Significance
Some of the most damaging tree diseases are caused by pathogens that induce cankers, a stem deformation often lethal. To investigate the cause of this adaptation, we sequenced the genomes of poplar pathogens that do and do not cause cankers. We found a unique cluster of genes that produce secondary metabolites and are co-activated when the canker pathogen is grown on poplar wood and leaves. The gene genealogy is discordant with the species phylogeny, showing a signature of horizontal transfer from fungi associated with wood decay. Furthermore, genes encoding hemicellulose-degrading enzymes are up-regulated on poplar wood chips, with some having been acquired horizontally. We propose that adaptation to colonize poplar woody stems is the result of acquisition of these genes.
Keywords: poplar pathogen, tree disease, fungal genomics, Septoria canker
Abstract
Some of the most damaging tree pathogens can attack woody stems, causing lesions (cankers) that may be lethal. To identify the genomic determinants of wood colonization leading to canker formation, we sequenced the genomes of the poplar canker pathogen, Mycosphaerella populorum, and the closely related poplar leaf pathogen, M. populicola. A secondary metabolite cluster unique to M. populorum is fully activated following induction by poplar wood and leaves. In addition, genes encoding hemicellulose-degrading enzymes, peptidases, and metabolite transporters were more abundant and were up-regulated in M. populorum growing on poplar wood-chip medium compared with M. populicola. The secondary gene cluster and several of the carbohydrate degradation genes have the signature of horizontal transfer from ascomycete fungi associated with wood decay and from prokaryotes. Acquisition and maintenance of the gene battery necessary for growth in woody tissues and gene dosage resulting in gene expression reconfiguration appear to be responsible for the adaptation of M. populorum to infect, colonize, and cause mortality on poplar woody stems.
Domestication of forest trees, in contrast to agricultural crops, has become prevalent only during the last few centuries and often encompasses a transition from wild, complex ecosystems to homogeneous and intensively managed plantations that are frequently composed of a single tree species (1). The recent ability to use modern genetic and genomic techniques with conventional breeding promises to speed up tree domestication (2). Poplar has emerged as an extremely versatile tree with natural attributes favorable to its domestication. The ease of vegetative propagation and the breeding of interspecific hybrids with broad adaptability, improved growth, and disease resistance has contributed to the widespread use of poplar for a variety of commercial products, including lumber, paper, and bioenergy feedstock (3). One of the challenges of poplar domestication has been the emergence of pathogens that were innocuous in their natural pathosystems, but can cause severe losses in plantations. Native poplars still vastly outnumber planted trees, and this large reservoir of a naturally coevolved pathosystem in close proximity to intensively managed clonal plantations could destabilize the host–pathogen equilibrium, leading to new disease epidemics (4).
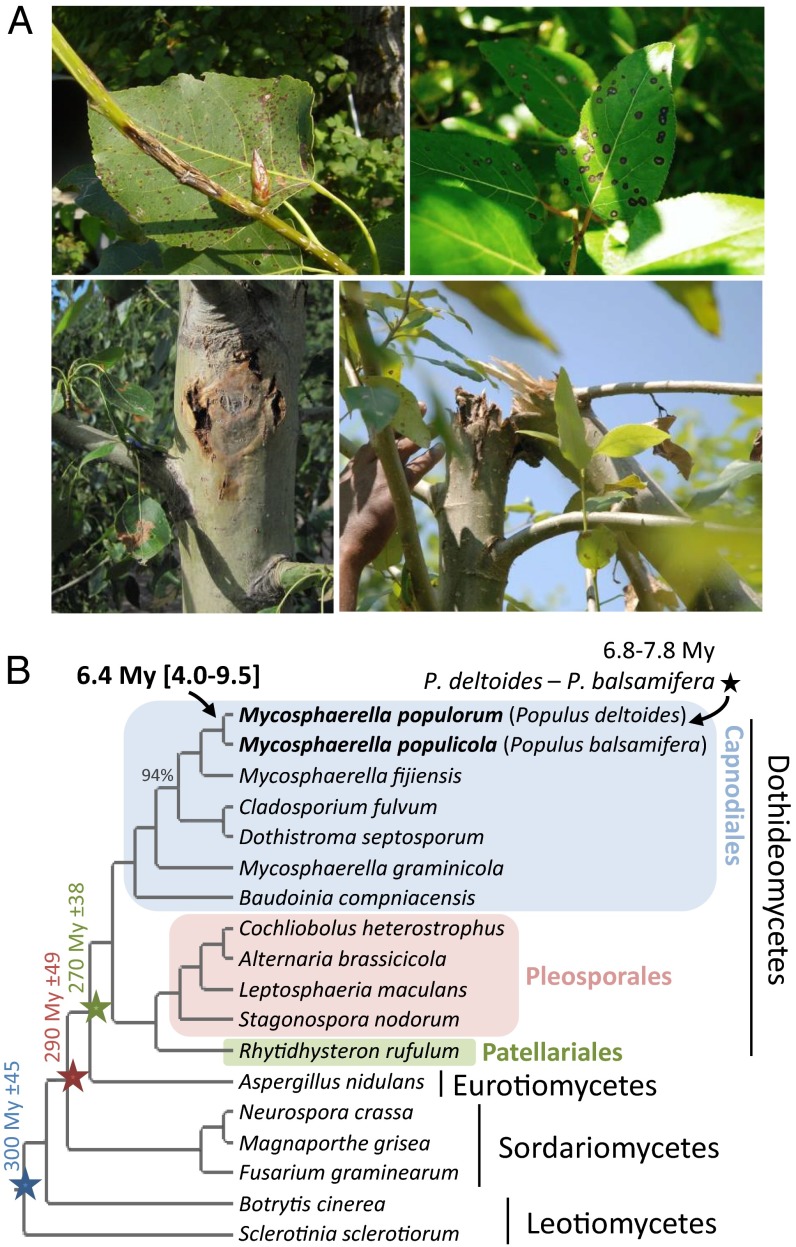
A native endemic fungus on northeastern and north-central North American poplars, Mycosphaerella populorum (anamorph = Sphaerulina musiva; class Dothideomycetes) occurs in natural stands of native Populus deltoides, causing necrotic foliar lesions, but rarely resulting in early defoliation (5). With the introduction of exotic poplar species at the beginning of the 20th century and the intensification of hybrid poplar cultivation in North America, M. populorum has emerged as a stem-infecting pathogen, causing stem cankers that lead to weakening and breakage of the tree trunk, often resulting in plantation failure (Fig. 1A) (6, 7). The pathogen can attack a broad range of susceptible hybrid poplars and has also expanded its geographic range. It has recently been reported, to our knowledge, for the first time west of the Rocky Mountains and in Argentina and Brazil (6–8). This disease has become the most important factor limiting poplar plantations in eastern North America and could threaten the poplar industry worldwide (6, 9).
