Significance
Gram-negative bacteria sense and respond to compromised outer-membrane assembly to maintain cell integrity. In Escherichia coli, outer-membrane stress is sensed via regulated intramembrane proteolysis, a universal signal-transduction system. The C-terminal peptides of unassembled outer-membrane proteins (OMPs) bind and activate DegS, a periplasmic protease that cleaves a transmembrane anti-sigma factor, increasing transcription of stress-response genes. The molecular mechanism by which OMPs allosterically activate DegS has been unclear. We show that OMP-peptide binding to DegS initiates a steric clash that breaks autoinhibitory interactions, resulting in proteolytic activation. This mechanism does not require strong sequence conservation. As proteases related to DegS play important roles in protein-quality control and regulation in most organisms, these studies provide an important step forward in understanding these enzymes.
Keywords: regulated intracellular proteolysis, allosteric regulation, outer-membrane proteins, stress sensor
Abstract
Escherichia coli senses envelope stress using a signaling cascade initiated when DegS cleaves a transmembrane inhibitor of a transcriptional activator for response genes. Each subunit of the DegS trimer contains a protease domain and a PDZ domain. During stress, unassembled outer-membrane proteins (OMPs) accumulate in the periplasm and their C-terminal peptides activate DegS by binding to its PDZ domains. In the absence of stress, autoinhibitory interactions, mediated by the L3 loop, stabilize inactive DegS, but it is not known how this autoinhibition is reversed during activation. Here, we show that OMP peptides initiate a steric clash between the PDZ domain and the L3 loop that results in a structural rearrangement of the loop and breaking of autoinhibitory interactions. Many different L3-loop sequences are compatible with activation but those that relieve the steric clash reduce OMP activation dramatically. Our results provide a compelling molecular mechanism for allosteric activation of DegS by OMP-peptide binding.
The Escherichia coli DegS protease is anchored to the periplasmic face of the inner membrane, where it functions to sense outer-membrane stress (1, 2). DegS is only activated when outer-membrane proteins (OMPs) accumulate in the periplasm as a consequence of stresses that compromise normal membrane insertion. The C-terminal peptides of these unassembled OMPs bind DegS and activate cleavage of the periplasmic portion of RseA (3), a transmembrane protein with a cytoplasmic domain that binds and inhibits the σE transcription factor (4). This cleavage event initiates a proteolytic cascade that ultimately releases σE to stimulate transcription of stress-response genes (5). Under nonstress conditions, OMPs do not accumulate in the periplasm, DegS cleavage of RseA is minimal, and σE remains bound to the cytoplasmic domain of RseA in an inactive state (1, 2).
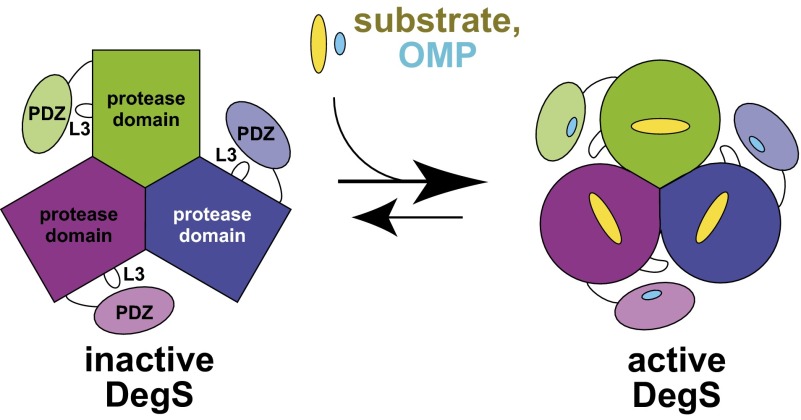
Each DegS subunit contains a membrane anchor, a trypsin-like protease domain, and a PDZ domain, which binds OMP C-terminal peptides (3). Crystal structures of DegS in active and inactive conformations reveal that the protease domains pack together to form a trimer with the PDZ domains located on the periphery (6, 7). Activation of DegS is generally well-described by a two-state allosteric model in which the preferential binding of both RseA and OMP peptides to the active conformation of the enzyme stabilizes this species in a positively cooperative manner (8) (Fig. 1). As expected for this model, OMP binding to a single PDZ domain can activate cis and trans protease domains (9). The most activating OMP peptide (YYF) increases DegS activity ∼1,000-fold, although other peptides activate to lesser extents because they bind active DegS less tightly or inactive DegS more tightly (8). Activation involves the rearrangement of a network of conserved residues at each subunit interface as well as the formation of a functional oxyanion hole at each catalytic site (6, 10, 11).
Fig. 1.
Cartoon depiction of the inactive and active states of the DegS trimer (subunits colored green, purple, and blue). The protease domains (darker colors) pack together at the center of the trimer and the PDZ domains (lighter colors) are on the periphery. The allosteric equilibrium between inactive and active DegS is controlled by the binding of OMP peptide (cyan ovals) to the PDZ domains and RseA substrate (yellow ovals) to the protease domains. The L3 loop of the protease domain makes autoinhibitory interactions with the PDZ domains that stabilize the inactive state and are broken upon OMP-peptide binding.
