Significance
In the mitogen-activated protein kinase (MAPK) pathways, N-terminal intrinsically disordered regulatory domains of the MAPK kinases (MKK) control signaling specificity by binding to their cognate MAPKs via docking sites carrying homologous recognition sequences. MKK7 activates the c-Jun N-terminal kinase (JNK) pathway and is the only MKK containing three motifs within its regulatory domain. Here we obtain a comprehensive picture of the structure, dynamics, affinity, stoichiometry, and kinetics of the MKK7–JNK signaling complex. Importantly, we show using a combination of X-ray crystallography and NMR exchange spectroscopy that the second docking site of MKK7 binds to JNK via two alternative binding modes, providing insight into the regulation of signaling specificity by short linear motifs.
Keywords: NMR, mitogen-activated protein kinase, intrinsically disordered protein, signaling, c-Jun N-terminal kinase
Abstract
Signaling specificity in the mitogen-activated protein kinase (MAPK) pathways is controlled by disordered domains of the MAPK kinases (MKKs) that specifically bind to their cognate MAPKs via linear docking motifs. MKK7 activates the c-Jun N-terminal kinase (JNK) pathway and is the only MKK containing three motifs within its regulatory domain. Here, we characterize the conformational behavior and interaction mechanism of the MKK7 regulatory domain. Using NMR spectroscopy, we develop an atomic resolution ensemble description of MKK7, revealing highly diverse intrinsic conformational propensities of the three docking sites, suggesting that prerecognition sampling of the bound-state conformation is not prerequisite for binding. Although the different sites exhibit similar affinities for JNK1, interaction kinetics differ considerably. Importantly, we determine the crystal structure of JNK1 in complex with the second docking site of MKK7, revealing two different binding modes of the docking motif correlating with observations from NMR exchange spectroscopy. Our results provide unique insight into how signaling specificity is regulated by linear motifs and, in general, into the role of conformational disorder in MAPK signaling.
Mitogen-activated protein kinases (MAPKs) are essential components of eukaryotic signal transduction networks that enable cells to respond appropriately to extracellular stimuli (1). The MAPK signaling pathways feature three sequentially acting protein kinases making up a signaling module: an MKKK (MAPK kinase kinase) that phosphorylates and thereby activates an MKK (MAPK kinase), which then activates the MAPK by phosphorylation. In mammalian organisms, four major MAPK pathways have been identified: extracellular-signal-regulated kinase (ERK1/2), ERK5, p38α/β/γ/δ, and c-Jun N-terminal kinase (JNK1/2/3) (2).
Seven human MKKs phosphorylate ERK, p38, and JNK according to the following selectivity scheme: MKK1/2 activate ERK1/2, MKK5 activates ERK5, MKK3/4/6 activate p38, and MKK4/7 activate JNK (3). Signaling specificity is controlled by intrinsically disordered N-terminal regulatory domains of the MKKs (ranging between 40 and 100 aa in length) that selectively bind to their cognate MAPKs (4–8). In the absence of these regulatory domains, phosphorylation of the downstream kinases is extremely inefficient (9, 10). Molecular recognition occurs through so-called docking sites in the regulatory domains composed of two to three basic residues followed by a short spacer of one to six residues and finally a hydrophobic-X-hydrophobic submotif. Crystal structures of ERK, p38, and JNK in complex with docking site peptides show that the basic residues contact the negatively charged common docking groove of the MAPKs, whereas the hydrophobic residues insert into hydrophobic pockets on the surface (Fig. S1A) (7, 11–16).
MKK7 activates the JNK pathway that primarily regulates stress and inflammatory responses. MKK7 is the only human MKK for which three putative docking sites (D1, D2, and D3) are present within its 100-aa regulatory domain (Fig. 1 A and B), posing a number of questions concerning the affinity, stoichiometry, and kinetics as well as the structure and dynamics of the MKK7–JNK signaling complex. Previously, it has been shown that all three putative docking sites are able to bind JNK, and it was proposed that binding occurs with moderate synergy (17).
Fig. 1.
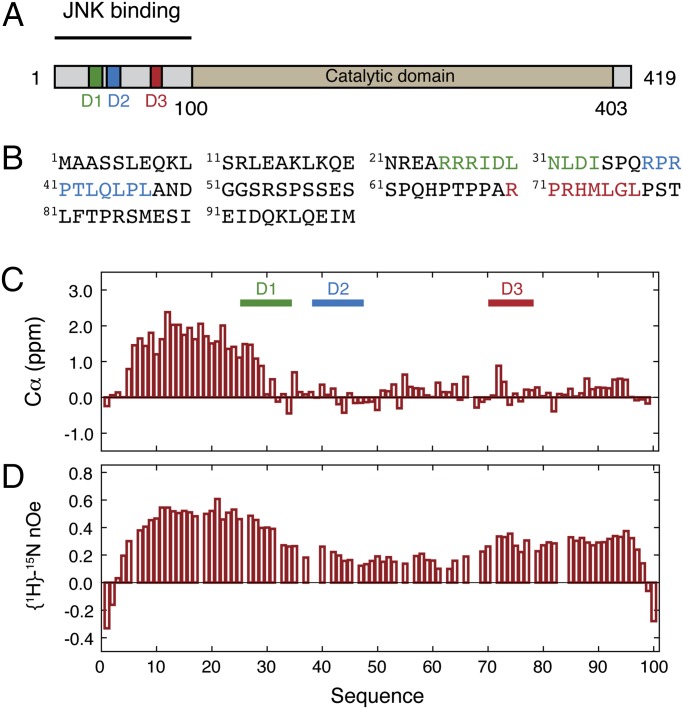
Secondary structure and dynamics in the regulatory domain of MKK7. (A) Domain organization of MKK7 indicating the position of the catalytic domain (beige) and the docking sites: D1 (green), D2 (blue), and D3 (red). (B) Primary sequence of the N-terminal regulatory domain of MKK7 with colors indicating the different docking sites: D1 (green), D2 (blue), and D3 (red). (C) Experimental secondary Cα chemical shifts obtained at 5 °C and pH 7.0. (D) Experimental 15N heteronuclear nOes measured at a 15N frequency of 60 MHz and 5 °C.
