Significance
Understanding emerging disease origins is important to gauge future human infection risks. This is particularly true for the various forms of the AIDS virus, HIV-1, which were transmitted to humans on four independent occasions. Previous studies identified chimpanzees in southern Cameroon as the source of the pandemic M group, as well as the geographically more restricted N group. Here, we show that the remaining two groups also emerged in southern Cameroon but had their origins in western lowland gorillas. Although group P has only been detected in two individuals, group O has spread extensively throughout west central Africa. Thus, both chimpanzees and gorillas harbor viruses that are capable of crossing the species barrier to humans and causing major disease outbreaks.
Keywords: AIDS, HIV-1, gorilla, SIVgor, zoonotic transmission
Abstract
HIV-1, the cause of AIDS, is composed of four phylogenetic lineages, groups M, N, O, and P, each of which resulted from an independent cross-species transmission event of simian immunodeficiency viruses (SIVs) infecting African apes. Although groups M and N have been traced to geographically distinct chimpanzee communities in southern Cameroon, the reservoirs of groups O and P remain unknown. Here, we screened fecal samples from western lowland (n = 2,611), eastern lowland (n = 103), and mountain (n = 218) gorillas for gorilla SIV (SIVgor) antibodies and nucleic acids. Despite testing wild troops throughout southern Cameroon (n = 14), northern Gabon (n = 16), the Democratic Republic of Congo (n = 2), and Uganda (n = 1), SIVgor was identified at only four sites in southern Cameroon, with prevalences ranging from 0.8–22%. Amplification of partial and full-length SIVgor sequences revealed extensive genetic diversity, but all SIVgor strains were derived from a single lineage within the chimpanzee SIV (SIVcpz) radiation. Two fully sequenced gorilla viruses from southwestern Cameroon were very closely related to, and likely represent the source population of, HIV-1 group P. Most of the genome of a third SIVgor strain, from central Cameroon, was very closely related to HIV-1 group O, again pointing to gorillas as the immediate source. Functional analyses identified the cytidine deaminase APOBEC3G as a barrier for chimpanzee-to-gorilla, but not gorilla-to-human, virus transmission. These data indicate that HIV-1 group O, which spreads epidemically in west central Africa and is estimated to have infected around 100,000 people, originated by cross-species transmission from western lowland gorillas.
AIDS is caused by HIV-1 and HIV-2, which are derived from a clade of lentiviruses [simian immunodeficiency viruses (SIVs)] found naturally in more than 40 species of nonhuman primates in sub-Saharan Africa (1, 2). These SIVs mostly fall into host-specific clades, but they have occasionally jumped species and spread successfully in new hosts. Of particular interest, chimpanzees (Pan troglodytes) acquired two distinct lineages of SIV from two different monkey species; all known strains of chimpanzee SIV (SIVcpz) are derived from a hybrid formed by recombination between these two viruses (3). The spread of this virus in chimpanzees appears to have occurred comparatively recently, because only two closely related subspecies in Central Africa are infected, whereas two other subspecies are not (4–8). Subsequently, strains of SIVcpz from one chimpanzee subspecies (Pan troglodytes troglodytes) have been subject to further transmission both to humans, leading to HIV-1, and to western gorillas (Gorilla gorilla), giving rise to gorilla SIV (SIVgor) (4, 9). The limited number of strains of SIVgor characterized so far form a single clade, but HIV-1 strains fall into four phylogenetically distinct groups, each of which must reflect a separate cross-species transmission from apes (1). These four zoonotic events have had very different outcomes. One gave rise to group M, the cause of the AIDS pandemic, which has infected more than 40 million people and spread across Africa and throughout the rest of the world. At the other extreme, group N and P viruses have only been found in small numbers of individuals from Cameroon: group N in fewer than 20 individuals (10) and group P in only two individuals (11, 12). Group O, although not nearly as prevalent as group M, has nonetheless caused a substantial epidemic. Although largely restricted to west central Africa, group O viruses have spread through Cameroon, Gabon, Nigeria, and other neighboring countries, and are estimated to have infected about 100,000 individuals (13, 14).
Molecular epidemiological studies of SIVcpz have shown that these viruses exhibit phylogeographic clustering, apparently largely due to major rivers and other barriers that limit the migration of wild chimpanzees (4–6). This clustering has allowed us to pinpoint the probable geographic origins of two of the human virus clades (4). Thus, strains of SIVcpz very closely related to HIV-1 group M have been found only in a small area in the southeast corner of Cameroon, implicating that region as the likely location of the chimpanzee-to-human transmission that gave rise to the AIDS pandemic. Similarly, strains of SIVcpz very closely related to HIV-1 group N have been found only in south-central Cameroon, pointing to chimpanzees in the Dja Forest as the source of this viral lineage. However, much less is known about the origins of the two other HIV-1 groups. Group O viruses are more closely related to SIVgor than to SIVcpz, but it is unclear whether the immediate precursor to the human viruses infected chimpanzees or gorillas (1, 9, 15). Group P viruses are even more closely related to strains of SIVgor (11, 12), indicating that they probably arose from a gorilla-to-human transmission, but the geographic location has not been defined.
Given the chimpanzee origin of pandemic HIV-1, previous studies have focused almost exclusively on characterizing SIVcpz in wild-living chimpanzees (4–7). To gain greater insight into the molecular epidemiology of SIVgor, and to learn more about the origins of HIV-1 groups O and P, we conducted extensive surveys of wild living gorillas. We screened western lowland gorillas across a large part of their geographic range in southern Cameroon and Gabon, and also sampled both lowland and mountain subspecies of the eastern gorilla (Gorilla beringei). Although eastern and western gorillas likely diverged before the origin of SIVgor, eastern gorillas may have independently acquired SIV, because they share their habitat with the second chimpanzee subspecies, the eastern chimpanzee (Pan troglodytes schweinfurthii), which is widely and commonly infected with SIVcpz (6). Finally, to gain insight into potential barriers to cross-species transfers, we investigated the role of the host restriction factor apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G) in inhibiting ape-to-ape, as well as ape-to-human, SIV transmission.
Results
SIVgor Infection Is Restricted to Western Lowland Gorillas in Southern Cameroon.
Previous molecular epidemiological studies of SIVgor were limited to field sites in Cameroon and the Democratic Republic of Congo (DRC) (16). To determine the prevalence and geographic distribution of this infection in western gorillas (G. gorilla), we collected additional fecal samples from southern Cameroon (n = 1,696) and extended our survey to 16 field sites in Gabon (n = 915) (Fig. 1 and Table S1). We also sampled eastern lowland gorillas (Gorilla beringei graueri) in the DRC (n = 103) and tested two different communities of mountain gorillas (G. beringei beringei) in the DRC and Uganda (n = 218). All fecal samples were examined for the presence of HIV cross-reactive antibodies using the INNO-LIA HIV I/II score confirmation test (Innogenetics), which contains HIV-1 and HIV-2 recombinant proteins and synthetic peptides coated as discrete lines on a nylon strip. This test was previously shown to detect SIVgor infection with greater than 90% sensitivity (16, 17) and has even uncovered more divergent SIV lineages from other nonhuman primate species (2). Of a total of 2,932 gorilla fecal samples tested, 70 reacted with at least one HIV-1 antigen. These samples came from four field sites, all located in southern Cameroon. Samples from the remaining 10 field sites in Cameroon and all sites in Gabon were INNO-LIA–negative, as were all samples from both subspecies of eastern gorillas.