Fig. 1.
M. populorum and M. populicola symptoms and divergence time estimate. (A, Top Left) M. populorum branch canker and leaf spots on a P. deltoides × P. trichocarpa clone. (Top Right) M. populicola leaf-spot symptoms on P. trichocarpa. (Bottom) M. populorum stem canker on P. deltoides × P. maximowiczii (Left) and P. deltoides × P. trichocarpa (Right) that caused the trunk to break at the canker. Photo provided by Harry Kope. (B) Maximum-likelihood phylogeny of M. populorum, M. populicola, and 16 other ascomycete fungi. All nodes received a bootstrap support of 100% except one, indicated with a 94% value. Colored stars: calibration points in million years (SI Appendix, Phylogenomic Analysis).
A sister species to this pathogen, Mycosphaerella populicola (anamorph = S. populicola; class Dothideomycetes), is also endemic on native poplars, causing a leaf spot symptom on Populus balsamifera and Populus trichocarpa, but has a much broader geographical distribution than M. populorum (6, 10). This pathogen is considered a lower threat to poplar plantations because it does not cause stem cankers under natural conditions or in plantations and it has a narrow host range (11).
To identify genetic factors underlying the canker symptom, we sequenced and compared the genomes of these two closely related pathogens with different natural host ranges and etiological characteristics. This provided a unique opportunity to contrast the evolutionary consequences of the adaptation of a tree pathogen to different hosts and the ability to gradually transition from natural to domesticated ecosystems. Our results show that the genome of M. populorum has evolved a broader battery of genes and has acquired genes through horizontal transfer that are absent in its sister species. These genes are enriched in functions that allow M. populorum to infect woody tissues.
Results and Discussion
Genome Sequences of the Two Poplar Pathogens.
The 29.3-Mb M. populorum genome is organized into 13 main chromosomes, 8 of which have telomeres on both ends and 5 a telomere at one end. The assembled genome has an additional 59 small scaffolds with an average length of 4.5 kb. A total of 10,233 genes was predicted, 10,179 of which were present on the 13 largest chromosomal assemblies (SI Appendix, Tables S1 and S2). These 13 chromosomes account for 99.5% of the gene space and 99.1% of the genome and represent the near-complete M. populorum genome. M. populorum chromosome sizes and numbers were confirmed by electrophoretic and cytological karyotyping (SI Appendix, Fig. S1 and Electrophoretic and Cytological Karyotypes). The slightly larger M. populicola genome (33.2 Mb) was composed of 502 scaffolds containing 9,739 predicted genes (SI Appendix, Tables S1 and S2) and showed an average nucleotidic similarity of 93.4% with the M. populorum genome (SI Appendix, Fig. S2A). The size difference between the two genomes (3.8 Mb) is explained mainly by a difference in repeat content (SI Appendix, Table S3 and De Novo Repeat Identification and Transposon Annotation).
Host–Pathogen Coevolution, Recent Speciation, and Macrosynteny.
We inferred a divergence time of 6.4 My (95% confidence interval of 4.0–9.5 My) between M. populorum and M. populicola (Fig. 1B). This overlaps with the estimated divergence time between their respective natural hosts, Populus deltoides and P. balsamifera/P. trichocarpa. The first fossils for members of the poplar sections Tacamahaca (i.e., balsam poplars including P. balsamifera and P. trichocarpa) and Aigeiros (cottonwoods, P. deltoides) were dated between 9 and 13 My ago (12), and divergence time between members of these sections ranged from 6.8 to 7.8 My (13). These estimations support the hypothesis of coevolution and cospeciation of these two species on their endemic hosts.
The genome organization has been conserved between these two poplar pathogens, reflecting their relatively recent speciation. The M. populorum and M. populicola genomes are highly collinear and macrosyntenic (SI Appendix, Fig. S2B), contrasting with the mesosynteny seen in other Mycosphaerella genome comparisons (14) (SI Appendix, Fig. S2C). The two poplar pathogens show a strong synteny with 171 (34%) repeat-masked M. populicola scaffolds aligned to the 13 repeat-masked M. populorum chromosomes. Most (95%) of the M. populicola scaffolds have a one-to-one relationship to a M. populorum single chromosome; i.e., there are no breakpoints. The orthologous gene content in these syntenic blocks is conserved, but their order and orientation often vary. M. populorum and M. populicola have a larger size of syntenic blocks (46 genes/block in the largest block) than M. populorum and Dothistroma septosporum (a pine pathogen), which diverged at least 87 Mya with only 10 genes in the largest block (14) (SI Appendix, Fig. S2C and Macro- and Microsynteny).
