Significance
Although poxvirus vectors are widely used in preclinical and clinical trials as candidate vaccines for multiple pathogens, how these vectors affect the host immune response is not clear. In this study, we developed a poxvirus vector based on the attenuated New York vaccinia virus (NYVAC), which is able to target a central host-cell signaling pathway, NFκB. In mice, the modified NYVAC acts on the immune system by increasing specific neutrophil migration via NFκB activation and in turn enhances CD8 T-cell responses to HIV antigens delivered by the viral vector. We show that these inherent properties define a mechanism for poxvirus-induced immune responses and offer novel approaches to vaccine vector design.
Keywords: vaccinia virus, neutrophils, NFκB, vaccine, HIV
Abstract
Neutrophils are antigen-transporting cells that generate vaccinia virus (VACV)-specific T-cell responses, yet how VACV modulates neutrophil recruitment and its significance in the immune response are unknown. We generated an attenuated VACV strain that expresses HIV-1 clade C antigens but lacks three specific viral genes (A52R, K7R, and B15R). We found that these genes act together to inhibit the NFκB signaling pathway. Triple ablation in modified virus restored NFκB function in macrophages. After virus infection of mice, NFκB pathway activation led to expression of several cytokines/chemokines that increased the migration of neutrophil populations (Nα and Nβ) to the infection site. Nβ cells displayed features of antigen-presenting cells and activated virus-specific CD8 T cells. Enhanced neutrophil trafficking to the infection site correlated with an increased T-cell response to HIV vector-delivered antigens. These results identify a mechanism for poxvirus-induced immune response and alternatives for vaccine vector design.
Although attenuated vaccinia virus (VACV) is a commonly used vector model for vaccine development (1), poxviruses have a sophisticated strategy to escape immune surveillance by expressing inhibitory molecules, which could compromise immune response and thus vaccine efficiency. VACV encodes several proteins involved in host immune evasion that limit virus recognition by innate immune cells such as neutrophils and could affect the virus’s ability to induce adaptive immunity (2).
During microbial invasion, neutrophils counteract infection immediately in a process termed microbial sterilization (3), for which they engulf extracellular pathogens and infected and apoptotic cells (4, 5), they produce reactive oxygen intermediates (6), they release lytic enzymes and antimicrobial peptides from their granules (3), and they generate neutrophil extracellular traps (7). Neutrophils can induce an adaptive immune response after migrating to the lymph nodes (8) and efficiently cross-prime naïve T cells (9). In the presence of a granulocyte macrophage–colony-stimulating factor (GM-CSF) (10), tumor necrosis factor (TNF)-α, interleukin (IL)-4 (11), or interferon (IFN)-γ (12), neutrophils can adopt an antigen-presenting cell (APC) phenotype, thus triggering T-cell activation. In the context of VACV infection, neutrophils generate virus-specific memory CD8 T cells, transporting antigens from the dermis to the bone marrow (BM) (13). It is nonetheless not known how VACV modulates neutrophil recruitment, the factors involved, or the significance in immune response triggering.
Proinflammatory cytokine/chemokine production by an infected cell, which signals the recruitment of neutrophils involved in the immune response, is limited by inhibition of signaling pathways such as nuclear factor kappa B (NFκB) (2). NFκB pathway activation depends on the virus’s capacity to interact with Toll-like receptors (TLRs). Viral pathogen-associated molecular patterns interact with TLR2 and TLR4 on the plasma membrane (14). Pathogen-associated molecular pattern binding induces TLR2 and TLR4 homo- or heterodimerization (15), followed by recruitment of Toll/IL-1 receptor (TIR) domain-containing adaptor proteins such as myeloid differentiation factor 88 (MyD88) and TIR domain-containing adapter protein (TIRAP)/MyD88 adaptor-like (MAL) or TIR domain-containing adapter-inducing IFNβ (TRIF) and TRIF-related adaptor molecule (TRAM) (14). In the MyD88-dependent and the TRIF-dependent pathways, activation of TNF receptor-associated factor 6 results in IκBα phosphorylation and degradation, allowing release of the NFκB complex, composed of p65/p50 or p65/p52 heterodimers, into the cell nucleus. NFκB then binds the κB site and regulates transcription of proinflammatory cytokine and chemokine genes (14).
VACV interacts with TLR2 (16) and TLR4 (17) and produces early direct or indirect viral inhibitors of the NFκB pathway such as A46 (18), A49 (19), A52 (18), B14 (20), C4 (21), E3 (22), K1 (23), K7 (24), M2 (25), and N1 (26). New York vaccinia virus (NYVAC), a highly attenuated VACV strain used as vaccine vector, lacks most of these proteins but encodes NFκB pathway inhibitors A52, K7, and B15 (corresponding to B14 in the Western Reserve strain) (2).
Given the interest in attenuated VACV vectors as vaccine candidates and their optimization as immunogens (1), we generated several NYVAC deletion mutants that express HIV-1 Envelope (Env) and group-specific antigen (Gag)–polymerase (Pol)–negative regulatory factor (Nef) clade C antigens (NYVAC-C) but lack specific genes to encode NFκB inhibitors. We show that A52, K7, and B15 viral proteins inhibit the NFκB pathway in macrophages and that infection with a virus that lacks these three inhibitors results in NFκB pathway activation. NFκB-dependent induction of cytokines and chemokines is enhanced in vitro and in vivo by NYVAC-C ΔA52RΔB15RΔK7R and is essential for specific neutrophil recruitment to the infection site. Neutrophils acquire features of APCs, activate antigen-specific T cells, and migrate to the draining lymph nodes and to the spleen, where they induce antigen-specific CD8 T-cell responses. These findings provide important insights into the mechanism of the poxvirus-induced immune response, which is relevant in vaccine vector design.
Results
Concomitant Deletion of A52R, B15R, and K7R Genes in NYVAC Leads to Enhanced NFκB Pathway Activation.
To define the immune modulatory role of the VACV viral genes that antagonize the NFκB pathway, we generated single, double, and triple deletion mutants for the viral genes encoding A52, K7, and B15 (Materials and Methods), using as a backbone the NYVAC-C vector expressing HIV-1 clade C antigens Env (gp120) as a cell-released product and Gag-Pol-Nef (GPN) as an intracellular polyprotein (27). With this targeted gene disruption, we generated the following NYVAC deletion mutants: NYVAC-C ΔB15R, NYVAC-C ΔA52R, NYVAC-C ΔK7R, NYVAC-C ΔB15RΔK7R, NYVAC-C ΔA52RΔB15R, NYVAC-C ΔA52RΔK7R, and NYVAC-C ΔA52RΔB15RΔK7R (also termed triple deletion mutant or NYVAC-C Δ3 for short). Gene deletion was confirmed by PCR, using primers annealed to flanking sequences of each gene (Fig. S1A). The gp120 and GPN HIV-1 proteins were expressed correctly by each deletion mutant, as confirmed by Western blot (Fig. S1B).
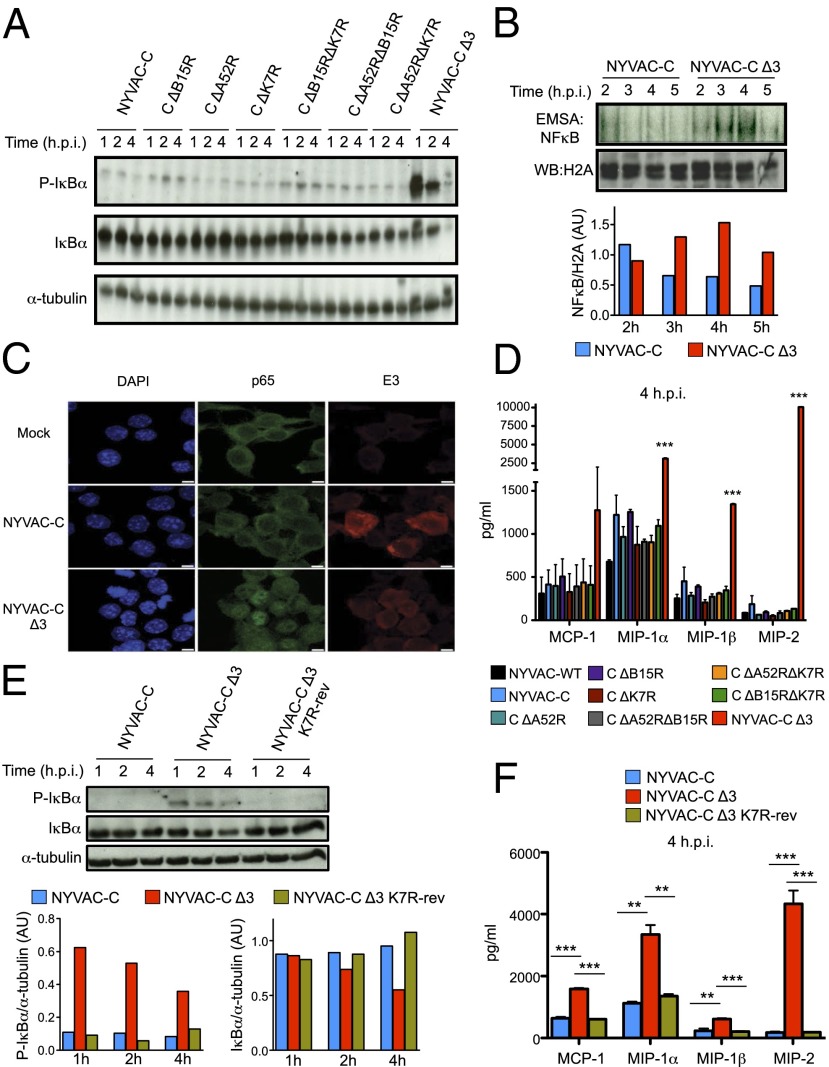
To determine the contribution of the NYVAC inhibitors to NFκB activation, we infected murine J774 macrophages with the different single, double, and triple NYVAC-C deletion mutants and performed a time-course assay to evaluate IκBα phosphorylation. We observed a clear increase in the level of IκBα phosphorylation in NYVAC-C Δ3-infected J774 cells compared with the parental or the other deletion mutants. As predicted, this phosphorylation was accompanied by enhanced IκBα degradation (Fig. 1A). Similar results between parental virus and NYVAC-C Δ3 were obtained following infection of human THP-1 monocytes differentiated into macrophages (Fig. S2A).