Fig. 1.
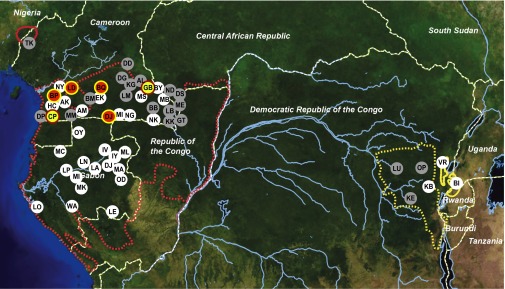
Geographic distribution of SIVgor in wild-living gorillas. Field sites are shown in relation to the ranges of western (G. gorilla, red) and eastern (G. beringei, yellow) gorillas, with subspecies indicated by broken (G. g. gorilla and G. b. graueri) and solid (G. g. diehli and G. b. beringei) lines. Forested areas are shown in dark green, whereas arid and semiarid areas are depicted in yellow and brown, respectively. Major lakes and rivers are shown in blue. Dashed white lines indicate national boundaries. Sites where SIVgor was detected in this study are highlighted in red (yellow border), with SIVgor-positive field sites reported previously shown in yellow (red border) (9, 16). White and gray circles indicate SIVgor-negative sites identified in this and previous studies, respectively (9, 16).
Analysis of 12S mitochondrial DNA sequences confirmed that the 70 antibody-positive fecal samples were all from western gorillas. Eighteen of these samples were too degraded to allow microsatellite analyses, but the remaining specimens represented 16 different individuals. Although the majority of INNO-LIA–positive samples cross-reacted with HIV-1 gp41 and/or p24 antigens, a subset exhibited strong cross-reactivity with all five HIV-1 antigens (Fig. 2). Because there was no evidence of false-positive reactivity, we tentatively classified all gorilla samples that reacted with at least one INNO-LIA antigen as SIVgor antibody-positive.
Fig. 2.
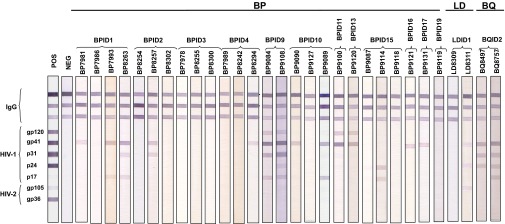
Detection of SIVgor antibodies in gorilla fecal samples. INNO-LIA banding patterns are shown, with molecular weight markers of HIV-1 and HIV-2 proteins indicated. IgG control lanes (n = 3) are shown on the top of each strip; plasma from HIV-1–infected (POS) and uninfected (NEG) humans were used for controls (two left lines). Fecal samples are grouped by individuals, with a two-letter code indicating the collection site of origin (Fig. 1).
Across the four sites with evidence of SIVgor infection, there was substantial variation in the number and proportion of INNO-LIA–positive samples. For example, at site BP, 48 (30%) of 161 samples were antibody-positive, corresponding to at least 10 infected individuals. In contrast, at site LD, located 50 km east of site BP, only a single antibody-positive sample was detected among almost 150 specimens tested. At site BQ, only nine (2%) of 435 samples were antibody-positive, and these samples were all from a single individual. Of note, this animal was not the same gorilla (BQ664) in which SIVgor infection was detected in 2004 (9). At site DJ, 12 (5%) of 237 samples were positive, corresponding to four gorillas. We estimated the prevalence of SIVgor infection for each field site based on the proportion of SIVgor-positive gorillas, but correcting for repeated sampling. Screening over 1,100 western lowland gorillas, we estimated an overall prevalence of 1.6% (95% confidence interval: 1.0–2.5%), ranging from less than 1% to over 20% at the four positive field sites (Table S1).
Field observations at site BP indicated that some of the samples came from members of two social groups. One group likely comprised at least 12 individuals based on nest counts, although genotyping identified only eight sampled gorillas, three of whom were SIVgor-positive. The second group consisted of at least 17 individuals based on nest counts and 16 individuals based on microsatellite analyses, seven of whom were antibody-positive. Thus, within each of these two social groups, around 40% of individuals were infected with SIVgor. At least one of these individuals (BP-ID4) may have represented a newly acquired infection, because among 12 samples collected over a 4-mo period, only the last one was positive (Table S2). Interestingly, 31 additional individuals from site BP, which appeared not to belong to either of these social groups, were all SIVgor-negative. At the other field sites where antibody-positive individuals were found (sites BQ, LD, and DJ), samples were collected mainly opportunistically and around feeding sites; thus, the size of the sampled social groups could not be estimated.
In addition to antibody detection, we tested all INNO-LIA–positive and –negative specimens from the same nesting and/or feeding site for the presence of SIVgor viral RNA using a real-time quantitative PCR (RT-qPCR) assay (18). Viral RNA was detected in 33 (58%) of 57 antibody-positive samples but also in 15 (7.4%) of 204 antibody-negative samples. In these specimens, SIVgor RNA copy numbers ranged from a few copies to more than 1,000 copies per milliliter of RNAlater-preserved (1:1 mixture; Ambion) fecal samples. Interestingly the RT-qPCR assay identified two additional SIVgor-infected gorillas, both of which were members of one of the two high-prevalence social groups at the BP site (Table S2). These individuals were likely sampled during acute infection before the onset of antibody responses, because 236 other INNO-LIA–negative samples collected both at SIVgor-positive (LD, n = 42; BQ, n = 69; CP, n = 37; DJ, n = 12) and SIVgor-negative (AK, n = 12; AM, n = 12; BY, n = 12; DD, n = 4; EK, n = 12; MB, n = 12; NY, n = 12) field sites (Fig. 1) were all RT-qPCR–negative.
High Genetic Diversity of SIVgor.
To examine the genetic diversity of SIVgor at the various collection sites, we used nested PCR to amplify viral sequences from antibody and/or RT-qPCR–positive samples. These analyses yielded SIVgor core protein (gag), polymerase (pol) and envelope (env) gene sequences from seven of the 16 antibody-positive gorillas, as well as from the two antibody-negative but RT-qPCR–positive animals (Table S2). Attempts to amplify viral sequences from the other INNO-LIA–positive individuals, including the single gorilla at site LD, were repeatedly unsuccessful (Table S2). Phylogenetic analyses showed that all of the newly identified SIVgor sequences were more closely related to each other, and to previously characterized SIVgor strains, than to SIVcpz (Fig. 3), indicating a single chimpanzee-to-gorilla transmission at the origin of the SIVgor lineage.
Fig. 3.
