Significance
Human color vision is tri-chromatic, with three opsins expressed in cone photoreceptors that are sensitive in the red, green, and blue region of the spectrum. As theories predict, such tri- or tetra-chromacy with three or four opsin genes is common among mammals, birds, and other animals, including insects. However, we discovered that dragonflies possess as many as 15–33 opsin genes that have evolved through dynamic gene multiplications and losses within the lineage of dragonflies. These opsin genes are differentially expressed between adult and larva, as well as between dorsal and ventral regions of adult compound eyes, which plausibly underpin the versatile behavioral and ecological adaptations of actively flying adults to aerial lifestyle and sedentary larvae to aquatic lifestyle.
Keywords: dragonfly, opsin, color vision, molecular evolution
Abstract
Dragonflies are colorful and large-eyed animals strongly dependent on color vision. Here we report an extraordinary large number of opsin genes in dragonflies and their characteristic spatiotemporal expression patterns. Exhaustive transcriptomic and genomic surveys of three dragonflies of the family Libellulidae consistently identified 20 opsin genes, consisting of 4 nonvisual opsin genes and 16 visual opsin genes of 1 UV, 5 short-wavelength (SW), and 10 long-wavelength (LW) type. Comprehensive transcriptomic survey of the other dragonflies representing an additional 10 families also identified as many as 15–33 opsin genes. Molecular phylogenetic analysis revealed dynamic multiplications and losses of the opsin genes in the course of evolution. In contrast to many SW and LW genes expressed in adults, only one SW gene and several LW genes were expressed in larvae, reflecting less visual dependence and LW-skewed light conditions for their lifestyle under water. In this context, notably, the sand-burrowing or pit-dwelling species tended to lack SW gene expression in larvae. In adult visual organs: (i) many SW genes and a few LW genes were expressed in the dorsal region of compound eyes, presumably for processing SW-skewed light from the sky; (ii) a few SW genes and many LW genes were expressed in the ventral region of compound eyes, probably for perceiving terrestrial objects; and (iii) expression of a specific LW gene was associated with ocelli. Our findings suggest that the stage- and region-specific expressions of the diverse opsin genes underlie the behavior, ecology, and adaptation of dragonflies.
Diverse animals have color vision, which enables efficient recognition of the environment, foods, enemies, mates, and so forth (1–3). Opsins, which belong to the subfamily of G protein-coupled transmembrane receptors and form visual pigments together with retinal chromophores, play key roles in animal photoreception (4, 5). Theories predict that a set of photoreceptors with several distinct spectral sensitivities is sufficient for encoding colors in the visible spectrum (6–8). Many birds and reptiles possess four genes encoding visual pigment opsins for covering a spectral range from 300 to 700 nm, whereas many diurnal mammals and insects have three opsin genes, losing a UV or red spectral end (2, 9). Recent accumulation of molecular and genomic data has uncovered some animals with more opsin genes than conventionally envisioned (10): for example, 8–9 genes in mosquitoes (11, 12), 8–11 genes in fish (13–18), and 10–33 genes in mantis shrimps (19, 20). The largest number of opsin genes was reported from the water flea Daphnia pulex, whose genome encodes 46 opsin genes, 27 of which belong to visual types based on molecular phylogeny (21). Whether and how so many opsins are involved in color perception, recognition, and discrimination in these animals is of physiological, ecological, and evolutionary interest (22).
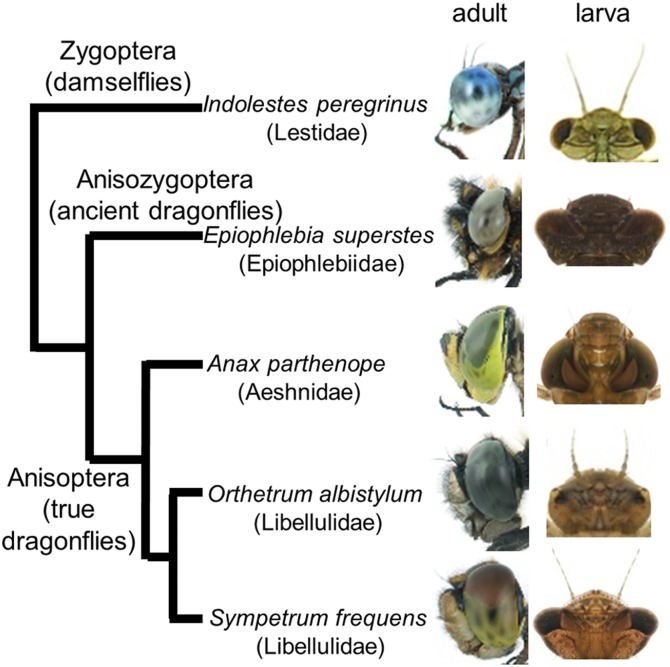
Dragonflies and damselflies (Insecta: Odonata) are colorful, diurnal, actively flying, and strongly dependent on visual sense. With auditory organs lacking and antennae reduced (23–25), dragonflies possess conspicuously large compound eyes, consisting of thousands of ommatidia (Figs. 1 and 2A). Dragonflies catch small prey in the air and form territories, wherein brightly colored males attack rival males and mate with dull-colored females (23, 26, 27). Previous morphological, spectrometric, electrophysiological, and transcriptomic studies have shown that the compound eyes of the dragonflies contain three to five classes of photoreceptors, with distinct spectral sensitivities covering the UV to red spectral range; the dorsal and ventral regions of the compound eyes are often differentiated both morphologically and physiologically (28–34).
Fig. 1.
Adult and larval eyes of zygopteran, anisozygopteran, and anisopteran dragonflies. Images courtesy of Akira Ozono.
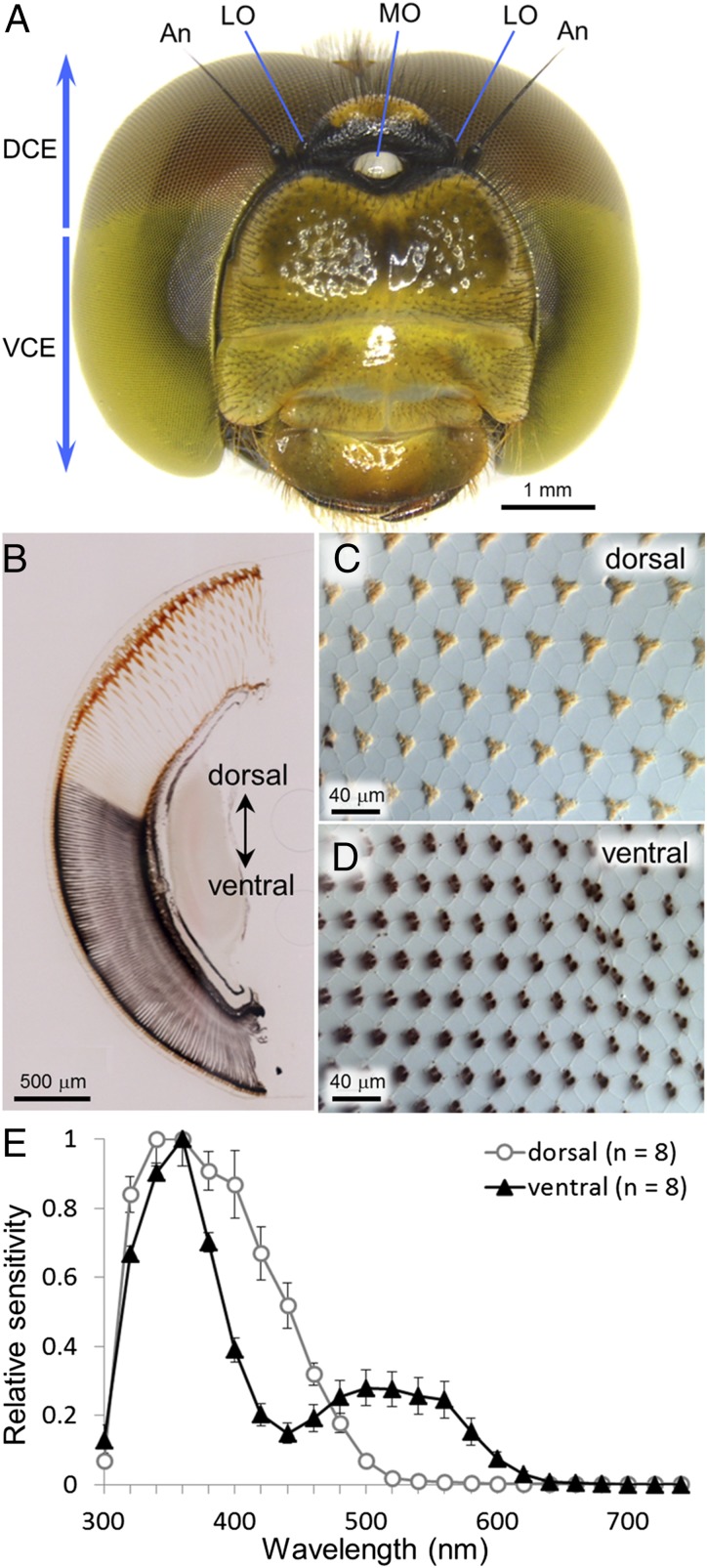
Fig. 2.