In the absence of OMP peptide, virtually all DegS molecules are inactive (3, 12). Two models have been proposed for OMP activation, each involving the L3 loop (residues 176–189) of the protease domain. This loop extends toward the PDZ domain and assumes different conformations in active and inactive DegS (6, 7). One model posits that PDZ-bound OMP peptides make specific contacts with the L3 loop to stabilize active DegS (6, 13). Several observations seem inconsistent with this model. For example, there are no conserved interactions between OMP peptides and the L3 loop in multiple crystal structures, and DegS∆PDZ has robust OMP-independent proteolytic activity (10, 12, 14). A second model posits that OMP-peptide binding destabilizes or breaks autoinhibitory contacts mediated by the L3 loop (8, 12). In inactive DegS, for example, several interactions are observed between the L3 loop and the PDZ domain that are absent in active DegS (6, 7). Consistent with an autoinhibitory role, DegS variants in which these interactions are removed have higher basal activity but can still be activated by OMP peptides (12).
Here, we provide strong evidence for a steric-clash mechanism in which OMP binding antagonizes autoinhibitory interactions, thereby activating DegS. Upon OMP binding, Met319 in the PDZ domain assumes a different rotamer to avoid a clash with bound peptide, and the M319A mutation reduces OMP activation dramatically (8). How this forced movement of Met319 might result in activation has not been clear, but the side chain of Asn182 in the L3 loop is reasonably close to Met319 in inactive DegS. Strikingly, replacement of Asn182 with Ala or Gly results in DegS variants that bind OMP peptides but show almost no activation. A wide variety of larger amino acids at this position support OMP-peptide activation in vitro and in vivo. We also probe the importance of additional L3-loop residues for autoinhibition and OMP activation. Our results provide a straightforward and compelling mechanism for allosteric activation of DegS in which Met319 in the PDZ domain must move to accommodate bound OMP peptide, forcing movement of Asn182 and the L3 loop, which breaks autoinhibitory interactions and allows DegS to assume the active conformation.
Results
Evidence for a Steric-Clash Model of Activation.
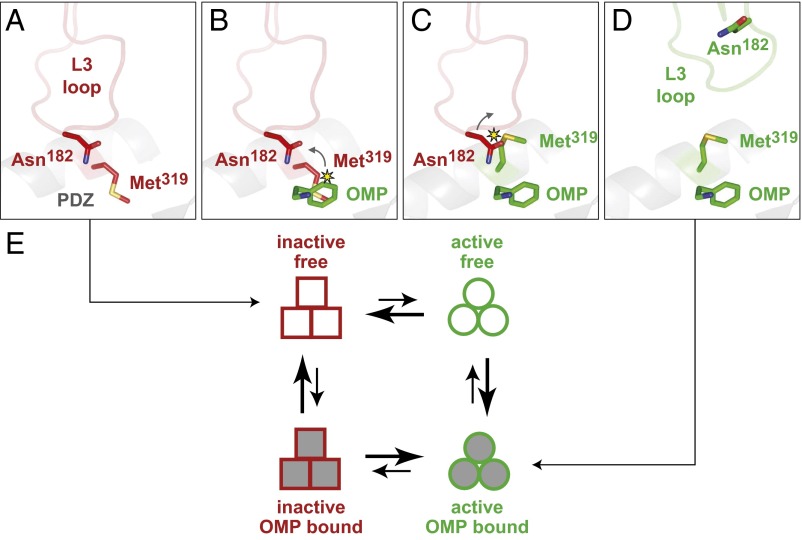
Met319 in the PDZ domain moves to avoid a steric clash with bound OMP peptide, contributing to DegS activation (8). Although no structures of OMP-bound inactive DegS have been solved, Asn182 is the L3-loop residue closest to the Met319 side chain in inactive DegS (Fig. 2A). Indeed, alignment of inactive and active DegS structures suggested that the Met319 rotamer in the OMP-bound active structure would clash with the side chain of Asn182 in inactive DegS but not in active DegS (Fig. 2 B–D). To avoid this clash, either or both side chains could adopt an unfavorable rotamer, or rearrangement of the L3 loop could occur. Either outcome would destabilize the autoinhibitory conformation of the L3 loop and protease domain relative to the active conformation, thereby altering the equilibrium constant relating the OMP-bound active and inactive conformations of DegS and leading to activation (Fig. 2E).
Fig. 2.
Alignment of inactive DegS and active DegS suggests a steric clash between a PDZ-domain residue and an L3-loop residue. (A) In inactive DegS (PDB ID: 4G0E, red), the Met319 side chain is part of the OMP-peptide binding pocket. (B) If OMP peptide bound to inactive DegS as it does to active DegS, then a clash between the Met319 side chain and the C-terminal phenylalanine of the OMP peptide would occur. (C) If the Met319 side chain in OMP-bound inactive DegS assumed the same rotamer it does in active DegS, then a clash with the Asn182 side chain in the L3 loop would occur. (D) In active DegS (PDB ID: 3GDS, green), the L3 loop assumes a completely different conformation in which Asn182 is distant from Met319. (E) Allosteric cycle relating peptide-free (open squares or circles) and peptide-bound (filled squares or circles) species of inactive (red) and inactive (green) DegS. For wild-type DegS in the absence of substrate, ∆G is 5.6 kcal/mol for the OMP-free inactive to active transition and 1.2 kcal/mol for the OMP-bound inactive to active transition (8).