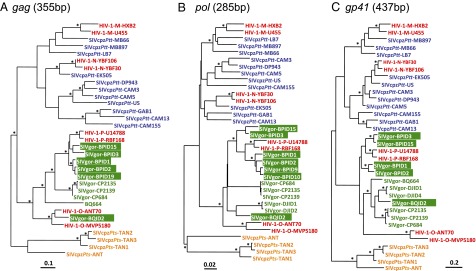
Evolutionary relationships across SIVcpz, SIVgor, and HIV-1 strains based on partial gene sequences. Phylogenetic trees were constructed using partial gag (A), pol (B), and env (C) sequences. Newly identified SIVgor strains (highlighted in green boxes) are compared with previously characterized SIVgor (green), SIVcpzPtt (blue), SIVcpzPts (orange), and HIV-1 (red) strains. Asterisks above branches correspond to nodes supported by bootstrap values over 70% from ML analyses and posterior probabilities over 0.90 from Bayesian analyses. (Scale bar: number of substitutions per site.)
Within the SIVgor clade, there was evidence of phylogeographic clustering; for example, all seven viruses from site BP formed a distinct clade as did previously characterized strains from sites CP and DJ. Interestingly, the two HIV-1 group P strains fell within the radiation of strains from the BP site, whereas the newly derived SIVgor strain from site BQ was closely related to HIV-1 group O in gag, but not in other genomic regions (Fig. 3).
Full-Length Genome Sequencing of SIVgor Strains Closely Related to HIV-1 Groups O and P.
To study the phylogenetic relationships of the SIVgor strains most closely related to HIV-1 groups O and P, we subjected three samples to whole-genome analysis. The concatenated fecal consensus sequences of SIVgor-BPID1, SIVgor-BPID15, and SIVgor-BQID2 were 9,029 bp, 9,012 bp, and 9,241 bp in length, respectively. All three viruses exhibited the same genomic organization as other members of the HIV-1/SIVcpz lineage, encoding a viral protein U (vpu) gene and nonoverlapping env and negative regulatory factor (nef) genes. Phylogenetic analyses of deduced Gag, Pol, and concatenated Env/Nef protein sequences showed that the two fully sequenced BP site strains, as well as two other BP site viruses (SIVgor-BPID2 and SIVgor-BPID3) for which Gag and/or Pol sequences were available, were very closely related to HIV-1 group P (Fig. 4). In contrast to the analyses based on short genomic fragments (Fig. 3), the single site BQ strain (SIVgor-BQID2) was found to cluster with HIV-1 group O in all three trees derived from full-length protein sequences (Fig. 4).
Fig. 4.
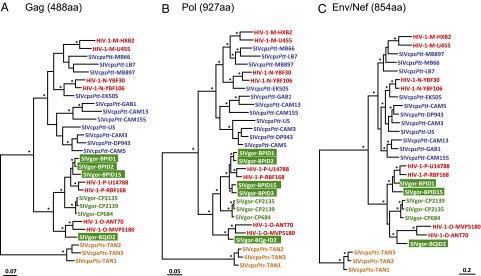
Evolutionary relationships across SIVcpz, SIVgor, and HIV-1 strains based on full-length protein sequences. Phylogenetic trees were constructed using complete Gag (A), Pol (B), and concatenated Env and Nef (C) protein sequences. Full-length genome sequences were obtained for SIVgor-BPID1, SIVgor-BPID15, and SIVgor-BQID2; complete gag and pol sequences were obtained for SIVgor-BPID2; and a complete pol sequence was obtained for SIVgor-BPID3. Other details are provided in Fig. 3.
The discordant positions of BQID2 in phylogenies derived from different genomic regions suggested a recombinant history. To examine the full-length sequences for evidence of recombination, the newly derived SIVgor sequences were compared with previously reported HIV-1, SIVcpz, and SIVgor genome sequences using similarity plot and bootstrap analyses for successive genomic regions, scanning along the alignment. SIVgor strains BPID1 and BPID15 consistently clustered with each other and with the two HIV-1 group P strains, across the entire genome (Fig. 5). The extent of divergence between these SIVgor and HIV-1 strains (i.e., 9.2%, 5.6%, and 18% in Gag, Pol, and Env proteins, respectively) is similar to the distances observed between HIV-1 groups M and N and their respective closest SIVcpzPtt relatives (4, 5). In contrast, the position of the BQID2 strain varied substantially and significantly among phylogenies derived from different genomic regions; recombination analyses suggested seven distinct regions, reflecting at least three, or perhaps four, different evolutionary histories (Fig. 5). For more than 70% of the genome, including gag, parts of pol (encoding the protease, the reverse transcriptase, and part of the integrase), the accessory genes (vif, vpr, and vpu), the 3′ end of env (gp41), and the nef gene, BQID2 was closely related to HIV-1 group O (Fig. 5A; trees a, c, and g). For a short region within pol, and for much of env, BQID2 clustered with other SIVgor strains, with the precise relationships varying among regions (Fig. 5A; trees b and d–f). These results indicate that the BQID2 genome represents a complex recombinant of multiple diverse SIVgor lineages, one of which is very closely related to HIV-1 group O. We calculated the overall genetic distance of HIV-1 group O from SIVgor-BQID2 by examining only genomic regions that clustered. The distances (0.161–0.172) were similar to those distances observed for the same genomic regions between HIV-1 group M (0.249–0.268) or group N (0.156–0.156) and their closest SIVcpz relatives.
Fig. 5.
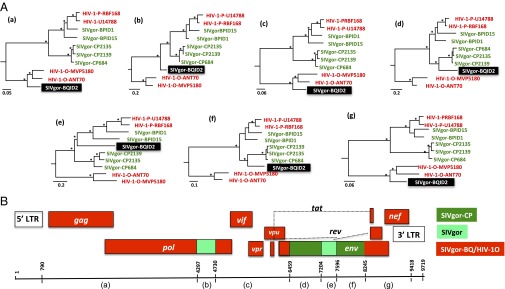
Mosaic genome structure of SIVgor-BQID2. (A) Phylogenies are shown for seven genomic regions (a–f) indicated below the genome diagram in B. Genomic regions in which SIVgor-BQID2 clusters with HIV-1 group O, or with SIVgor from site CP, are shown in red and dark green, respectively. (B) In two other regions (light green), BQID2 is not specifically closely related to other strains. Bootstrap values over 70% from ML analyses are shown with an asterisk. (Scale bar: number of substitutions per site.)
Overall, the analysis of full-length SIVgor genomes provided evidence for three distinct SIVgor clades: one comprising viruses from site CP in southwest Cameroon, a second comprising viruses from site BP in western Cameroon, and a third represented by the virus from site BQ in south-central Cameroon. The close relationships of HIV-1 groups P and O to the BP and BQ strains of SIVgor, respectively, indicate that gorillas were the source of both HIV-1 clades, and point to likely geographic locations of the cross-species transmission events.
Resistance of Gorilla APOBEC3G to SIVcpz Vif-Mediated Degradation.
