Significance
The present work extends redox-based change in enzyme activity to the TCA cycle of plant mitochondria. Thioredoxin (TRX) was found to regulate the activity of enzymes of the mitochondrial cycle (succinate dehydrogenase and fumarase) and of an enzyme associated with it (ATP-citrate lyase) by modulating thiol redox status. A combination of experiments based on mutant and carbon isotope labeling analyses provides evidence that flux through this pathway is coordinately modulated by TRX at the enzyme level of both mitochondria and cytosol. The results provide in vivo confirmation of earlier in vitro results and further show that mitochondria resemble plastids in using TRX and redox status to regulate the main carbon flux pathway of the organelle.
Keywords: Arabidopsis, redox regulation, thioredoxin TCA cycle regulation, citric acid cycle regulation, ATP-citrate lyase
Abstract
Plant mitochondria have a fully operational tricarboxylic acid (TCA) cycle that plays a central role in generating ATP and providing carbon skeletons for a range of biosynthetic processes in both heterotrophic and photosynthetic tissues. The cycle enzyme-encoding genes have been well characterized in terms of transcriptional and effector-mediated regulation and have also been subjected to reverse genetic analysis. However, despite this wealth of attention, a central question remains unanswered: “What regulates flux through this pathway in vivo?” Previous proteomic experiments with Arabidopsis discussed below have revealed that a number of mitochondrial enzymes, including members of the TCA cycle and affiliated pathways, harbor thioredoxin (TRX)-binding sites and are potentially redox-regulated. We have followed up on this possibility and found TRX to be a redox-sensitive mediator of TCA cycle flux. In this investigation, we first characterized, at the enzyme and metabolite levels, mutants of the mitochondrial TRX pathway in Arabidopsis: the NADP-TRX reductase a and b double mutant (ntra ntrb) and the mitochondrially located thioredoxin o1 (trxo1) mutant. These studies were followed by a comparative evaluation of the redistribution of isotopes when 13C-glucose, 13C-malate, or 13C-pyruvate was provided as a substrate to leaves of mutant or WT plants. In a complementary approach, we evaluated the in vitro activities of a range of TCA cycle and associated enzymes under varying redox states. The combined dataset suggests that TRX may deactivate both mitochondrial succinate dehydrogenase and fumarase and activate the cytosolic ATP-citrate lyase in vivo, acting as a direct regulator of carbon flow through the TCA cycle and providing a mechanism for the coordination of cellular function.
As in animals and aerobic microorganisms (1, 2), the tricarboxylic acid (TCA) cycle of plant mitochondria is composed of a set of eight enzymes that oxidize pyruvate and malate formed in the cytosol to CO2 and NADH (3). The CO2 is released and the NADH is oxidized by the electron transport chain for the generation of ATP. Recent years have witnessed major advances in our understanding of the cycle in plants, including its different modes of operation and properties of its constituent enzymes (4–6). We also now understand a great deal about the physiological role, kinetic features, and transcriptional and posttranslational regulation of enzymes participating in the cycle.
In addition to these studies, experiments have focused on functional interactions taking place between mitochondria and the other organelle that generates energy in plant cells, namely, the chloroplast (7, 8). The results suggest that the two compartments are tightly linked by regulatory mechanisms acting at the levels of the gene and interorganellar metabolite transport (9–13). Further, a long-standing body of evidence indicates that the cycle is regulated by a feedback regulator in animals, bacteria, and plants (14–16). However, despite decisive progress made in our understanding of the cycle (4–6), the nature of the regulator remains unknown. In the current work, we have addressed this question.
Recent advances have identified a candidate for this elusive role: thioredoxin (TRX), a regulatory protein widely distributed throughout the three domains of life. TRX harbors a dithiol active site and regulates target enzymes by thiol/disulfide exchange, and a TRX-dependent reduction system in plant mitochondria was identified as early as 2000 (17, 18). The first clue to its identity as the mitochondrial regulator emerged from proteomics studies in which members of the TCA cycle of plant mitochondria were found to bind TRX (13, 19). Supporting evidence came from pharmacologically based approaches that linked activation of anapleurotic fluxes of the TCA cycle to redox changes, compatible with a role for TRX (20). The reduced TRX needed for this type of regulation could be supplied by NADPH and the flavoenzyme NADP-TRX reductase (NTR); both proteins are present in mitochondria (18, 21). However, aside from the alternative oxidase (AOX) (19, 22) and citrate synthase (CS) (23, 24), TRX has not been functionally linked to the regulation of enzymes of the TCA cycle or other processes in plant mitochondria. Thus, although TRX has long been proposed to be involved in regulating CS, and thereby TCA cycle activity (25), a clear indication that such regulation is acting in vivo is lacking.