Morphology, anatomy and spectral sensitivity of adult compound eyes of Sympetrum frequens. (A) Frontal view of adult head. An, antenna; DCE, dorsal region of compound eye; LO, lateral ocellus; MO, median ocellus; VCE, ventral region of compound eye. (B) Unstained dorso-ventral section of a compound eye. (C) Unstained superficial section of the dorsal region of a compound eye, wherein photoreceptor cells and/or screening pigment cells accumulate orange pigment. (D) Unstained superficial section of the ventral region of a compound eye, wherein photoreceptor cells and/or screening pigment cells accumulate dark purple pigment. (E) Spectral sensitivity of the dorsal and ventral regions of adult eyes measured by electroretinography. Each symbol indicates the mean value of four dark-adapted individuals with SE (n = 8).
Here we report an extreme diversity of opsin genes in dragonflies: our comprehensive RNA sequencing of adult and larval eyes of 12 dragonfly species, representing 11 families, unveiled that as many as 15–33 opsin genes, of which 11–30 are visual opsin genes, have evolved in the lineage of dragonflies through dynamic gene multiplications and losses. Interestingly, expression patterns of the opsin genes exhibited striking differences between the adult stage and the larval stage, and also between the dorsal region and the ventral region of adult compound eyes, which unveil the previously unrecognized relevance of evolutionary diversification and spatiotemporal expression of opsin genes to ecological adaptation of the actively flying diurnal insects with keen visual sense.
Results
Identification of 20 Opsin Genes in Dragonflies of the Family Libellulidae.
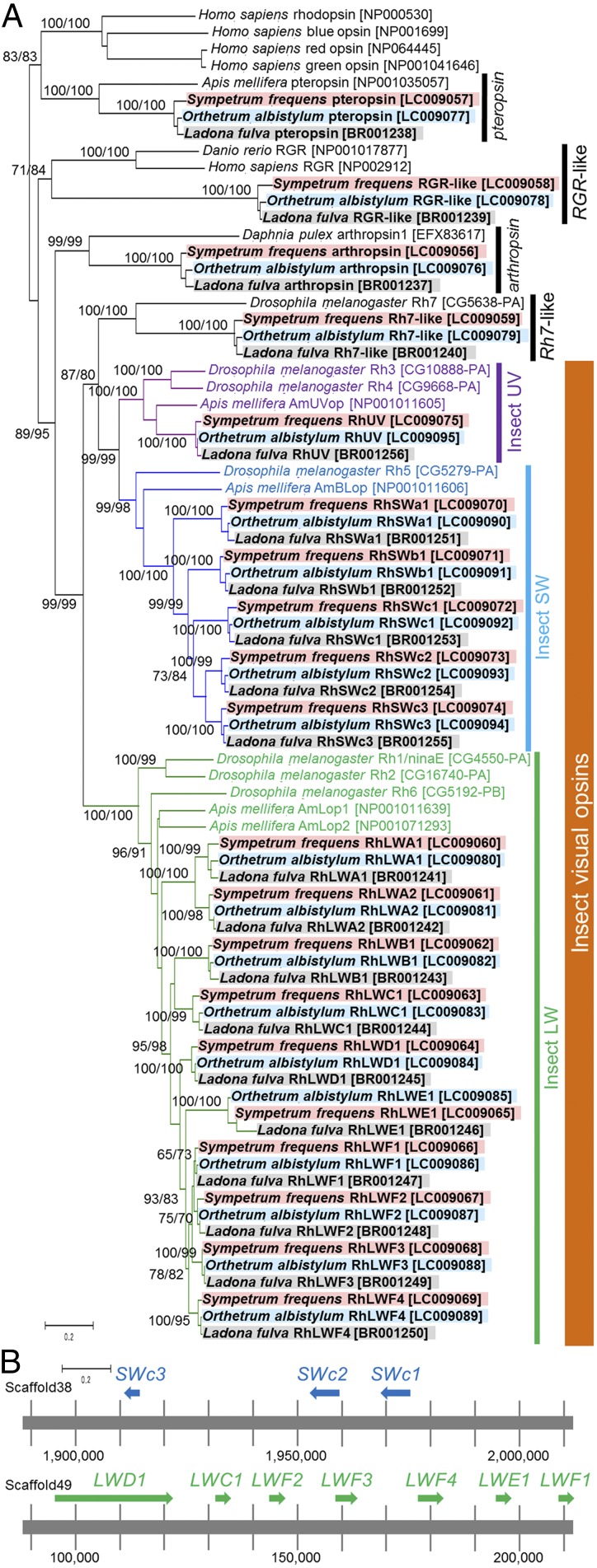
The Libellulidae is among the largest of dragonfly families, embracing over 1,000 species in the world (35, 36). First, we surveyed the visual transcriptomics of the red dragonfly Sympetrum frequens (Libellulidae) by RNA sequencing of the dorsal region of adult eyes, the ventral region of adult eyes, the adult head region including ocelli, and the larval whole head (Figs. 1 and 2A). Over 43 million HiSeq 100 base paired-end reads were subjected to de novo assembly (Table S1), which yielded 96,828 contigs of 201 bp or larger. Using known opsin genes of insects and other animals as queries, we obtained 60 opsin gene-like contigs. Because these contigs contained many partial or chimeric sequences, we carefully checked and manually corrected each of the contig sequences using Integrative Genomics Viewer (37) and experimentally verified them by RT-PCR and DNA sequencing. We also added over 5 million MiSeq 300 base paired-end reads (Table S1), which facilitated determination of full-length opsin gene sequences. In this way, a total of 20 opsin genes were identified from S. frequens: 1 pteropsin-type gene, 1 retinal G protein-coupled receptor (RGR)-like gene, 1 arthropsin type gene, 1 rhodopsin7 (Rh7)-like gene, 1 UV type (UV) gene, 5 short-wavelength type (SW) genes, and 10 long-wavelength type (LW) genes (Fig. 3A and Fig. S1; accession nos. LC009056–LC009075). Next, we performed RNA sequencing analysis of the white-tailed skimmer dragonfly Orthetrum albistylum (Libellulidae) in the same way, wherein 67 million HiSeq reads and 2.4 million MiSeq reads (Table S1) yielded the same set of 20 opsin genes (Fig. 3A; accession nos. LC009076–LC009095). Finally, we inspected the recently released draft genome data of the scarce chaser dragonfly Ladona fulva (Libellulidae) (BCM-HGSC: I5K, GenBank accession no. APVN01000000), identified the same set of 20 opsin genes (Fig. 3A; accession nos. BR001237–BR001256), and confirmed no other opsin genes in the genome. On the genome of L. fulva, three SW opsin genes (SWc1-c3) and seven LW opsin genes (LWC1, LWD1, LWE1, and LWF1–F4) were located in tandem, respectively (Fig. 3B).
Fig. 3.
Identification of 20 opsin genes in three dragonflies of the family Libellulidae. (A) A neighbor-joining phylogeny of 20 opsin genes each from S. frequens (red shade), O. albistylum (blue shade), and L. fulva (gray shade) inferred from 869 aligned amino acid sites. On each node, bootstrap values are indicated in the order of neighbor-joining method/maximum-likelihood method. Accession numbers or annotation IDs are shown in brackets. Classification of the opsin genes is indicated on the right side. Insect visual opsin genes are highlighted in purple for UV type, in blue for SW type, and in green for LW type, respectively. (B) Gene clusters of SW and LW opsin genes on the genome of L. fulva.
Diversity and Conservation of Opsin Genes in Libellulid Dragonflies.