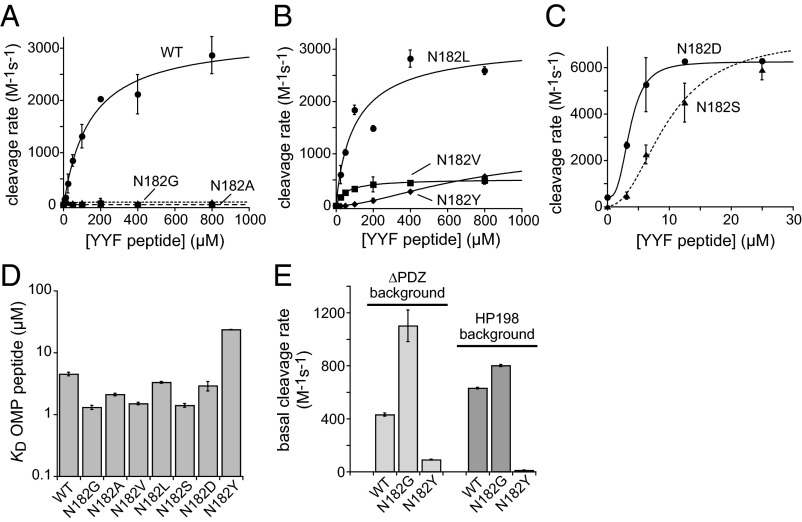
This steric-clash model predicts that the size of the residue-182 side chain but not its specific chemical identity should be an important determinant of activation. To test this prediction, we constructed DegS mutants with glycine, alanine, serine, valine, aspartic acid, leucine, and tyrosine at position 182 and purified these enzymes. For each variant and wild-type DegS, we assayed the rate of cleavage of a sub-KM concentration of a 35S-labeled RseA fragment by measuring accumulation of an acid-soluble radioactive product over a range of YYF peptide concentrations (Fig. 3 A–C). In support of a steric-clash model, the two mutants with the smallest side chains (N182G and N182A) showed maximal levels of YYF-dependent activity that were at least 50-fold lower than wild-type DegS (Fig. 3A). This extremely poor activation did not result from a failure to bind OMP peptide, as N182G and N182A DegS bound a fluorescent OMP decapeptide with KD’s slightly tighter than wild-type DegS (Fig. 3D and Table 1). To test whether the N182G mutation might reduce activity of the DegS protease domain, we combined it with the ∆PDZ mutation, which allows robust OMP-independent cleavage. The purified N182G DegS∆PDZ variant was more active than the parental DegS∆PDZ enzyme in this assay (Fig. 3E). We also combined the N182G mutation with the H198P mutation, which stabilizes active DegS and increases OMP-independent basal activity (8, 10). The N182G/H198P double mutant had slightly higher basal cleavage activity than H198P DegS (Fig. 3E). These results show that the very low level of OMP-activated cleavage observed for the N182G mutant is a consequence of a failure to respond normally to peptide binding.
Fig. 3.
Enzymatic activity and OMP-binding of DegS variants with amino acid substitutions for Asn182 in the L3 loop. (A–C) Cleavage of RseA (100 µM) by wild-type (WT) DegS or variants (1 µM trimer) was assayed in the presence of different concentrations of the YYF OMP peptide. Values are averages ± SD (n = 2–4). Curves were fit to the equation y = basal + Amax* [YYF]h/(Kacth + [YYF]h). Fitted values of Kact, the Hill constant (h), and Amax were 140 µM, 1.0, and 3260 M-1s−1 (WT); 23 µM, 1.45, and 49 M-1s−1 (N182G); 105 µM, 0.92, and 3140 M-1s−1 (N182L); 55 µM, 0.96, and 270 M-1s−1 (N182V); 725 µM, 1.7, and 1040 M-1s−1 (N182Y); 3.6 µM, 3.0, and 5880 M-1s−1 (N182D); and 9.7 µM, 2.2, and 7,270 M-1s−1 (N182S). Reliable fitted values were not obtained for N182A. (D) KDs for binding of DegS and variants to a fluorescent OMP decapeptide are shown on a log scale. (E) Second-order rate constants for RseA cleavage by DegS variants in ∆PDZ or H198P backgrounds were determined in the absence of OMP peptide. Values in D and E are averages ± SEM (n = 2–4).
Table 1.