In this study, we characterize the structure and dynamics of the regulatory domain of MKK7 at atomic resolution by using NMR spectroscopy, providing an ensemble model of the full-length kinase. We study the interaction between MKK7 and JNK1 by NMR and isothermal titration calorimetry (ITC), providing a comprehensive picture of the affinity, stoichiometry, and kinetics of the JNK1–MKK7 complex. Furthermore, we determine the crystal structure of JNK1 in complex with the second MKK7 docking site. By combining these data with experimental NMR measurements of the same complex, we provide insight into the dynamic nature of the interaction between the two proteins, suggesting conformational exchange between distinct MKK7 binding modes. Our results support the conclusion that MKK7 forms a highly dynamic complex with JNK1 that relies on multisite interactions to control signaling specificity. The atomic resolution description of full-length MKK7 obtained here extends our view beyond the folded, catalytic domains of the MAPK family and highlights potential new avenues for the discovery of drugs targeting this protein family.
Results
Structure and Dynamics of the Regulatory Domain of MKK7.
We obtained the complete NMR backbone resonance assignment of the regulatory domain of MKK7 (Fig. S2A). The 13Cα secondary chemical shifts reveal that MKK7 contains helical propensity at its N terminus (residues 5–30), whereas the remaining residues have secondary chemical shifts close to zero indicating a near random coil state (Fig. 1C). We also measured {1H}-15N heteronuclear Overhauser enhancement (nOe) showing that residues 5–30 have reduced flexibility compared with the remainder of the chain, coinciding with the residual helical structure in this region (Fig. 1D). The central region separating D2 and D3 shows increased dynamics on the picosecond to nanosecond time scale due to the presence of a large number of serine and glycine residues.
Structural Ensemble Description of MKK7.
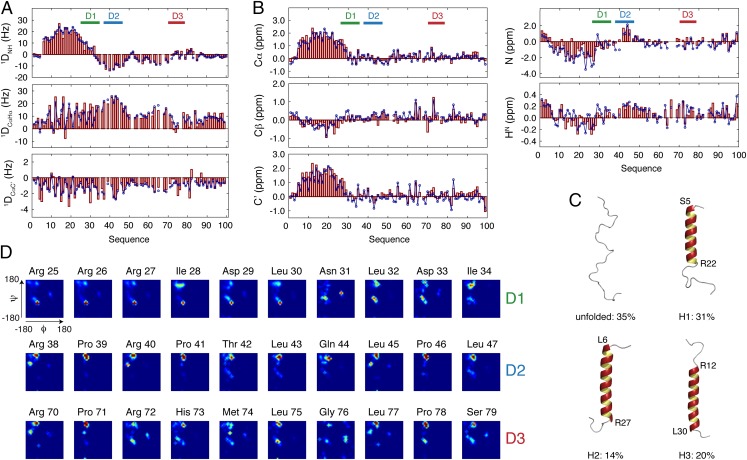
To obtain detailed information about the site-specific conformational sampling of the three docking sites in their prerecognition states, the experimental chemical shifts were complemented with measurements of multiple residual dipolar couplings (RDCs) (18). The experimental 1DNH, 1DCαHα, and 1DCαC′ RDCs display “dipolar waves” at the N terminus of MKK7, providing evidence for exchange between a limited number of specific α-helices (Fig. 2A) (19–22). Analysis of experimental 13Cα chemical shifts and 1DNH, 1DCαHα, and 1DCαC′ RDCs from residues 5–30 by using a minimal ensemble approach (22, 23), combined with the genetic algorithm ASTEROIDS (24–26), reveals that this sequence samples an ensemble of three specific helical conformers (H1, S5-R22; H2, L6-R27; H3, R12-L30) in exchange with a completely disordered form of the protein (Fig. 2 A–C). The selected helices are all preceded by serine residues (S4, S5, and S11), indicating that the main stabilization mechanism is N-capping nucleation (27).
Fig. 2.