Fig. 1.
Deletion of A52R, B15R, and K7R genes induces robust NFκB activation. (A) Phosphorylated and total IκBα forms analyzed by Western blot (WB) in J774 mouse macrophages infected with NYVAC-C or NYVAC-C deletion mutants (5 PFUs per cell) for 1, 2, and 4 h. α-tubulin was used as the internal loading control. (B) J774 nuclear extracts incubated with 32P-end–labeled NFκB probe and assayed by EMSA for DNA binding complexes at 2, 3, 4, and 5 h postinfection with NYVAC-C or NYVAC-C Δ3. Histone H2A was used as the internal loading control for WB. Bars show the ratio of NFκB probe bands to histone H2A bands, quantified using ImageJ. (C) Confocal microscopy of mock-, NYVAC-C–, or NYVAC-C Δ3-infected J774 cells (3 h postinfection). Cells were stained with antibodies to E3 VACV protein (red), p65 cell protein (green), and DAPI probe to detect DNA (blue). (Scale bars, 5 μm.) (D) Concentrations of chemokines MCP-1, MIP-1α, MIP-1β, and MIP-2 at 4 h postinfection, quantified by immunoassay in supernatants of J774 cells infected as indicated. (E) Phosphorylated and total IκBα forms analyzed by WB in J774 mouse macrophages infected with NYVAC-C, NYVAC-C Δ3, or NYVAC-C Δ3 K7R-rev (5 PFUs per cell) for 1, 2, and 4 h. α-tubulin was used as the internal loading control. Bars show the ratio of phospho- or total IκBα bands to α-tubulin bands, quantified with ImageJ. (F) Concentrations of chemokines MCP-1, MIP-1α, MIP-1β, and MIP-2 at 4 h postinfection, quantified by immunoassay in supernatants of NYVAC-C–, NYVAC-C Δ3-, or NYVAC-C Δ3 K7R-rev–infected J774 macrophages. Values show mean ± SEM of duplicates, representative of three independent experiments. ***P < 0.001, **P < 0.01.
To confirm that increased IκBα phosphorylation in triple deletion mutant-infected cells enhanced NFκB activity, we used electrophoretic mobility shift assay (EMSA) to analyze NFκB binding to its consensus binding sequence motif and an immunofluorescence assay to detect p65 translocation from the cytoplasm to the nucleus. EMSA indicated marked NFκB pathway activation in NYVAC-C Δ3-infected macrophages, with a twofold increase from 3 to 5 h postinfection compared with NYVAC-C–infected macrophages (Fig. 1B), which was confirmed by immunofluorescence of p65 migration from the cytoplasm to the nucleus (Fig. 1C). These results suggest that these genes (A52R, K7R, and B15R) must be removed concomitantly from the viral vector to induce robust pathway activation following VACV infection.
A variety of cytokines and chemokines are known NFκB target genes. We therefore analyzed their secretion in supernatants of J774 macrophages infected with the various deletion mutants. Compared with parental and other deletion mutant viruses, only NYVAC-C Δ3-infected cells showed significantly increased production of cytokines such as TNF-α and IL-6 at 24 h postinfection (Fig. S2B) and of chemokines such as macrophage inflammatory proteins (MIPs)-1α, -1β, and -2 at 4 h postinfection (Fig. 1D). We observed a similar chemokine/cytokine profile in primary peritoneal macrophages infected with the NYVAC-C deletion mutants (Fig. S2C). These data indicate enhanced NFκB-dependent chemokine and cytokine secretion after macrophage infection with the NYVAC-C Δ3.
To demonstrate that only one of the three inhibitory molecules is sufficient to abolish NFκB activation, we generated the revertant NYVAC-C Δ3 K7R-rev virus, in which the K7R gene was reinserted in the HA locus (Fig. S1C); expression was confirmed by RT-PCR (Fig. S1D). IκBα phosphorylation, degradation, and chemokine secretion levels in NYVAC-C Δ3 K7R-rev–infected J774 macrophages resembled those of the same cells infected with NYVAC-C (Fig. 1 E and F).
Increased in Vitro and in Vivo Neutrophil Migration After NYVAC-C Δ3 Infection.
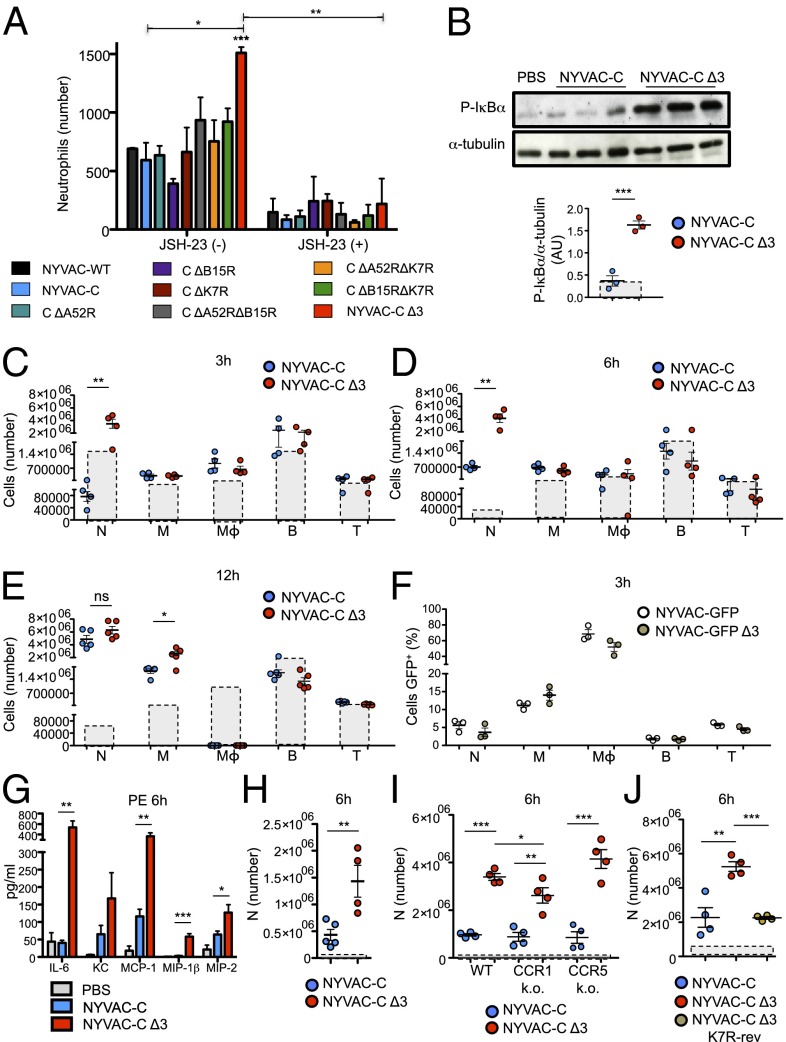
Because chemokines such as MCP-1, MIP-1α, -1β, and -2 or keratinocyte chemoattractant (KC) are essential for recruitment of several cell types, we tested whether a specific cell subset responds to this chemokine–cytokine mixture. We used an in vitro migration assay in which total murine BM cells were allowed to migrate after stimulation with supernatants of the different NYVAC-infected (4 h) J774 macrophages. Only neutrophils (Ly6G+CD11b+) exposed to NYVAC-C Δ3-infected macrophage supernatants showed significantly more migration compared with the parental virus or the other deletion mutants (Fig. 2A). Migrated neutrophils showed the same percentages of early and late apoptotic cells when stimulated with supernatants of NYVAC-C– and NYVAC-C Δ3-infected J774 macrophages (Fig. S3A).
Fig. 2.
Increased neutrophil recruitment after NYVAC-C Δ3 infection. (A) Absolute number of BM neutrophils that migrated after stimulation with supernatants of virus-infected J774 cells (4 h). J774 cells were untreated or pretreated with the p65 inhibitor JSH-23 (30 μM). (B) Phosphorylated IκBα form analyzed by WB in PECs of BALB/c mice at 1 h after injection with PBS or 107 PFUs of NYVAC-C or NYVAC-C Δ3. α-tubulin was used as the internal loading control. Each point represents the ratio of phospho-IκBα to α-tubulin bands in individual mice, as quantified by ImageJ. Boxes with dashed lines indicate the ratio for PBS-injected mice. Absolute numbers of neutrophils (N), monocytes (M), macrophages (Mϕ), B cells (B), and T cells (T) in the peritoneal cavity of BALB/c mice at 3 h (C), 6 h (D), and 12 h (E) after injection of 107 PFUs of NYVAC-C or NYVAC-C Δ3. Boxes with dashed lines indicate absolute numbers for PBS-injected mice. (F) Percentages of GFP+ neutrophils (N), monocytes (M), macrophages (Mϕ), B (B), and T cells (T) in the peritoneal cavity of NYVAC-GFP– or NYVAC-GFP Δ3-injected (107 PFUs) mice at 3 h postinfection. (G) Cytokine/chemokine levels at 6 h postinfection in peritoneal exudates of PBS-, NYVAC-C–, or NYVAC-C Δ3- (107 PFUs) injected mice (n = 5 per group). (H) Absolute number of neutrophils (N) in the peritoneal cavity of mice 6 h after injection of NYVAC-C– or NYVAC-C Δ3-peritoneal exudates at 6 h postinfection. Boxes with dashed lines indicate total numbers in PBS-peritoneal exudate-injected mice. (I) Absolute numbers of neutrophils (N) in the peritoneal cavity of C57/BL6J WT, CCR1 KO, and CCR5 KO mice at 6 h after injection with PBS or 107 PFUs of NYVAC-C or NYVAC-C Δ3. Boxes with dashed lines indicate absolute numbers of cells in PBS-injected mice. (J) Absolute numbers of neutrophils (N) in the peritoneal cavity of BALB/c mice at 6 h after injection with 107 PFUs of NYVAC-C, NYVAC-C Δ3, or NYVAC-C Δ3 K7R-rev. Boxes with dashed lines indicate absolute numbers of cells in PBS-injected mice. Graphs show mean ± SEM; each point represents an individual mouse. Data are representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
In a similar experiment, we used a supernatant of J774 cells pretreated with JSH-23, an inhibitor of p65 translocation to the nucleus (28), and then infected it with the NYVAC virus. The number of neutrophils that migrated toward supernatants of JSH-23–treated NYVAC-C Δ3-infected cells was significantly lower than those that migrated to supernatants of untreated NYVAC-C Δ3-infected cells and was similar to the other NYVAC-C deletion mutants (Fig. 2A). These data show that NFκB activation underlies the differences in neutrophil migration induced by the triple deletion mutant virus and its parental strain.