Properties of DegS and variants with L3-loop mutations
| Enzyme | Basal activity, M-1s−1 | YYF-stimulated activity, M-1s−1 | KD for OMP decapeptide, µM |
| DegS | <10 | 2,300 ± 230 | 4.5 ± 0.4 |
| N182G | <10 | 74 ± 35 | 1.3 ± 0.1 |
| N182A | <10 | 24 ± 5 | 2.1 ± 0.2 |
| N182V | <10 | 390 ± 170 | 1.5 ± 0.1 |
| N182L | <10 | 1,500 ± 150 | 3.3 ± 0.3 |
| N182S | <10 | 5,600 ± 760 | 1.4 ± 0.1 |
| N182D | 410 ± 50 | 6,100 ± 610 | 2.9 ± 0.6 |
| N182Y | <10 | 560 ± 56 | 24 ± 2 |
| L181A | 93 ± 25 | 1,500 ± 150 | 0.6 ± 0.06 |
| ∆183–187 | 20 ± 10 | 24 ± 4 | 0.3 ± 0.07 |
| L3-Gly5 | 480 ± 210 | 680 ± 160 | 0.4 ± 0.05 |
| H198P | 630 ± 63 | 17,000 ± 2,200 | 2.9 ± 0.4 |
| N182G/H198P | 800 ± 80 | 16,000 ± 1,600 | 0.7 ± 0.07 |
| N182Y/H198P | <10 | 15,000 ± 1,500 | 14 ± 1.4 |
| L181/H198P | 4,700 ± 470 | 17,000 ± 1,700 | 0.1 ± 0.05 |
| ∆183–187/H198P | 3,300 ± 900 | 3,500 ± 670 | 0.4 ± 0.04 |
| L3-Gly5/H198P | 11,000 ± 1,100 | 12,000 ± 2,500 | 0.2 ± 0.03 |
| ∆PDZ | 430 ± 43 | — | — |
| N182G/∆PDZ | 1,100 ± 120 | — | — |
| N182Y/∆PDZ | 88 ± 8 | — | — |
YYF-stimulated cleavage of RseA was measured in the presence of 200 µM YYF tripeptide for most variants and 800 µM YYF for N182Y. There is no binding site for OMP peptide in the ∆PDZ variants. Values reported are averages ± SEM (n = 2–7) or 10% of average, whichever is larger.
YYF peptide stimulated RseA cleavage by the remaining residue-182 mutants to maximal activities higher than wild type (N192D; N182S), similar to wild type (N182L), or lower than wild type (N182V; N182Y) (Fig. 3 B and C). As the maximal activities of these mutant did not scale in any simple fashion with the size of the residue 182 side chain, additional factors must also influence activation. Such factors could include altering the equilibrium between the active and inactive conformations of DegS either directly or by changing the extent to which OMP peptide binding favors these conformations. Relative to wild-type DegS, for example, the N182D mutant had higher unstimulated activity, higher maximal YYF-stimulated activity, and was half-maximally activated at lower concentrations of YYF peptide (Fig. 3C). These phenotypes are similar to other DegS mutants that destabilize inactive DegS (12). Indeed, Asp182 would be close to Asp320 in the PDZ domain of inactive DegS, and thus unfavorable electrostatic interactions should decrease autoinhibition and shift the conformational equilibrium to favor active DegS. By contrast, the N182Y mutant required much higher concentrations of YYF peptide to observe stimulation (Fig. 3B) and bound OMP peptide more weakly than wild-type DegS or the other residue-182 mutants (Fig. 3D). These results would be expected if Tyr182 alters the effects of OMP binding (modeling suggests that Tyr182 could pack against Met319) and/or stabilizes inactive DegS. In support of the latter model, introducing the N182Y mutation into the H198P or ∆PDZ DegS enzymes reduced unstimulated RseA cleavage substantially (Fig. 3E). These results show that interactions mediated by different residue-182 side chains in the L3 loop can affect unstimulated activity and activation by OMP peptides both positively and negatively.
Testing the Clash Model in Vivo.
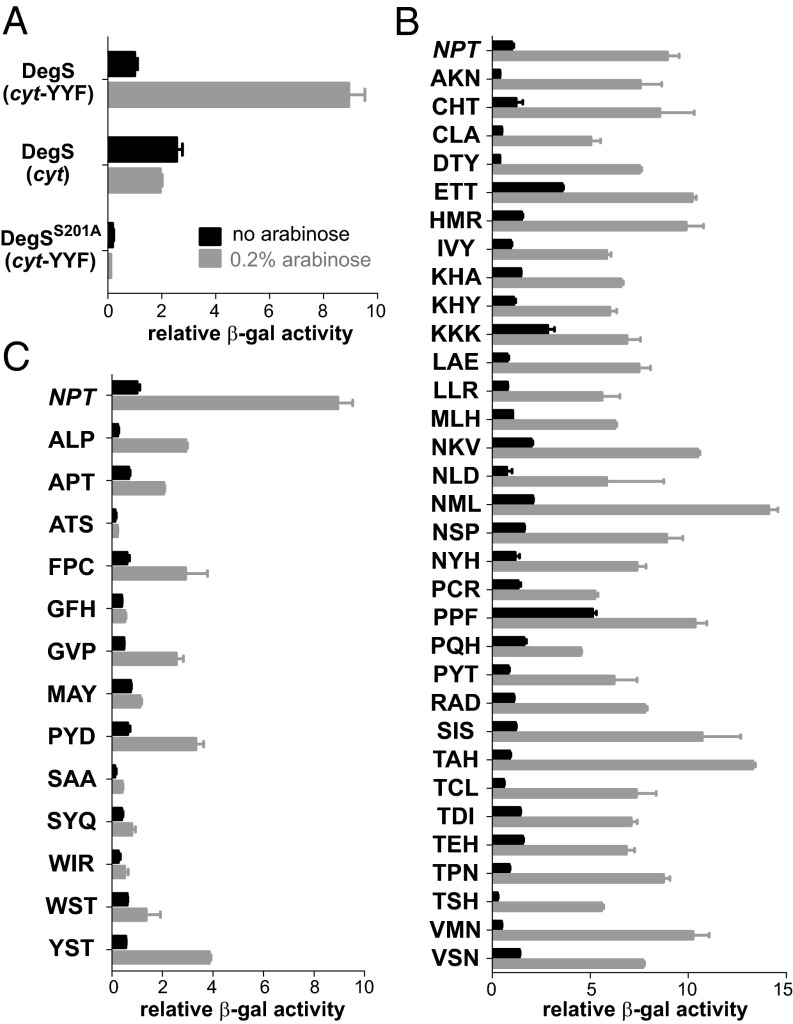
If a steric clash between the side chains of residue 182 and Met319 is a major determinant of OMP activation, then many different L3-loop sequences should be compatible with activation. To test this possibility, we randomized the codons for residues 182–184 and tested a set of variants for their ability to respond to OMP stimulation in vivo. For these studies, we expressed DegS variants from a plasmid in a ∆degS strain of E. coli carrying a σE-dependent chromosomal lacZ reporter (15). To assay OMP dependence, the test strain contained a second plasmid expressing a fusion of cytochrome b562 to a C-terminal sequence ending in YYF (cyt-YYF) under arabinose control (5). When wild-type DegS was expressed in the test strain, addition of 0.2% arabinose resulted in the expected induction of β-gal activity when the cyt-YYF protein was present but not when an otherwise identical protein lacking the C-terminal YYF sequence was present (Fig. 4A). An inactive DegS variant bearing the S201A mutation in the catalytic triad resulted in minimal β-galactosidase activity with or without arabinose induction of cyt-YYF (Fig. 4A).