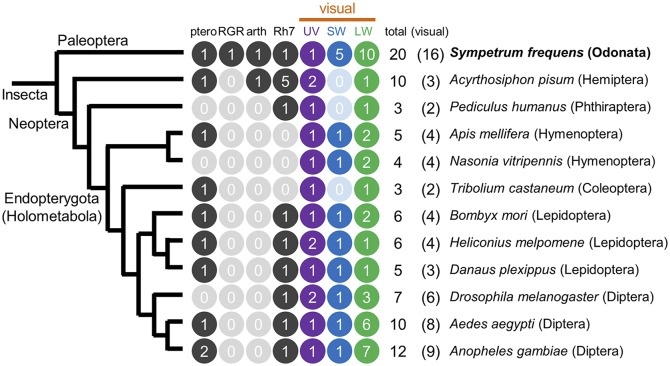
Molecular phylogenetic analysis revealed that the 20 opsin genes of S. frequens, O. albistylum, and L. fulva formed compact and distinct 20 monophyletic clusters (Fig. 3A), indicating that all of the 20 genes already existed in the common ancestor of the libellulid dragonflies. Of these, the 16 genes of UV, SW, and LW types have been classified as insect visual opsins (38) (Fig. 3A and Fig. S1), of which five SW opsin genes (SWa1, SWb1, and SWc1–c3), two LW opsin genes (LWA1 and LWA2), and eight LW opsin genes (LWB1, LWC1, LWD1, LWE1, and LWF1–F4), respectively, formed clusters distinct from previously identified insect opsin genes (Fig. 3A and Fig. S1). The 20 opsin genes in the dragonflies were strikingly larger in number than the opsin genes in the other insects whose genomes had been determined (Fig. 4). These patterns suggest that opsin gene multiplications occurred after divergence of the dragonflies from other insect groups.
Fig. 4.
Opsin genes of the red dragonfly S. frequens in comparison with those encoded in the genomes of diverse insects. Numbers of opsin genes of pteropsin type (ptero), RGR-like (RGR), arthropsin type (arth), Rh7-like (Rh7), UV type, SW type, and LW type are mapped on the insect phylogeny (75). Numbers of insect visual opsins (visual) are highlighted by purple for UV type, blue for SW type, and green for LW type.
Developmental and Regional Differentiation of Dragonfly Eyes.
Compound eyes of dragonflies are markedly different between adult and larva (Fig. 1). In adult dragonflies, the dorsal region of compound eyes is structurally different from the ventral region (32, 39). In S. frequens, for example, the dorsal ommatidia have larger facets and orange screening pigment, whereas the ventral ommatidia have smaller facets and dark purple screening pigment (Fig. 2 A–D). Such structural differences seem likely to entail functional differentiation, which can be verified, at least in part, by electroretinographic recording of spectral sensitivity (32). In S. frequens, the dorsal eye region was sensitive to a SW range with a peak at 360 nm, whereas the ventral eye region was sensitive to a wider wavelength range with two peaks at 360 nm and 520 nm (Fig. 2E). It is of great interest how these developmental, regional, structural, and functional differentiations in the compound eyes are relevant to as many as 20 opsin genes.
Stage- and Region-Specific Expression of Opsin Genes.
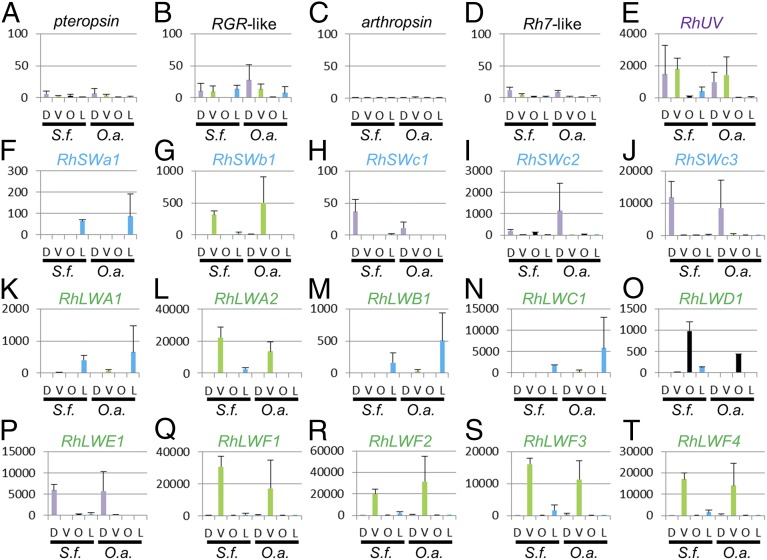
RNA sequencing revealed that, in both S. frequens and O. albistylum, expression patterns of the visual opsin genes exhibited striking differences between the adult stage and the larval stage, and also between the dorsal region and the ventral region of adult eyes (Fig. 5). Although one SW opsin gene (SWa1) and three LW opsin genes (LWA1, LWB1, and LWC1) were specifically expressed in larva (Fig. 5 F, K, M, and N), the other 12 visual opsin genes were mainly expressed in adult (Fig. 5 E, G–J, L, and O–T). In adult compound eyes, three SW opsin genes (SWc1, SWc2, and SWc3) and one LW opsin gene (LWE1) were specifically expressed in the dorsal region (Fig. 5 H–J and P), whereas one SW opsin gene (SWb1) and five LW opsin genes (LWA2 and LWF1–F4) were predominantly expressed in the ventral region (Fig. 5 G, L, and Q–T). The only UV opsin gene was expressed in both the dorsal and ventral regions of adult eyes (Fig. 5E). Notably, one LW opsin gene (LWD1) was specifically expressed in the adult ocelli (Fig. 5O). Nonvisual opsin genes, pteropsin, RGR-like, arthropsin, and Rh7-like, were scarcely expressed in the larval and adult visual organs (Fig. 5 A–D), which is concordant with their presumed nonvisual functions inferred from the phylogenetic analysis (Fig. 3A and Fig. S1). These results indicate that in S. frequens, the dorsal and the ventral regions of adult eyes are differentiated not only morphologically (Fig. 2 A–D) and physiologically (Fig. 2E), but also biochemically in terms of expression profiles of the diverse visual opsin genes (Fig. 5 E–T). Moreover, it should be noted that expression profiles of the visual opsin genes in O. albistylum were almost the same as those in S. frequens (Fig. 5), suggesting that the dorso-ventrally differentiated expression of the visual opsin genes is conserved among the libellulid dragonflies.
Fig. 5.
Expression levels of 20 opsin genes in adult and larval visual organs of S. frequens and O. albistylum. (A–D) Nonvisual opsin genes. (E) Visual opsin gene of UV type. (F–J) Visual opsin genes of SW type. (K–T) Visual opsin genes of LW type. D, dorsal region of adult eyes; L, larval whole head; O, adult head region containing ocelli; O.a., O. albistylum; S.f., S. frequens; V, ventral region of adult eyes. The numbers indicate FPKM values.
Diversity and Evolutionary Dynamics of Opsin Genes in the Odonata.
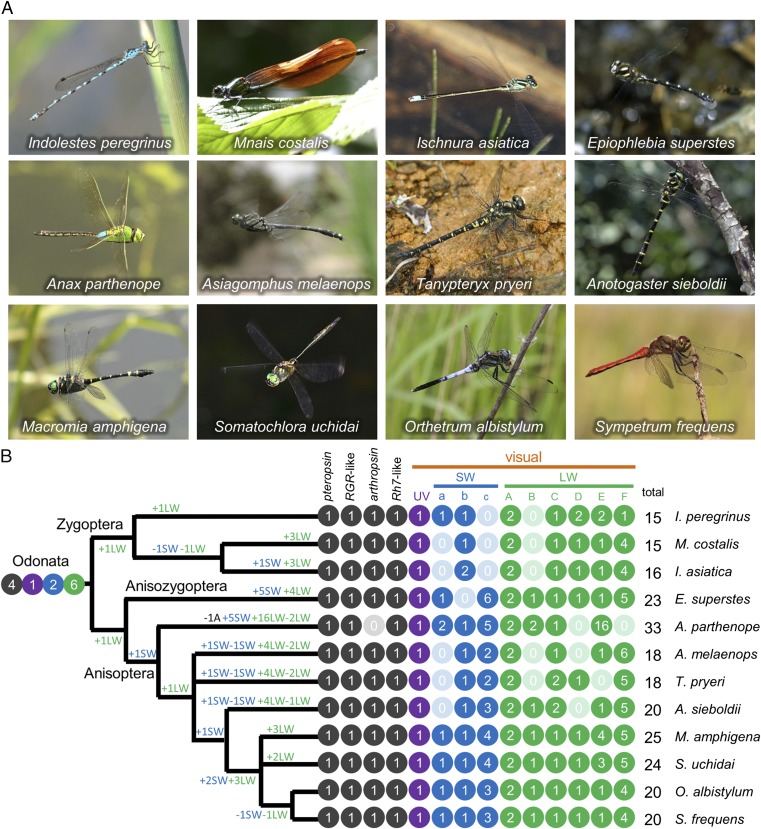
How and when have so many opsin genes emerged in the evolutionary course of dragonflies? Are the stage- and region-specific expression patterns of the diverse opsin genes also found in other dragonflies? To address these questions, in addition to S. frequens and O. albistylum (Libellulidae), we conducted the RNA sequencing analyses on 10 dragonfly species representing 10 different dragonfly families: Somatochlora uchidai (Corduliidae), Macromia amphigena (Macromiidae), Anotogaster sieboldii (Cordulegastridae), Tanypteryx pryeri (Petaluridae), Asiagomphus melaenops (Gomphidae), Anax parthenope (Aeshnidae), Epiophlebia superstes (Epiophlebiidae), Ischnura asiatica (Coenagrionidae), Mnais costalis (Calopterygidae), and Indolestes peregrinus (Lestidae) (Fig. 6A and Table S1). The first six species belong to the suborder Anisoptera (true dragonflies), the last three species belong to the suborder Zygoptera (damselflies), and E. supertes belongs to the suborder Anisozygoptera (ancient dragonflies; sometimes integrated to the Anisoptera) (Figs. 1 and 6B) (34, 35). Although all visual opsin genes from these 10 species were obtained as full-length, some nonvisual opsin genes (pteropsin, arthropsin, or Rh7-like) from several species were partial, probably because of low expression levels of these genes (Dataset S1). In this way, we obtained presumably complete sets of opsin genes for 12 dragonfly species representing 11 families (Fig. 6B and Dataset S1; accession numbers LC009056–LC009302). The total number of opsin genes varied considerably among the dragonflies, ranging from 15 (M. costalis and I. peregrinus) to 33 (A. parthenope). All of the species consistently possessed the four nonvisual opsin genes and the UV opsin gene as single-copies, except for A. parthenope lacking arthropsin gene. The number of SW opsin genes ranged from 1 (M. costalis) to 8 (A. parthenope); so did the number of LW opsin genes from 8 (I. peregrinus) to 21 (A. parthenope). Molecular phylogenetic analysis of the opsin genes (Fig. S2) and mapping of presence/absence of the opsin genes on the dragonfly phylogeny (Fig. 6B) unveiled the dynamic evolutionary trajectories of SW and LW opsin genes with repeated multiplications and losses in the evolutionary course of the dragonflies. It was estimated that the common ancestor of the extant dragonflies possessed at least 13 opsin genes consisting of 4 nonvisual, 1 UV, 2 SW, and 6 LW types (Fig. 6B).
