community succession in a salt marsh chronosequence: Evidences for temporal niche partitioning. ISME J. 2014;8(10):1989–2001. doi: 10.1038/ismej.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chase JM. Stochastic community assembly causes higher biodiversity in more productive environments. Science. 2010;328(5984):1388–1391. doi: 10.1126/science.1187820. [DOI] [PubMed] [Google Scholar]
- 49.McGenity TJ, Gemmell RT, Grant WD, Stan-Lotter H. Origins of halophilic microorganisms in ancient salt deposits. Environ Microbiol. 2000;2(3):243–250. doi: 10.1046/j.1462-2920.2000.00105.x. [DOI] [PubMed] [Google Scholar]
- 50.Oren A. Microbial life at high salt concentrations: Phylogenetic and metabolic diversity. Saline Syst. 2008;4:2. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oren A. Halophilic Microorganisms and Their Environments. Kluwer; Dordrecht, The Netherlands: 2002. [Google Scholar]
- 52.Morrissey EM, Gillespie JL, Morina JC, Franklin RB. Salinity affects microbial activity and soil organic matter content in tidal wetlands. Glob Change Biol. 2014;20(4):1351–1362. doi: 10.1111/gcb.12431. [DOI] [PubMed] [Google Scholar]
- 53.Herlemann DP, et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5(10):1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chase JM. Community assembly: When should history matter? Oecologia. 2003;136(4):489–498. doi: 10.1007/s00442-003-1311-7. [DOI] [PubMed] [Google Scholar]
- 55.Bianchi TS. The role of terrestrially derived organic carbon in the coastal ocean: A changing paradigm and the priming effect. Proc Natl Acad Sci USA. 2011;108(49):19473–19481. doi: 10.1073/pnas.1017982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen SL, et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci USA. 2012;109(42):E2846–E2855. doi: 10.1073/pnas.1207574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martiny JB, Eisen JA, Penn K, Allison SD, Horner-Devine MC. Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci USA. 2011;108(19):7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.