Here, we provide evidence that TRX acts to regulate the TCA cycle via a single mechanism controlling carbon flow through the entire pathway that has long remained mysterious. It now appears that mitochondria resemble chloroplasts as an organelle in which TRX plays a primary role in linking redox to the regulation of fundamental processes of the organelle.
Results
Extraplastidial TRX Function Is Important for Seed and Root Growth.
To investigate the potential role of TRX in regulating the mitochondrial TCA cycle, we deemed it important to determine the growth properties of the previously characterized NADP-TRX reductase a and b double mutant (ntra ntrb) double-KO plant that lacks two NTR isoforms localized in cytosol and mitochondria (26). In addition, we isolated a T-DNA insertion mutant in the thioredoxin o1 (trxo1) gene (At2g35010) encoding a mitochondrial TRX from the Salk collection (SALK 042792) (27) (Fig. S1A). The insertion in the TRXo1 gene was mapped to the first intron and resulted in KO of gene expression (Fig. S1 B and C). In an early experiment, we followed germination and seedling establishment under standard growth conditions in which 7- to 10-d-old plants were transferred to long-day conditions (16 h light/8 h dark). Shoot material was harvested from 2- to 8-wk-old plants, and the fresh weight (FW) was determined at harvest. Relative to WT, a significant reduction in FW was observed for both the ntra ntrb double-KO and trxo1 plants up to the fourth week (Table S1). After this time, no further difference in shoot FW was observed in the ntra ntrb mutant, whereas the shoot FW was significantly higher in trxo1 after 6 wk.
To gain a more complete perspective of the mutants, we characterized seeds with respect to germination efficiency and seedling establishment. To this end, radicle emergence was scored daily for a week in replicate experiments carried out in the presence and absence of 20 mM sucrose (Fig. S2 A and B). In the presence of sucrose, there was a clear deceleration in the rate of germination in both mutants, whereas in its absence, only the ntra ntrb double-KO plants displayed a reduced rate of germination. This point notwithstanding, the vast majority of mutant seedlings overcame these seemingly deleterious phenotypes in both cases and were able to establish photosynthetically active seedlings. In related experiments with roots, growth was reproducibly reduced by up to 30% in the ntra ntrb double-KO plants compared with WT plants with or without sucrose as found earlier (26). By contrast, trxo1 displayed similar root growth to WT under both conditions (Fig. S2 C and D).
Mutations of ntra ntrb and trxo1 alter extractable activities of the TCA cycle and affiliated enzymes.
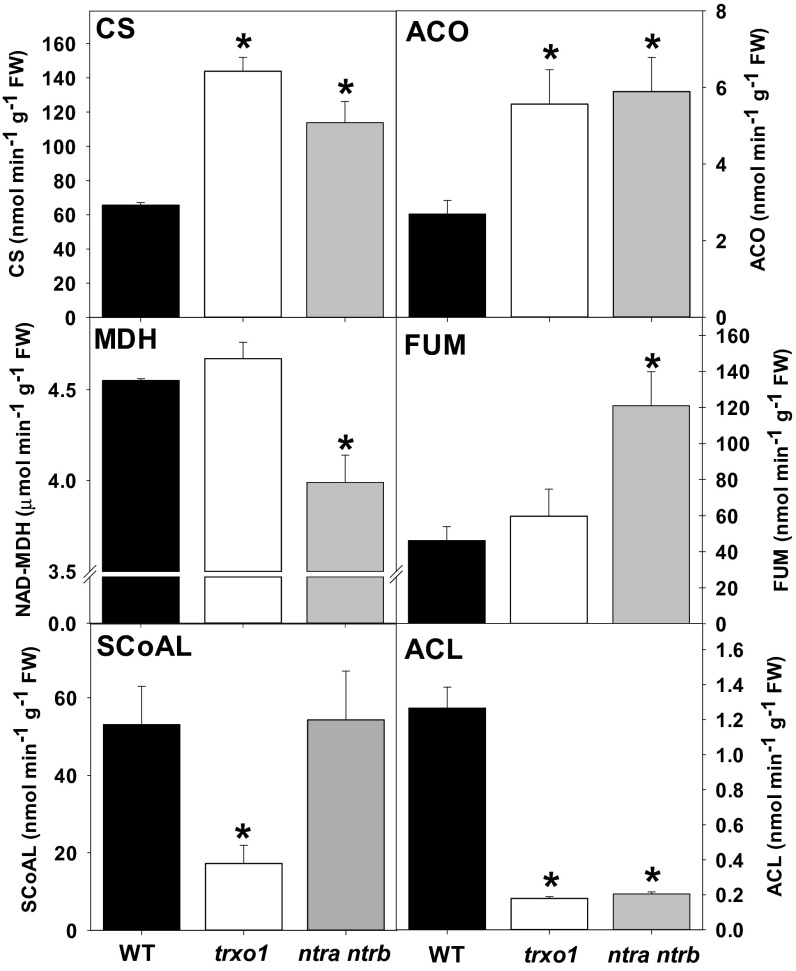