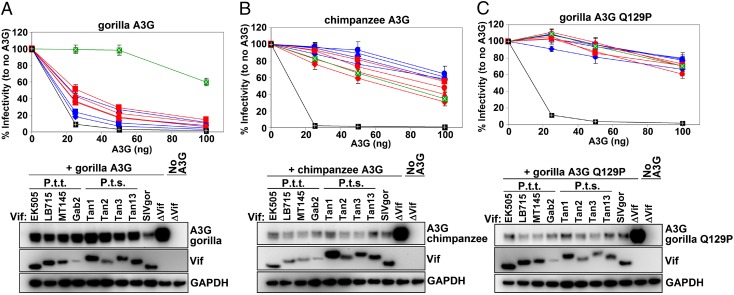
Although each of the four groups of HIV-1 resulted from a different cross-species transmission event, all known SIVgor strains, including the nine strains newly characterized here, form a monophyletic lineage within the SIVcpz radiation. Thus, SIVgor appears to have resulted from a single initial transmission event, which is surprising, given that central chimpanzees and western gorillas are sympatric over a large area and SIVcpz infection is widespread among central chimpanzees (4, 5). Furthermore, although SIVcpz is similarly common among eastern chimpanzees (6), we have found no evidence of transmission to sympatric eastern gorillas. This raised the question of whether host restriction factors are limiting chimpanzee-to-gorilla transmission. One potential restriction factor is APOBEC3G (A3G), which is normally counteracted by the Vif protein of HIV-1 and SIVcpz, leading to its degradation. However, we had previously observed that a Pro (P)-to-Gln (Q) change at site 129 in the gorilla A3G protein confers resistance to Vif-mediated degradation by some HIV-1 and SIVcpz strains (19). To examine this result further, we tested the anti-A3G activity of a larger panel of Vif proteins from both SIVcpzPtt (from P. t. troglodytes) and SIVcpzPts (from P. t. scheinfurthii) strains. Specifically, we transfected 293T cells with a Vif-minus (NL4-3 ΔVif) HIV-1 molecular clone, as well as increasing amounts of WT chimpanzee (129P), WT gorilla (129Q), or mutant gorilla (Q129P) A3G-expressing plasmids in the presence of different SIVcpz and SIVgor Vif variants, and then determined the effect on viral infectivity and A3G degradation (Fig. 6). The results show that only SIVgor Vif was able to counteract gorilla A3G efficiently, whereas the Vif proteins from eight different SIVcpz strains were inactive or only minimally active (Fig. 6A). This was the case despite the fact that these same SIVcpz Vif proteins were fully active against the chimpanzee A3G (Fig. 6B). Most importantly, replacing the Q at position 129 with a P rendered the gorilla A3G protein completely sensitive to all Vif variants tested (Fig. 6C), indicating that the difference in susceptibility was controlled by a single residue. Western blots of transfected 293T cell lysates confirmed these results, showing that the gorilla A3G was only efficiently degraded in the presence of the SIVgor Vif, and not the various SIVcpz Vif variants (Fig. 6A, Right), whereas chimpanzee and mutant gorilla A3G was efficiently degraded by all Vif proteins (Fig. 6 B and C, Right). Using a much larger set of SIVcpz strains, we thus confirmed that gorilla A3G is resistant to SIVcpz Vif-mediated degradation, resulting in potent viral restriction. To ensure that the gorilla gene used for the analyses in Fig. 6 did not represent an unusual A3G variant, we used PCR to amplify exon 3 of the A3G gene from four additional SIVgor-positive fecal samples (one from each of the BP, BQ, DJ, and CP sites; Fig. 1). The results revealed that the four SIVgor-infected gorillas were all homozygous for a Q codon at position 129 of their A3G gene.
Fig. 6.
Gorilla A3G is resistant to degradation by SIVcpz but not SIVgor Vif proteins. Plasmids expressing vif genes from SIVcpzPtt (red), SIVcpzPts (blue), and SIVgor (green) were cotransfected with increasing quantities (0, 25, 50, and 100 ng) of WT gorilla A3G (A), WT chimpanzee A3G (B), and the gorilla A3G mutant Q129P (C), along with a vif-deficient HIV-1 molecular clone (NL4-3 ΔVif) into 293T cells. Two days after transfection, viral supernatants were used to infect TZM-bl reporter cells (a HeLa-derived cell line that constitutively expresses CD4, CCR5, and CXCR4 cell surface receptors) and infectious particle release was measured. (Upper) Virus infectivity (y axis) plotted in the presence of increasing quantities (x axis) of the various Vif expression plasmids (infectivity in the absence of Vif is shown in black) is depicted. Values represent averages (with SDs) from three different transfections. (Lower) Western blots of the corresponding 293T cell lysates (transfected with 50 ng of A3G), probed for A3G and Vif expression (GAPDH represents the loading control), are depicted.
Discussion
Cross-species transmissions of SIVcpz from chimpanzees have given rise to SIVgor in gorillas and HIV-1 in humans. Comparatively little is known about SIVgor; thus, we investigated its prevalence, host range, geographic distribution, and genetic diversity. Our results indicate that SIVgor is much less common and widespread than SIVcpz in chimpanzees, with infection restricted to a small fraction of communities of western lowland gorillas in southern Cameroon. Moreover, there is only evidence for a single chimpanzee-to-gorilla transmission event. Nonetheless, SIVgor strains exhibit considerable genetic diversity, indicating that this virus has infected gorillas for longer than HIV-1 has infected humans. Most interestingly, among the newly characterized strains, we identified two distinct SIVgor lineages that are very closely related to HIV-1 groups O and P, indicating that gorillas were the immediate source of both of these human viruses.
SIVgor in Gorillas Is Less Common and Widespread than SIVcpz in Chimpanzees.
Gorillas are classified into two species, eastern and western, that are estimated to have split about 100,000 y ago (20); both comprise two subspecies: western lowland (Gorilla gorilla gorilla) and Cross River (Gorilla gorilla diehli) gorillas in west-central Africa and eastern lowland (G. beringei graueri) and mountain (G. b. beringei) gorillas in east-central Africa (21). Eastern and western lowland gorillas are sympatric with eastern and central chimpanzees, respectively, and both chimpanzee subspecies are commonly and widely infected with SIVcpz. However, we have found evidence of SIVgor infection only in western lowland gorillas. We tested more than 100 (n = 115) mountain gorillas at two different field sites (Table S2) from a population that is thought to number now only about 900 individuals (22), and we found all to be SIVgor-negative. Similarly, we found no evidence of SIVgor infection in eastern lowland gorillas in this and a previous study (16). Finally, a previous survey of Cross River gorillas in northwest Cameroon also failed to identify SIVgor infection, although only a small number of samples were examined (16). Thus, combining all data, we have screened a total of 5,793 samples from ∼2,500 western and 250 eastern gorillas at 55 different field sites, covering a large proportion of the geographic range of both species (Fig. 1); yet, we identified SIVgor-positive gorillas only at six sites, all located in southern Cameroon.
Overall, we found fewer than 2% of western lowland gorillas to be infected with SIVgor, which is much lower than the estimates of SIVcpz infection rates for central (6%) or eastern (13%) chimpanzees (4, 6, 16). Although it is possible that some fecal samples were too degraded to yield positive results, this possibility alone cannot explain the low SIVgor detection rates in western gorillas or the lack of infection in eastern gorillas. The INNO-LIA assay is highly cross-reactive and has detected antibodies from divergent SIV infections in the past (2). Thus, it is unlikely that lack of cross-reactivity resulted in false-negative results for large numbers of gorillas, including individuals that may have acquired more divergent SIVcpzPts strains. Indeed, in a previous study (16), we used a Western blot approach that detects both SIVcpzPtt and SIVcpzPts infections with high (92%) sensitivity (4, 6) to screen eastern gorillas and also failed to detect SIVgor infection. Finally, 236 INNO-LIA–negative samples from four sites where SIVgor was detected, as well as from seven sites where SIVgor was absent, were all RT-qPCR–negative. Thus, it is unlikely that methodological differences or technical shortcomings have led to a gross underestimation of the prevalence and distribution of SIVgor infections.