Fig. 6.
Opsin genes in diverse dragonflies. (A) Adult dragonflies representing 12 species and 11 families examined in this study. (B) Numbers of opsin genes of pteropsin type, RGR-like, arthropsin type, Rh7-like, UV type, SW type, and LW type mapped on the dragonfly phylogeny (76). Numbers of insect visual opsins (visual) are highlighted by purple for UV type, blue for SW type, and green for LW type. SW genes are further categorized into groups a, b, and c; so are LW genes into groups A, B, C, D, E, and F (also see Fig. S2). Estimated gains and losses of the opsin genes in the evolutionary course of the dragonflies are indicated on the branches.
Expression Patterns of SW Opsin Genes in Diverse Dragonflies.
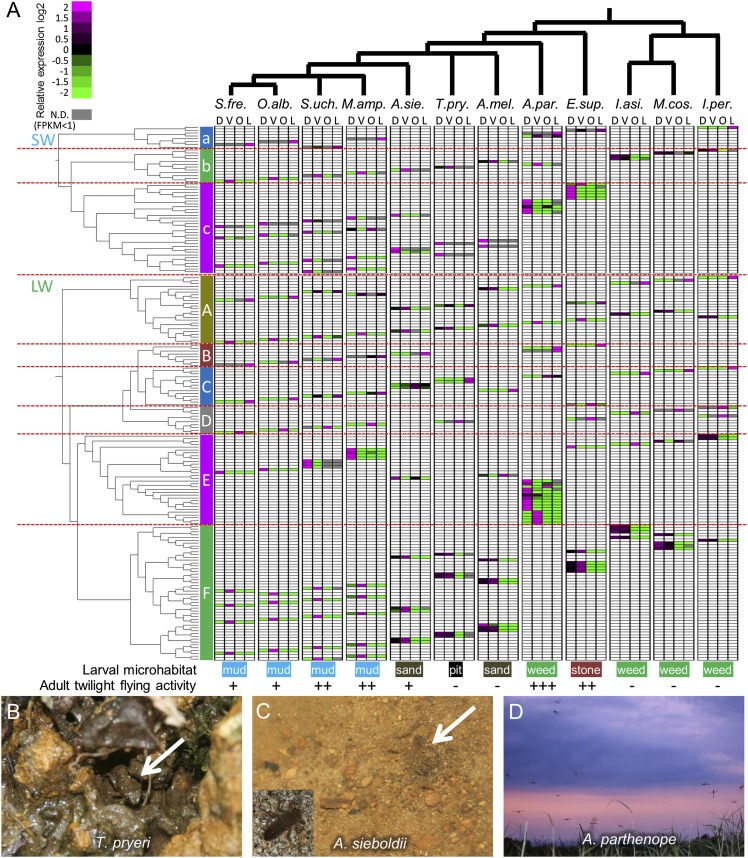
The diverse SW opsin genes of the 12 dragonfly species were categorized into three groups—a, b and c—on the basis of their phylogenetic placements and expression patterns (Figs. 6B and 7A and Fig. S2A). The group a genes were expressed mainly in larvae, whereas the group b and group c genes were expressed mainly in adults (Fig. 7A and Dataset S1). The group a, group b, and group c genes were absent in five species (A. sieboldii, T. pryeri, A. melaenops, I. asiatica, and M. costalis), one species (E. superstes), and three species (I. asiatica, M. costalis, and I. peregrinus), respectively. The group a gene was specifically duplicated in A. parthenope, as was the group b gene in I. asiatica. The group c genes varied in copy number, ranging from 0 through 2, 3, 4, and 5 to 6 (E. superstes) copies (Figs. 6B and 7A).
Fig. 7.
Expression profiles of opsin genes in diverse dragonflies. (A) Expression levels of visual opsin genes in adult and larval visual organs of 12 dragonfly species. Phylogenetic relationship of the dragonflies is shown on the top: A.mel., Asiagomphus melaenops; A.par., Anax parthenope; A.sie., Anotogaster sieboldii; E.sup., Epiophlebia superstes; I.asi., Ischnura asiatica; I.per., Indolestes peregrinus; M.amp., Macromia amphigena; M.cos., Mnais costalis; O.alb., Orthetrum albistylum; S.fre., Sympetrum frequens; S.uchi., Somatochlora uchidai; T.pry., Tanypteryx pryeri. Molecular phylogeny of SW-type opsin genes is shown on the upper left, where a, b, and c indicate the SW gene groups a, b, and c (see also Fig. S2A). Molecular phylogeny of LW-type opsin genes is shown on the lower left, where A, B, C, D, E, and F indicate the LW gene groups A, B, C, D, E and F (see also Fig. S2B). Gene expression levels are displayed by heat map presentation in connection to the dragonfly phylogeny and the opsin gene phylogenies, in which D, V, O, and L indicate expression levels in dorsal region of adult eyes, ventral region of adult eyes, adult head region containing ocelli, and larval whole head, respectively. In the color spectrum bar (Upper Left), magenta and green indicate positive and negative values relative to the average FPKM per gene per dragonfly species, whereas gray indicates no detectable expression with FPKM < 1. On the bottom, ecological traits for each dragonfly species are indicated according to refs. 40 and 41. For larval microhabitat: mud is crawling, or burrowing in muddy water bottom of open lowland ponds, marsh, rice paddies, or rivers; sand is burrowing in sandy water bottom of mountain streams or small rivers under dense tree cover; pit is digging pits on wet mountain slopes under forest cover; stone is hiding under stones in rocky water bottom of mountain streams under forest cover; and weed is hanging on water plants in open ponds or rivers. For adult twilight flying activity: +++, very active; ++, active; +, moderate; –, not observed. (B) A pit-dwelling larva of T. pryeri (arrow). (C) A bottom-burrowing larva of A. sieboldii (arrow); Inset shows an exposed larva. (D) Adults of A. parthenope in twilight flight.
Expression Patterns of LW Opsin Genes in Diverse Dragonflies.