To determine whether infection with the triple mutant virus also leads to increased neutrophil migration in vivo, BALB/c mice were infected by i.p. injection of 107 plaque-forming units (PFUs) of NYVAC-C or NYVAC-C Δ3. Peritoneal exudate cells (PECs) were collected 1 h postinfection to study NFκB activation. In PECs of NYVAC-C Δ3-infected mice, IκBα phosphorylation was significantly higher than in PECs of parental virus-infected mice (Fig. 2B). At 3 and 6 h postinfection, mice that received the triple deletion mutant virus showed a significant increase in neutrophils (Ly6G+CD11b+) in the peritoneal cavity compared with NYVAC-C–infected mice (Fig. 2 C and D). There were no differences in monocytes (M), macrophages (Mϕ), B cells (B), or T cells (T) (Fig. 2 C and D), which indicated that in the first 6 h of infection, the triple deletion mutant virus promotes neutrophil migration to the infection site before migration of the other hematopoietic cells. After 12 h, we detected no difference in neutrophil migration, but triple deletion mutant virus-infected mice showed a significant increase in monocytes compared with NYVAC-C–infected mice; resident macrophages were absent for both viruses (Fig. 2E). The infection kinetics indicates that neutrophil migration precedes monocyte migration in NYVAC-C Δ3-infected mice.
To rule out the possibility that increased neutrophil recruitment was caused by differences in virus infective capacity, we replaced the HIV-1 antigen cassette with the GFP gene to generate NYVAC-GFP and NYVAC-GFP Δ3 viruses, which were injected (107 PFUs; i.p.) into BALB/c mice. At 3 h postinfection, comparison of NYVAC-GFP Δ3- and NYVAC GFP-injected mice showed similar percentages of GFP+ cells in PEC types (neutrophils, monocytes, macrophages, and B and T cells; Fig. 2F). These percentages indicate that both viruses have similar in vivo infective capacities and that macrophages, which constitute ∼20% of the total peritoneal cell yield, are the most susceptible cell type to NYVAC infection (∼80%). The absolute numbers of GFP+ cells indicated that only GFP+ neutrophils were significantly higher in NYVAC-GFP Δ3- than in NYVAC-GFP–infected mice (Fig. S3B), due to a significant increase in neutrophil migration in NYVAC-GFP Δ3- compared with NYVAC-GFP–infected mice (Fig. S3C).
The peritoneal exudates of NYVAC-C Δ3-infected mice (6 h postinfection) showed significantly higher levels of cytokines such as IL-6 and chemokines such as MCP-1, MIP-1β, and MIP-2 compared with those of NYVAC-C–infected mice (Fig. 2G). We detected no difference in IL-8 secretion between these two groups (Fig. S3D). To demonstrate that the neutrophils were recruited by increased cytokine/chemokine secretion at the infection site, peritoneal exudate pools of 6-h NYVAC-C– and NYVAC-C Δ3-infected mice, free of PECs and viruses, were injected i.p. into mice, and neutrophils in the peritoneal cavity were analyzed at 6 h postinfection. Total neutrophils in NYVAC-C Δ3-peritoneal exudate-injected mice were significantly higher compared with those in NYVAC-C–peritoneal exudate-injected mice (Fig. 2H). These data indicate that enhanced neutrophil recruitment was dependent on cytokine/chemokines produced during the infection.
To determine the chemokine(s) involved in increased neutrophil migration in NYVAC-C Δ3-infected mice, we performed the same experiment in C57/BL6J CCR1 and CCR5 knockout (KO) mice, which are deficient in the CCR1 and CCR5 receptors involved in binding the chemokines MIP and MCP. Total neutrophil number in the peritoneal cavity of NYVAC-C Δ3-infected CCR1 KO mice was significantly lower compared with NYVAC-C Δ3-infected wild-type (WT) mice, although not as low as in NYVAC-C–infected CCR1 KO mice (Fig. 2I). There were no differences between CCR5 KO and WT mice (Fig. 2I). These data suggest that the increased neutrophil migration is mediated partially by CCR1 and not by CCR5.
To demonstrate that the enhanced neutrophil migration in NYVAC-C Δ3-infected mice is NFκB-dependent, we infected mice with NYVAC-C Δ3 K7R-rev and analyzed PEC neutrophils at 6 h postinfection. The revertant virus induced a significant decrease in neutrophil number compared with the NYVAC-C Δ3 virus and resembled the value for the NYVAC-C virus (Fig. 2J).
Enhanced Neutrophil Trafficking After NYVAC-C Δ3 Infection.
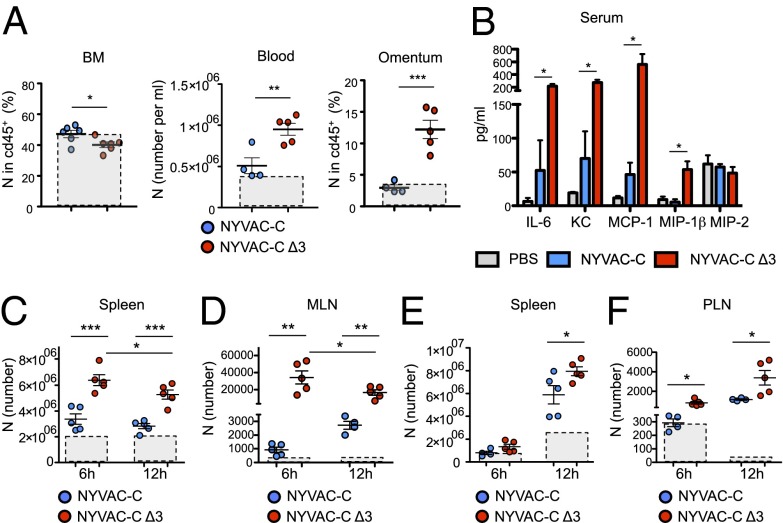
To determine how neutrophil trafficking after NYVAC-C Δ3 infection occurs, we quantified neutrophils in BM, spleen, lymph nodes, peripheral blood, and omentum. We included omentum in this analysis as it is the primary site of neutrophil exudation and mediates recruitment from circulation to the peritoneal cavity. NYVAC-C Δ3-infected mice had significantly fewer neutrophils in BM but more neutrophils in blood and a larger percentage of neutrophils in omentum than NYVAC-C–infected mice (Fig. 3A). These changes were the result of increased IL-6, KC, MCP-1, and MIP-1β production in sera of triple deletion mutant-infected mice compared with those infected with the parental strain (Fig. 3B). No differences were detected in IL-8 levels between the two groups (Fig. S3E). Compared with neutrophils, at 6 h NYVAC-C Δ3-infected mice had significantly more monocytes (CD115+) in the blood than NYVAC-C–infected mice, but we found no difference between the two groups in the percentage of monocytes in BM or omentum (Fig. S3F).
Fig. 3.
Increased neutrophil trafficking after NYVAC-C Δ3 infection. (A) Percentages of neutrophils in BM and omentum, and total number of neutrophils in blood at 6 h postinfection, from NYVAC-C– or NYVAC-C Δ3-injected mice. Boxes with dashed lines indicate absolute numbers of cells in PBS-injected mice. (B) Cytokine/chemokine levels at 6 h postinfection in serum of PBS-, NYVAC-C–, or NYVAC-C Δ3-injected mice (n = 5 per group). Shown are total neutrophil numbers in spleen (C) and MLNs (D) at 6 and 12 h postinfection, from NYVAC-C– or NYVAC-C Δ3-injected mice, as well as the total neutrophil numbers in spleen (E) and PLNs (F) at 6 and 12 h postintramuscular infection, from NYVAC-C– or NYVAC-C Δ3-injected mice. Boxes with dashed lines indicate absolute numbers of cells in PBS-injected mice. Graphs show mean ± SEM; each point represents an individual mouse. Data are representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
In the secondary lymphoid organs such as the spleen and mediastinal lymph nodes (MLNs), we found significantly more neutrophils in NYVAC-C Δ3- than in NYVAC-C–infected mice (Fig. 3 C and D). In MLNs and in the spleen of NYVAC-C Δ3-infected mice, the absolute number of neutrophils was significantly reduced from 6 to 12 h postinfection (Fig. 3 C and D). We also detected a significant increase in neutrophil death from 6 to 12 h in the spleens of NYVAC-C Δ3- compared with NYVAC-C–infected mice (Fig. S3G), as well as a clear trend (P = 0.07) in increased MLN neutrophil death (Fig. S3H). This increase could explain the significant reduction in spleen and MLN neutrophils from 6 to 12 h.
To test a common human immunization route, BALB/c mice also received intramuscular injections of NYVAC-C or NYVAC-C Δ3 viruses. NYVAC-C Δ3 induced a significant neutrophil increase in the spleen at 12 h postinfection (Fig. 3E) and in draining popliteal lymph nodes (PLNs) at 6 and 12 h postinfection (Fig. 3F) compared with NYVAC-C.
These data indicate that neutrophil migration occurs in a specific time window and is enhanced after NYVAC-C Δ3 infection. Neutrophils abandon BM, enter the blood stream, and through the omentum reach the infection site, where they encounter infected cells; they subsequently leave toward secondary lymphoid organs to initiate an adaptive immune response.
Neutrophils Act as APCs After NYVAC-C Δ3 Infection.