Ensemble description of the regulatory domain of MKK7 on the basis of chemical shifts and RDCs. (A) Comparison of experimental RDCs (red) with values back-calculated (blue) from a selected ASTEROIDS ensemble comprising 200 conformers. (B) Comparison of experimental chemical shifts (red) with values back-calculated (blue) from a selected ASTEROIDS ensemble. (C) Model of the N terminus of MKK7 in terms of a minimal ensemble of cooperative α-helices in exchange with a completely disordered form of the protein. For simplicity, each ensemble is represented by a single structure and the contribution from each ensemble is indicated as a percentage. (D) Ramachandran maps for residues in the three docking sites D1, D2, and D3 derived from five independent runs of ASTEROIDS selecting against experimental chemical shifts and RDCs. Red indicates high population, whereas blue corresponds to negligible population.
In a second step, we carried out an ensemble selection of the entire regulatory domain on the basis of chemical shifts (13Cα, 13Cβ, 13C’, 15N, and 1HN) and RDCs (1DNH, 1DCαHα, and 1DCαC′). Ensembles comprising 200 conformers were selected from a large pool of structures containing the already described conformational helical equilibrium at the N terminus, giving rise to excellent agreement with experimental data (Fig. 2 A and B). Importantly, the derived ensembles are also capable of reproducing independent (passive) data (4DHNHα RDCs), testifying to the validity of the ensembles (Fig. S2B).
These representative ensembles provide amino acid-specific insight into the conformational sampling of the three docking sites in their prerecognition states (Fig. 2D and Fig. S2C). Although the D1 docking site displays enhanced α-helical sampling compared with statistical coil distributions, the other two sites, D2 and D3, populate more extended regions of Ramachandran space. In particular, residues within the D2 docking site oversample PPII conformations. We note that the 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of the isolated regulatory domain superimposes on that of full-length MKK7, showing that the conformational sampling of the regulatory domain derived here is conserved in the context of the full-length kinase, allowing us to propose an ensemble model of full-length MKK7 (Fig. S3).
Stoichiometry and Affinity of the MKK7–JNK1 Complex.
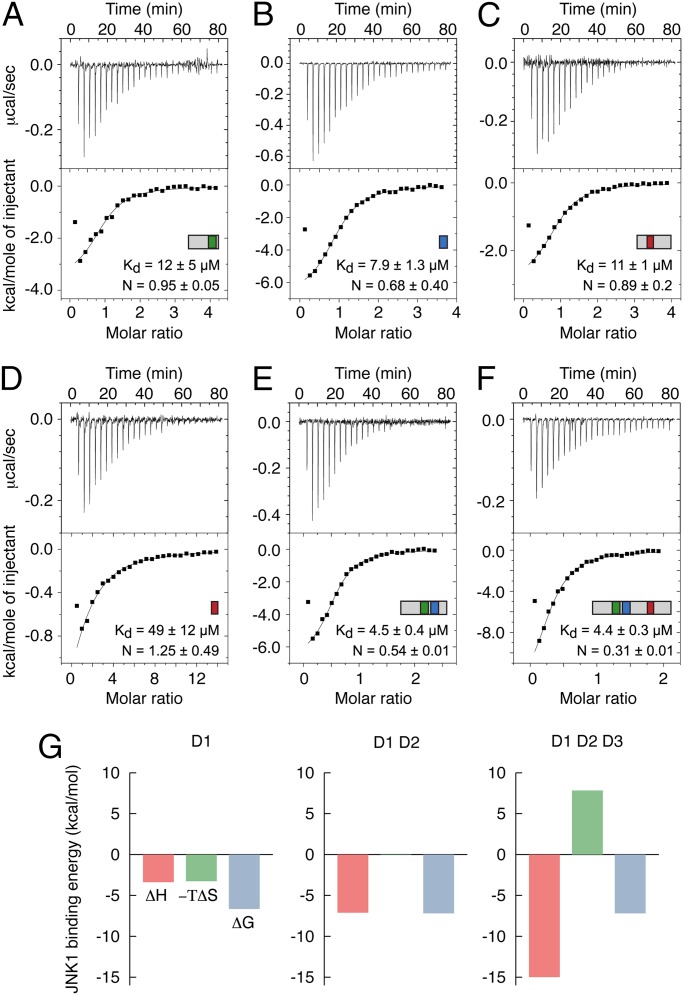
Isothermal titration calorimetry (ITC) was used to determine the affinity and stoichiometry of the MKK7–JNK1 interaction. The ITC measurements reveal that all docking sites bind with similar affinity in the low micromolar range with D2 (Kd = 7.9 μM) as the strongest binder followed by D3 (Kd = 11 μM) and D1 (Kd = 12 μM) (Fig. 3 A–C). We note that the peptide corresponding to the canonical D3 site shows a lower affinity (Kd = 49 μM; Fig. 3D) than the longer construct containing an extended D3 site (Kd = 11 μM; Fig. 3C), suggesting potential binding contributions from residues outside the canonical docking site.
Fig. 3.