Fig. 4.
Levels of β-galactosidase expressed from a σE-dependent lacZ reporter were used to assess the basal and OMP-stimulated RseA-cleavage activities of DegS and L3-loop mutants in vivo. Cytochrome b562 with a YYF C-terminal tail (cyt-YYF) or no tail (cyt) was expressed from an arabinose-inducible promoter. Black and gray bars represent β-gal levels in the absence and presence of 0.2% arabinose, respectively. (A) Control experiments showed that arabinose-mediated expression of cyt-YYF but not cyt stimulate wild-type DegS activity. Expression of cyt-YYF did not stimulate catalytically inactive DegSS201A. (B) β-gal levels with or without arabinose induction of cyt-YYF for DegS variants with different amino acids at residues 182–184. All variants in this panel support arabinose-induced activity at least fourfold higher than the basal activity of wild-type DegS (NPT). (C) DegS variants from the residue 182–184 library with arabinose-induced activity less than fourfold higher than the basal activity of wild-type DegS (NPT). Values in all panels are averages ± SEM (n = 2–8). DegS variants in B and C are designated by the one-letter code for the amino acids at residues 182, 183, and 184.
We assayed β-gal levels in strains transformed with 45 DegS variants containing L3-loop mutations with and without arabinose induction of cyt-YYF. Of these DegS variants, 32 supported moderate to strong β-gal induction in the presence of arabinose (fourfold or more over the wild-type basal activity; Fig. 4B). Among these variants, 15 different amino acids were represented at each randomized position (residue 182, ACDEHIKLMNPRSTV; residue 183, ACDEHIKLMPQSTVY; residue 184, ADEFHIKLNPRSTVY). Thus, the sequence determinants for DegS function and OMP activation at these L3-loop positions are extremely relaxed. Based on the failure of the N182A mutant to be strongly activated by YYF in vitro, we were surprised to find that a variant with an AKN184 sequence seemed fully active in vivo. By contrast and in line with results in vitro, the N182A variant (APT184) displayed weak induction in the cellular assay (Fig. 4C). For AKN184, it seems likely that Lys183 or Asn184 compensates for Ala182 and restores OMP responsiveness.
For 13 DegS variants, either no or low levels of β-gal induction were observed (Fig. 4C). In six cases, these results were expected because residue 182 was a glycine, alanine, or tyrosine (GFH184, GVP184, APT184, ATS184, ALP184, and YST184). Position 182 was a tryptophan or phenylalanine in three additional variants in this category (FPC184, WIR184, and WST184), suggesting that any aromatic residue at this position can stabilize inactive DegS. Notably, no variants in the strongly inducing class had an aromatic residue at position 182. The reason for poor induction by the remaining variants in this class (PYD184, MAY184, SAA184, and SYQ184) is less clear. These mutations may allow the L3 loop to adopt an alternative autoinhibitory conformation that alleviates the clash between Met319 and the residue-182 side chain, stabilize inactive DegS by other mechanisms, or reduce intracellular levels of the mutant proteins because of increased proteolysis or decreased mRNA stability.
Role of Additional L3 Loop Residues in Autoinhibition and OMP Activation.