As previously shown for SIVcpz-infected chimpanzees, SIVgor infection rates varied considerably among field sites, with some gorilla communities reaching prevalences of 20% or higher (16, 17). Moreover, within two nesting groups, up to 40% of group members were infected, indicating efficient spread between individuals belonging to the same social unit. Interesting, gorillas from the same field site (BP) that did not belong to these social groups, were not SIVgor-infected, suggesting that group interactions (or lack thereof) play an important role in intercommunity virus spread. In general, the observations of varying prevalence rates between different primate species is not surprising; for example, at least 50% of sooty mangabeys (Cercocebus atys), red-capped mangabeys (Cercocebus torquatus), and western red colobus monkeys (Piliocolobus badius) are infected with SIV, compared with only about 1% of greater spot-nosed monkeys (Cercopithecus nictitans) or mustached monkeys (C. cephus) (2).
SIVgor Infection in Gorillas Is Restricted to Southern Cameroon.
Although high prevalence rates within certain gorilla social groups may reflect transmissions from acutely infected mating partners, who are likely more infectious than chronically infected individuals (23), the geographic distribution of SIVgor is nevertheless puzzling. Confirmed infections in western lowland gorillas are spread over a distance of at least 400 km but are restricted to the northern part of their range in southern Cameroon (Fig. 1). It is unclear whether the current distribution is related to geographic barriers or whether it is perhaps the consequence of an overall population decline. Over recent decades, the number of wild-living gorilla populations, some of which may have harbored SIVgor, has declined rapidly because of increased human presence, hunting, and habitat loss due to deforestation, as well as infectious disease outbreaks, including Ebola (24, 25). It is also possible that SIVgor is pathogenic in gorillas, because SIVcpz infection is in chimpanzees (23, 26). Thus, SIVgor could have once been present across a larger geographic area but may have led to the extinction of certain gorilla populations. SIVcpz has been shown to have a substantive negative impact on the health, reproduction, and survival of chimpanzees in the wild, and has caused the decline of at least one chimpanzee community (25, 27). Thus, studies on additional infected and uninfected gorilla populations are required to determine the impact of SIVgor on gorilla survival (17). Alternatively, it is possible that following its introduction, SIVgor may have not yet spread beyond southern Cameroon. The home ranges of gorilla groups are small compared with chimpanzees, and their movements, as inferred from gene flow, are influenced by both major and smaller rivers, perhaps more so than the migration of chimpanzees (28–30). It has been estimated that SIVgor sequence distances reflect at least 100–200 y of diversification (15). The date of the most recent common ancestor of SIVgor could be much older than this estimate, given the apparent time dependence of lentivirus molecular clocks (31, 32). Nonetheless, it is possible that the introduction of SIVgor into gorillas has been too recent for this infection to have spread to a larger geographic area.
SIVgor Resulted from a Single Introduction of SIVcpz from Sympatric Chimpanzees.
All newly characterized SIVgor sequences fall within the previously identified radiation of SIVgor strains, indicating that SIVgor has emerged only once. Chimpanzees and gorillas have overlapping habitats and often feed in the same fruit trees (33–35). Sharing the same habitat leads to direct and indirect contacts, which have resulted in the cross-species transmission of other pathogens, such as the agents of anthrax, Ebola, and hepatitis B (15, 36–38). Transmission of SIV would require physical encounters between the two species, possibly involving biting or other contact with infected blood or body fluids. Although such incidences have not been reported, they would need to occur only rarely to allow SIV transmission. We thus reasoned that other factors are likely responsible for the apparently low rate of transmission between these sympatric species. Previous studies have highlighted the importance of host restriction factors as a barrier to SIV cross-species transmission (39). Among these host restriction factors, A3G has been reported to be one of the most effective (40–42). In fact, gorilla A3G is resistant to HIV-1 and SIVcpz Vif proteins (Fig. 6), providing a possible explanation for why SIVcpz has not crossed between chimpanzees and gorillas more often. Although all gorillas we have examined are homozygous at the site in A3G conferring resistance to SIVcpz, extensive A3G sequence diversity has been observed in other primate species (e.g., rhesus macaques, sooty mangabeys, African green monkeys) (40, 41). A3G polymorphism could render certain individuals more susceptible to transmissions of SIVcpz, allowing the virus to gain a foothold in gorillas (19). It is also possible that the spread of SIVgor selected for an increased frequency of the A3G 129Q mutation in gorillas, but we note that 129Q is also found in eastern gorillas (20), which are not naturally SIVgor-infected, suggesting that this mutation spread before the divergence of the two species.
SIVgor from Gorillas Is the Precursor of HIV-1 Groups O and P in Humans.
A total of 56 SIVgor-infected gorillas have now been identified in this and previous studies. Partial env or pol sequences have been obtained for about half of them (9, 16, 17), although only small subgenomic fragments were amplified for most cases. Nonetheless, available sequences indicate greater similarity among SIVgor strains from the same field site than among strains from different field sites. Phylogeographic clustering was particularly evident at the BP site, where all seven viruses form a distinct clade (Fig. 3). HIV-1 group P viruses fall within the radiation of the SIVgor strains from site BP, strongly suggesting that this rare form of HIV-1 originated in this region of western Cameroon.
The newly characterized SIVgor strain from site BQ resolves the question of whether HIV-1 group O resulted from a chimpanzee-to-human or gorilla-to-human transmission (1). SIVgor-BQID2 is closely related to HIV-1 group O viruses across most of its genome, and its phylogenetic position (Fig. 5) indicates that this lineage was transmitted from chimpanzees to gorillas before the onward transmission to humans. The mosaic nature of the SIVgor-BQID2 genome indicates that at some point in the past, prevalence rates must have been sufficiently high to allow coinfection of the same individual with diverse SIVgor lineages. Although we cannot be precise about the location of the source population of HIV-1 group O, we can conclude that gorillas transmitted SIVgor to humans on at least two occasions.
SIVgor Has Adapted to Spread Efficiently in the Human Population.
The four HIV-1 groups have different virological and epidemiological histories. Only HIV-1 group M spread globally and is responsible for the current HIV-1 pandemic. Infections with group N and P strains are very rare and largely restricted to Cameroon (43), but group O is the second most widespread HIV-1 lineage. Group O viruses, which currently account for up to 1% of all HIV-1 infections in Cameroon (44), also spread to other countries, including some outside of Africa. Indeed, the earliest (albeit retrospectively) documented AIDS cases in Europe were identified in a Norwegian sailor and his family, who became infected in the 1960s with viruses later shown to be group O (45). Group O infections are most prevalent in west and central Africa. In addition to Cameroon, group O viruses have been found in Chad, Gabon, Niger, Nigeria, Senegal, and Togo (13). In Nigeria, group O reactive sera were detected in about 1% of HIV-positive samples (13). Because of the size of the AIDS epidemic in that country, this finding extrapolates to tens of thousands of group O infections in Nigeria alone. The natural history of group O infection appears to be similar to the natural history of group M infection (43), and it is thus likely that a roughly similar number of people have died as are currently infected. Therefore, it is reasonable to estimate that HIV-1 group O may have infected around 100,000 people (14).