Reduced seed germination rates and root growth are features commonly described for a deficiency in TCA cycle activity (28, 29). The observed morphological phenotypes, however, are not, on their own, convincing evidence of reduced activity of this pathway, particularly in light of the very mild phenotypes displayed by the lack of mitochondrial trxo1. For this reason, we directly assessed the total maximal catalytic activity of a range of metabolic enzymes in desalted leaf extracts. The chloroplast enzymes ribulose-1,5-biphosphate carboxylase oxygenase (Rubisco), NADP-dependent malate dehydrogenase (MDH), ADP-Glc pyrophosphorylase (AGPase), and NADP-GAPDH showed only minor activity differences in mutants compared with WT plants (Table S2), thereby confirming that mutation effects are independent of chloroplasts and consistent with their being confined to the cytosol/mitochondria (ntra ntrb) and to the mitochondria (trxo1). By contrast, enzymes of the TCA cycle, or associated with citrate metabolism, were significantly altered, with a clear increase in fumarase (FUM) activity in the ntra ntrb double mutant and a dramatic decrease (up to 80%) in ATP-citrate lyase (ACL) activity in both mutants (Fig. 1). These changes were coupled with an increase in the activity of CS and aconitase (ACO) in both mutants, whereas succinyl-CoA ligase (SCoAL) and mitochondrial/cytosolic NAD+-dependent MDH decreased in trxo1 and in ntra ntrb, respectively (Fig. 1).
Fig. 1.
Enzyme activities in whole-leaf extract of WT, trxo1 mutant, and ntra ntrb double mutant. Activities (nmol⋅min−1⋅g−1 of FW) were determined in leaf material harvested at the end of the day from 4-wk-old plants before bolting. Data presented are mean ± SEM (n = 6). Asterisks indicate values significantly different from WT by the Student’s t test (*P < 0.05).
In addition to the striking effect that the perturbation of the mitochondrial TRX system has on enzymes of or associated with the TCA cycle, these results suggest that TRX promotes differential redox regulation of these enzymes. Given that the catalytic activity of TRX is dependent on redox-active Cys residues in the target proteins, we next aligned the sequences of all TCA cycle enzymes and ACL from Arabidopsis thaliana with their homologs from plants, animals, and microorganisms to determine whether these enzymes have conserved Cys residues. We found two conserved Cys residues in the sequences of CS, isocitrate dehydrogenase (IDH), FUM, MDH, and succinate dehydrogenase (SDH); three conserved Cys residues in the sequences of SCoAL; and five conserved Cys residues in the sequences of ACO (Dataset S1 A–G). Three of the conserved Cys residues shown in yellow in ACO are essential for binding the FeS cluster (Dataset S1B). Arabidopsis isoforms ACL-A and ACL-B have one and two conserved Cys residues across all species included in the alignment, respectively. ACL-B presented one Cys residue that is conserved in all species except the gorilla. Additionally, ACL-A and ACL-B have five and six conserved Cys residues, respectively, but this conservation does not extend beyond the plant kingdom (Dataset S1 H–I). The enzyme activity data, together with the presence of conserved Cys residues in almost all of the TCA cycle enzymes, are consistent with their being potential TRX targets. However, the presence of conserved Cys residues could also be related to other posttranslational modifications, such as glutathionylation and nitrosylation (30). Furthermore, these data alone are not sufficient to determine whether the observed effects are mediated at the transcriptional, translational, or posttranslational level.