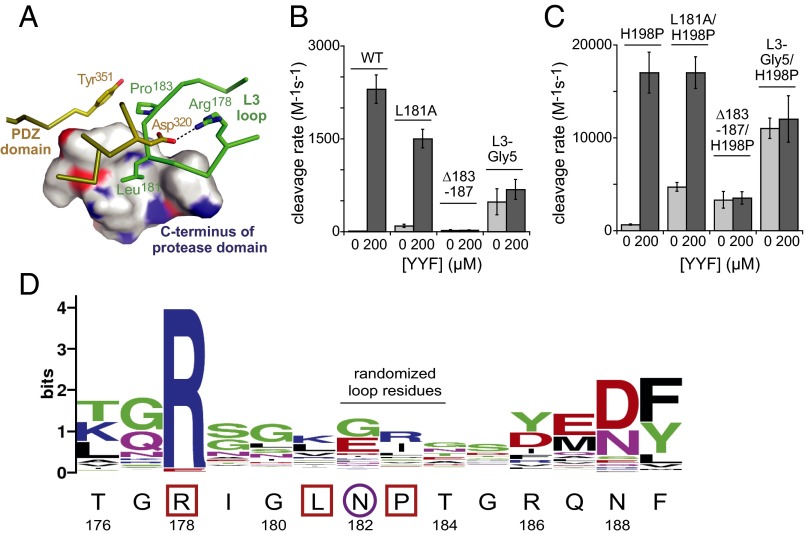
Some interactions that appear to be autoinhibitory do not involve the PDZ domain. Only in inactive DegS, for example, does Leu181 in the L3 loop pack into a hydrophobic pocket formed by side chains from the C-terminal segment of the protease domain (6, 7) (Fig. 5A). To test whether the Leu181 side chain contributes to autoinhibition, we constructed and purified L181A and L181A/H198P variants and assayed cleavage of RseA and OMP-peptide binding (Fig. 5 B and C and Table 1). The basal activity of L181A DegS was ∼80-fold higher than wild-type DegS and the basal activity of L181A/H198P DegS was ∼fivefold higher than H198P DegS, supporting a role for Leu181 in autoinhibition. YYF peptide enhanced cleavage by L181A and L181A/H198P to rates similar to the parental enzymes. The L181A mutation resulted in stronger OMP-peptide binding by a factor of ∼sevenfold in the wild-type background and ∼40-fold in the H198P background (Table 1). Tighter OMP binding to the L181A variants is expected if the free-energy gap between the inactive and active conformations is reduced in L181A DegS and becomes even smaller in L181A/H198P DegS, as OMP-binding affinity is weakened by the cost of the coupled inactive to active transition (8, 12).
Fig. 5.
The L3 loop. (A) Autoinhibitory interactions involving L3-loop residues (green) stabilize the inactive conformation of DegS (PDB ID: 4RR1). The side chain of Leu181 packs into a hydrophobic pocket formed by Leu240, Ile244, Lys247, and Val254 from the C-terminal part of the protease domain, and a hydrogen bond between the Arg256 side chain and the carbonyl oxygen of Leu181 helps stabilize these interactions. In addition, Arg178 makes a salt-bridge with Asp320 in the PDZ domain, and Pro183 packs against Tyr351 in the PDZ domain. (B) Basal and YYF-stimulated cleavage activities of wild-type DegS and variants with L3-loop mutations. (C) Basal and YYF-stimulated cleavage activities of H198P DegS and variants with L3-loop mutations. (D) Most residues in the L3 loop are poorly conserved in an alignment of prokaryotic and eukaryotic proteases homologous to DegS. The wild-type DegS sequence is shown below the WebLogo figure (19). L3-loop residues that mediate DegS autoinhibition are boxed; residue 182, which is critical for linking OMP binding to allosteric activation, is circled. Asn182 is flanked by residues that make autoinhibitory contacts, restricting its ability to avoid a clash upon OMP binding to inactive DegS.
We also constructed variants in which residues 183–187 of the L3 loop were deleted (∆183–187) or mutated en bloc to glycines (L3-Gly5) in wild-type and H198P backgrounds. Both mutants had increased basal activities compared with the parental enzymes, but these activities were not stimulated significantly by OMP peptide (Fig. 5C and Table 1). The activities of the ∆183–187 and L3-Gly5 mutants were lower in the wild-type than H198P backgrounds, indicating that the protease domains of both mutants still equilibrate between inactive and active conformations. These mutants were less active than their OMP-activated parents, however, suggesting residual autoinhibition and/or reduced stability of the active conformation relative to the inactive conformation. The latter possibility is plausible, as Arg178 in the L3 loop plays a role in stabilizing the active conformation of DegS (10). The ∆183–187 and L3-Gly5 mutations increased the affinity of OMP-decapeptide binding ∼10-fold (Table 1). Again, this strengthening of binding is expected if autoinhibitory interactions removed by both loop mutations keep the PDZ domain in a conformation with relatively weak OMP-peptide affinity. In combination, these results indicate that the wild-type L3 loop plays important roles both in maintaining DegS in an inactive state and in mediating OMP activation.
Discussion
The side-chain rotamer of Met319 in the PDZ domain changes between inactive DegS and OMP-bound active DegS, relieving a clash that would otherwise occur with the side chain of the C-terminal phenylalanine of the OMP peptide (8). As all OMP peptide variants known to activate DegS strongly have a C-terminal phenylalanine (6, 12, 14), as do most E. coli outer-membrane proteins (3), this OMP-dependent repositioning of Met319 is very likely to be common to all activating events. Consistent with an important role for this rotamer rearrangement, the M319A mutation resulted in stronger binding of OMP peptide but very low levels of activation. However, these studies did not answer the question of how OMP-mediated repositioning of Met319 transmits an activating signal to the protease domain. Based on modeling, we hypothesized that the repositioned Met319 side chain would clash with the side chain of Asn182 in the L3 loop, unless one or both of these side chains and/or the loop adopted unfavorable conformations that destabilize the autoinhibitory conformation and shift the equilibrium to favor active DegS. In support of this model, we find that mutants with small glycine or alanine side chains at position 182 bind OMP peptide normally but show almost no activation. For these mutants, we propose that repositioning of the Met319 side chain in OMP-bound DegS results in very mild destabilization of the autoinhibited conformation (perhaps to avoid minor clashes with other atoms of the L3 loop) and thus causes little activation.