Reduced Gene Content in M. populorum and M. populicola.
The genomes of M. populorum and M. populicola have the smallest gene content (10,233 and 9,739 protein models predicted, respectively) (SI Appendix, Table S2) within the 18 Dothideomycetes fungal genomes analyzed recently (14). It is unlikely that the reduced gene sets in these poplar pathogens result from incomplete genome coverage and/or assembly; the M. populorum and M. populicola assemblies encompass 96.0% and 94.4% of the Core Eukaryotic Genes Mapping Approach (15) eukaryotic gene space, respectively.
A total of 8,548 homologous genes between the two poplar pathogens was clustered in groups of two to six proteins (SI Appendix, Clusters of Homologous Proteins). Within this core gene set, a large number of lineage-specific genes are present, with ∼7% (719 genes in M. populorum and 749 in M. populicola) of their respective gene content shared exclusively between the two poplar pathogens. We found that 8.3% (850) of the gene models in M. populorum and 7.2% (703) of those in M. populicola were unique; i.e., they did not have homologs in the other fungal genomes available in the National Center for Biotechnology (NCBI) nonredundant database (SI Appendix, Fig. S3A). Only ∼9% of the unique genes encode putative secreted proteins, and only 4–5% carry known functional domains (SI Appendix, Fig. S3B). Many of these unique genes represent putative transcription factors as inferred from zinc-ion binding and nucleic-acid–binding gene ontology terms. Similar lineage-specific enrichments in transcription factors have been found in the fungal obligate biotrophs Melampsora larici-populina and Puccinia graminis (16).
To further study gene family expansions and contractions in these two genomes, we compared them to 10 Dothideomycetes genomes, resulting in an analysis of 16,909 OrthoMCL gene families (SI Appendix, Fig. S3C). A large majority (∼99.5%) of these gene families were contracted in the two poplar pathogens, with only 32 (0.18%) of them expanded in M. populorum and 23 (0.13%) in M. populicola; 34 (0.2%) additional OrthoMCL gene families were expanded in both fungi (SI Appendix, Fig. S3D; Dataset S1). More than 91% of the genes included within these expanded gene families had RNA-seq transcript support, 66.2% with gene-count values (fragments per kilobase of exon per million fragments mapped) up to 10.0 (Dataset S1). Strikingly, among these expanded gene families, those encoding peptide and amino acid transporters are significantly overrepresented (SI Appendix, Fig. S3E). This suggests that these two poplar pathogens have diversified their potential for nitrogen assimilation, as observed in other obligate biotrophs (16, 17).
Gene Content and Expression in M. populorum and M. populicola Reflect Differences in Etiology and Pathogenicity.
The high similarity in genome organization and gene number observed between M. populorum and M. populicola raise the possibility that differences in the etiology of the diseases caused by these pathogens are due to subtle contrasts in gene dosage (i.e., the number of copies of a given gene) and expression. To investigate whether this could explain canker formation in M. populorum, we analyzed gene content and expression of gene families known to be involved in interaction with plant hosts, including those coding for secondary metabolism proteins, plant cell-wall–degrading enzymes, and transporters. We compared RNA-seq expression profiles of both fungi grown on artificial media enriched with host-derived carbon sources (poplar leaf extract or poplar wood chips). Inoculation of host-derived media has been widely used to efficiently mimic conditions occurring during host infection and colonization by fungal pathogens (18). Even if artificial media does not completely mimic the structure and dynamics of the nutritional environment that exists in planta (18, 19), we expect that our simplified model of growth on poplar-enriched artificial media can provide a convenient way to compare factors involved in leaf and woody tissue invasion, breakdown, and assimilation.
All gene families examined had more copies in M. populorum than in M. populicola. Furthermore, expanded gene families systematically showed a higher number of genes that were up-regulated in M. populorum when interacting with host compounds, suggesting a gene dosage effect probably acting at the level of gene expression (SI Appendix, Fig. S4A).
A unique horizontally transferred M. populorum nonribosomal peptide synthetase-polyketide synthase hybrid cluster strongly induced by poplar.
A total of 31 and 29 secondary metabolite clusters were predicted in M. populorum and M. populicola, respectively. This is slightly higher than any other fungi sequenced in the Capnodiales (14, 20). In addition to the 26 syntenic clusters present in the two genomes, five M. populorum [one nonribosomal peptide synthetase (NRPS), two polyketide synthases (PKS), two NRPS–PKS hybrids] and three M. populicola (three PKS) clusters were lineage specific (SI Appendix, Clusters of secondary metabolite genes).