The diverse LW opsin genes of the 12 dragonfly species were similarly categorized into six groups: A, B, C, D, E, and F (Figs. 6B and 7A and Fig. S2B). All species retained two copies of the group A genes: one copy was highly expressed in adult, whereas the other copy was predominantly expressed in larva or weakly expressed in both adult and larva. The group B genes were preferentially expressed in larva, absent in five species (T. pryeri, A. melaenops, I. asiatica, M. costalis, and I. peregrinus) and duplicated in one species (A. parthenope). The group C genes also tended to be expressed in larva, being duplicated in A. sieboldii and T. pryeri. The group D genes were single-copied, except for loss in three species (A. sieboldii, A. melaenops, and A. parthenope) and duplication in one species (I. peregrinus), and expressed in association with the head region containing ocelli. Interestingly, in the dragonfly species lacking the group D gene, a group C gene and a group E gene were expressed in the head region containing ocelli (Fig. 7A), presumably compensating for the loss of the group D gene. The group E genes were mostly expressed in the dorsal region of adult eyes, and drastically varied in copy number: absent in T. pryeri, 1 copy in 7 species, 2 copies in I. peregrinus, 3 copies in S. uchidai, 4 copies in M. amphigena, and as many as 16 copies in A. parthenope. In contrast, the group F genes were preferentially expressed in the ventral region of adult eyes, and usually multiple-copied: four copies in three species, five copies in five species, and six copies in A. melaenops, but only one copy in I. peregrinus and absent in A. parthenope. Notably, in T. pryeri lacking the group E genes, multiple group F genes were expressed not only in the ventral region but also in the dorsal region of adult eyes. In A. parthenope lacking the group F genes, some of the multiplied group E genes exhibited predominant expression in the ventral region of adult eyes. These patterns highlight compensatory evolution of regional gene expression patterns mediated by dynamic gene losses and multiplications in the dragonflies. Meanwhile, in zygopteran damselflies I. asiatica, M. costalis, and I. peregrinus, the region-specific expression patterns of the group E and group F genes were obscure (Figs. 6B and 7A and Dataset S1).
Discussion
Diversity of Opsin Genes in Dragonflies.
In this study, we identified a strikingly large number of opsin genes in diverse dragonflies, ranging from 15 to 33 opsin genes in total, of which 11–30 are visual opsin genes (Fig. 6B). The opsin genes in dragonflies are overwhelmingly larger in number than those in other insects (Fig. 4), and rivaling 14–33 visual opsin genes in mantis shrimps (20) and 27 visual opsin genes in the water flea (21). Theoretically, a set of photoreceptors with several distinct spectral sensitivities is sufficient for encoding colors in the visible spectrum (6–8), which leads to the following questions: Why are so many opsin genes required for some animals like mantis shrimps and dragonflies? What roles do these opsins play in their color vision? In mantis shrimps, the diverse opsin genes contribute to spectrally enrich their unique compound eyes that appear to function as a scanning photodetector array. As many as 12 photoreceptor types, each sampling a narrow wavelength range and in total covering from deep UV to far red, are arranged on their eyes in a highly ordered manner, especially in the color-sensitive midband region. With rapid scanning eye movement, mantis shrimps are able to detect and recognize diverse colors quickly, although their wavelength discrimination capability is relatively poor (22). Needless to say, dragonflies as terrestrial insects are quite different from mantis shrimps as oceanic crustaceans not only taxonomically but also ecologically and behaviorally. Molecular phylogenetic analysis clearly showed that multiplication of the visual opsin genes in dragonflies has occurred independently of those in mantis shrimps and the water flea (Fig. 8). Our data are suggestive of the hypothesis that the evolution of the diverse opsin genes in dragonflies is relevant to the ecological niche division between adult and larval dragonflies, and also to spatiotemporal heterogeneity of light conditions for adult dragonflies, as discussed below.
Fig. 8.
Phylogenetic relationship of opsin genes of insects (red shade), mantis shrimps (yellow shade), water flea (blue shade), and human. A neighbor-joining phylogeny inferred from 979 aligned amino acid sites is shown. On each node, bootstrap values are indicated in the order of neighbor-joining method/maximum-likelihood method. Accession numbers or annotation IDs are in brackets. Classification of the opsin genes is indicated on the right side.
Ecological Relevance of Adult-Larva Differentiation of Opsin Gene Expression.
Compound eyes of dragonflies are markedly different between adult and larva (Fig. 1), which should be relevant to their distinct ecological niches: adults actively fly in the air whereas larvae are sedentary on the water bottom (26). We found that visual opsin genes expressed in larval compound eyes are distinct in repertoire, fewer in number, and LW-skewed in comparison with those in adult eyes (Figs. 5 E–T and 7A), which presumably reflects less visual dependence and LW-skewed light condition for aqueous larval dragonflies. In this context, it may be notable that dragonfly species whose larval lifestyle entails particularly limited exposure to light, such as T. pryeri digging pits on wet mountain slopes (Fig. 7B) and A. sieboldii and A. melaenops burrowing in the sandy bottom of small mountain streams (Fig. 7C), where dense tree cover usually exists, tend to lack SW gene expression in larval compound eyes (Fig. 7A and Dataset S1) (26, 40, 41).
Functional Dorso-Ventral Differentiation of Compound Eyes in Adult Dragonflies at Molecular Level.
The sun in the sky is the major source of light under natural conditions. Hence, the light from above, which is dominated by direct radiation from the light source, tends to be of high intensity and SW-skewed, whereas the light from below, which is mostly a result of reflecting and scattering objects, such as plants, water surfaces, and soil, tends to be of lower intensity and LW-skewed (42). This vertical heterogeneity of light conditions is ubiquitous across terrestrial and aquatic ecosystems, which have probably prompted the evolution of dorso-ventrally differentiated eyes known from diverse animals (39). The most striking cases are found in water surface dwellers, like whirligig beetles and four-eyed fish, whose dorsal half and ventral half of eyes are anatomically separated and functionally specialized for aerial vision and aquatic vision, respectively (17, 43). Dorso-ventral structural differentiation of compound eyes has been reported not only in dragonflies but also in diverse actively flying or swarming insects, such as black flies (44), march flies (45), mayflies (46, 47), owlflies (48), butterflies (49), and others (39, 50). Similar dorso-ventrally differentiated compound eyes are also known from planktonic crustaceans, such as hyperiid amphipods, euphausiid krills, and mysid shrimps (51, 52). Here we demonstrate that, in diverse dragonflies, the dorsal and ventral regions of adult eyes exhibit not only morphological and physiological differences (Fig. 2) (32, 39), but also striking differentiation at the molecular level in that the majority of 11–30 visual opsin genes are, respectively, expressed in dorso-ventrally distinct patterns (Figs. 5 and 7A), which must be relevant to the ecology of the strongly visual adult dragonflies.
Ecological Relevance of Dorso-Ventral Differentiation of Opsin Gene Expression in Adult Compound Eyes.
The general expression patterns of the visual opsin genes in compound eyes of adult dragonflies are summarized as follows: (i) in true dragonflies (Anisoptera including Anisozygoptera), the SW and LW opsin genes expressed in the dorsal region are distinct from those expressed in the ventral region; (ii) in the dorsal region, multiple SW opsin genes and one or a few LW opsin genes are preferentially expressed; (iii) conversely, in the ventral region one or a few SW opsin genes and multiple LW opsin genes are preferentially expressed; whereas (iv) in contrast, in damselflies (Zygoptera) the dorso-ventrally differentiated expression patterns of the SW and LW opsin genes are not obvious (Figs. 5 and 7A). These patterns suggest that, in true dragonflies, the dorsal and ventral regions of adult eyes are tuned to perceive SW light and LW light, respectively. Such regional specialization of adult eyes seems to make sense in the light of ecological and behavioral aspects of the dragonflies. In the daytime, male dragonflies, which are often brightly colored, make a territory at an open space around riverside, pond, forest edge, or tree canopy, patrol and defend the territory against conspecific males, and chase and attempt to mate with conspecific females (26). On the other hand, although all dragonflies are diurnal, territorial, and resting in the nighttime, true dragonflies, but not damselflies, generally exhibit an array of characteristic behaviors called “twilight flight” just before and after sunset (Fig. 7 A and D) (26). In the twilight hour, numerous small insects, such as midges and flies, form swarming groups, many nocturnal insects like moths start flying, and many true dragonfly species actively fly and efficiently catch these small insects at dusk (Fig. 7D) (26, 40, 41). In the daytime, territorial males are looking down on the environment and paying attention to conspecific dragonflies that are usually approaching from surroundings, where LW-skewed light signals reflected from the environmental objects should be perceived by the ventral or lateral region of their compound eyes. In the twilight time, on the other hand, the environmental light becomes dimmer and SW-skewed (42), where foraging dragonflies are probably recognizing the silhouette of small flying preys against SW-skewed light from the sky by the dorsal ommatidia. This hypothesis is certainly speculative, but can account for the general expression pattern of the visual opsin genes in compound eyes of adult dragonflies in accordance with their ecology and behavior. It is notable that dragonfly species that are particularly active in twilight tend to express more SW opsin genes in the dorsal region of compound eyes (Fig. 7 A and D). Biochemical and molecular genetic approaches to spectral sensitivity of the respective opsin genes, and also electrophysiological and neuroethological approaches to photoreceptive characteristics of the dorsal and ventral regions of adult eyes, will be important for testing the hypothesis in future studies. In this context, it may be relevant that dorso-ventrally differentiated expression patterns of several opsin genes have been identified in compound eyes of butterflies (9, 53–55), and SW-sensitive vision has been shown to be advantageous for twilight-active animals (56–58).