ITC measurements of JNK1 binding to MKK7 constructs. (A) Binding of JNK1 to residues 1–42 of MKK7. (B) Binding of JNK1 to residues 37–48 of MKK7. (C) Binding of JNK1 to residues 48–100 of MKK7. (D) Binding of JNK1 to residues 69–78 of MKK7. (E) Binding of JNK1 to residues 1–55 of MKK7. (F) Binding of JNK1 to residues 1–100 of MKK7. In A–F, representative ITC data are shown with raw injection heats (Upper) and the corresponding specific binding isotherms (Lower). All ITC measurements were carried out at 20 °C with JNK1 in the sample cell, and the data were fit to a model corresponding to n independent binding sites. The final values correspond to averages of at least two independent measurements. (G) Dissection of binding free energies into the enthalpic and entropic contributions for the interaction of JNK1 with MKK7 constructs containing D1 only, D1 and D2, and all three docking sites.
ITC provides a clear picture of the stoichiometry of the MKK7–JNK1 complex, revealing a 1:1 complex for the construct containing only D1 (Fig. 3A), a 1:2 complex for the construct containing both D1 and D2 (Fig. 3E), and finally a 1:3 complex for the full-length regulatory domain containing all three docking sites (Fig. 3F). Furthermore, the binding of several JNK1 molecules to MKK7 is supported by the correlated increase in binding enthalpy with increasing number of binding sites (Fig. 3G). ITC measurements thus confirm that all MKK7 docking sites are equally binding competent and that MKK7 can simultaneously accommodate three JNK1 molecules.
Characterization of the MKK7–JNK1 Complex by NMR.
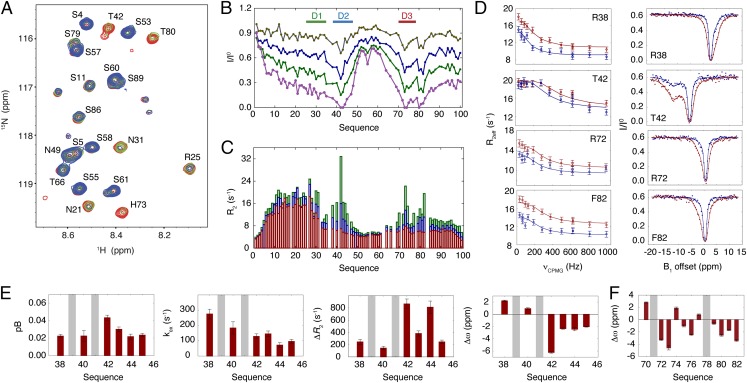
The interaction of MKK7 with JNK1 was further monitored by using NMR at 5 °C. Although, chemical shift changes in the 1H-15N HSQC spectrum of MKK7 were small upon addition of JNK1, large changes in signal intensities were observed, leading to the disappearance of a number of resonances, even at substoichiometric amounts of JNK1 (Fig. 4A). The intensity profile of MKK7 shows a decrease in signal intensities localized at the three JNK1 docking sites with D2 and D3 showing the clearest binding signatures (Fig. 4B). The intensities also decrease toward the C terminus, in agreement with previous NMR studies of p38α showing that residues outside the canonical docking sites contact the MAPK surface (28).
Fig. 4.
Interaction of the regulatory domain of MKK7 with JNK1 by NMR. (A) Region of the 1H-15N HSQC spectra of MKK7 in the absence (red) and presence of 28% (green) and 69% (blue) JNK1. (B) Intensity profile of MKK7 with increasing amounts of JNK1: 8% (gray), 14% (blue), 28% (green), and 69% (magenta). (C) R2 relaxation rates at 600 MHz and 5 °C of MKK7 in the absence of JNK1 (red) and presence of increasing amounts of JNK1: 14% (blue), 28% (green). (D) RD (Left) and CEST profiles (Right) of selected residues within MKK7 obtained upon addition of 10% (molar ratio) of JNK1. The RD experiments were recorded at 1H frequencies of 600 MHz (blue) and 800 MHz (red), whereas the CEST experiments were recorded at a 1H frequency of 700 MHz for two B1 saturating fields of 22 Hz (blue) and 44.5 Hz (red). Lines correspond to fits of the profiles to a two-site exchange model. (E) Simultaneous analysis of RD and CEST data for each residue within D2. The parameters presented are the following: The population (pB) of MKK7 in complex with JNK1, the exchange rate (kex), the difference in effective R2 relaxation rates (ΔR2), and the difference in 15N chemical shifts (Δω) between the free and bound forms of MKK7. (F) Difference in 15N chemical shifts between the free and bound form of MKK7 for residues within D3 obtained from a simultaneous analysis of RD and CEST data for all residues.
The gradual disappearance of the NMR resonances of MKK7 at substoichiometric amounts of JNK1 indicates contributions to the line widths from conformational exchange occurring on the microsecond to millisecond time scale between free and JNK1-bound forms of MKK7. The 15N R2 rates measured on the visible resonances at selected points during the chemical shift titration increase only slightly upon addition of JNK1 (Fig. 4C), indicating that the exchange between free and bound forms of MKK7 is slow to intermediate on the chemical shift time scale. We also repeated the titration of the regulatory domain of MKK7 with JNK1 at 20 °C (the temperature at which the ITC was performed), showing similar features as the 5 °C spectra (Fig. S4).