Metabolite profiling.
Having established that the activity of enzymes associated with mitochondrial energy metabolism was affected in at least one of the two mutants under study, we next determined whether the mutants showed alterations in their metabolite profiles. For this purpose, we measured the levels of starch, sugars, and nitrate at the end of the day in illuminated leaves of mutants and WT, as well as a broad range of primary and secondary metabolites, by established methods (31, 32). Primary metabolites showing pronounced changes are listed in Table 1, whereas secondary metabolites and primary metabolites showing less pronounced changes are listed in Table S3. We observed a significant decrease in starch in the ntra ntrb mutant coupled with a doubling of the glucose and fructose content (Table 1). Of these changes, only the increase in fructose was seen in the trxo1 mutant, which otherwise resembled the WT. Similarly, only the ntra ntrb mutant displayed significant decreases in nitrate levels (Table 1). GC-MS data also revealed increases in Asp, Asn, Gln, Glu, and Phe, as well as considerable increases in malate and citrate in both the trxo1 and ntra ntrb mutants. In addition, the levels of Ala, putrescine, and succinate were exclusively increased in the ntra ntrb mutant, whereas pyruvate was increased in the trxo1 mutant only. Pro and Ser were exclusively decreased in the trxo1 mutant, and the levels of Gly and Thr were lower in the trxo1 mutant but higher in the ntra ntrb than in WT (Table 1). No other changes were observed in amino acids or minor sugars, sugar alcohols, or polyamines measured by GC-MS (Table S3), with the exception of trehalose, which was enhanced in the ntra ntrb mutant but not in the trxo1 mutant (Table S3). We next evaluated the levels of the major flavonols and glucosinolates by liquid chromatography-MS. These analyses revealed large changes in flavonoids (kaempferol and anthocyanin), phenylpropanoids, and the majority of glucosinolates and indole glucosinolates (Table S3), thus confirming and extending previous observations (33). Altogether, these results are consistent with a reprogramming of the main pathways of primary and secondary metabolism to maintain a balanced metabolism. Although the mutation effects are striking, the mechanism responsible for the changes cannot be identified with the data. We therefore conducted a series of isotope redistribution experiments to complement the other results and help with the interpretation.
Table 1.
Content of metabolites showing significant differences between WT, trxo1 mutant, and ntra ntrb double mutant
| Metabolite | WT | trxo1 | ntra ntrb |
| Enzymatic | |||
| Starch | 21.2 ± 3.0 | 25.9 ± 3.8 | 12.1 ± 2.7 |
| Fructose | 8.3 ± 0.9 | 19.9 ± 1.5 | 22.4 ± 2.3 |
| Glc | 3.5 ± 1.1 | 3.4 ± 0.4 | 7.7 ± 0.6 |
| Nitrate | 31.2 ± 2.9 | 28.5 ± 0.8 | 23.0 ± 1.3 |
| GC-MS | |||
| Asp | 1.00 ± 0.1 | 1.44 ± 0.1 | 2.03 ± 0.3 |
| Asn | 1.00 ± 0.2 | 2.24 ± 0.4 | 1.95 ± 0.2 |
| Gly | 1.00 ± 0.1 | 0.64 ± 0.1 | 1.49 ± 0.1 |
| Thr | 1.00 ± 0.1 | 0.78 ± 0.0 | 1.28 ± 0.1 |
| Pro | 1.00 ± 0.2 | 0.48 ± 0.1 | 1.17 ± 0.1 |
| Ser | 1.00 ± 0.1 | 0.76 ± 0.0 | 1.03 ± 0.0 |
| Gln | 1.00 ± 0.2 | 2.28 ± 0.6 | 3.25 ± 0.3 |
| Glu | 1.00 ± 0.1 | 1.59 ± 0.1 | 1.76 ± 0.7 |
| Ala | 1.00 ± 0.1 | 0.95 ± 0.0 | 1.40 ± 0.1 |
| Pyruvate | 1.00 ± 0.1 | 1.40 ± 0.1 | 1.40 ± 0.2 |
| Malate | 1.00 ± 0.2 | 1.65 ± 0.2 | 1.88 ± 0.3 |
| Citrate | 1.00 ± 0.1 | 2.55 ± 0.6 | 1.72 ± 0.2 |
| Succinate | 1.00 ± 0.0 | 0.95 ± 0.1 | 1.29 ± 0.0 |
| Glc | 1.00 ± 0.2 | 0.84 ± 0.1 | 1.91 ± 0.2 |
| Phe | 1.00 ± 0.1 | 1.20 ± 0.0 | 1.44 ± 0.1 |
| Putrescine | 1.00 ± 0.1 | 1.16 ± 0.1 | 1.57 ± 0.2 |
Metabolites were determined at end of the day as described in Materials and Methods. Starch, fructose, Glc, and nitrate were measured through the enzymatic method and are presented as millimoles per kilogram of FW. Other metabolites were measured through GC-MS. The GC-MS data were normalized with the values obtained for ribitol (internal standard) and the FW used for the extraction (∼100 mg). The data presented are normalized with respect to the mean response calculated for WT plants. Data presented are mean ± SEM (n = 6). Values in bold and underlined type indicate values significantly different from WT by the Student’s t test at 5% (P < 0.05).