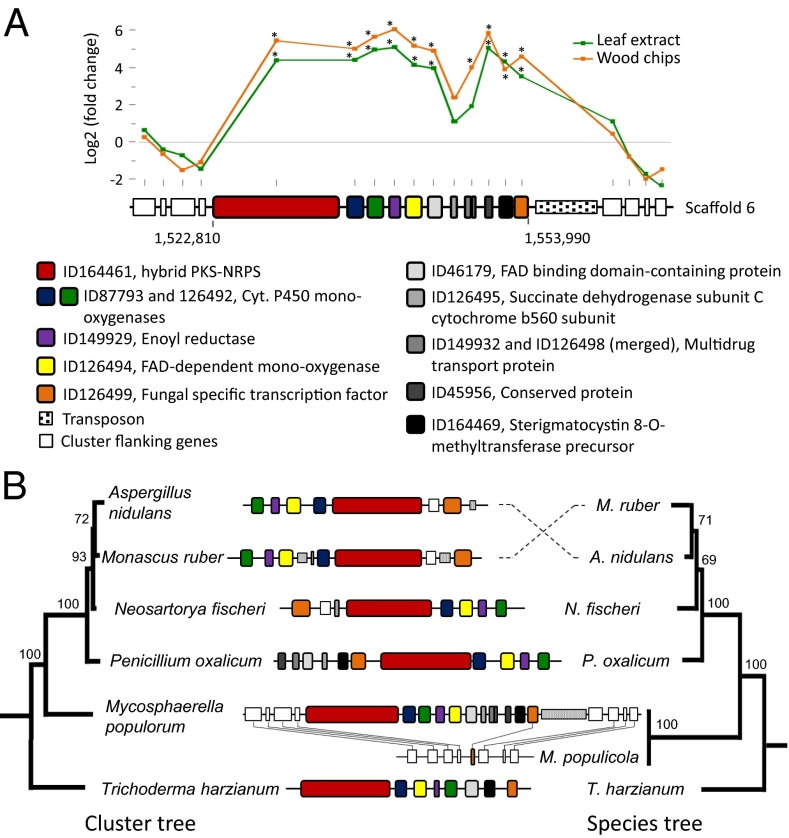
One of the two unique NRPS–PKS clusters found on M. populorum scaffold 6 showed a strong induction pattern during growth on media amended with poplar tissues. This cluster is ∼20- and ∼40-fold overexpressed for the leaf-extract and wood-chip treatments, respectively, compared with the control (skim milk) (SI Appendix, Table S4; Fig. 2A). This shows that the expression of this gene cluster is coregulated following induction by poplar leaves or wood. Homologs of these genes were also present in the M. populicola and other Dothideomycetes genomes, but were not organized in a coregulated gene cluster as seen in M. populorum.
Fig. 2.
Chaetoglobosin-like cluster unique to M. populorum. (A) Transcript abundance comparison (log2-fold change ratio) between the control medium and the poplar leaf-extract (green) and the poplar wood-chips (orange) media for the 11 genes of the cluster; in contrast with other genes of scaffold 6 in M. populorum, log2-fold values of 8 genes located outside the cluster at the 5′- and 3′-ends are given. Asterisk indicates a significant change in gene expression (Cuffdiff q-value < 0.01) between the leaf-extract and the wood-chips treatment compared with the control (skim milk). (B) Maximum-likelihood (ML) phylogeny of cluster genes (3,707 variable amino acid sites from the five congruent datasets corresponding to the chaetoglobosin-like proteins) and ML species phylogeny (five congruent protein datasets and 1,535 variable amino acid sites) for the five taxa most closely related to the M. populorum protein clusters (i.e., P1 group; see SI Appendix, Origin and evolutionary history of the chaetoglobosin-like gene cluster in M. populorum). Structure and conservation of the cluster is given with the legend used for Fig. 2A; lines between genes indicate homolog pairs between M. populorum and M. populicola. The chaetoglobosin-like cluster is absent from M. populicola and all other members of the Dothideomycetes.
Secondary metabolite cluster identification with antiSMASH (21) indicated that the first part of the predicted M. populorum cluster (five key genes and one gene coding for a fungal-specific transcription factor) was homologous to the Penicillium expansum chaetoglobosin biosynthesis cluster (22) (Fig. 2A). RNAi silencing of the P. expansum hybrid NRPS–PKS gene completely eliminated chaetoglobosin production (23). Chaetoglobosins are known to have phytotoxic, antifungal, and antibacterial activities, which may benefit the fungus during host interaction and while fending off competitors (24, 25). As chaetoglobosins are produced by fungi exhibiting different lifestyles—broad host range (P. expansum), narrow host range (Diplodia maydis), and endophytes (Chaetomium spp.)—they presumably serve different roles in these diverse interactions.
Similarity searches against all publicly available fungal genomes revealed that the full cluster is shared only with the ascomycetes Penicillium oxalicum (synonym: Penicillium decumbens). P. oxalicum is a saprophytic fungus commonly associated with wood decay and possesses improved lignocellulose degradation abilities (26). The striking conservation in synteny and gene sequence as well as the sporadic distribution of the full cluster in only two groups of the fungal phylogeny (Dothideomycetes and Eurotiales) raises the possibility that this cluster was acquired through horizontal gene transfer (HGT) (Fig. 2B). To test this hypothesis, individual phylogenies were reconstructed for all of the 11 M. populorum cluster genes and their respective homologs identified across different fungal genomes. Most of the M. populorum secondary metabolite cluster proteins were found nested within a group of Eurotiomycetes fungi, including P. oxalicum, as expected under the hypothesis that the cluster was transferred from a Eurotiomycetes ancestor through a horizontal transfer event (Fig. 2B; Dataset S2; SI Appendix, Origin and evolutionary history of the chaetoglobosin-like gene cluster in M. populorum). This unique and highly expressed cluster could have been transferred from fungi with wood-decaying abilities to early Mycosphaerella and may contribute to the ability of M. populorum to infect woody tissues, an activity missing in M. populicola.
Enzyme arsenal facilitates wood colonization.
Microscopy analyses of young stems and branches indicated that infection by M. populorum results in wood colonization (7, 27, 28). Fungal growth was not limited to the cortex, periderm, and phloem, but also invaded and expanded within xylem elements. Hyphae of M. populorum were observed growing in the cell lumens of xylem vessel members and fibers (28), suggesting that differences in the ability to infect woody tissues could be related to wood cell-wall–degrading gene families. The genes targeting plant cell-wall components found in the two poplar pathogens could ensure at least a partial cellulose digestion and possibly allow these pathogens to use cellulose as a carbon source during host penetration (14) [SI Appendix, Fig. S4B and Carbohydrate-active enzymes (CAZy); Dataset S3A].