Identification of an Ocellus-Specific Opsin Gene.
In 9 of 12 of the dragonfly species we examined, a LW opsin gene of the group D exhibited a characteristic expression pattern associated with ocelli (Figs. 5O and 7A). In the remaining three species in which the group D LW opsin gene was absent, either a group C LW opsin gene or a group E LW opsin gene was preferentially expressed in association with ocelli (Fig. 7A and Dataset S1). These results strongly suggest that the LW opsin genes comprise the principal visual pigments in ocelli of the dragonflies. Previous studies have shown that ocelli of dragonflies perceive not only intensity but also directionality of UV and green light with crude form vision, thereby resolving horizontally extended features like the horizon for attitude adjustment during flight (59–62), in which the ocellus-specific LW opsin genes must play a pivotal role. Opsin genes specifically expressed in ocelli have also been identified in the fruit fly Drosophila melanogaster (63) and the cricket Gryllus bimaculata (64).
Evolutionary Dynamics of Diverse Opsin Genes in Dragonflies.
Our molecular phylogenetic and gene expression analyses unveiled the following evolutionary aspects of the diverse opsin genes in dragonflies: (i) ancient multiplication of the opsin genes in the Odonata, which gave rise to at least four nonvisual opsin genes and nine visual opsin genes of one UV, two SW, and six LW types; (ii) diversification of the extant dragonflies with retaining the large number of opsin genes in general; and (iii) meanwhile, repeated local multiplications and losses of SW and LW opsin genes during the dragonfly diversification (Fig. 6B). On the grounds that many of the SW and LW opsin genes are located on the dragonfly genome in tandem (Fig. 3B), it seems likely that unequal crossing over is the principal genetic mechanism underlying the dynamic multiplications and losses of the opsin genes (65, 66). Another notable aspect of the evolutionary dynamics is highlighted by acquisition of compensational expression patterns associated with losses of some visual opsin genes during the dragonfly evolution: for example, (i) in A. sieboldii, A. melaenops, and A. parthenope, loss of the ocellus-specific LW opsin gene of the group D accompanied ocellus-associated expression of other LW opsin genes of either group C or group E; (ii) in T. pryeri, loss of the group E LW opsin genes, which are generally expressed in the dorsal region of adult eyes, entailed novel expression patterns of multiple group F LW opsin genes in the entire region of adult eyes; and (iii) in A. parthenope, loss of the group F opsin genes, which are generally expressed in the ventral region of adult eyes, were presumably compensated for by highly multiplied group E LW opsin genes, some of which acquired preferential expression in the ventral region of adult eyes (Fig. 7A). These evolutionary patterns provide a dynamic perspective as to how multiplied visual opsin genes have contributed to the functional diversity, genetic redundancy, and evolutionary robustness of color vision in dragonflies. How these diverse opsin genes are functioning in the dragonfly’s visual organs deserves future studies. Conventionally, it has been envisaged that, on the basis of vertebrate studies, each photoreceptor cell expresses a single opsin gene, and multiple photoreceptor cells expressing several different opsin genes with different absorption spectra constitute a retina for color vision (30, 31, 33, 34). However, recent studies have identified coexpression of multiple opsin genes in a single photoreceptor cell in compound eyes of butterflies and other arthropods (67–70), raising the possibility that such opsin coexpression may enrich the photoreceptor diversity among invertebrates including dragonflies.
Conclusion and Perspective.
We humans have a RGB trichromatic system based on three opsin proteins expressed in cone photoreceptors (71). Neuroethological studies revealed that most animals are either di-, tri-, or tetra-chromats (8). Theories predict that several classes of photoreceptors with distinct spectral sensitivities are sufficient for encoding colors (6–8). Actually, many vertebrates and invertebrates possess three, four, or five genes encoding visual pigment opsins (2, 8, 9). However, our discovery of up to 30 visual opsin genes functioning in visual organs of dragonflies highlights that trichromacy with several opsins is a minimal requirement and the molecular basis of animal’s color vision can potentially be much more complex. For dragonflies, their visual environments are spatiotemporally partitioned into the aquatic light condition for larval eyes, the SW-skewed aerial light condition for the dorsal region of adult eyes, and the LW-skewed environmental light condition for the ventral region of adult eyes. These regions express different sets of visual opsins generated through dynamic gene multiplications. Plausibly, although speculative, the highly multiplied opsin genes may play substantial roles in ecological adaptation of dragonflies via finely tuned visual systems for better thriving in this colorful world.
Materials and Methods
Insects.
Table S1 lists the dragonfly samples, an adult and a larva per species, subjected to RNA sequencing analyses. For histology and electroretinography, adult insects of S. frequens were collected at Tsukuba, Ibaraki, Japan.
Histology and Image Acquisition.
Compound eyes were carefully dissected from adult insects in 4% (wt/vol) paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.5) (CB), and fixed in the same fixative for 15 min at room temperature. After a brief wash with CB, the tissues were dehydrated through a graded water-acetone series, infiltrated with propylene oxide two times for 15 min each, embedded in Quetol 812, cut into 26-µm sections using a microtome (Microm HM 355S), and mounted on glass slides using Mountquick (Daido Sangyo). The sections were photographed using a digital camera (Leica DFC420) attached to a light microscope (Leica DM2500) equipped with an Apo 10× oil-immersion objective. The images at different focal depths were automatically stacked and aligned using the e-Tiling software (v3.8, Mitani Corporation).
Electrophysiology.
Different regions of adult eyes were examined for spectral sensitivity by electroretinography. An adult dragonfly, whose wings and legs had been removed, was mounted on a stage using beeswax so that its lateral side was fixed upward. A small hole was made either on the dorsal or ventral cornea to insert a recording electrode. A silver wire was inserted into the head capsule as the reference electrode. After the insect was set in a Faraday cage, a glass micropipette filled with tap water was inserted into the retina through the corneal hole. Light stimuli were provided by a xenon lamp (500 W) through a series of interference filters with 20-nm serial steps from 300 nm to 740 nm. Photon flux of each monochromatic light was measured using a radiometer (Model-470D, Sanso) and attenuated to equal density (1.0 × 1012 photons per square centimeter per second) using an optical wedge. Electroretinograms were recorded through an amplifier (MEZ-7200 Nihonkohen) connected to a computer via a MP-150 AD converter (BIOPAC). The light stimulus was applied through a liquid light guide set close to the compound eye. After fine-tuning of the position of the light guide by applying dim white light, the insect was kept in the dark for 15 min. After the dark adaptation, responses to monochromatic light flashes of 50-ms duration with 10-s intervals from short to long wavelength, and in the reverse direction, were recorded. This bidirectional stimulation protocol was repeated at least twice, thereby yielding four or more spectral responses. The response-light intensity (V-logI) function with a range of three log units was recorded at the wavelength that gave the maximal response. The V-logI data were fitted to the Naka–Rushton function, V/Vmax = In/(In + Kn), where I is the stimulus intensity, V is the response amplitude, Vmax is the maximum response amplitude, K is the stimulus intensity eliciting 50% of Vmax, and n is the exponential slope. We then converted the spectral response into spectral sensitivity, which is the reciprocal of the stimulus intensity required for a criterion response.
RNA Sequencing and Opsin Gene Assembly.
For all of the dragonfly species examined, total RNA samples were extracted from freshly dissected dorsal side of adult eyes, ventral side of adult eyes, adult head region around ocelli, and whole larval head using RNAiso plus (Takara) and RNeasy mini columns (Qiagen). For several species, the whole adult head was also subjected to RNA sample preparation. Using 1 µg total RNA per sample as template, cDNA libraries were constructed using TruSeq RNA Sample Preparation Kits v2 (Illumina), and the libraries were sequenced by HiSeq 2000, HiSeq 2500 or MiSeq (Illumina). Sequence data were deposited in the DNA Data Bank Japan Sequence Read Archive (accession numbers are shown in Table S1). The raw reads were subjected to de novo assembling using the Trinity program (72) implemented in the MASER pipeline (cell-innovation.nig.ac.jp/public/contents/service.html#pf_maser). After automatic assembling, we checked and corrected each of the opsin gene sequences manually using Integrative Genomics Viewer (37), and confirmed the revised sequences by RT-PCR and DNA sequencing. After revising the opsin gene sequences, sequence read mapping was performed using the BWA-mem software (73) implemented in the MASER pipeline, whereby transcript expression levels were estimated to calculate fragments per kilobase of exon per million (FPKM) values. Opsin genes of L. fulva were obtained by tBLASTn search against the draft genome sequence (APVN01000000) (https://www.hgsc.bcm.edu/).