To characterize the conformational exchange between free and JNK1-bound forms of MKK7, we measured 15N relaxation dispersion (RD) (29) and chemical exchange saturation transfer (CEST) experiments (30) of MKK7 containing 10% (molar ratio) of JNK1. Both experiments reveal exchange on the microsecond to millisecond time scale for residues located in and around the docking sites D2 and D3 (Fig. 4D). No RD is detected in free MKK7, showing that the observed dispersion is associated with JNK1 binding. The RD and CEST data were analyzed simultaneously assuming a two-site exchange system for each individual residue within D2 (Fig. S5). The fitted population of JNK1-bound MKK7 from this analysis ranged between 2.2 and 4.3% (Fig. 4E), and the associated exchange rates vary from 275 to 70 s−1 over the D2 site with the fastest rates observed for the group of positively charged arginines, whereas the slowest rates are observed for the hydrophobic residues within the docking site (Fig. 4E). These observations suggest that the residues L43, L45, and L47 function as anchors of the docking site, whereas the charged residues R38 and R40 establish less well-defined contacts with the surface of JNK1. We note that large ΔR2 values between free and JNK1-bound MKK7 are obtained from the analysis of the CEST experiments (Fig. 4E). These rates exceed the expected values for the size of the MKK7–JNK1 complex by an order of magnitude and can only be consistent with conformational exchange contributions to R2 within the bound form of MKK7. These contributions could report on exchange between different bound conformations of the MKK7 peptide occurring on the microsecond to millisecond time scale.
The RD and CEST profiles observed within D3 did not allow a reliable per-residue analysis of the data. We therefore performed a simultaneous analysis by using a two-state model of all residues within D3 to a common population and exchange rate with chemical shift changes varying over the individual residues (Fig. 4F and Fig. S6). The analysis gives an exchange rate of 707 ± 34 s−1 and a population of bound MKK7 of 0.8%. Clear dispersion profiles are observed for the residues T80, L81, and F82 that are located outside the canonical D3 docking site (R70-L77), confirming their involvement in the binding of JNK1. The common kinetics exhibited by these additional sites indicates that they bind in a concerted fashion together with the rest of the D3 motif. The extension of the binding motif at the level of D3 is also reflected in the decrease in signal intensities of MKK7 measured for increasing JNK1 concentrations (Fig. 4B), as well as in the difference observed in the measured dissociation constants by ITC of the canonical D3 docking site peptide compared with the protein construct containing the extended docking site (Fig. 3 C and D). In contrast to D2 and D3, the kinetics associated with binding of JNK1 to D1 could not be derived from the RD and CEST experiments. NMR experiments carried out by using a shorter construct of the regulatory domain containing only the D1 docking site demonstrate that D1 binds JNK1 under these conditions (Fig. S7).
To evaluate the effect of the presence of the catalytic domain of MKK7 on the interaction with JNK1, we carried out an NMR titration of full-length MKK7 with JNK1 (Fig. S3C). An intensity profile is observed that is almost identical to the profile obtained for the isolated regulatory domain (Fig. 4B). Importantly, this titration demonstrates that D3 can bind JNK1 despite its close proximity to the catalytic domain of MKK7.
Crystal Structure of the JNK1–MKK7D2 Complex.
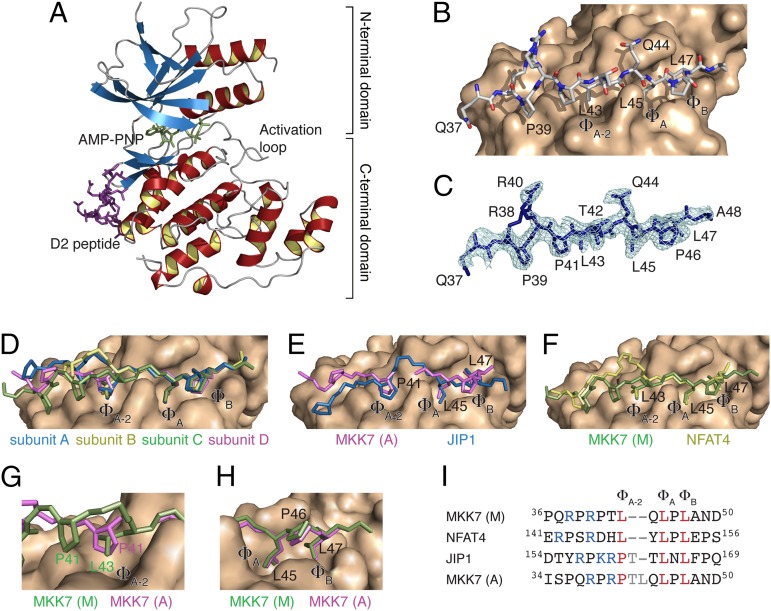
To obtain insight into the structural basis of the interaction between JNK1 and MKK7, we solved the crystal structure of JNK1 to 2.4 Å resolution in complex with a peptide derived from the D2 docking site of MKK7 (QRPRPTLQLPLA) and the ATP analog AMP-PNP (Fig. 5A and Table S1). Four JNK1 molecules and four peptides are present in the asymmetric unit of the crystal, where subunits A, B, and C adopt a similar interlobe angle and have only partially visible activation loops. An analysis of the domain motions by using DynDom (31) shows that subunit D exhibits a 10° rotation inwards compared with the other subunits, which is accompanied by an ordering of the activation loop (Fig. S8A) (11, 12). The density and temperature factors, however, indicate that the activation loop and residues 178–192 likely adopt multiple conformations (Fig. S8 B and C).