Selective retention of horizontally acquired genes from exchanges between fungi as well as from prokaryote donors have recently been shown to play an important role in the reconfiguration of carbohydrate metabolism of filamentous plant and animal associated fungi (29–31). Several gene families related to hemicellulose and pectin degradation and fungal cell-wall modification are significantly enriched in the canker pathogen, M. populorum (SI Appendix, Fig. S4B). None of these genes arose from a recent lineage-specific gain in M. populorum, as they were found in other Dothideomycetes lineages. Instead, they were most likely inherited from ancient gene duplications before the M. populorum/M. populicola split, followed by a loss in the M. populicola lineage (Dataset S4). Eleven of these genes had significant similarities with bacterial genes, suggesting a possible ancient prokaryote origin (Dataset S4). Phylogenetic approaches showed that two genes potentially involved in cellulose degradation, including an Aryl-alcohol/glucose oxidase, were probably inherited in Dothideomycetes fungi through a horizontal gene transfer from a bacterial donor (SI Appendix, Fig. S5 and HGT of CAZy and AA genes). Additionally, one gene encoding a putatively secreted cutinase showed a clear footprint of HGT from a Basidiomycota donor to ascomycete fungi, including members of the genus Fusarium and M. populorum (SI Appendix, Fig. S5 A–D and HGT of CAZy and AA genes).
The two poplar pathogens also encode several homologs of ligninolytic enzymes and lytic polysaccharide mono-oxygenases [Auxiliary Activity (AA) enzyme families] (32), including peroxidases and glucose/methanol/choline oxidoreductases. More than half of those (61% in M. populorum and 63% in M. populicola) are predicted to be secreted (Dataset S3B). Secreted multicopper oxidases are considered to be “attack” enzymes, involved in melanin and lignin depolymerization, that contribute to the reduction of host lignification in response to pathogen infection (33). M. populicola has a similar content of secreted laccase genes as M. populorum, except for a gene encoding a vanillyl-alcohol oxidase (Dataset S3B). Within the Dothideomycetes genomes, genes encoding vanillyl-alcohol oxidases have been gained only by the canker pathogen M. populorum and the pine needle pathogen D. septosporum (Dataset S4) (14, 20). An additional difference between the two poplar pathogens is an enrichment in genes encoding chloroperoxidases with a total of 16 gene models in M. populorum vs. 10 in M. populicola (Dataset S3B). Several plant-pathogenic ascomycetes that also occur as leaf litter-inhabiting saprophytes use extracellular chloroperoxidases to catalyze lignin chlorination, whereby the major structures in lignin are not only chlorinated, but also cleaved by these enzymes (34).
Expression of the ligno-cellulolytic enzyme arsenal and carbon assimilation.
We observed a major difference in the gene expression profile of the two poplar pathogens that is correlated with the ability of M. populorum to partially degrade lignocellulose compounds and grow in woody tissues (Fig. 3A). Genes encoding carbohydrate active enzymes are more differentially expressed in M. populorum grown on the wood-chip medium compared with M. populicola. Twenty-one percent of the 214 genes encoding carbohydrate-active [including AA (32)] enzymes in M. populorum were significantly up-regulated during growth on wood-chip medium relative to the skim-milk and the leaf-extract media; only 7.5% were down-regulated (Fig. 3B). By contrast, only 4.2% of the 194 carbohydrate active enzymes were significantly up-regulated in M. populicola but 14.1% were down-regulated (Fig. 3B). Differential regulation thus appears to represent a key functional contrast between these two pathogens during colonization of wood.
Fig. 3.
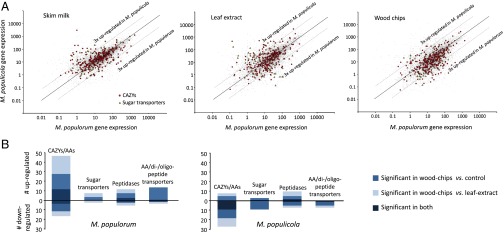
Comparison of gene expression in M. populorum and M. populicola during growth on an artificial medium enriched with skim milk (control), poplar leaf extract, and wood chips. (A) Log2-fold change ratio of the gene expression values between one-to-one orthologs of M. populorum and M. populicola (mean of samples from 1, 2, and 7 days of infection as replicates). Genes encoding carbohydrate active enzymes (CAZy) and sugar transporters are highlighted with red circles and yellow triangles, respectively. (B) Number of genes encoding CAZy, sugar transporters, peptidases, and nitrogen transporters that are significantly up- and down-regulated (with Cuffdiff q-value < 0.05) when M. populorum (Left) and M. populicola (Right) were grown on the wood-chip medium.
A large majority of up-regulated transcripts encode enzymes targeting the three most important carbohydrates in hardwood: cellulose, xylan, and pectin. The two genes encoding lytic polysaccharide mono-oxygenase enzymes (family AA9; formerly GH61) are strongly up-regulated when M. populorum was grown on the wood-chip medium (SI Appendix, Fig. S6). Enzymes of this AA9 family act in concert with classical cellulases to enhance lignocellulose degradation by oxidatively cleaving glycosidic bonds on the surface of cellulose (35). Activation of this partial cellulosic system in M. populorum is complemented by the up-regulation of full xylanolytic machinery. β-Xylosidases, α-arabinofuranosidases, and α-methylglucuronidases are part of the degradation machinery that is capable of the complete hydrolysis of xylan, the major polysaccharide component of hemicellulose accounting for 15–30% of the cell-wall content in poplar (3). The same cellulosic and xylanolytic systems in M. populicola were reduced (one β-xylosidase, one α-d-galactosidase, and one α-l-fucosidase missing) and not up-regulated under the wood-chip treatment (Datasets S3A and S4). In vitro measurement of the xylanase, β-glucosidase, and cellobiohydrolase activities using pNP-coupled glycosides confirmed the activation of these wood-degradation abilities in M. populorum grown on wood-chip medium compared with M. populicola (SI Appendix, Fig. S7 and Enzymatic Activities: Strains and Experiments). Up-regulation of genes encoding enzymes involved in cellulose, xylan, and pectin degradation correlates well with growth profiles on artificial media enriched for these complex molecules, as M. populorum grows better than M. populicola on media enriched with these products (SI Appendix, Fig. S8 and Growth Profiling on Artificial Media).