Phylogenetic Analysis.
Deduced amino acid sequences of opsin genes were aligned using the Clustal W program implemented in the program package MEGA6 (74). Molecular phylogenetic analyses were conducted by neighbor-joining method and maximum-likelihood method using MEGA6. Bootstrap values were obtained by 1,000 resamplings.
Supplementary Material
Acknowledgments
We thank Makoto Machida, Tooru Oono, Osamu Shimbori, and Hiroyuki Futahashi for insect samples, and Akira Ozono for photos of dragonfly heads in Fig. 1. This work was supported by the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research Grants 23780058 and 26711021 (to R.F.) and 21247009 and 26251036 (to K.A.), and the Cooperative Research Grant of the Genome Research for BioResource, NODAI Genome Research Center, Tokyo University of Agriculture (to R.F., R.K.-M., and S.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Bank Japan Read Archive, www.ddbj.nig.ac.jp (accession nos. BR001237–BR001256, LC009056–LC009302, DRA001714, DRA001715, DRA001718, DRA001744, DRA001755–DRA001758, DRA001761, DRA001779, DRA001780, DRA001792, DRA001793, DRA002343–DRA002347, DRA002352, DRA002354, DRA002390, DRA002393, DRA002395, DRA002455–DRA002461, DRA002482, DRA002485–DRA002487, DRA002489–DRA002492, DRA002494–DRA002499, DRA002761–DRA002764, DRA002766–DRA002769, DRA002771, DRA002773–DRA002781, DRA002790–DRA002791, DRA002806).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424670112/-/DCSupplemental.
References
- 1.Jacobs GH. Comparative Color Vision. Academic; New York: 1981. [Google Scholar]
- 2.Kelber A, Vorobyev M, Osorio D. Animal colour vision—Behavioural tests and physiological concepts. Biol Rev Camb Philos Soc. 2003;78(1):81–118. doi: 10.1017/s1464793102005985. [DOI] [PubMed] [Google Scholar]
- 3.Osorio D, Vorobyev M. A review of the evolution of animal colour vision and visual communication signals. Vision Res. 2008;48(20):2042–2051. doi: 10.1016/j.visres.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Terakita A. The opsins. Genome Biol. 2005;6(3):213. doi: 10.1186/gb-2005-6-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci. 2009;364(1531):2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barlow HB. What causes trichromacy? A theoretical analysis using comb-filtered spectra. Vision Res. 1982;22(6):635–643. doi: 10.1016/0042-6989(82)90099-2. [DOI] [PubMed] [Google Scholar]
- 7.Maloney LT. Evaluation of linear models of surface spectral reflectance with small numbers of parameters. J Opt Soc Am A. 1986;3(10):1673–1683. doi: 10.1364/josaa.3.001673. [DOI] [PubMed] [Google Scholar]
- 8.Cronin TW, Johnsen S, Marshall NJ, Warrant EJ. Visual Ecology. Princeton Univ Press; Princeton: 2014. [Google Scholar]
- 9.Briscoe AD. Reconstructing the ancestral butterfly eye: Focus on the opsins. J Exp Biol. 2008;211(Pt 11):1805–1813. doi: 10.1242/jeb.013045. [DOI] [PubMed] [Google Scholar]
- 10.Porter ML, et al. Shedding new light on opsin evolution. Proc Biol Sci. 2012;279(1726):3–14. doi: 10.1098/rspb.2011.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill CA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298(5591):176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 12.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316(5832):1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chinen A, Hamaoka T, Yamada Y, Kawamura S. Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics. 2003;163(2):663–675. doi: 10.1093/genetics/163.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto Y, Fukamachi S, Mitani H, Kawamura S. Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes) Gene. 2006;371(2):268–278. doi: 10.1016/j.gene.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Owens GL, Windsor DJ, Mui J, Taylor JS. A fish eye out of water: Ten visual opsins in the four-eyed fish, Anableps anableps. PLoS ONE. 2009;4(6):e5970. doi: 10.1371/journal.pone.0005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laver CR, Taylor JS. RT-qPCR reveals opsin gene upregulation associated with age and sex in guppies (Poecilia reticulata)—A species with color-based sexual selection and 11 visual-opsin genes. BMC Evol Biol. 2011;11:81. doi: 10.1186/1471-2148-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owens GL, Rennison DJ, Allison WT, Taylor JS. In the four-eyed fish (Anableps anableps), the regions of the retina exposed to aquatic and aerial light do not express the same set of opsin genes. Biol Lett. 2012;8(1):86–89. doi: 10.1098/rsbl.2011.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagi R, et al. Correlation between nuptial colors and visual sensitivities tuned by opsins leads to species richness in sympatric Lake Victoria cichlid fishes. Mol Biol Evol. 2012;29(11):3281–3296. doi: 10.1093/molbev/mss139. [DOI] [PubMed] [Google Scholar]
- 19.Porter ML, Bok MJ, Robinson PR, Cronin TW. Molecular diversity of visual pigments in Stomatopoda (Crustacea) Vis Neurosci. 2009;26(3):255–265. doi: 10.1017/S0952523809090129. [DOI] [PubMed] [Google Scholar]
- 20.Porter ML, et al. The evolution of complexity in the visual systems of stomatopods: Insights from transcriptomics. Integr Comp Biol. 2013;53(1):39–49. doi: 10.1093/icb/ict060. [DOI] [PubMed] [Google Scholar]
- 21.Colbourne JK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331(6017):555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoen HH, How MJ, Chiou TH, Marshall J. A different form of color vision in mantis shrimp. Science. 2014;343(6169):411–413. doi: 10.1126/science.1245824. [DOI] [PubMed] [Google Scholar]
- 23.Tillyard BJ. The Biology of Dragonflies. Cambridge Univ Press; London: 1917. [Google Scholar]
- 24.Yager DD. Structure, development, and evolution of insect auditory systems. Microsc Res Tech. 1999;47(6):380–400. doi: 10.1002/(SICI)1097-0029(19991215)47:6<380::AID-JEMT3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 25.Cocroft RB, Rodríguez RL. The behavioral ecology of insect vibrational communication. Bioscience. 2005;55(4):323–334. [Google Scholar]
- 26.Corbet PS. Dragonflies, Behavior and Ecology of Odonata. Cornell Univ Press; Ithaca, NY: 1999. [Google Scholar]
- 27.Córdoba-Aguilar A. Dragonflies and Damselflies: Model Organisms for Ecological and Evolutionary Research. Oxford Univ Press; London: 2008. [Google Scholar]
- 28.Autrum H, Kolb G. Spektrale Empfindlichkeit einzelner Sehzellen der Aeschniden. Z Vgl Physiol. 1968;60(4):450–477. [Google Scholar]
- 29.Eguchi E. Fine structure and spectral sensitivities of retinular cells in the dorsal sector of compound eyes in the dragonfly, Aeschna. Z Vgl Physiol. 1971;71(2):201–218. [Google Scholar]
- 30.Meinertzhagen IA, Menzel R, Kahle G. The identification of spectral receptor types in the retina and lamina of the dragonfly Sympetrum rubicundulum. J Comp Physiol. 1983;151(3):295–310. [Google Scholar]
- 31.Yang EC, Osorio D. Spectral sensitivities of photoreceptors and lamina monopolar cells in the dragonfly, Hemicordulia tau. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1991;169(6):663–669. [Google Scholar]
- 32.Labhart T, Nilsson DE. The dorsal eye of the dragonfly Sympetrum: Specializations for prey detection against the blue sky. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1995;176(4):437–453. [Google Scholar]
- 33.Bybee SM, Johnson KK, Gering EJ, Whiting MF, Crandall KA. All the better to see you with: A review of odonate color vision with transcriptomic insight into the odonate eye. Org Divers Evol. 2012;12(3):241–250. [Google Scholar]
- 34.Huang SC, Chiou TH, Marshall J, Reinhard J. Spectral sensitivities and color signals in a polymorphic damselfly. PLoS ONE. 2014;9(1):e87972. doi: 10.1371/journal.pone.0087972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silsby J. Dragonflies of the World. Smithonian Institution Press; Washington, DC: 2001. [Google Scholar]
- 36.Dijkstra KDB, et al. The classification and diversity of dragonflies and damselflies (Odonata) Zootaxa. 2013;3703(1):36–45. [Google Scholar]