By contrast, we find that DegS variants with serine, aspartic acid, valine, leucine, and tyrosine at position 182 can be activated by OMP peptide, as expected if these side chains also clash with Met319 in OMP-bound DegS and stabilize the active conformation. Indeed, when we screened a random library in which residues 182–184 of the L3 loop were randomly mutagenized, many different combinations of side chains at all three positions were compatible with robust OMP-mediated activation of DegS in vivo. Consistently, the central portion of the L3 loop shows very little sequence conservation in bacterial and metazoan DegS homologs (Fig. 5D). These results support a simple activation model based on steric incompatibility. An alternative proposal that activation results from stabilizing interactions between the penultimate side chain of bound OMP peptide and the L3 loop (6, 13) is inconsistent with other studies (8, 12, 14) and with our current results, as the Asn182 side chain is ∼10 Å from the nearest atoms of bound OMP peptide. Are clash mechanisms also likely to operate in other Gram-negative bacteria, where unassembled OMPs could also activate DegS signaling? In most DegS homologs from Gram-negative proteobacteria, the residue corresponding to 319 is methionine or leucine, whereas position 182 is typically serine or asparagine, supporting a clash mechanism similar to that used by E. coli DegS. However, a subset of these DegS homologs have methionine or leucine at 319 but glycine at position 182. A clash mechanism might still operate in these enzymes if their L3 loops adopted a somewhat different conformation in which residues other than 182 clashed with a repositioned 319 side chain.
OMP peptide does not activate DegS variants bearing a deletion (∆183–187) in the L3 loop or with residues 183–187 changed to glycines (L3-Gly5), despite the presence of the wild-type Asn182. The cleavage activities of these variants are still increased substantially by the H198P mutation, which stabilizes functional DegS, indicating that they still equilibrate between inactive and active enzyme conformations. For these OMP-unresponsive mutants, we suggest that changes in the L3-loop conformation prevent a clash between Met319 and Asn182 in OMP-bound inactive DegS and thus uncouple OMP binding from activation. This model is supported by the increased affinities of these variants for OMP peptide.
In addition to mediating OMP-peptide activation, the side chain of residue 182 can influence the equilibrium between the inactive and active conformations of DegS. For example, our results indicate that N182Y mutation stabilizes the inactive conformation of DegS, whereas the N182D mutation destabilizes this conformation. Interestingly, the N182Y mutation also decreases the affinity of DegS for OMP peptide, probably by restricting movement of Met319. Thus, the identity of the position-182 side chain can affect the intrinsic allosteric equilibrium of DegS as well as its ability to respond to OMP peptide.
Our current work, in combination with previous structural and biochemical studies (6–8, 12, 14), provides strong support for a model in which the L3 loop and PDZ domain play major roles in autoinhibition of proteolytic activity in OMP-free DegS and in triggering activation in OMP-bound DegS. In inactive DegS, interactions between residues in the L3 loop and residues in the PDZ or protease domains are clearly autoinhibitory. For example, we find that the L181A, N182D, ∆183–187, and L3-Gly5 mutations in the L3 loop all substantially increase the cleavage activity of DegS in the absence of OMP peptide. Previous studies show that the P183A mutation in the L3 loop, the D320A mutation in the PDZ domain (which eliminates a salt bridge with Arg178 in the L3 loop), and deletion of the PDZ domain also increase the basal activity of DegS (12). Thus, interactions between the L3 loop and the OMP-free PDZ domain or protease domain stabilize inactive DegS, whereas conformational rearrangements needed to accommodate the binding of OMP peptides to the PDZ domain of inactive DegS destabilize these autoinhibitory interactions, shifting the equilibrium to favor the active DegS conformation.
Materials and Methods
Protein Expression and Purification.
Variants of E. coli DegS (residues 27–355) with an N-terminal His6 tag in place of the wild-type membrane anchor and a 35S-labeled variant of the periplasmic domain of E. coli RseA (residues 121–216) were expressed and purified as described (12). Synthetic YYF peptide (GenScript) was purified by HPLC.
Enzymatic and Biochemical Assays.
Cleavage and binding assays were performed at room temperature in buffer containing 150 mM sodium phosphate (pH 8.3), 380 mM NaCl, 10% (vol/vol) glycerol and 4 mM EDTA following published protocols (12). Briefly, cleavage assays were performed by incubating 1 µM trimeric DegS or variants with or without YYF tripeptide (typically at a concentration of 200 µM) and three sub-KM concentrations of 35S-RseA. At different times, samples were quenched trichloroacetic acid (TCA) to a final concentration of 8.6% (wt/vol) and incubated on ice for 20 min. After centrifugation, acid-soluble radioactive cleavage products were quantified by scintillation counting. Second-order rate constants were calculated by taking the slope of the plot of initial velocity versus substrate concentration. For peptide-activation assays, 1 µM trimeric DegS or variants were incubated with different concentrations of YYF peptide and 100 µM 35S-RseA. Samples were quenched in TCA, incubated, centrifuged, and quantified as described above. Cleavage rates were determined by dividing the initial velocity by the substrate and enzyme concentrations.