Hemicellulose and pectin accessory esterases are another set of overexpressed genes in M. populorum grown on wood-chip medium that may play an important role in the colonization of woody tissues. These enzymes are involved in xylan de-acetylation and include carboxylesterase, rhamnogalacturonan acetylesterase, and feruloyl esterase enzymes (Dataset S3A). Feruloyl esters are found in cell walls of poplar stem and branch tissues, forming complex cross-links between linear polysaccharide chains such as xylan, lignin, and structural proteins and act as a physical barrier hindering microbial degradation of polysaccharide polymers (36). Xylan esterases working in concert with xylanases are required for effective biodegradation of xylan (37). As observed in M. populorum, the coexpression of xylanases and deactylating esterases facilitates this canker-causing pathogen in the degradation of xylan as opposed to M. populicola.
Along with the differential up-regulation of plant cell-wall–degrading enzymes in M. populorum, transmembrane sugar transporter genes are also activated; these may be required for the assimilation of the products from the degradation activities (Fig. 3). Of the 49 sugar transporters found in the M. populorum genome, the transcription of 8 (16.3%) was significantly up-regulated during growth on the wood-chip treatment, whereas only 2 (4.1%) were significantly down-regulated (Fig. 3B). An opposite trend was observed for M. populicola, with only three sugar transporters (of 41; 7.3%) being significantly up-regulated and nine (21.9%) down-regulated (Fig. 3B).
The mobilization of starch from the host and the synthesis of fungal-specific products such as α-glucan were also up-regulated in the wood-chip treatment for M. populorum (SI Appendix, Stealth pathogenicity and the CAZy GH13 family). A gene cluster homologous to the α-glucan cluster found in other ascomycete fungi (38, 39) showed a tissue-dependent coexpression and a strong up-regulation in the canker pathogen M. populorum when grown on the wood-chips medium, whereas in M. populicola this cluster was up-regulated only on the leaf-extract medium (SI Appendix, Fig. S9 and Stealth pathogenicity and the CAZy GH13 family). This subtle difference in host tissue recognition between the two poplar pathogens also hints at an increased potential of M. populorum to interact with woody tissues.
Nitrogen assimilation via proteolysis.
Nitrogen constitutes an important limiting factor for fungal growth in woody tissues (40, 41). The unique protein degradation/assimilation pathway active in the canker pathogen M. populorum allows it to harvest nitrogen from wood, giving it access to a niche in a woody environment. Plant pathogen genomes are generally enriched in secreted trypsin and subtilisin-like peptidases (MEROPS subfamilies S01 and S08) that play a role in pathogenicity and direct degradation of plant cell-wall components, such as hydroxyproline-rich glycoproteins (42, 43). An underrepresentation of these subfamilies in M. populorum and M. populicola is balanced by an increase in tripeptidyl-peptidases (S53) and amino-peptidases (M28) (SI Appendix, Tables S5 and S6 and Peptidases). Another abundant peptidase family in the M. populorum and M. populicola genomes is the atypical secreted oligo-peptidases (M03B) (SI Appendix, Tables S5 and S6). These oligo-peptidases display a signal peptide, whereas the fungal M03B peptidases are generally not secreted and are actively involved in the degradation of oligo-peptides in the intracellular compartment (SI Appendix, Fig. S10A). Furthermore, genes encoding these atypical secreted oligo-peptidases are expressed by both M. populorum and M. populicola in the leaf-extract and wood-chip treatments with a high up-regulation pattern induced by the leaf-extract medium for M. populorum (SI Appendix, Fig. S10B). On the other hand, the nitrate and nitrite assimilation systems were down-regulated in M. populorum growing on wood-chip medium. This was counteracted by an up-regulation of the di- and tripeptidyl peptidases, which helps the fungus to derive N nutrition through the protein degradation pathway. However, the transcriptional control of the nitrate/nitrite assimilation pathway in M. populicola was not repressed during its interaction with the host tissues (SI Appendix, Fig. S10C). This shows that the usual path of nitrogen assimilation (nitrate/nitrite uptake) seen in a wide range of fungi (from saprophytic to endophytic and necrotrophic fungi) has been abandoned by M. populorum in favor of nitrogen uptake via the protein degradation pathway.
A similar trend was observed for transporters involved in small peptide and protein uptake (Fig. 3B). Both M. populorum and M. populicola show an expansion of oligo-peptide transporters and amino acid transporters that may be required for the assimilation of proteolysis degradation products (SI Appendix, Fig. S3E; Dataset S5). Peptide and amino acid transporter genes that are transcriptionally up-regulated by M. populorum on the wood-chip medium are likely to be involved in the transport of small peptides that are released by the action of peptidases induced under the same experimental conditions (SI Appendix, Table S7). This implies that M. populorum has the potential to activate a unique protein degradation/assimilation pathway during growth on wood-chip medium, suggesting a reliance on proteolysis for nutrition while growing in contact with wood.