Efficient spread of a zoonotic pathogen in the human population depends on a combination of viral, host, and environmental factors. The most recent common ancestors of HIV-1 groups M and O are both estimated to have existed around 1920 (46, 47), whereas the lower levels of diversity seen in group N, and especially in group P, indicate that they emerged more recently. Coalescent studies suggest that groups O and M underwent similar rates of exponential growth until about 1960, and only since then has the spread of group M far outstripped the spread of group O (47). Importantly, groups M and O have undergone independent adaptations to the human host. In both lineages, Met at position 30 in the Gag matrix protein was replaced by a basic residue, Arg (48), which enhances viral replication in human lymphoid tissue (49). Both lineages also had to acquire resistance to the potent restriction factor tetherin: SIVcpz and SIVgor antagonize tetherin via their Nef proteins, but the tetherin motif that these proteins target was deleted in a human ancestor. In group M viruses, Vpu adapted to acquire antitetherin activity (50), whereas in the ancestor of group O, the Nef protein evolved to use a different target within tetherin (14). The fact that group O viruses have not spread even more widely in the human population is thus unlikely to be due to a lack of adaptation to the human host, but may simply reflect the absence of epidemiological opportunity during the early stages of the pandemic expansion of AIDS starting a little over 50 y ago. Clearly, SIVs from both chimpanzees and gorillas have the capacity to spread efficiently in the human host. Thus, it will be important to continue to monitor humans for primate lentiviruses and to study the viral and host factors that govern cross-species infection and onward transmission.
Materials and Methods
Sample Collection and Study Sites.
Fecal samples were collected between July 2009 and June 2013 from wild western lowland gorillas (G. g. gorilla) in Cameroon and Gabon, eastern lowland gorillas (G. beringei graueri) in the DRC, and mountain gorillas (G. b. beringei) in the DRC and Uganda. Most samples were collected around night nests and feeding sites, but also opportunistically (16, 17, 22, 51). Samples were preserved in RNAlater (Ambion), kept at ambient temperature in the field for a maximum of 3 wk and then stored at −20 °C or −80 °C. Field information included the global positioning system position and condition of fecal samples, as well as the number and age of gorilla nests.
Noninvasive Detection of SIVgor.
All gorilla fecal samples were first tested for the presence of HIV-1 cross-reactive antibodies using the INNO-LIA HIV I/II score confirmation test (Innogenetics) as previously described (9, 16). An RT-qPCR assay, previously shown to detect HIV-1 groups M, N, O, and P; SIVcpzPtt; SIVcpzPts; and SIVgor in fecal samples (18), was used to screen all antibody-positive, as well as some antibody-negative, fecal samples. Viral RNA was extracted using the NucliSens Magnetic Extraction Kit (BioMerieux) (17). To characterize individual SIVgor strains, nested RT-PCR was used to amplify partial env (315–692 bp) and pol (285 bp) sequences (4, 9, 15–17). In addition, diagnostic gp41 (155 bp) and gag (408 bp) fragments were amplified using newly designed primer sets (Table S3). To confirm their species origin, antibody-positive samples and a subset of negative samples were subjected to mitochondrial 12S sequence analyses (9, 52). To determine the number of sampled individuals, microsatellite analysis was performed on all samples collected at the same nesting sites. Gorilla group sizes were estimated based on the number of night nests (53). Fecal samples collected less than 20 m from the nests were considered to belong to the same group (16, 17). Samples were genotyped at seven loci using multiplex PCR (D18s536, D4s243, D10s676, D9s922 and D2s1326, D2s1333, D4s1627). For gender determination, a region of the amelogenin gene was amplified (4). Homozygous loci were amplified at least four times to minimize allelic dropout. Matching samples were given a consensus identification number and genotype (Table S2).
SIVgor Prevalence.
SIVgor prevalence rates were estimated for each field site based on the proportion of SIVgor-positive gorillas as determined by microsatellite analysis, taking into account repeated sampling per mission and on consecutive missions. Previous studies of western and eastern lowland gorillas indicated that each individual was sampled ∼1.8 and 1.9 times, respectively, and that a maximum of 50% of animals were resampled on consecutive missions (16, 17, 51). The prevalence was thus calculated by multiplying the number of samples with the coefficient [(n × 1.8)/(1 + (n − 1) × 0.50], with n representing the number of missions; 95% confidence limits were calculated as described (ww3.ac-poitiers.fr/math/prof/resso/cali/ic_phrek.html) (54).
Generation of Full-Length SIVgor Sequences.
Whole-genome sequences of SIVgor were generated by amplifying partially overlapping subgenomic fragments from fecal viral RNA as previously described (4, 5, 15). Ambiguous sites in sequence chromatograms were resolved as reported (4, 5, 15).
Phylogenetic Analyses.
Nucleotide and protein sequences were aligned using Muscle (55) and the online version of MAFFT (mafft.cbrc.jp/alignment/server/) with minor manual adjustments. Sites that could not be unambiguously aligned were automatically (Gblocks; phylogeny.lirmm.fr/phylo_cgi/index.cgi) excluded and manually checked. Proteome sequences were generated by joining deduced Gag, Pol, Env, and Nef amino acid sequences; the carboxy termini of Gag sequences that overlapped with Pol sequences were excluded. Newly derived partial and full-length SIVgor sequences were compared with previously published SIVgor; SIVcpz; and HIV-1 group M, N, O, and P reference sequences. Phylogenies were inferred using maximum likelihood (ML) and Bayesian methods. ML trees were constructed using PhyML version 3.1 for nucleotide sequence and version LG4X for amino acid sequences, and branch support was evaluated with 1,000 bootstrap replicates for each tree (56, 57); only values higher than 70% were considered meaningful. Bayesian inference of phylogeny was performed using MrBayes3.2 (58). For nucleotide alignments, 10,000 trees were sampled with Markov Chain Monte Carlo (MCMC) algorithms under the GTR + G + I model with six rate categories within two runs totaling 20,000,000 generations. Estimated sample sizes for all parameters were >200, indicating that the two runs converged. For protein alignments, a mixed model of amino acid substitution was used and MCMC runs were performed until the runs converged (10–20 million generations). For both nucleotide and protein alignments, the burn-in was set to 25%. Posterior probabilities ≥0.95 were considered significant.
Recombination Analysis.
Full-length SIVgor sequences were analyzed using bootscan and similarity plots with SimPlot version 3.5.1 and Recco version 0.93 (59, 60). Bootscan analyses examined neighbor-joining trees with a 400-bp window and 20-bp increments. Breakpoints were inferred using SimPlot and Recco, and verified by visual inspection. The recombinant patterns were subsequently confirmed by phylogenetic tree analysis of nucleotide sequences using ML methods implemented in PhyML version 3.1 (56). Branch support was evaluated with the nonparametric bootstrap method, and 1,000 bootstrap replicates were performed for each tree. Only values higher than 70% were considered significant.