OMP-peptide binding assays were performed by titrating increasing concentrations of DegS or variants against 100 nM fluorescein-DNRDGNVYYF and monitoring changes in fluorescence anisotropy (excitation 480 nm; emission 520 nm) after correction for protein scattering. Binding curves were fit using nonlinear least-square algorithms implemented in Prism (GraphPad).
Construction of L3 Mutant Library.
A degS plasmid with an engineered silent SalI site midgene was amplified and digested with SalI and BamHI restriction enzymes. Primers were constructed to amplify a portion of degS containing this SalI site at the 5′ end and a native BamHI site at the 3′ end. The latter plasmid contained an NNS NNS NNS sequence (N = A, C, G, T; S = C, G) in place of the wild-type sequence for codons 182–184. This amplified DNA was cut with SalI and BamHI and ligated into the cut vector described above. The ligation reaction was transformed into E. coli DH5α, and colonies were selected for plasmid isolation and sequencing.
β-Galactosidase Assay.
σE activity was measured by monitoring β-galactosidase (β-gal) expression from a chromosomal σE -dependent lacZ reporter gene in Φλ(rpoHP3::lacZ), as described (5, 15–17). Strain CAG33315 (14) was transformed with plasmid BA182 (3) containing cyt-YYF in pBAD33. That strain was further transformed with plasmids containing DegS variants in a pEXT20 vector. Strains were grown at 30 °C in ampicillin (100 µg/mL) and chloramphenicol (34 µg/mL) overnight, diluted 1:150 into the wells of a 96-well assay plate (Corning), and allowed to grow to midlog phase with or without 0.2% arabinose in the presence of the same concentrations of ampicillin/chloramphenicol. The plate was then incubated on ice to arrest growth, a final OD600 was measured, and cells were lysed with PopCulture + rLysozyme (EMD Millipore, Novagen). Lysate was added to an ortho-nitrophenyl-β-galactoside (ONPG) solution and OD420 was monitored for 1 h. The slope of this line, minus a blank, was taken as the β-galactosidase activity, which was normalized to the activity of the strain containing the wild-type DegS plasmid grown in the absence of arabinose. For the no-stress-signal control, plasmid pBA175, which contains cyt instead of cyt-YYF, was transformed into CAG33315, and this strain was transformed with a wild-type DegS plasmid.
Sequence Alignment.
The five closest DegS homologs from each phylum of bacteria as well as the 43 closest homologs in metazoans and four Homo sapiens HtrA proteases were aligned in ClustalW2 (18). Alignments at positions 176–189 were collected and submitted to WebLogo (19) for visualization.
Modeling.
Modeling and preparation of structural figures were performed using the PyMOL Molecular Graphics System (Schrödinger, LLC).
Acknowledgments
We thank Monica Guo for advice, strains, and plasmids. This work was supported by NIH Grant AI-16892. T.A.B. is an employee of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Ades SE. Regulation by destruction: Design of the sigmaE envelope stress response. Curr Opin Microbiol. 2008;11(6):535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Guo MS, Gross CA. Stress-induced remodeling of the bacterial proteome. Curr Biol. 2014;24(10):R424–R434. doi: 10.1016/j.cub.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113(1):61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 4.Campbell EA, et al. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol Cell. 2003;11(4):1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 5.Chaba R, et al. Signal integration by DegS and RseB governs the σE-mediated envelope stress response in Escherichia coli. Proc Natl Acad Sci USA. 2011;108(5):2106–2111. doi: 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117(4):483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 7.Zeth K. Structural analysis of DegS, a stress sensor of the bacterial periplasm. FEBS Lett. 2004;569(1-3):351–358. doi: 10.1016/j.febslet.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Sohn J, Sauer RT. OMP peptides modulate the activity of DegS protease by differential binding to active and inactive conformations. Mol Cell. 2009;33(1):64–74. doi: 10.1016/j.molcel.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mauldin RV, Sauer RT. Allosteric regulation of DegS protease subunits through a shared energy landscape. Nat Chem Biol. 2013;9(2):90–96. doi: 10.1038/nchembio.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohn J, Grant RA, Sauer RT. Allostery is an intrinsic property of the protease domain of DegS: Implications for enzyme function and evolution. J Biol Chem. 2010;285(44):34039–34047. doi: 10.1074/jbc.M110.135541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Regt AK, et al. A conserved activation cluster is required for allosteric communication in HtrA-family proteases. Structure. 2015;23(3):517–526. doi: 10.1016/j.str.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131(3):572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 13.Hasselblatt H, et al. Regulation of the sigmaE stress response by DegS: How the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress. Genes Dev. 2007;21(20):2659–2670. doi: 10.1101/gad.445307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohn J, Grant RA, Sauer RT. OMP peptides activate the DegS stress-sensor protease by a relief of inhibition mechanism. Structure. 2009;17(10):1411–1421. doi: 10.1016/j.str.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli σ(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-σ factor. Genes Dev. 1999;13(18):2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Plainview, NY: 1972. [Google Scholar]
- 17.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of σE, an Escherichia coli heat-inducible σ-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7(12B):2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 18.Larkin MA, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]