Isotope redistribution through the TCA cycle and associated pathways.
We next sought to obtain more direct evidence that these pathways were altered in vivo. Despite the fact that a protocol has recently been established for estimating metabolic fluxes in intact plants following the supplying of 13CO2 to illuminated leaves (34), this approach labels mitochondrial pools very slowly, and therefore is not currently suitable for the system under study. For this reason, we adopted an approach frequently used in other studies (35, 36); that is, we performed independent feedings of 13C-Glc, 13C-malate, and 13C-pyruvate for a period of 4 h. This procedure was used for the evaluation of unidirectional rates of carbon exchange. This approach is important because changes in metabolic fluxes can occur without changes in the metabolite pool sizes of pathway intermediates, rendering it facile to underestimate or even possible to miss important mechanisms of metabolic regulation on the basis of steady-state metabolite levels alone. Such approaches also allow quantitative evaluation of a broad range of metabolic pathways without the need for laborious (and potentially inaccurate) chemical fractionation procedures commonly used in the estimation of fluxes following incubation with radiolabeled substrates.
For these experiments, the leaves were snap-frozen in liquid nitrogen and the metabolites were extracted, derivatized, and evaluated by a specially modified GC-MS protocol (37). Following [U-13C]-Glc feeding, label enrichment in heavy isotope peaks (i.e., enhanced heavy isotope labeling with respect to the control treatment of the same genotype) was observed in a total of 13 metabolites (listed in Table S4), including increased accumulation in malate and decreased accumulation in Pro in both the ntra ntrb mutant and the trxo1 mutant, reflecting an enhanced flow of 13C to malate via respiratory metabolism and a decreased flow to Pro, respectively (Fig. 2A). The label accumulation in Asp (Fig. 2A) and raffinose (Table S4) following 13C-Glc feeding was dramatically decreased, whereas the label accumulation in Ala and succinate was increased in the ntra ntrb mutant but not in the trxo1 mutant (Fig. 2A and Table S4). Following 13C-malate feeding, label enrichment was seen in only eight metabolites (Table S4); however, the trends in accumulation in Ala, Asp, and succinate were conserved (Fig. 2B). Similarly, following 13C-pyruvate feeding, very similar differences were observed in the labeling of Ala and Asp compared to the results obtained following 13C-malate or 13C-Glc feeding (Fig. 2C). In addition, in this experiment, significantly increased labeling was observed in Val (only in the ntra ntrb mutant) and decreased in Ser (in both mutants) (Fig. 2C). Interestingly, a 13C-enrichment in malate under 13C-Glc and a 13C-decrease in Pro (under 13C-Glc) and Ser (under 13C-pyruvate) were conserved in both mutants (Fig. 2 A and C), with all results from these experiments, furthermore, in close agreement with the observed changes in the steady-state levels (Table 1). Most saliently, there was an accumulation of label in succinate following feeding of either 13C-Glc or 13C-malate, as well as an increase in label redistributed to Ala and a decrease in label redistributed to Asp in the ntra ntrb double mutant observed following incubation with any of the three substrates supplied.
Fig. 2.