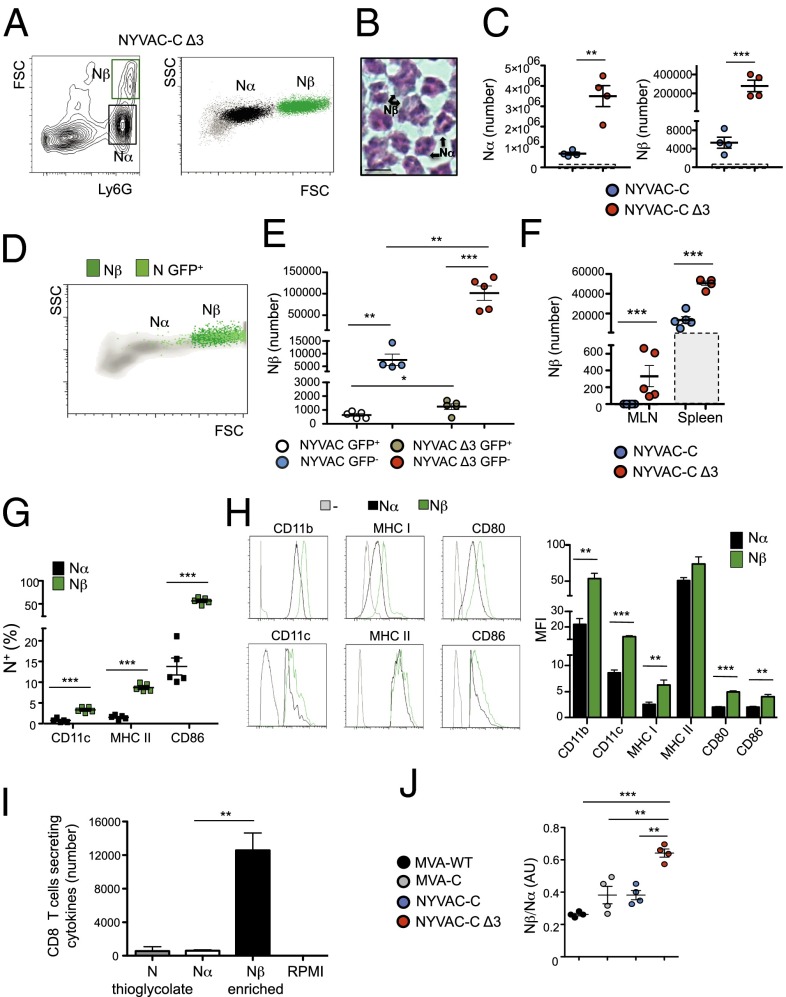
We next characterized the neutrophil population recruited to the peritoneum after infection. Using flow cytometry, we found two populations that differed in cell size and complexity, which we termed Nα and Nβ (Fig. 4A). To evaluate the morphology of Nα and Nβ populations, we sorted Ly6G+ cells. Both populations had clear neutrophil-like morphology, but Nβ cells were larger, more lobulated, and more hypersegmented than Nα cells (Fig. 4B). In PECs, the Nα subset showed a fourfold increase, whereas the Nβ subset showed a 20-fold increase in NYVAC-C Δ3-infected compared with parental virus-infected mice (Fig. 4C).
Fig. 4.
Nβ neutrophil population with APC features is involved in specific activation of CD8 T cells after NYVAC-C Δ3 infection. (A) FACS plots determined by forward scatter (FSC) and Ly6G, and side scatter (SSC) and FSC of PEC from NYVAC-C Δ3-injected mice. Nα (black) and Nβ (green) neutrophil subsets are shown as dot plots. (B) Hematoxylin/eosin staining of peritoneal neutrophils presorted for Ly6G+. (Scale bar, 10 μm.) (C) Absolute number of Nα and Nβ neutrophils in the peritoneal cavity at 6 h postinfection in NYVAC-C– or NYVAC-C Δ3- (107 PFUs) injected mice. Boxes with dashed lines indicate total number in PBS-injected mice. Graphs show mean ± SEM; each point represents an individual mouse. (D) Nβ (dark green) and infected neutrophils (N GFP+; light green) from PECs of NYVAC-GFP Δ3-infected mice. (E) Absolute numbers of Nβ GFP+/− neutrophils in NYVAC-C (107 PFUs)/NYVAC-GFP– (107 PFUs) or NYVAC-C Δ3 (107 PFUs)/NYVAC-GFP Δ3- (107 PFUs) infected mice. Graphs show mean ± SEM; each point represents an individual mouse. (F) Absolute numbers of Nβ neutrophils in MLNs and spleen at 6 h postinfection of NYVAC-C– or NYVAC-C Δ3-injected mice. Boxes with dashed lines indicate absolute number in PBS-injected mice. Graphs show mean ± SEM; each point represents an individual mouse. (G) Percentages of Nα (black) and Nβ (green) neutrophils positive for CD11c, MHC II, and CD86 markers. Graphs show mean ± SEM; each square represents an individual mouse. (H) Layouts and median fluorescence intensity (MFI) of indicated markers in Nα (black) and Nβ (green) neutrophils at 6 h postinfection in NYVAC-C Δ3-injected mice. In gray is the isotype control. Columns show mean ± SEM of five mice. (I) Total numbers of CD8 T cells per spleen secreting IFN-γ and/or TNF-α and/or IL-2 after stimulation with thioglycolate-induced neutrophils, with virus-induced Nα, with virus-induced Nβ-enriched, or with RPMI. Columns show mean ± SEM of triplicates. (J) Nβ:Nα ratio in peritoneal cavity at 6 h postinfection in MVA-WT–, MVA-C–, NYVAC-C–, or NYVAC-C Δ3- (107 PFUs) injected mice. Graphs show mean ± SEM; each point represents an individual mouse. Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
We tested whether acquisition of the Nβ profile depended on direct virus infection of neutrophils or on the cytokine/chemokine milieu produced after infection. Mice were initially infected with NYVAC-C or NYVAC-C Δ3 to induce neutrophil recruitment to the peritoneal cavity and subsequently injected with NYVAC-GFP or NYVAC-GFP Δ3 to infect the migrated neutrophils. Most GFP+ neutrophils in mice infected with the parental or triple deletion mutant virus had a Nβ-like profile (Fig. 4D), indicating that Nβs were more susceptible to the infection than Nα cells. The majority of Nβ neutrophils were GFP− in both mouse groups (Fig. 4E), which indicated that neutrophil viral infection was not directly responsible for Nβ subset generation. As predicted, the Nβ population (GFP+ and GFP−) was higher in the NYVAC-C Δ3- than in NYVAC-C–infected mice (Fig. 4E). To demonstrate that Nβ and Nα were recruited by increased cytokine/chemokine secretion at the infection site, peritoneal exudate pools of 6 h NYVAC-C– and NYVAC-C Δ3-infected mice, free of PECs and viruses, were injected i.p. into mice; neutrophils in the peritoneal cavity were analyzed at 6 h postinfection. The absolute numbers of Nα and Nβ neutrophils in NYVAC-C Δ3-peritoneal exudate-injected mice were significantly higher than those of NYVAC-C–peritoneal exudate-injected mice (Fig. S4A).
Examination of Nβ neutrophils in the spleen and MLNs of NYVAC-C Δ3-infected mice showed significantly more Nβ cells than in NYVAC-C–infected mice (Fig. 4F).
To characterize Nα and Nβ neutrophil phenotype in NYVAC-C Δ3-infected mice, we analyzed the expression of several surface markers including the CD11b activation marker involved in neutrophil adhesion and migration, MHC class I and class II molecules used for presentation of antigenic determinants to T cells, the CD11c dendritic cell (DC) marker, and CD80/CD86 APC costimulatory markers involved in T-cell activation. All neutrophils expressed CD11b, MHC class I, and CD80 markers. The percentage of Nβ neutrophils that expressed MHC class II, CD11c, or CD86 was significantly higher than Nα cells (Fig. 4G). Moreover, levels of all surface markers were higher in Nβ than in Nα neutrophils, with significant differences in most cases (Fig. 4H). These results indicate that Nβs have a more APC-like profile than Nα neutrophils.
To demonstrate the APC potential of Nα and Nβ neutrophils, both populations were sorted for Ly6G+ (purity, >96%) and cell size and then incubated 16 h with isolated spleen CD8 T cells (purity, >97%; ratio, 1:20) from mice that received i.p. injections of NYVAC or PBS 90 d earlier. Ly6G+ neutrophils (purity, >96%) sorted from i.p. thioglycolate-injected mice were used as controls. We measured antigen-specific CD8 T-cell activation by intracellular cytokine staining (ICS) of IL-2, IFN-γ, and TNF-α. Nα cells alone did not induce antigen-specific CD8 T activation (Fig. 4I): The number of cytokine-secreting CD8 T cells was similar to thioglycolate-induced neutrophils (<0.01% of total CD8 T cells). The Nβ-enriched fraction induced a significant increase in CD8 T-cell activation compared with Nα alone (Fig. 4I). The CD8 T-cell response in PBS-injected mice was subtracted for each group; this response was <0.01% of total CD8 T cells, indicating that the neutrophils did not activate naïve CD8 T cells. These data indicate that Nα cells alone are unable to activate CD8 T cells and that the presence of Nβ cells is sufficient to activate antigen-specific CD8 T cells, suggesting that Nβs have an APC role.
Given the distinct roles of Nα and Nβ in CD8 T-cell activation, we analyzed the Nβ/Nα ratio in the peritoneal cavity of poxvirus-infected mice. In NYVAC-C Δ3-infected mice, this ratio was significantly higher than in NYVAC-C–infected mice, and also compared with MVA-WT– and MVA-C–infected mice (Fig. 4J). The total neutrophil percentage in MVA-WT–, MVA-C–, or NYVAC-C Δ3-infected mice was similar and significantly higher than in NYVAC-C–infected mice (Fig. S4B).
NYVAC-C Δ3 Enhances the HIV-Specific T-Cell Response Through Neutrophil Migration.
To study the ability of NYVAC-C Δ3 to induce a specific T-cell response to HIV-1 antigens, we used a DNA intramuscular prime/poxvirus i.p. boost approach. This heterologous immunization protocol is more immunogenic than a homologous combination in activating T-cell responses to HIV-1 antigens (27).