Resistance Testing of Gorilla A3G to Degradation by SIVcpz and SIVgor Vif.
A ΔVif replication-competent molecular clone (NL4-3) was obtained from the AIDS Research and Reference Reagent Program. C-terminally FLAG-tagged expression plasmids of SIVcpzPts (TAN1 and TAN2), SIVcpzPtt (EK505 and LB715), and SIVgor (CP2139) vif genes; a ΔVif-GFP control construct; and HA-tagged chimpanzee and gorilla A3G expression plasmids have been described (19). Vif alleles from SIVcpzPtt MT145 (4), SIVcpzPtt GAB2 (61, 62), SIVcpzPts TAN3 (63), and SIVcpzPts TAN13 (63) were amplified from full-length molecular clones and cloned into the pCRV1 Vif-expression plasmid (19). Increasing amounts of HA-tagged A3G (0, 25, 50, and 100 ng) and Vif-expression (or ΔVif-GFP for control) plasmids (50 ng) were cotransfected with NL4-3 ΔVif (500 ng) in 293T cells. The total amount of DNA for each transfection was kept identical by substituting the A3G-expression plasmid with a GFP-expression plasmid. Supernatants were collected 48 h after transfection and used to infect 1 × 104 TZM-bl cells in 96-well plates, and infectivity was assessed by measuring β-gal activity (Applied Biosystems). Average relative infectivity values and SDs were calculated using data from three independent transfections. For A3G degradation analysis, transfected 293T cells were lysed, separated on 10% SDS-polyacrylamide gels (Invitrogen), transferred to PVDF membranes (Pierce), and probed with rabbit anti-HA polyclonal (Sigma) and rabbit anti-FLAG monoclonal (Sigma) antisera. GAPDH was detected with a mouse primary antibody (Santa Cruz Biotechnology). Membranes were incubated with HRP-conjugated secondary antibodies (Sigma), developed with SuperSignal West Femto (Pierce), and detected using a FluorChem E imaging system (Protein Simple).
GenBank Accession Numbers.
Full-length and partial SIVgor sequences are available under GenBank accession nos. KP004989–KP004991 and KP004992–KP004999, respectively.
Supplementary Material
Acknowledgments
We thank the staff and SIV team of Projet PRESICA (Prévention du Sida au Cameroun), Jacob Willie, Donald Mbohli, and Marcel Salah for sample collection and logistical support in Cameroon; the Cameroonian Ministries of Health, Environment and Forestry, and Research for permission to collect samples in Cameroon; the Ministry of Water and Forests of Gabon, Philippe Engandja, Alain-Prince Okouga, Martine Koné and the Mikongo Conservation Center for logistical support in Gabon; Sabrina Locatelli, Frank Kirchhoff, Daniel Sauter, and Peter Sudmant for helpful discussions; and Coralie Sigounios, Alexandra Meyer, and Shivani Sethi for assistance with preparation of figures and manuscript submission. The study was supported by grants from the NIH (Grants R37 AI50529, R01 AI 058715, P30 AI 045008, R37 AI 066998, R01 AI064001, and AI 089246), the Agence Nationale de Recherches sur le SIDA (ANRS), France (ANRS 12125, ANRS 12182, ANRS 12555, and ANRS 12325), the Centre International de Recherches Médicales de Franceville, and the Institut de Recherche pour le Développement. L.E. and V.B. received a PhD grant from Sidaction and Fonds de dotation Pierre Bergé. M.D. received a PhD grant from the Brazilian PDSE-CAPES (Programa de Doutorado Sanduíche no Exterior from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) “Scholarship Program Doctoral Sandwich Abroad.”
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP004989–KP004991 and KP004992–KP004999).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502022112/-/DCSupplemental.
References
- 1.Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1(1):a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Locatelli S, Peeters M. Cross-species transmission of simian retroviruses: How and why they could lead to the emergence of new diseases in the human population. AIDS. 2012;26(6):659–673. doi: 10.1097/QAD.0b013e328350fb68. [DOI] [PubMed] [Google Scholar]
- 3.Bailes E, et al. Hybrid origin of SIV in chimpanzees. Science. 2003;300(5626):1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 4.Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313(5786):523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Heuverswyn F, et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368(1):155–171. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, et al. Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J Virol. 2012;86(19):10776–10791. doi: 10.1128/JVI.01498-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santiago ML, et al. SIVcpz in wild chimpanzees. Science. 2002;295(5554):465. doi: 10.1126/science.295.5554.465. [DOI] [PubMed] [Google Scholar]
- 8.Switzer WM, et al. The epidemiology of simian immunodeficiency virus infection in a large number of wild- and captive-born chimpanzees: Evidence for a recent introduction following chimpanzee divergence. AIDS Res Hum Retroviruses. 2005;21(5):335–342. doi: 10.1089/aid.2005.21.335. [DOI] [PubMed] [Google Scholar]
- 9.Van Heuverswyn F, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444(7116):164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 10.Delaugerre C, De Oliveira F, Lascoux-Combe C, Plantier JC, Simon F. HIV-1 group N: Travelling beyond Cameroon. Lancet. 2011;378(9806):1894. doi: 10.1016/S0140-6736(11)61457-8. [DOI] [PubMed] [Google Scholar]
- 11.Plantier J-C, et al. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15(8):871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 12.Vallari A, et al. Confirmation of putative HIV-1 group P in Cameroon. J Virol. 2011;85(3):1403–1407. doi: 10.1128/JVI.02005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peeters M, et al. Geographical distribution of HIV-1 group O viruses in Africa. AIDS. 1997;11(4):493–498. doi: 10.1097/00002030-199704000-00013. [DOI] [PubMed] [Google Scholar]
- 14.Kluge SF, et al. Nef proteins of epidemic HIV-1 group O strains antagonize human tetherin. Cell Host Microbe. 2014;16(5):639–650. doi: 10.1016/j.chom.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takehisa J, et al. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol. 2009;83(4):1635–1648. doi: 10.1128/JVI.02311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neel C, et al. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J Virol. 2010;84(3):1464–1476. doi: 10.1128/JVI.02129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etienne L, et al. Noninvasive follow-up of simian immunodeficiency virus infection in wild-living nonhabituated western lowland gorillas in Cameroon. J Virol. 2012;86(18):9760–9772. doi: 10.1128/JVI.01186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Etienne L, et al. Single real-time reverse transcription-PCR assay for detection and quantification of genetically diverse HIV-1, SIVcpz, and SIVgor strains. J Clin Microbiol. 2013;51(3):787–798. doi: 10.1128/JCM.02792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letko M, et al. Vif proteins from diverse primate lentiviral lineages use the same binding site in APOBEC3G. J Virol. 2013;87(21):11861–11871. doi: 10.1128/JVI.01944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499(7459):471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butynski TM. In: Great Apes and Humans: The Ethics of Coexistence. Beck BB, et al., editors. Smithsonian Institution Press; Washington, DC: 2001. [Google Scholar]