Fig. 5.
Crystal structure of JNK1 in complex with the D2 docking site peptide. (A) Crystal structure of JNK1 in complex with the ATP analog AMP-PNP (green) and the D2 peptide (magenta). (B) Structural basis of the interaction between the D2 peptide (sticks) and JNK1 (surface) as observed in chain C (main binding mode). (C) Simulated annealed omit electron density map contoured at 1σ of the D2 peptide of chain C. (D) Superposition of the four MKK7 docking site peptides on the surface of JNK1. The MKK7 peptides are shown as sticks with side chains displayed for residues P41, L43, L45, P46, and L47. (E) Superposition of the MKK7 (magenta, alternative binding mode) and JIP1 (blue) docking site peptides on the surface of JNK1. (F) Superposition of the MKK7 (green, main binding mode) and NFAT4 (yellow) docking site peptides on the surface of JNK1. (G) Zoom of the ΦA-2 hydrophobic pocket. (H) Zoom of the ΦA and ΦB hydrophobic pockets. (I) Sequence alignment of the docking site motif of MKK7 with that of NFAT4 and JIP1. Two alignments are shown for MKK7 corresponding to the main (M) and alternative (A) binding mode observed in the crystal structure.
The MKK7 peptide inserts into three pockets (denoted ΦA-2, ΦA, and ΦB) on the surface of JNK1 (Fig. 5 B and C). In subunits A, B, and C, the peptide is anchored to the hydrophobic pockets via L43, L45, and L47, whereas the positively charged arginines make looser and less well-defined contacts with the surface of JNK1 (Fig. 5D). Interestingly, an alternative binding mode is observed in subunit D, where R40 contacts D326 and E329 on the surface of JNK1 and P41, L45, and L47 occupy the three pockets, resulting in increased flexibility at the level of T42 and L43 and consequently missing electron density for these two residues (Fig. 5 G and H). In general, the main (M) binding mode of the MKK7 peptide resembles that of the docking site motif of the JNK substrate NFAT4, whereas the alternative (A) binding mode resembles the binding mode of the docking site motif of the scaffold protein JIP1 (Fig. 5 E, F, and I). The fact that both conformations are observed within the crystal structure suggests that the MKK7 peptide is able to switch between these two alternative binding modes.
Discussion
In recent years, it has become increasingly clear that cellular signaling relies on intrinsically disordered domains that play key roles in controlling specificity and compartmentalization. Insight into the molecular basis of cellular signaling therefore requires the characterization of these highly flexible domains and their functional complexes. Indeed, as demonstrated in the case of phosphatase PTP1B, understanding the dynamic behavior of intrinsically disordered signaling proteins provides potent tools for drug discovery (32). In this study, we have obtained, to our knowledge, the first atomic resolution characterization of the structure and interaction kinetics of a disordered domain of the MKK family. Using a multidisciplinary approach, we have combined NMR, X-ray crystallography, and ITC to obtain a comprehensive picture of the affinity, stoichiometry, kinetics, structure, and dynamics of the MKK7–JNK1 signaling complex.
Analysis of experimental NMR data in terms of a dynamic ensemble reveals the remarkable complexity of intrinsic conformational sampling present in the regulatory domain of MKK7. The N terminus possesses residual helical structure, potentially constituting a molecular recognition element for the upstream dual leucine zipper kinase (DLK) that has been shown to interact with the disordered domain of MKK7 via its leucine zippers (33, 34). The helical region of MKK7 is predicted with high confidence to form a coiled coil (Fig. S9A), and as the regulatory domain does not homodimerize (Fig. S9B), we therefore speculate that this motif mediates heterodimerization with the leucine zipper region of DLK. In general, the regulatory domains of the MKK family may play a more important role in binding to upstream MKKKs than previously thought, as also exemplified by the identification of the MEKK3–MKK5 (35) and MEKK2–MKK7 (36) complexes.
The N-terminal helix also overlaps with the first docking motif, so that the intrinsic local backbone sampling of D1 is dominated by α-helical conformations, whereas D3 exhibits propensities resembling a random coil state (Fig. 2D). By contrast the D2 site exhibits elevated propensity to populate extended (PPII) conformations in the prerecognition state. Analysis of dihedral angle distributions in crystal structures of JNK, ERK, and p38 reveals an overall tendency of docking site motifs to adopt PPII or extended conformations in complex with their cognate kinases (Fig. S1B). The different intrinsic conformational propensities within the docking sites of MKK7 therefore suggest that prerecognition sampling of the bound-state conformation is not a prerequisite for binding.
The ITC studies show that JNK1 employs a multisite interaction mechanism, visiting all of the MKK7 docking sites with almost equal affinities. The RD and CEST experiments mainly probe the rate of complex dissociation, koff. The obtained off rate of 707 ± 34 s−1 for the D3 docking site is faster than that observed for the D2 site, exhibiting a value of 137 ± 8 s−1 (simultaneous analysis of all residues equivalent to the D3 analysis resulting in a population of 3.3%). Although the affinities of the three interactions (D1–D3) are similar, neither CEST nor RD is observed for the D1 site, clearly indicating a different kinetic regime for this site.