Conclusions.
Our comparative genomic analyses show that relatively few genomic changes—acquisition of novel functions via HGT, gene family expansion, and gene expression reprogramming—could enable a tree pathogen to exhibit novel disease etiology. Genomic innovation through trans-specific exchange of genetic material is proposed as a significant driver of adaptation to infect woody tissues in the canker pathogen M. populorum. Gene arsenal dosage observed across all gene families examined suggests that after divergence from M. populicola a rapid reorganization of the protein machinery in M. populorum enabled it to use poplar woody tissues. This can have major impacts on tree domestication because it transforms an innocuous coevolved pathogen into one that causes severe mortality and economic damage to plantation forestry.
Materials and Methods
Fungal Strains, Genome Sequencing, and Annotation.
The M. populorum SO2202 (NCBI taxonomy ID: 692275) and M. populicola p0202b (NCBI taxonomy ID: 1136490) strains sequenced in this study were deposited in the culture collection of the Centraalbureau voor Schimmelcultures (CBS) (CBS_133246 and CBS_133247). The M. populorum genome was sequenced at the Joint Genome Institute on both the Roche 454 and Illumina Solexa platforms. The M. populicola genome was sequenced on the IBIS platform (Laval University) with Roche 454 technology.
Illumina RNA-Seq Analysis.
RNA-seq data were generated with an Illumina (GAII) sequence from poly(A+) mRNA. Treatments consisted of M. populorum and M. populicola grown on YNB-minimal liquid media (HIMEDIA) enriched with skim milk, poplar leaf extract, or wood chips. Fungal material was harvested 1, 2, and 7 d after inoculation. Poly(A+) mRNA extraction, library construction, and postrun analyses are described in SI Appendix, Illumina RNA-Seq Experiments.
Detailed materials and methods for all experiments and results are shown in SI Appendix and Datasets S1–S7.
Supplementary Material
Acknowledgments
This work was supported by the Genomic Research and Development Initiative of Natural Resources Canada, Genome Canada, and Genome BC Project 2112. The work conducted by the US Department of Energy Joint Genome Institute was supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231. E.M. was supported by a grant from The Netherlands Organization for Scientific Research and The Netherlands Genomics Initiative 93511035 (to R.P.d.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GeneBank database (accession nos. AIDU01000000, AEFD01000000, and SRX759472). The annotated genomes are available at genome.jgi-psf.org/Sepmu1 and genome.jgi-psf.org/Seppo1. Fungal cultures have been deposited in the CBS strains collection, www.cbs.knaw.nl/Collections/Biolomics.aspx?Table=CBS%20strain%20database (accession nos. CBS_133246 and CBS_133247).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424293112/-/DCSupplemental.
References
- 1.Michon G, De Foresta H. Agroforests: Pre-domestication of forest trees or true domestication of forest ecosystems? Neth J Agric Sci. 1997;45(4):451–462. [Google Scholar]
- 2.Boerjan W. Biotechnology and the domestication of forest trees. Curr Opin Biotechnol. 2005;16(2):159–166. doi: 10.1016/j.copbio.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Sannigrahi P, Ragauskas AJ, Tuskan GA. Poplar as a feedstock for biofuels: A review of compositional characteristics. Biofuels Bioprod Biorefining. 2010;4(2):209–226. [Google Scholar]
- 4.Newcombe G, Bradshaw HDJ. Quantative trait loci confering resistance in hybrid poplar to Septoria populicola, the cause of leaf spot. Can J Bot. 1996;26(11):1943–1950. [Google Scholar]
- 5.Waterman AM. Canker of hybrid poplar clones in the United States, caused by Septoria musiva. Phytopathology. 1946;35:148–156. [Google Scholar]
- 6.Feau N, Mottet M-J, Périnet P, Hamelin RC, Bernier L. Recent advances related to poplar leaf spot and canker caused by Septoria musiva. Can J Plant Pathol. 2010;32(2):122–134. [Google Scholar]
- 7.Weiland JE, Stanosz JC, Stanosz GR, Pathology P, Madison W. Prediction of long-term canker disease damage from the responses of juvenile poplar clones to inoculation with Septoria musiva. Plant Dis. 2003;87(12):1507–1514. doi: 10.1094/PDIS.2003.87.12.1507. [DOI] [PubMed] [Google Scholar]
- 8.Dos Santos AF, Machado EB, Stanosz GR, Smith DR. First report of Septoria musiva in poplar in Brazil. Trop Plant Pathol. 2010;35(1):52–53. [Google Scholar]
- 9.Ares A, Gutierrez L. Selection of poplar clones for the Lower Valley of the Colorado River, Argentina. Forestry. 1996;69(1):75–82. [Google Scholar]
- 10.Feau N, Hamelin RC, Bernier L. Attributes and congruence of three molecular data sets: Inferring phylogenies among Septoria-related species from woody perennial plants. Mol Phylogenet Evol. 2006;40(3):808–829. doi: 10.1016/j.ympev.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Zalasky H. Stem and leaf spot infections caused by Septoria musiva and Septoria populicola on poplar seedlings. Phytoprotection. 1978;59(1):43–50. [Google Scholar]
- 12.Eckenwalder JE. Systematics and evolution of Populus. In: Bradshaw H, Heilman PHT, editors. Biology of Populus and Its Implications for Management and Conservation. National Research Council Research Press; Ottawa: 1996. pp. 7–32. [Google Scholar]
- 13.Rajora OP, Zsuffa L. Allozyme divergence and evolutionary relationships among Populus deltoides, P. nigra, and P. maximowiczii. Genome. 1990;33(1):44–49. [Google Scholar]