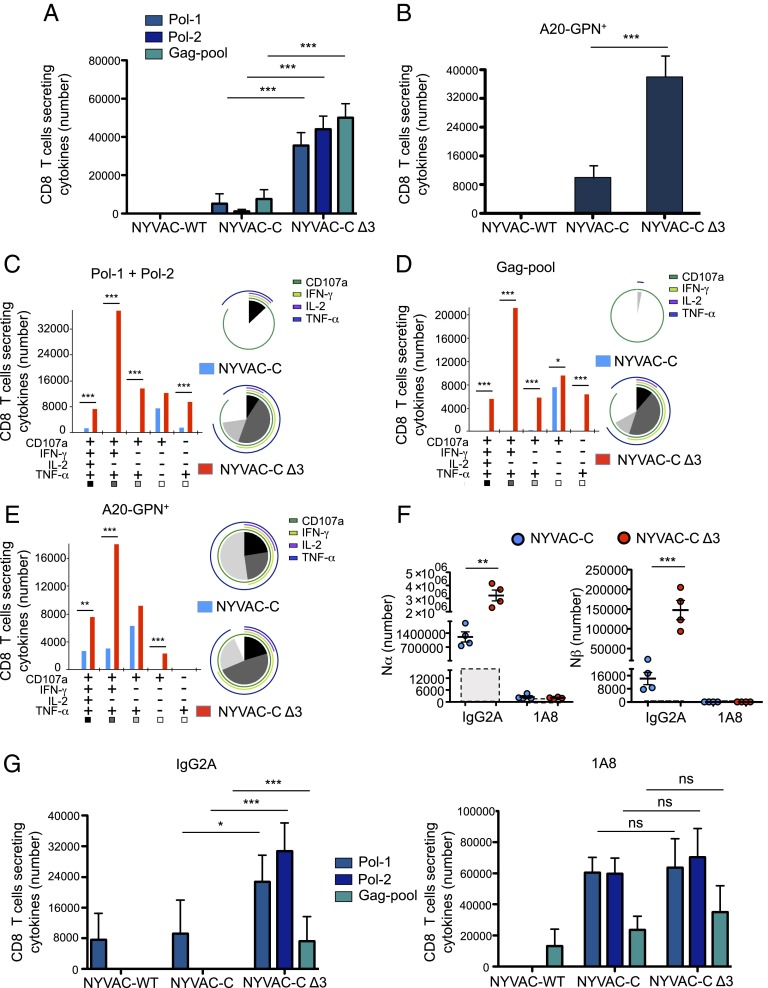
BALB/c mice were immunized (Materials and Methods), and adaptive T-cell immune responses were measured by ICS of spleen cells. To study the HIV-1–specific T-cell response, splenocytes from infected mice were stimulated with HIV-1 Env-1, Pol-1, or Pol-2 peptides, which are the most immunogenic MHC class I-restricted cytotoxic T lymphocytes (CTLs) peptides of HIV clade C in BALB/c mice (29) or with a pool of overlapping Gag peptides. Alternatively, to study CD8 and CD4 T-cell responses to Env and GPN antigens, splenocytes were pulsed with A20 cells previously nucleofected with the mammalian expression plasmids pcDNA-gp120 (Env) or pcDNA-GPN. To determine the functional profile of CD4 and CD8 T cells, we measured levels of IFN-γ, TNF-α, and IL-2 as well as of lysosomal-associated membrane protein-1 (LAMP-1 or CD107a) as a surrogate marker for induction of killing. We determined the magnitude of the T-cell response as the number of T cells that expressed IFN-γ and/or TNF-α and/or IL-2 and/or CD107a and the polyfunctionality of the response as T cell’s capacity to express more than one of these activation markers (13).
NYVAC-C Δ3 induced a significantly higher specific CD8 T-cell response to HIV intracellular antigens than NYVAC-C, both when Pol and Gag antigens were analyzed separately (Fig. 5A) and when the overall GPN response was studied (Fig. 5B). There were no significant differences between both viruses in the magnitude of the specific CD8 T-cell response to the Env extracellular antigen, using either Env-1 peptide (Fig. S5A) or A20-ENV+ (Fig. S5B). Neither virus induced a GPN-specific CD4 T-cell response when stimulated with A20-GPN+. We detected no differences in Env-specific CD4 T-cell response with A20-Env+ between two-virus–infected mouse groups (Fig. S5C).
Fig. 5.
NYVAC-C Δ3 enhances the magnitude and polyfunctionality of the CD8 T-cell response to HIV-1 Gag–Pol antigens. Vaccine-induced HIV-1–specific CD8 T-cell response in mice (n = 4 per group) infected with 107 PFUs of NYVAC-WT, NYVAC-C, or NYVAC-C Δ3. The response was measured 11 d after the last immunization, after splenocyte stimulation with HIV-1 peptides/pools or with A20 GPN+. Total value (magnitude) is the sum of total CD8 T cells per spleen that secrete IFN-γ and/or TNF-α and/or IL-2 and/or CD107a. (A) Magnitude of Pol-1–, Pol-2–, or Gag pool-specific CD8 T-cell response. Graphs show mean ± CI. (B) Magnitude of CD8 T-cell response to A20 GPN+. Graphs show mean ± CI. (C) Functional profile of adaptive Pol-1 + Pol-2–specific CD8 T cells. (D) Functional profile of adaptive Gag pool-specific CD8 T cells. (E) Functional profile of adaptive GPN-specific CD8 T cells. Combinations of responses (x axis) and total numbers of functionally distinct cell populations (y axis) are shown. Responses are grouped and color-coded based on the number of functions. Pie chart colors indicate the percentage of cytokine-producing cells based on number of functions (inside) and the different activation markers (outside). (F) Absolute number of Nα and Nβ neutrophils in the peritoneal cavity at 6 h postinfection in NYVAC-C– or NYVAC-C Δ3- (107 PFUs) injected and IgG2A-pretreated or 1A8-pretreated mice. Boxes with dashed lines indicate absolute number in PBS-injected mice. Graphs show mean ± SEM; each point represents an individual mouse. (G) Vaccine-induced HIV-1–specific CD8 T-cell response in mice (n = 4 per group) infected with 107 PFUs NYVAC-WT, NYVAC-C, or NYVAC-C Δ3 and IgG2A-pretreated or 1A8-pretreated. The response was measured 11 d after the last immunization, after splenocyte stimulation with HIV-1 peptides/pools. Total value (magnitude) is the sum of total CD8 T cells per spleen that secrete IFN-γ and/or TNF-α and/or IL-2 and/or CD107a. Graphs show mean ± CI. Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
The quality of the Gag and Pol responses, defined as cytokine production and cytotoxic potential, showed that compared with the parental strain, the triple deletion mutant induced a marked increase in the CTL polyfunctional profile (Fig. 5 C and D). The CD8 T-cell subset that produced IFN-γ, TNF-α, and CD107a was the most representative population induced with Pol and Gag peptides (Fig. 5 C and D) and with A20-GPN+ (Fig. 5E) in triple deletion mutant-infected mice. This CD8 T-cell subset marks the difference in the CTL polyfunctional profile between two-virus–infected mice (Fig. 5 C–E, pie charts). These results indicate that the triple deletion mutant enhances the magnitude and the polyfunctional profile of specific CD8 T cells to HIV-1 GPN intracellular antigens.
We also used a DNA intramuscular prime/poxvirus intramuscular boost to immunize and measured adaptive CD8 T-cell immune responses by ICS in spleen cells. NYVAC-C Δ3 induced a significantly higher specific CD8 T-cell response to HIV-1 Pol antigen than NYVAC-C; no significant difference was found between two groups in the response to Gag antigen (Fig. S5D).
To test whether neutrophils are involved in the enhanced HIV-specific CTL response, we depleted these cells in vivo using anti-Ly6G mAb 1A8 administered 24 h before virus boost (30). Nα and Nβ populations were depleted similarly in 1A8-pretreated/virus-infected mice compared with isotype control IgG2A-pretreated/virus-infected mice (Fig. 5F). The difference in magnitude of the Pol-1–, Pol-2–, and Gag pool-specific CD8 T-cell responses between viruses was greatly reduced in 1A8-pretreated mice (Fig. 5G) compared with the significant differences in IgG2A-pretreated mice between two groups (Fig. 5G).
The increase in HIV antigen-responsive CD8 T cells in mAb 1A8-pretreated/NYVAC-C Δ3-infected mice compared with mAb IgG2a-pretreated/NYVAC-C Δ3-infected mice might be due to the greater monocyte activation (based on CD11b and CD62L overexpression) in 1A8- than in IgG2a-pretreated mice (Fig. S5E). Monocyte numbers in 1A8-pretreated/NYVAC-C Δ3-infected mice were significantly lower compared with IgG2A-pretreated/NYVAC-C Δ3-infected mice, suggesting neutrophil-dependent monocyte migration (Fig. S5F). We found no differences in DC activation markers (CD80, CD86) or in DC number between the two groups (Fig. S5 G and H). These data suggest that neutrophils are involved in the increase in HIV-1 Gag- and Pol-specific CD8 T-cell responses after infection with the triple deletion mutant and that other immune cells induce higher antigen-specific CD8 T-cell responses in the nonphysiological absence of neutrophils.
Discussion
Here we define the biological contribution of the deletion of three VACV inhibitors of the NFκB signaling pathway in the NYVAC genome. The A52, K7, and B15 early proteins, members of the B-cell lymphoma-2 family (31, 32), are involved in suppression of host immune responses (2). During infection, A52 and K7 interact with TNF receptor-associated factor 6 and inhibit its kinase activation capacity (24, 33), whereas B15 binds the IκB kinase complex to inhibit IκBα phosphorylation and degradation (20). In this study, we focused on the A52, K7, and B15 proteins; using single, double, and triple deletion mutants, we demonstrate that the presence of only one of the three inhibitory molecules is sufficient to abolish NFκB activation, ruling out synergy between these proteins as a mode of action. We establish that combined deletion of A52R, K7R, and B15R is necessary for efficient triggering of the NFκB pathway and neutrophil recruitment.
Neutrophils treated with GM-CSF and/or other cytokines can up-regulate MHC class II and the costimulatory molecules CD80/CD86 (APC markers) and promote T-cell activation (11, 12). Neutrophils can acquire macrophage (34) or dendritic phenotypes (10), and such hybrid neutrophil populations with APC-like properties participate in adaptive immune responses (10). In the context of a tumor, the neutrophil subsets generated in the cytokine/chemokine environment can affect tumor growth by influencing CD8 T-cell activation (35). In our study, during NYVAC-C Δ3 infection, we observed recruitment of Nα and Nβ neutrophil subsets as a consequence of the cytokine/chemokine profile produced. Nβ neutrophils are more lobulated, larger, and morphologically more complex; display an enhanced activation profile; have higher levels of APC markers (CD11c, CD80, and CD86); and have greater capacity to induce antigen-specific T-cell activation than Nα cells.