Analysis of the CEST data displays large ΔR2 values between the free and bound states of MKK7 for residues within D2 (Fig. 4E). As mentioned above, this observation suggests that the peptide of MKK7 experiences conformational exchange on the microsecond to millisecond timescale when bound to JNK1. This finding correlates with the crystal structures, in which we observed two different binding modes of D2 on the surface of JNK1 (Fig. 5 E and F). It is possible that the conformational exchange contributions in the complex probed by the CEST experiments correspond to the switching between the two alternative binding modes observed in the crystal structures, in particular the highest level of detectable dynamics within the complex is observed for T42 which lies between P41 and L43 that occupy pocket ΦA-2 in the two alternative binding modes (Fig. 5G). The two binding modes of the D2 peptide are associated with a 10° clamping motion of the N- and C-terminal lobes of JNK1 (Fig. S8A). The two different conformations resemble active- and auto-inhibited forms of JNK1, suggesting that the changes in structure resulting from the two different peptide-binding modes are related to kinase function (Fig. S8D) (37).
Conclusion
In conclusion, this study provides unique insight into the conformational behavior, interaction mechanisms, and possible role of the intrinsically disordered regulatory domain of MKK7 in the JNK signaling pathway. Distinct intrinsic conformational propensities in the JNK1 interaction sites are associated with different interaction kinetics potentially resulting from different apparent binding mechanisms. We also obtain direct evidence for the flexible nature of the bound state of the D2 site, combining observations derived from crystallography concerning available binding modes, and NMR concerning exchange between conformations within the bound state. More generally this first atomic resolution molecular description of full-length MKK7 paves the way for future studies of interactions with other cellular partners, in particular noncognate kinases, providing insight into the molecular mechanisms underpinning signaling specificity in the MAPK pathways.
Materials and Methods
Expression and purification protocols of MKK7 and JNK1, experimental details of ITC and NMR experiments, and ensemble selection procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work used the platforms of the Grenoble Instruct Centre (Integrated Structural Biology Grenoble; Unité Mixte de Service 3518) with support from the French Infrastructure for Integrated Structural Biology (ANR-10-INSB-05-02) and Grenoble Alliance for Integrated Structural and Cell Biology (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. Financial support is acknowledged from the French Agence Nationale de la Recherche (ANR) through ANR Jeunes Chercheuses et Jeunes Chercheurs ProteinDisorder (M.R.J.), ANR MALZ TAUSTRUCT (M.B.), and ANR ComplexDynamics (M.B.). J.K. is a fellow of the Marie Curie action “IDPbyNMR” of the European Commission (Contract 264257).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of JNK1 in complex with the second MKK7 docking site have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4UX9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419528112/-/DCSupplemental.
References
- 1.Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: A 10-year update. Physiol Rev. 2012;92(2):689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 2.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 3.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc Natl Acad Sci USA. 1997;94(14):7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanoue T, Nishida E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol Ther. 2002;93(2-3):193–202. doi: 10.1016/s0163-7258(02)00188-2. [DOI] [PubMed] [Google Scholar]
- 5.Bardwell L. Mechanisms of MAPK signalling specificity. Biochem Soc Trans. 2006;34(Pt 5):837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardwell AJ, Frankson E, Bardwell L. Selectivity of docking sites in MAPK kinases. J Biol Chem. 2009;284(19):13165–13173. doi: 10.1074/jbc.M900080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garai Á, et al. Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Sci Signal. 2012;5(245):ra74. doi: 10.1126/scisignal.2003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peti W, Page R. Molecular basis of MAP kinase regulation. Protein Sci. 2013;22(12):1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. The MKK7 gene encodes a group of c-Jun NH2-terminal kinase kinases. Mol Cell Biol. 1999;19(2):1569–1581. doi: 10.1128/mcb.19.2.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming Y, et al. Synergistic activation of stress-activated protein kinase 1/c-Jun N-terminal kinase (SAPK1/JNK) isoforms by mitogen-activated protein kinase kinase 4 (MKK4) and MKK7. Biochem J. 2000;352(Pt 1):145–154. [PMC free article] [PubMed] [Google Scholar]
- 11.Chang CI, Xu BE, Akella R, Cobb MH, Goldsmith EJ. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol Cell. 2002;9(6):1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 12.Heo YS, et al. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 2004;23(11):2185–2195. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure. 2006;14(6):1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Liu S, Sun J-P, Zhou B, Zhang Z-Y. Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3. Proc Natl Acad Sci USA. 2006;103(14):5326–5331. doi: 10.1073/pnas.0510506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hom RK, et al. Design and synthesis of disubstituted thiophene and thiazole based inhibitors of JNK. Bioorg Med Chem Lett. 2010;20(24):7303–7307. doi: 10.1016/j.bmcl.2010.10.066. [DOI] [PubMed] [Google Scholar]