- 14.Ohm RA, et al. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 2012;8(12):e1003037. doi: 10.1371/journal.ppat.1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parra G, Bradnam K, Ning Z, Keane T, Korf I. Assessing the gene space in draft genomes. Nucleic Acids Res. 2009;37(1):289–297. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duplessis S, et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci USA. 2011;108(22):9166–9171. doi: 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330(6010):1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 18.Tan KC, Ipcho SVS, Trengove RD, Oliver RP, Solomon PS. Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology. Mol Plant Pathol. 2009;10(5):703–715. doi: 10.1111/j.1364-3703.2009.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solomon PS, Tan K-C, Oliver RP. The nutrient supply of pathogenic fungi: A fertile field for study. Mol Plant Pathol. 2003;4(3):203–210. doi: 10.1046/j.1364-3703.2003.00161.x. [DOI] [PubMed] [Google Scholar]
- 20.de Wit PJGM, et al. The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. 2012;8(11):e1003088. doi: 10.1371/journal.pgen.1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blin K, et al. antiSMASH 2.0: A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41(Web Server issue):W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen B, Smedsgaard J, Frisvad JC. Penicillium expansum: Consistent production of patulin, chaetoglobosins, and other secondary metabolites in culture and their natural occurrence in fruit products. J Agric Food Chem. 2004;52(8):2421–2428. doi: 10.1021/jf035406k. [DOI] [PubMed] [Google Scholar]
- 23.Schümann J, Hertweck C. Molecular basis of cytochalasan biosynthesis in fungi: Gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. J Am Chem Soc. 2007;129(31):9564–9565. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- 24.Von Wallbrunn C, Luftmann H, Bergander K, Meinhardt F. Phytotoxic chaetoglobosins are produced by the plant pathogen Calonectria morganii (anamorph Cylindrocladium scoparium) J Gen Appl Microbiol. 2001;47(1):33–38. doi: 10.2323/jgam.47.33. [DOI] [PubMed] [Google Scholar]
- 25.Qin J-C, et al. Bioactive metabolites produced by Chaetomium globosum, an endophytic fungus isolated from Ginkgo biloba. Bioorg Med Chem Lett. 2009;19(6):1572–1574. doi: 10.1016/j.bmcl.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Liu G, et al. Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS ONE. 2013;8(2):e55185. doi: 10.1371/journal.pone.0055185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin R, LeBoldus JM. The infection biology of Sphaerulina musiva: Clues to understanding a forest pathogen. PLoS ONE. 2014;9(7):e103477. doi: 10.1371/journal.pone.0103477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiland JE, Stanosz GR. The histology of hybrid poplar clones inoculated with Septoria musiva. Plant Dis. 2007;91(12):1524–1530. doi: 10.1094/PDIS-91-12-1524. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Vallvé S, Romeu A, Palau J. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol Biol Evol. 2000;17(3):352–361. doi: 10.1093/oxfordjournals.molbev.a026315. [DOI] [PubMed] [Google Scholar]
- 30.Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2010;26(1):5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Richards TA, Leonard G, Soanes DM, Talbot NJ. Gene transfer into the fungi. Fungal Biol Rev. 2011;25(2):98–110. [Google Scholar]
- 32.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6(1):41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edens WA, Goins TQ, Dooley D, Henson JM. Purification and characterization of a secreted laccase of Gaeumannomyces graminis var. tritici. Appl Environ Microbiol. 1999;65(7):3071–3074. doi: 10.1128/aem.65.7.3071-3074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz-Bermúdez P, Srebotnik E, Hammel KE. Chlorination and cleavage of lignin structures by fungal chloroperoxidases. Appl Environ Microbiol. 2003;69(8):5015–5018. doi: 10.1128/AEM.69.8.5015-5018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beeson WT, Phillips CM, Cate JHD, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012;134(2):890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- 36.Gou J-Y, Park S, Yu X-H, Miller LM, Liu C-J. Compositional characterization and imaging of “wall-bound” acylesters of Populus trichocarpa reveal differential accumulation of acyl molecules in normal and reactive woods. Planta. 2008;229(1):15–24. doi: 10.1007/s00425-008-0799-9. [DOI] [PubMed] [Google Scholar]
- 37.Gou J-Y, et al. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell. 2012;24(1):50–65. doi: 10.1105/tpc.111.092411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujikawa T, et al. Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 2012;8(8):e1002882. doi: 10.1371/journal.ppat.1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marion CL, Rappleye CA, Engle JT, Goldman WE. An alpha-(1,4)-amylase is essential for alpha-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol Microbiol. 2006;62(4):970–983. doi: 10.1111/j.1365-2958.2006.05436.x. [DOI] [PubMed] [Google Scholar]
- 40.Bolton MD, Thomma BPHJ. The complexity of nitrogen metabolism and nitrogen-regulated gene expression in plant pathogenic fungi. Physiol Mol Plant Pathol. 2008;72(4-6):104–110. [Google Scholar]
- 41.Dickson RE. Carbon and allocation in trees. Ann Sci. 1989;46(suppl.):631s–647s. [Google Scholar]
- 42.Carlile AJ, et al. Characterization of SNP1, a cell wall-degrading trypsin, produced during infection by Stagonospora nodorum. Mol Plant Microbe Interact. 2000;13(5):538–550. doi: 10.1094/MPMI.2000.13.5.538. [DOI] [PubMed] [Google Scholar]
- 43.Di Pietro A, et al. Molecular characterization of a subtilase from the vascular wilt fungus Fusarium oxysporum. Mol Plant Microbe Interact. 2001;14(5):653–662. doi: 10.1094/MPMI.2001.14.5.653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.