MVA induces NFκB activation (23) and robust neutrophil recruitment after intranasal (36) or intradermal infection (37). We demonstrate that, after i.p. infection, the percentage of neutrophils in MVA-WT–, MVA-C–, or NYVAC-C Δ3-infected mice was similar and that the Nβ/Nα ratio is significantly higher in NYVAC-C Δ3- than in MVA-infected mice. The NYVAC Δ3 vector might thus offer an advantage in generating antigen-specific CD8 T-cell responses compared with the MVA vector.
Attenuated VACV vectors are considered vaccine candidates; specifically, NYVAC, MVA, and ALVAC poxvirus strains are used against emerging infectious diseases and cancer in humans (1). For HIV/AIDS VACV vectors, several strategies have been developed to improve immunogenicity to HIV antigens, such as use of costimulatory molecules, administration of heterologous vectors, and deletion of immunomodulatory viral genes (38). These approaches have yielded promising results in primates and elicited protection after challenge with simian immunodeficiency virus (39), although effectiveness was limited in the RV144 phase III HIV/AIDS human clinical trial (40). In this context, NYVAC-C Δ3 could be considered a valid prototype for future vaccines due to its ability to activate NFκB, to induce specific neutrophil migration, and to enhance CD8 T-cell immune responses to Gag and Pol antigens.
Gag and Pol are the best conserved HIV-1 proteins (41) and are able to shift the CTL response from variable Env epitopes to the conserved Gag and Pol epitopes in the first years of HIV-1 infection (42). In untreated chronic HIV-1–infected individuals, a Gag CTL response correlates with lower HIV viral loads, decreasing HIV viremia (43); furthermore, it correlates with decreasing viremia in early HIV-1–infected patients with suspension of retroviral therapy (42). A prophylactic vaccine that induces a Gag CTL response was recently shown to control simian immunodeficiency virus infection (44). Because the Gag/Pol-specific CD8 T-cell response in NYVAC-C Δ3-infected mice is mainly polyfunctional compared with that of NYVAC-C and because most human HIV nonprogressors preferentially maintain highly functional HIV-specific CD8 T cells (45), the NYVAC-C Δ3 vaccinia vector might constitute a promising approach for prophylactic and therapeutic treatment. By triggering enhanced NFκB activation and specific neutrophil recruitment, the triple deletion mutant could offer a considerable advantage over current NYVAC-based vectors being tested in phase I prophylactic and therapeutic clinical trials (46).
In contrast to Gag and Pol responses, NYVAC-C Δ3 does not induce a significant increase in the CD8 T-cell response to Env compared with NYVAC-C. This difference in antigen response probably depends on Env extracellular secretion compared with Gag–Pol intracellular production. Our data suggest greater neutrophil involvement in the engulfment of VACV-infected cells that express intracellular Gag and Pol than of a secreted antigen such as Env, which could be sequestered by macrophages normally found in the peritoneal cavity.
Neutrophils reportedly have difficulties in priming CD4 T cells compared with CD8 T cells after vaccinia infection (13); in our model, we observe no CD4 T-cell response to GPN in NYVAC-C– or in NYVAC-C Δ3-infected mice.
After migrating to the lymph nodes, neutrophils can compete with classic APCs (DCs, macrophages) to present antigens (47), and a direct interaction between antigen-pulsed neutrophils and CD8 T cells has also been proposed (9). In contrast, VACV-infected neutrophils cannot induce a CD8 T-cell proliferative response in BM in the absence of myeloid APCs (13). When mice were NYVAC-GFP– or NYVAC-GFP Δ3-infected, we did not detect GFP+-infected neutrophils in secondary lymphoid organs, which would suggest indirect neutrophil infection. Based on this evidence, we speculate that antigen transport results from neutrophil uptake of apoptotic cells in which the antigen has already been processed. The neutrophils that transport processed antigens are able to prime CD8 T cells directly, which then respond/proliferate specifically.
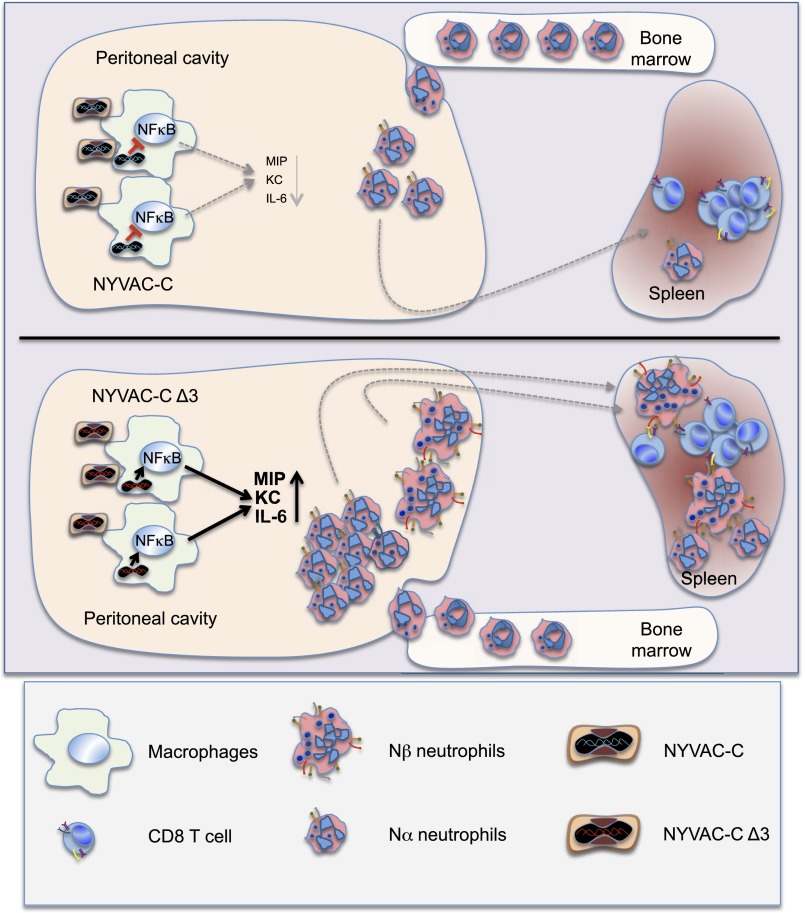
In summary, here we identify a mechanism by which the VACV vector NYVAC lacking A52R, K7R, and B15R genes triggers selective immune responses (Fig. 6). Infection with NYVAC-C Δ3 led to activation of the NFκB signaling pathway in vitro and in vivo, with increased cytokine/chemokine expression followed by specific neutrophil activation and recruitment. This enhanced neutrophil recruitment during infection correlated with an increased CD8 T-cell adaptive immune response specific for heterologous antigens. Further analysis of neutrophil populations identified an activated Nβ cell subtype with APC function that migrated to MLNs and to the spleen. Based on these inherent properties, the NYVAC triple deletion mutant demonstrates a new mechanism for poxvirus-induced immune response that provides a novel basis for virus vaccine vector design.
Fig. 6.
NYVAC-C Δ3 capacity to increase neutrophil peritoneal recruitment and spleen migration compared with NYVAC-C.
Materials and Methods
Mice and Injections.
BALB/c and C57/BL6J mice (6–8 wk old) were purchased from Harlan. C. Combadière (INSERM, Paris) provided C57/BL6J CCR1 KO mice, and S. Mañes (Centro Nacional de Biotecnología, Madrid) provided C57/BL6J CCR5 KO mice. NYVAC-WT and NYVAC-C vectors were provided by Sanofi Pasteur, and their biological properties have been described (27). For the heterologous DNA prime/NYVAC boost immunization protocol to assay NYVAC virus immunogenicity, mice received 100 μg DNA-C (50 μg plasmid construct pcDNA-CN54GP120 + 50 μg plasmid construct pcDNA-CN54GPN) or 100 μg DNA-ϕ (100 μg plasmid construct pcDNA) by the intramuscular route (27). Plasmids were purified using the Endofree Plasmid Mega Kit (Qiagen). After 2 wk, mice were immunized i.p. or i.m. with 107 PFUs of NYVAC-WT, NYVAC-C, or NYVAC-C Δ3. Mice immunized with sham DNA (DNA-ϕ) followed by a NYVAC-WT boost were used as controls. For in vivo neutrophil depletion, mice were injected i.v. with 200 μg 1A8 mAb (BioXcell) or 200 μg Isotype Control IgG2A mAb (Abcam) 24 h before the boost immunization. Animal studies were approved by the Ethical Committee of Animal Experimentation of the Centro Nacional de Biotecnologia/Consejo Superior de Investigaciones Cientificas in accordance with national and international guidelines and the Royal Decree (RD 1201/2005) (permits 12013 and 13013).
Cells.
African green monkey kidney cells (BSC-40; American Type Culture Collection) and primary chicken embryo fibroblast (CEF) cells (Intervet) were grown in DMEM supplemented with 10% (vol/vol) FCS. Human monocytic THP-1 cells (ATCC), murine macrophage J774.G8 cells (ATCC), and A20 cells (ATCC) were cultured in RPMI medium 1640 (Sigma-Aldrich) supplemented with 50 μM 2-mercaptoethanol and 10% (vol/vol) FCS. THP-1 cells were differentiated into macrophages by treatment with 5 ng/mL phorbol 12-myristate 13-acetate (Sigma-Aldrich) for 24 h before use. Murine macrophage cells were obtained from peritoneal exudates 4 d after i.p. administration of 1 mL 9% (wt/vol) casein; total cells were cultured (12 h) in RPMI 1640 with 10% (vol/vol) FCS, and nonadherent cells were removed. Cells were cultured in a humidified air 5% (vol/vol) CO2 atmosphere at 37 °C.
Viruses.