- 37.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koyanagi M, Nagata T, Katoh K, Yamashita S, Tokunaga F. Molecular evolution of arthropod color vision deduced from multiple opsin genes of jumping spiders. J Mol Evol. 2008;66(2):130–137. doi: 10.1007/s00239-008-9065-9. [DOI] [PubMed] [Google Scholar]
- 39.Land MF. Variations in the structure and design of compound eyes. In: Stavenga DG, Hardie RC, editors. Facets of Vision. Springer; Heidelberg: 1989. pp. 90–111. [Google Scholar]
- 40.Sugimura M, Ishida S, Kojima K, Ishida K, Aoki T. Dragonflies of the Japanese Archipelago in Color. Hokkaido Univ Press; Sapporo, Japan: 2001. [Google Scholar]
- 41.Ozono A, Kawashima I, Futahashi R. Dragonflies of Japan. Bunichi-Sogo Syuppan; Tokyo: 2012. [Google Scholar]
- 42.Coemans MA, Vos Hzn JJ, Nuboer JF. The relation between celestial colour gradients and the position of the sun, with regard to the sun compass. Vision Res. 1994;34(11):1461–1470. doi: 10.1016/0042-6989(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 43.Blagodatski A, et al. Under- and over-water halves of Gyrinidae beetle eyes harbor different corneal nanocoatings providing adaptation to the water and air environments. Sci Rep. 2014;4:6004. doi: 10.1038/srep06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirschfeld K. The visual system of the fly: Physiological optics and functional anatomy as related to behaviour. In: Schmitt FO, Worden FG, editors. The Neurosciences 4th Study Program. MIT Press; Cambridge, MA: 1979. pp. 297–310. [Google Scholar]
- 45.Zeil J. Sexual dimorphism in the visual system of flies; The compound eyes and neural superposition in Bibionidae (Diptera) J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1983;150(3):379–393. [Google Scholar]
- 46.Horridge GA. The ommatidium of the dorsal eye of cloeon as a specialization for photoreisomerization. Proc R Soc Lond B Biol Sci. 1976;193(1110):17–29. doi: 10.1098/rspb.1976.0028. [DOI] [PubMed] [Google Scholar]
- 47.Wolburg-Buchholz K. The dorsal eye of Chloeon dipterum (Ephemeroptera). A light- and electron microscopical study. Z Naturforsch C. 1976;31c:335–336. [Google Scholar]
- 48.Schneider L, Draslar K, Langer H, Gogala M, Schlecht P. Feinstruktur und Schirmpigment-Eigenschaften der Ommatidien des Doppelauges von Ascalaphus (Insecta, Neuroptera) Cytobiologie. 1978;16(2):274–307. [Google Scholar]
- 49.Stavenga DG, Kinoshita M, Yang EC, Arikawa K. Retinal regionalization and heterogeneity of butterfly eyes. Naturwissenschaften. 2001;88(11):477–481. doi: 10.1007/s001140100268. [DOI] [PubMed] [Google Scholar]
- 50.Dietrich W. Die Facettenaugen der Dipteren. Z Wiss Zool. 1909;92(3):465–539. [Google Scholar]
- 51.Land MF. Optics and vision in invertebrates. In: Autrum H, editor. Handbook of Sensory Physiology vol VII/6B. Springer; Heidelberg: 1981. pp. 471–592. [Google Scholar]
- 52.Nilsson DE. The transparent compound eye of Hyperia (Crustacea): Examination with a new method for analysis of refractive index gradients. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1982;147(3):339–349. [Google Scholar]
- 53.Kitamoto J, Sakamoto K, Ozaki K, Mishina Y, Arikawa K. Two visual pigments in a single photoreceptor cell: Identification and histological localization of three mRNAs encoding visual pigment opsins in the retina of the butterfly Papilio xuthus. J Exp Biol. 1998;201(Pt 9):1255–1261. doi: 10.1242/jeb.201.9.1255. [DOI] [PubMed] [Google Scholar]
- 54.Awata H, Wakakuwa M, Arikawa K. Evolution of color vision in pierid butterflies: Blue opsin duplication, ommatidial heterogeneity and eye regionalization in Colias erate. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2009;195(4):401–408. doi: 10.1007/s00359-009-0418-7. [DOI] [PubMed] [Google Scholar]
- 55.Ogawa Y, et al. Coexpression of three middle wavelength-absorbing visual pigments in sexually dimorphic photoreceptors of the butterfly Colias erate. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2012;198(12):857–867. doi: 10.1007/s00359-012-0756-8. [DOI] [PubMed] [Google Scholar]
- 56.Lall AB, Strother GK, Cronin TW, Seliger HH. Modification of spectral sensitivities by screening pigments in the compound eyes of twilight-active fireflies (Coleoptera: Lampyridae) J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1988;162(1):23–33. doi: 10.1007/BF01342700. [DOI] [PubMed] [Google Scholar]
- 57.Cronin TW, Järvilehto M, Weckström M, Lall AB. Tuning of photoreceptor spectral sensitivity in fireflies (Coleoptera: Lampyridae) J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2000;186(1):1–12. doi: 10.1007/s003590050001. [DOI] [PubMed] [Google Scholar]
- 58.Melin AD, Moritz GL, Fosbury RA, Kawamura S, Dominy NJ. Why aye-ayes see blue. Am J Primatol. 2012;74(3):185–192. doi: 10.1002/ajp.21996. [DOI] [PubMed] [Google Scholar]
- 59.Stange G, Stowe S, Chahl JS, Massaro A. Anisotropic imaging in the dragonfly median ocellus: A matched filter for horizon detection. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2002;188(6):455–467. doi: 10.1007/s00359-002-0317-7. [DOI] [PubMed] [Google Scholar]
- 60.van Kleef J, James AC, Stange G. A spatiotemporal white noise analysis of photoreceptor responses to UV and green light in the dragonfly median ocellus. J Gen Physiol. 2005;126(5):481–497. doi: 10.1085/jgp.200509319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berry RP, Stange G, Warrant EJ. Form vision in the insect dorsal ocelli: An anatomical and optical analysis of the dragonfly median ocellus. Vision Res. 2007;47(10):1394–1409. doi: 10.1016/j.visres.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 62.van Kleef J, Berry R, Stange G. Directional selectivity in the simple eye of an insect. J Neurosci. 2008;28(11):2845–2855. doi: 10.1523/JNEUROSCI.5556-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pollock JA, Benzer S. Transcript localization of four opsin genes in the three visual organs of Drosophila; RH2 is ocellus specific. Nature. 1988;333(6175):779–782. doi: 10.1038/333779a0. [DOI] [PubMed] [Google Scholar]
- 64.Henze MJ, Dannenhauer K, Kohler M, Labhart T, Gesemann M. Opsin evolution and expression in arthropod compound eyes and ocelli: Insights from the cricket Gryllus bimaculatus. BMC Evol Biol. 2012;12:163. doi: 10.1186/1471-2148-12-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohno S. Evolution by Gene Duplication. Springer; New York: 1970. [Google Scholar]
- 66.Zhang J. Evolution by gene duplication: An update. Trends Ecol Evol. 2003;18(6):292–298. [Google Scholar]
- 67.Arikawa K, Mizuno S, Kinoshita M, Stavenga DG. Coexpression of two visual pigments in a photoreceptor causes an abnormally broad spectral sensitivity in the eye of the butterfly Papilio xuthus. J Neurosci. 2003;23(11):4527–4532. doi: 10.1523/JNEUROSCI.23-11-04527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmeling F, et al. Opsin expression, physiological characterization and identification of photoreceptor cells in the dorsal rim area and main retina of the desert locust, Schistocerca gregaria. J Exp Biol. 2014;217(Pt 19):3557–3568. doi: 10.1242/jeb.108514. [DOI] [PubMed] [Google Scholar]
- 69.Sison-Mangus MP, Bernard GD, Lampel J, Briscoe AD. Beauty in the eye of the beholder: The two blue opsins of lycaenid butterflies and the opsin gene-driven evolution of sexually dimorphic eyes. J Exp Biol. 2006;209(Pt 16):3079–3090. doi: 10.1242/jeb.02360. [DOI] [PubMed] [Google Scholar]
- 70.Rajkumar P, Rollmann SM, Cook TA, Layne JE. Molecular evidence for color discrimination in the Atlantic sand fiddler crab, Uca pugilator. J Exp Biol. 2010;213(Pt 24):4240–4248. doi: 10.1242/jeb.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: The genes encoding blue, green, and red pigments. Science. 1986;232(4747):193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 72.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN]
- 74.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Misof B, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346(6210):763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 76.Futahashi R. A revisional study of Japanese dragonflies based on DNA analysis. Tombo. 2014;56:57–59. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.