- 22.Roy J, et al. Challenges in the use of genetic mark-recapture to estimate the population size of Bwindi mountain gorillas (Gorilla beringei beringei) Biol Conserv. 2014;180:249–261. [Google Scholar]
- 23.Keele BF, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460(7254):515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh PD, et al. Catastrophic ape decline in western equatorial Africa. Nature. 2003;422(6932):611–614. doi: 10.1038/nature01566. [DOI] [PubMed] [Google Scholar]
- 25.Bermejo M, et al. Ebola outbreak killed 5000 gorillas. Science. 2006;314(5805):1564. doi: 10.1126/science.1133105. [DOI] [PubMed] [Google Scholar]
- 26.Etienne L, et al. Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms. Retrovirology. 2011;8:4. doi: 10.1186/1742-4690-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudicell RS, et al. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 2010;6(9):e1001116. doi: 10.1371/journal.ppat.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bermejo M. Home-range use and intergroup encounters in western gorillas (Gorilla g. gorilla) at Lossi forest, North Congo. Am J Primatol. 2004;64(2):223–232. doi: 10.1002/ajp.20073. [DOI] [PubMed] [Google Scholar]
- 29.Doran-Sheehy DM, Greer D, Mongo P, Schwindt D. Impact of ecological and social factors on ranging in western gorillas. Am J Primatol. 2004;64(2):207–222. doi: 10.1002/ajp.20075. [DOI] [PubMed] [Google Scholar]
- 30.Fünfstück T, et al. The genetic population structure of wild western lowland gorillas (Gorilla gorilla gorilla) living in continuous rain forest. Am J Primatol. 2014;76(9):868–878. doi: 10.1002/ajp.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Worobey M, et al. Island biogeography reveals the deep history of SIV. Science. 2010;329(5998):1487. doi: 10.1126/science.1193550. [DOI] [PubMed] [Google Scholar]
- 32.Sharp PM, Simmonds P. Evaluating the evidence for virus/host co-evolution. Curr Opin Virol. 2011;1(5):436–441. doi: 10.1016/j.coviro.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 33.Morgan D, Sanz C. Chimpanzee feeding ecology and comparisons with sympatric gorillas in the Goualougo Triangle, Republic of Congo. In: Hohmann G, Robbins M, Boesch C, editors. Feeding Ecology in Apes and Other Primates. Cambridge Univ Press; Cambridge, U.K.: 2006. pp. 97–122. [Google Scholar]
- 34.Stanford C, Nkurunungi JB. Behavioral ecology of sympatric chimpanzees and gorillas in Bwindi Impenetrable National Park, Uganda: Diet. Int J Primatol. 2003;24(4):901–918. [Google Scholar]
- 35.Head J, Boesch C, Makaga LØ, Robbins M. Sympatric chimpanzees (Pan troglodytes troglodytes) and gorillas (Gorilla gorilla gorilla) in Loango National Park, Gabon: Dietary composition, seasonality, and intersite comparisons. Int J Primatol. 2011;32(3):755–775. [Google Scholar]
- 36.Leendertz FH, et al. Anthrax in Western and Central African great apes. Am J Primatol. 2006;68(9):928–933. doi: 10.1002/ajp.20298. [DOI] [PubMed] [Google Scholar]
- 37.Walsh PD, Breuer T, Sanz C, Morgan D, Doran-Sheehy D. Potential for Ebola transmission between gorilla and chimpanzee social groups. Am Nat. 2007;169(5):684–689. doi: 10.1086/513494. [DOI] [PubMed] [Google Scholar]
- 38.Lyons S, et al. Species association of hepatitis B virus (HBV) in non-human apes; Evidence for recombination between gorilla and chimpanzee variants. PLoS ONE. 2012;7(3):e33430. doi: 10.1371/journal.pone.0033430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8(1):55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Krupp A, et al. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS Pathog. 2013;9(10):e1003641. doi: 10.1371/journal.ppat.1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Compton AA, Hirsch VM, Emerman M. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe. 2012;11(1):91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Compton AA, Emerman M. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog. 2013;9(1):e1003135. doi: 10.1371/journal.ppat.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mourez T, Simon F, Plantier JC. Non-M variants of human immunodeficiency virus type 1. Clin Microbiol Rev. 2013;26(3):448–461. doi: 10.1128/CMR.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vessière A, et al. Diagnosis and monitoring of HIV-1 group O-infected patients in Cameroun. J Acquir Immune Defic Syndr. 2010;53(1):107–110. doi: 10.1097/QAI.0b013e3181b97ec1. [DOI] [PubMed] [Google Scholar]
- 45.Jonassen TO, et al. Sequence analysis of HIV-1 group O from Norwegian patients infected in the 1960s. Virology. 1997;231(1):43–47. doi: 10.1006/viro.1997.8510. [DOI] [PubMed] [Google Scholar]
- 46.Worobey M, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455(7213):661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faria NR, et al. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346(6205):56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wain LV, et al. Adaptation of HIV-1 to its human host. Mol Biol Evol. 2007;24(8):1853–1860. doi: 10.1093/molbev/msm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bibollet-Ruche F, et al. Efficient SIVcpz replication in human lymphoid tissue requires viral matrix protein adaptation. J Clin Invest. 2012;122(5):1644–1652. doi: 10.1172/JCI61429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauter D, et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6(5):409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gray M, et al. Genetic census reveals increased but uneven growth of a critically endangered mountain gorilla population. Biol Conserv. 2013;158:230–238. [Google Scholar]
- 52.van der Kuyl AC, Kuiken CL, Dekker JT, Goudsmit J. Phylogeny of African monkeys based upon mitochondrial 12S rRNA sequences. J Mol Evol. 1995;40(2):173–180. doi: 10.1007/BF00167111. [DOI] [PubMed] [Google Scholar]
- 53.Tutin CG, Parnell R, White LT, Fernandez M. Nest building by lowland gorillas in the Lopé Reserve, Gabon: Environmental influences and implications for censusing. Int J Primatol. 1995;16(1):53–76. [Google Scholar]
- 54.Newcombe RG. Confidence Intervals for Proportions and Related Measures of Effect Size. CRC Press; London: 2012. [Google Scholar]
- 55.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 57.Dang CC, Lefort V, Le VS, Le QS, Gascuel O. ReplacementMatrix: A web server for maximum-likelihood estimation of amino acid replacement rate matrices. Bioinformatics. 2011;27(19):2758–2760. doi: 10.1093/bioinformatics/btr435. [DOI] [PubMed] [Google Scholar]
- 58.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lole KS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maydt J, Lengauer T. Recco: Recombination analysis using cost optimization. Bioinformatics. 2006;22(9):1064–1071. doi: 10.1093/bioinformatics/btl057. [DOI] [PubMed] [Google Scholar]
- 61.Janssens W, et al. Phylogenetic analysis of a new chimpanzee lentivirus SIVcpz-gab2 from a wild-captured chimpanzee from Gabon. AIDS Res Hum Retroviruses. 1994;10(9):1191–1192. doi: 10.1089/aid.1994.10.1191. [DOI] [PubMed] [Google Scholar]
- 62.Bibollet-Ruche F, et al. Complete genome analysis of one of the earliest SIVcpzPtt strains from Gabon (SIVcpzGAB2) AIDS Res Hum Retroviruses. 2004;20(12):1377–1381. doi: 10.1089/aid.2004.20.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takehisa J, et al. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J Virol. 2007;81(14):7463–7475. doi: 10.1128/JVI.00551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.