The poxvirus strains used included the genetically attenuated VACV-based vector NYVAC-WT and the recombinant NYVAC-C (27). Deletion mutants were constructed using Discosoma sp. (ds)Red2 and red shifted (rs)GFP as fluorescent markers. BSC-40 cells were infected with 0.01 PFU per cell of NYVAC-C (1 h) and transfected with pGem-RG-A52R-wm, pGem-RG-B15R-wm, or pGem-RG-K7R-wm plasmids using Lipofectamine (Invitrogen) to generate single, double, and triple deletion mutants. Deletion mutants were selected from progeny virus by consecutive rounds of plaque purification as described (48). NYVAC-GFP and NYVAC-GFP Δ3 were constructed using rsGFP as a fluorescent marker and β-galactosidase as a reporter gene. BSC-40 cells were infected with 0.01 PFU per cell of NYVAC-C or NYVAC-C Δ3 (1 h) and transfected with VACV insertional plasmid vector pLZAW1-LEO (49) to replace the HIV cassette with the GFP gene. Recombinant viruses were selected from progeny virus by consecutive rounds of plaque purification as reported (27). NYVAC-C Δ3 Κ7-rev was constructed using the K7R gene and β-glucuronidase as the reporter gene. BSC-40 cells were infected with 0.01 PFU per cell of NYVAC-C Δ3 (1 h) and transfected with VACV insertional plasmid vector pCAR-2/K7R to insert the K7R gene into the HA locus. Recombinant viruses were selected from progeny virus by consecutive rounds of plaque purification as reported (50). In vitro virus infections were performed with 2% (vol/vol) FCS. All viruses were grown in primary CEF cells, similarly purified through two 36% (wt/vol) sucrose cushions, and virus titers were determined by immunostaining plaque assay in BSC-40 cells as described (51).
Plasmids.
The plasmid transfer vectors pGem-RG-A52R-wm, pGem-RG-B15R-wm, and pGem-RG-K7R-wm, used for deletion of A52R, B15R, and K7R ORF from the NYVAC-C genome, respectively, were obtained by sequential cloning of A52R, B15R, and K7R recombination flanking sequences into the plasmid pGem-Red-GFP-wm as described (52). The plasmid transfer vector pCAR-2/K7R used for the insertion of the K7R gene was obtained by cloning the K7R sequence into the plasmid pCAR-2 (patent WO2007132052 A1).
The NYVAC genome was used as the template to amplify the left flank, the repeated left flank and the right flank of the three genes, or the sequence of K7R; the oligonucleotides used are listed in Table S1. The resulting plasmids pGem-RG-A52R-wm, pGem-RG-B15R-wm, pGem-RG-K7R-wm, and pCAR-2/K7R were confirmed by DNA sequence analysis.
PCR.
Viral DNA was extracted by the SDS-proteinase K-phenol method. Primers LF´A52R-Eco and RFA52R-Bam (spanning A52R flanking regions), LF´B15R-Aat and RFB15R-Bam (spanning B15R flanking regions), and LFK7R-Aat and fi-FIL-BR (TTATAGGATCCCTCCAGGAGAAAG) (spanning K7R flanking regions) were used for PCR analysis of A52R, B15R, and K7R loci, respectively. Primers TK-L and TK-R were used for PCR analysis of the TK (thymidine kinase) locus as described (53) to confirm the replacement of the HIV cassette with the GFP gene. Primers HA1 (GTCACGTGTTACCACGCA) and HA2-NYVAC–reverse (CCGAGTAAGGCATTAGG) were used for PCR analysis of the HA locus to confirm insertion of the K7R gene.
RT-PCR.
Total RNA was extracted using RNeasy Mini Kit (Qiagen) and was treated with DNase I recombinant, RNase-free. Subsequently, first-strand cDNA was synthesized with Oligo (dT)12–18 primers (Invitrogen) using SuperScriptIII Reverse transcriptase (Invitrogen) following the manufacturer’s protocol, and cDNA was used for PCR amplification. The primers K7RinternalFwd-Bam and K7RinternalRev-Not used for amplification of K7R cDNA were the same as for cloning and are described in Table S1.
Western Blot.
BSC-40 cells were lysed in Laemmli buffer, and extracts were fractionated by 8% (vol/vol) SDS/PAGE and analyzed by Western blot using rabbit polyclonal anti-gp120 (Centro Nacional de Biotecnología), -gag p24 (ARP 432, NIBSC, Centralised Facility for AIDS Reagents), or -E3 antibodies (Centro Nacional de Biotecnología). J774.G8 and THP-1 cells were lysed in Cell Lysis Buffer (Cell Signaling) supplemented with PMSF, and extracts were fractionated by 10% (vol/vol) SDS/PAGE and analyzed by Western blot using anti-phospho IκBα, -IκBα, and -tubulin (Cell Signaling). Nuclear extracts were obtained using a Nuclear/Cytosol Fractionation Kit (BioVision Research) and analyzed by Western blot using anti-H2A mAb (Cell Signaling).
EMSAs.
For gel shift assays, nuclear extracts were incubated with 0.5 ng 32P-end–labeled double-stranded oligonucleotide probe 5′-AGTTGAGGGGACTTTCCCAGGC-3′, which has NFκB binding sites. The binding reaction was performed in binding buffer with 1.5 mg poly (dI-dC), and binding products were resolved by electrophoresis.
Immunofluorescence.
Cells were washed with PBS, fixed with 4% (vol/vol) paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 20% (vol/vol) FCS. Cells were coincubated with polyclonal anti-p65 (Santa Cruz) and -E3 antibodies (Centro Nacional de Biotecnología). Bound primary antibodies were detected with AlexaFluor488 or 594-conjugated antibodies (Invitrogen). Cell nuclei were DAPI-stained (Sigma). The model of the microscope was LEICA TCS SP5 multispectral confocal system; type, magnification, and numerical aperture of the objective lenses were HCX PL APO lambda blue 63.0×/1.4 oil immersion; the temperature was 25 °C; the imaging medium was Prolong Gold (Invitrogen); and the acquisition software was Leica LASAF version 2.6.0.
Cytokine/Chemokine Analysis.
Sera and peritoneal washes from PBS- or virus-injected mice, as well as supernatants of mock- or virus-infected J774 and peritoneal macrophages, were assayed to detect cytokines and chemokines in multiplex analyses using LuminexXMAP technology. IL-8 levels from sera, peritoneal washes, and supernatants of J774 macrophages were quantified by IL-8 ELISA Kit (MyBioSource).
In Vitro Migration Assay.
Migration assays were performed in 6.5-mm-diameter transwell dishes (Corning Costar) with 3-μm pore filters. Murine BM cells were added to the upper chamber, and supernatants of mock- or virus-infected J774 cells were added to the bottom chamber. Migration in medium alone served as the negative control. After 3 h of incubation, cells in the medium of the bottom chamber were collected and passed for 1 min at a high flow rate in a LSRII cytometer (BD). The number of cell events was counted, and remaining cells in the tube were stained with anti-Ly6G (1A8) and -CD11b (M1/70) (BD).
Apoptosis Assay.
The annexin V binding assay was performed in combination with propidium iodide (PI) to monitor the integrity of the neutrophil membrane.
Hematoxylin/Eosin Staining.
Ly6G+ cells were fixed with acetone for 10 min and then stained.
Peptides.
The HIV-1 peptides Env-1 (sequence, PADPNPQEM), Pol-1 (LVGPTPVNI), and Pol-2 (YYDPSKDLI) were described as H-2d–restricted CTL epitopes (29) and were provided by the CNB-CSIC Proteomics Service. The HIV-1 Gag pool (60 peptides of Gag-1 + 61 peptides of Gag-2) and Nef pool (49 peptides) were provided by the EuroVacc Foundation and have been described (27); they span the entire HIV-1 Gag and Nef regions included in the immunogens as consecutive 15 mers with 11-amino acid overlaps.
Nucleofection Assay.
A20 cells were nucleofected using 4D-Nucleofector (Lonza) and the Amaxa SF Cell Line Kit (Lonza). Cells were nucleofected with 6 μg pcDNA-CN54gp120 or pcDNA-CN54GPN; pcDNA-ϕ was used as the negative control. At 24 h postnucleofection, A20 cells were used to restimulate splenocytes from infected mice (1:10, A20–splenocyte ratio).
Cell Isolation.
CD8+ T cells were purified from spleen using the Dynabeads FlowComp Mouse CD8 Kit (Invitrogen). Ly6G+ cells were sorted using MoFlo XDP sorter (Beckman Coulter).
Flow Cytometry.
For ICS, erythrocyte-depleted splenocytes were rested overnight and resuspended in RPMI 1640 with 10% (vol/vol) FCS and 1 μg/mL Golgiplug (BD), monensin (eBioscience), and anti-CD107a (1D4B) (BD). After restimulation with peptides or A20 cells [6 h, 37 °C, 5% (vol/vol) CO2], splenocytes were stained for surface markers with anti-CD3 (145-2C11), -CD4 (GK1.5), and -CD8 (53-6.7) (all from BD); fixed; permeabilized (Cytofix/Cytoperm Kit; BD); and stained intracellularly with anti–IL-2 (JES6-5H4), –IFN-γ (XMG 1.2), and –TNF-α (MP6-X722) (all from BD). PECs, obtained after injection of cold PBS into previously infected mice, were counted and stained with anti-Ly6G (1A8), -CD3 (145-2C11), -CD11b (M1/70), -CD19 (1D3), -CD80 (16-10A1), -CD86 (GL-1), -MHC class I (H-2Kd; SF1-1.1), -MHC class II (1-A/1-E; 2G9) (all from BD), -CD45 (30-F11; Biolegend), -CD11c (N418; eBioscience), -F4/80 (BM8, eBioscience), and -CD62L (MEL-14, Beckman Coulter). Peripheral blood, BM, omentum, spleen, and lymph nodes cells were stained with anti-LyG (1A8), -CD45 (30-F11), -CD11b (M1/70), and -CD115 (AFS98, eBioscience). The dead cells were stained using the violet LIVE/DEAD stain kit (Invitrogen) in all cytometry analyses.
Cells were acquired using a GALLIOS (Beckman Coulter) or LSRII (BD) flow cytometer, and data analyses were performed with FlowJo software version 8.5.3 (Tree Star). Boolean combinations of single functional gates were created with FlowJo to determine the frequency of each response based on all possible combinations of cytokines or of differentiation marker expression.
Statistical Analysis.
For statistical analysis of CD8 T-cell response to HIV-1 Gag–Pol antigens, we used an approach that corrects measurements for medium response and allows calculation of confidence intervals (CIs) and P values of hypothesis tests (50). Only antigen response values significantly larger than the corresponding RPMI are shown. Background levels (splenocytes in RPMI) were subtracted from all values used to allow analysis of proportional representation of responses. Analysis and presentation of distribution were performed using SPICE version 5.1, downloaded from exon.niaid.nih.gov. For statistical analysis of T-cell responses to HIV-1 Env antigen, the Mann–Whitney t test was used. For statistical analysis of cytokine/chemokine expression and in vitro neutrophil migration, one-way ANOVA was applied to compare all viruses used. Student’s t test was used for other analyses to establish the differences between two groups.