- 16.Ma W, et al. Phosphorylation of DCC by ERK2 is facilitated by direct docking of the receptor P1 domain to the kinase. Structure. 2010;18(11):1502–1511. doi: 10.1016/j.str.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Ho DT, Bardwell AJ, Grewal S, Iverson C, Bardwell L. Interacting JNK-docking sites in MKK7 promote binding and activation of JNK mitogen-activated protein kinases. J Biol Chem. 2006;281(19):13169–13179. doi: 10.1074/jbc.M601010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozenne V, et al. Mapping the potential energy landscape of intrinsically disordered proteins at amino acid resolution. J Am Chem Soc. 2012;134(36):15138–15148. doi: 10.1021/ja306905s. [DOI] [PubMed] [Google Scholar]
- 19.Fieber W, Kristjansdottir S, Poulsen FM. Short-range, long-range and transition state interactions in the denatured state of ACBP from residual dipolar couplings. J Mol Biol. 2004;339(5):1191–1199. doi: 10.1016/j.jmb.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 20.Mohana-Borges R, Goto NK, Kroon GJA, Dyson HJ, Wright PE. Structural characterization of unfolded states of apomyoglobin using residual dipolar couplings. J Mol Biol. 2004;340(5):1131–1142. doi: 10.1016/j.jmb.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Jensen MR, Blackledge M. On the origin of NMR dipolar waves in transient helical elements of partially folded proteins. J Am Chem Soc. 2008;130(34):11266–11267. doi: 10.1021/ja8039184. [DOI] [PubMed] [Google Scholar]
- 22.Jensen MR, et al. Quantitative conformational analysis of partially folded proteins from residual dipolar couplings: Application to the molecular recognition element of Sendai virus nucleoprotein. J Am Chem Soc. 2008;130(25):8055–8061. doi: 10.1021/ja801332d. [DOI] [PubMed] [Google Scholar]
- 23.Jensen MR, et al. Intrinsic disorder in measles virus nucleocapsids. Proc Natl Acad Sci USA. 2011;108(24):9839–9844. doi: 10.1073/pnas.1103270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nodet G, et al. Quantitative description of backbone conformational sampling of unfolded proteins at amino acid resolution from NMR residual dipolar couplings. J Am Chem Soc. 2009;131(49):17908–17918. doi: 10.1021/ja9069024. [DOI] [PubMed] [Google Scholar]
- 25.Jensen MR, Salmon L, Nodet G, Blackledge M. Defining conformational ensembles of intrinsically disordered and partially folded proteins directly from chemical shifts. J Am Chem Soc. 2010;132(4):1270–1272. doi: 10.1021/ja909973n. [DOI] [PubMed] [Google Scholar]
- 26.Salmon L, et al. NMR characterization of long-range order in intrinsically disordered proteins. J Am Chem Soc. 2010;132(24):8407–8418. doi: 10.1021/ja101645g. [DOI] [PubMed] [Google Scholar]
- 27.Serrano L, Fersht AR. Capping and alpha-helix stability. Nature. 1989;342(6247):296–299. doi: 10.1038/342296a0. [DOI] [PubMed] [Google Scholar]
- 28.Francis DM, et al. Structural basis of p38α regulation by hematopoietic tyrosine phosphatase. Nat Chem Biol. 2011;7(12):916–924. doi: 10.1038/nchembio.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen DF, Vallurupalli P, Kay LE. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem B. 2008;112(19):5898–5904. doi: 10.1021/jp074793o. [DOI] [PubMed] [Google Scholar]
- 30.Vallurupalli P, Bouvignies G, Kay LE. Studying “invisible” excited protein states in slow exchange with a major state conformation. J Am Chem Soc. 2012;134(19):8148–8161. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 31.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins. 1998;30(2):144–154. [PubMed] [Google Scholar]
- 32.Krishnan N, et al. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat Chem Biol. 2014;10(7):558–566. doi: 10.1038/nchembio.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holzman LB, Merritt SE, Fan G. Identification, molecular cloning, and characterization of dual leucine zipper bearing kinase. A novel serine/threonine protein kinase that defines a second subfamily of mixed lineage kinases. J Biol Chem. 1994;269(49):30808–30817. [PubMed] [Google Scholar]
- 34.Mooney LM, Whitmarsh AJ. Docking interactions in the c-Jun N-terminal kinase pathway. J Biol Chem. 2004;279(12):11843–11852. doi: 10.1074/jbc.M311841200. [DOI] [PubMed] [Google Scholar]
- 35.Glatz G, Gógl G, Alexa A, Reményi A. Structural mechanism for the specific assembly and activation of the extracellular signal regulated kinase 5 (ERK5) module. J Biol Chem. 2013;288(12):8596–8609. doi: 10.1074/jbc.M113.452235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura K, Johnson GL. Noncanonical function of MEKK2 and MEK5 PB1 domains for coordinated extracellular signal-regulated kinase 5 and c-Jun N-terminal kinase signaling. Mol Cell Biol. 2007;27(12):4566–4577. doi: 10.1128/MCB.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laughlin JD, et al. Structural mechanisms of allostery and autoinhibition in JNK family kinases. Structure. 2012;20(12):2174–2184. doi: 10.1016/j.str.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.