Supplementary Material
Acknowledgments
We are grateful to Victoria Jiménez for tissue culture, Raquel Castañera for help with animals, M. Carmen Moreno-Ortiz for Luminex and cell sorter assistance, Sylvia Gutiérrez for help with confocal microscopy, Catherine Mark for editorial assistance, and Marcos Malumbres for his critical review of the manuscript. M.D.P. received a Formación en Investigación en Salud PhD fellowship, and E.M.-P. received a Formación del Profesorado Universitario PhD fellowship from the Spanish Ministries of Health and Education. The research was supported by Spanish Grants SAF-2008-02036 and Red de Sida Royal Decree 12/0017/0038 and in part by a grant from the PTVDC (Poxvirus T Cell Vaccine Discovery Consortium) Bill and Melinda Gates Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424341112/-/DCSupplemental.
References
- 1.Gómez CE, Nájera JL, Krupa M, Perdiguero B, Esteban M. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr Gene Ther. 2011;11(3):189–217. doi: 10.2174/156652311795684731. [DOI] [PubMed] [Google Scholar]
- 2.Smith GL, et al. Vaccinia virus immune evasion: Mechanisms, virulence and immunogenicity. J Gen Virol. 2013;94(Pt 11):2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 3.Nathan C. Neutrophils and immunity: Challenges and opportunities. Nat Rev Immunol. 2006;6(3):173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 4.Sørensen OE, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97(12):3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto Y, Moki T, Takizawa T, Shiratsuchi A, Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J Immunol. 2007;178(4):2448–2457. doi: 10.4049/jimmunol.178.4.2448. [DOI] [PubMed] [Google Scholar]
- 6.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33(5):657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 8.Chtanova T, et al. Dynamics of neutrophil migration in lymph nodes during infection. Immunity. 2008;29(3):487–496. doi: 10.1016/j.immuni.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beauvillain C, et al. Neutrophils efficiently cross-prime naive T cells in vivo. Blood. 2007;110(8):2965–2973. doi: 10.1182/blood-2006-12-063826. [DOI] [PubMed] [Google Scholar]
- 10.Matsushima H, et al. Neutrophil differentiation into a unique hybrid population exhibiting dual phenotype and functionality of neutrophils and dendritic cells. Blood. 2013;121(10):1677–1689. doi: 10.1182/blood-2012-07-445189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oehler L, et al. Neutrophil granulocyte-committed cells can be driven to acquire dendritic cell characteristics. J Exp Med. 1998;187(7):1019–1028. doi: 10.1084/jem.187.7.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fanger NA, et al. Activation of human T cells by major histocompatability complex class II expressing neutrophils: Proliferation in the presence of superantigen, but not tetanus toxoid. Blood. 1997;89(11):4128–4135. [PubMed] [Google Scholar]
- 13.Duffy D, et al. Neutrophils transport antigen from the dermis to the bone marrow, initiating a source of memory CD8+ T cells. Immunity. 2012;37(5):917–929. doi: 10.1016/j.immuni.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 14.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29(2):182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Delaloye J, et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009;5(6):e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Hutchens MA, et al. Protective effect of Toll-like receptor 4 in pulmonary vaccinia infection. PLoS Pathog. 2008;4(9):e1000153. doi: 10.1371/journal.ppat.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowie A, et al. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc Natl Acad Sci USA. 2000;97(18):10162–10167. doi: 10.1073/pnas.160027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansur DS, et al. Poxvirus targeting of E3 ligase β-TrCP by molecular mimicry: A mechanism to inhibit NF-κB activation and promote immune evasion and virulence. PLoS Pathog. 2013;9(2):e1003183. doi: 10.1371/journal.ppat.1003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen RA, Ryzhakov G, Cooray S, Randow F, Smith GL. Inhibition of IkappaB kinase by vaccinia virus virulence factor B14. PLoS Pathog. 2008;4(2):e22. doi: 10.1371/journal.ppat.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ember SW, Ren H, Ferguson BJ, Smith GL. Vaccinia virus protein C4 inhibits NF-κB activation and promotes virus virulence. J Gen Virol. 2012;93(Pt 10):2098–2108. doi: 10.1099/vir.0.045070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myskiw C, Arsenio J, van Bruggen R, Deschambault Y, Cao J. Vaccinia virus E3 suppresses expression of diverse cytokines through inhibition of the PKR, NF-kappaB, and IRF3 pathways. J Virol. 2009;83(13):6757–6768. doi: 10.1128/JVI.02570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shisler JL, Jin XL. The vaccinia virus K1L gene product inhibits host NF-kappaB activation by preventing IkappaBalpha degradation. J Virol. 2004;78(7):3553–3560. doi: 10.1128/JVI.78.7.3553-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schröder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008;27(15):2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gedey R, Jin XL, Hinthong O, Shisler JL. Poxviral regulation of the host NF-kappaB response: The vaccinia virus M2L protein inhibits induction of NF-kappaB activation via an ERK2 pathway in virus-infected human embryonic kidney cells. J Virol. 2006;80(17):8676–8685. doi: 10.1128/JVI.00935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maluquer de Motes C, et al. Inhibition of apoptosis and NF-κB activation by vaccinia protein N1 occur via distinct binding surfaces and make different contributions to virulence. PLoS Pathog. 2011;7(12):e1002430. doi: 10.1371/journal.ppat.1002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez CE, et al. Generation and immunogenicity of novel HIV/AIDS vaccine candidates targeting HIV-1 Env/Gag-Pol-Nef antigens of clade C. Vaccine. 2007;25(11):1969–1992. doi: 10.1016/j.vaccine.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 28.Shin HM, et al. Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-kappaB without affecting IkappaB degradation. FEBS Lett. 2004;571(1-3):50–54. doi: 10.1016/j.febslet.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 29.Wild J, et al. Preclinical evaluation of the immunogenicity of C-type HIV-1-based DNA and NYVAC vaccines in the Balb/C mouse model. Viral Immunol. 2009;22(5):309–319. doi: 10.1089/vim.2009.0038. [DOI] [PubMed] [Google Scholar]
- 30.Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol. 2008;83(1):64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- 31.Graham SC, et al. Vaccinia virus proteins A52 and B14 share a Bcl-2-like fold but have evolved to inhibit NF-kappaB rather than apoptosis. PLoS Pathog. 2008;4(8):e1000128. doi: 10.1371/journal.ppat.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalverda AP, et al. Poxvirus K7 protein adopts a Bcl-2 fold: Biochemical mapping of its interactions with human DEAD box RNA helicase DDX3. J Mol Biol. 2009;385(3):843–853. doi: 10.1016/j.jmb.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 33.Harte MT, et al. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J Exp Med. 2003;197(3):343–351. doi: 10.1084/jem.20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araki H, et al. Reprogramming of human postmitotic neutrophils into macrophages by growth factors. Blood. 2004;103(8):2973–2980. doi: 10.1182/blood-2003-08-2742. [DOI] [PubMed] [Google Scholar]
- 35.Fridlender ZG, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 2009;16(3):183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann MH, et al. Modified vaccinia virus Ankara triggers chemotaxis of monocytes and early respiratory immigration of leukocytes by induction of CCL2 expression. J Virol. 2009;83(6):2540–2552. doi: 10.1128/JVI.01884-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abadie V, et al. Original encounter with antigen determines antigen-presenting cell imprinting of the quality of the immune response in mice. PLoS ONE. 2009;4(12):e8159. doi: 10.1371/journal.pone.0008159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gómez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum Vaccin Immunother. 2012;8(9):1192–1207. doi: 10.4161/hv.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barouch DH, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013;155(3):531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rerks-Ngarm S, et al. MOPH-TAVEG Investigators Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 41.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8(8):619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang OO, Daar ES, Ng HL, Shih R, Jamieson BD. Increasing CTL targeting of conserved sequences during early HIV-1 infection is correlated to decreasing viremia. AIDS Res Hum Retroviruses. 2011;27(4):391–398. doi: 10.1089/aid.2010.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiepiela P, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 44.Iwamoto N, et al. Control of simian immunodeficiency virus replication by vaccine-induced Gag- and Vif-specific CD8+ T cells. J Virol. 2014;88(1):425–433. doi: 10.1128/JVI.02634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harari A, et al. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J Exp Med. 2008;205(1):63–77. doi: 10.1084/jem.20071331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang CW, Strong BS, Miller MJ, Unanue ER. Neutrophils influence the level of antigen presentation during the immune response to protein antigens in adjuvants. J Immunol. 2010;185(5):2927–2934. doi: 10.4049/jimmunol.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perdiguero B, et al. Deletion of the vaccinia virus gene A46R, encoding for an inhibitor of TLR signalling, is an effective approach to enhance the immunogenicity in mice of the HIV/AIDS vaccine candidate NYVAC-C. PLoS ONE. 2013;8(9):e74831. doi: 10.1371/journal.pone.0074831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Pilato M, et al. New vaccinia virus promoter as a potential candidate for future vaccines. J Gen Virol. 2013;94(Pt 12):2771–2776. doi: 10.1099/vir.0.057299-0. [DOI] [PubMed] [Google Scholar]
- 50.Nájera JL, Gómez CE, García-Arriaza J, Sorzano CO, Esteban M. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS ONE. 2010;5(6):e11406. doi: 10.1371/journal.pone.0011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramírez JC, Gherardi MM, Esteban M. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: Virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J Virol. 2000;74(2):923–933. doi: 10.1128/jvi.74.2.923-933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kibler KV, et al. Improved NYVAC-based vaccine vectors. PLoS ONE. 2011;6(11):e25674. doi: 10.1371/journal.pone.0025674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gómez CE, et al. Head-to-head comparison on the immunogenicity of two HIV/AIDS vaccine candidates based on the attenuated poxvirus strains MVA and NYVAC co-expressing in a single locus the HIV-1BX08 gp120 and HIV-1(IIIB) Gag-Pol-Nef proteins of clade B. Vaccine. 2007;25(15):2863–2885. doi: 10.1016/j.vaccine.2006.09.090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.