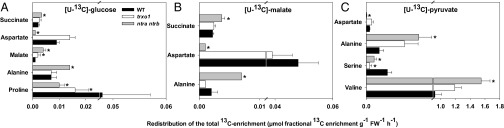
Redistribution of the 13C label enrichment in selected metabolites following incubation of leaves from WT, trxo1 mutant, and ntra ntrb double-mutant plants. Fully expanded leaves of 6-wk-old plants were harvested at the middle of the light period and fed via the petiole with [U-13C]-Glc (A), [U-13C]-malate (B), or [U-13C]-pyruvate (C) solution. Data presented are mean ± SEM (n = 6). Asterisks indicate values significantly different from WT by the Student’s t test at 5% (*P < 0.05).
In vitro confirmation and complementation of the effects of TRX on TCA cycle and associated enzymes.
To confirm that altered TCA cycle and associated enzyme activities in the trxo1 and ntra ntrb mutants were due to inactivation of the TRX system, we performed protein-based complementation studies with cell-free extracts. To this end, we conducted enzyme assays for all enzymes of the TCA cycle and the citrate-metabolizing enzyme ACL in the presence of recombinant NTRB alone or in combination with recombinant TRXo1 or TRXh2, a mitochondrial/cytosolic TRX (38). Given that NTRB alone was not able to modify the activities of the enzymes analyzed here, the results are shown only as the effect of the addition of TRXo1 and TRXh2.
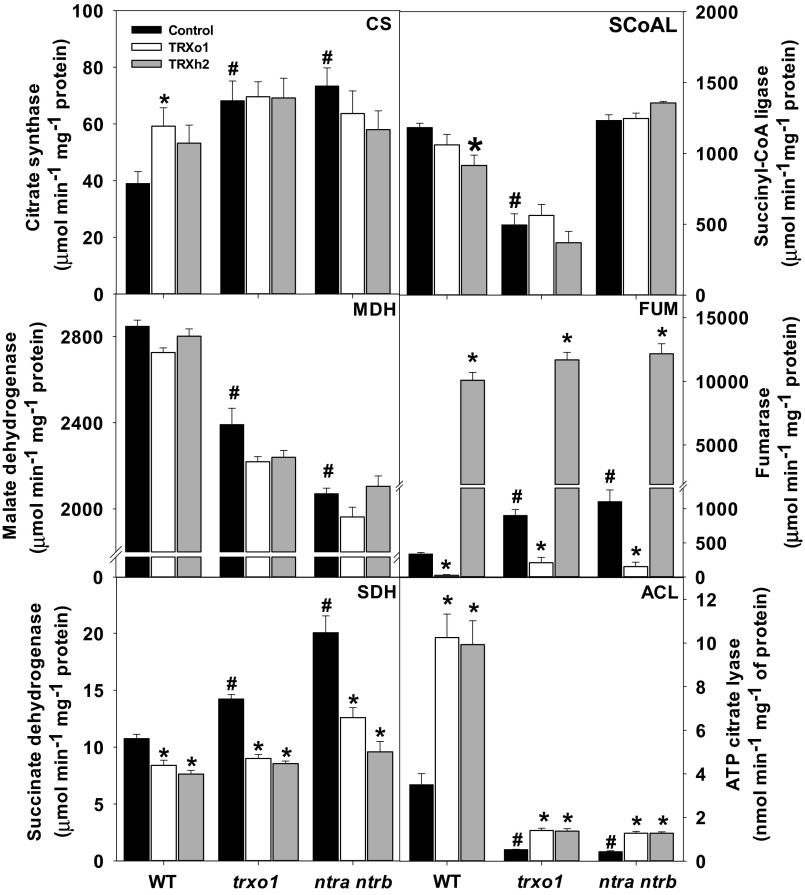
The complementation experiments were carried out using preparations from either isolated mitochondria (for the TCA cycle enzymes; details are provided in SI Materials and Methods) or whole leaves (for the cytosolic enzyme ACL). As seen in Fig. 3, the complementation experiments generally confirmed the previous results based on enzyme assays obtained with leaf extracts shown in Fig. 1. These assays enabled us to determine whether the differences observed in the activity of TCA cycle enzymes in leaf extracts (Fig. 1) are related to the mitochondrial isoforms. As observed in Fig. 1, the activity of FUM and MDH was much higher and lower, respectively, in mitochondrial preparations of the ntra ntrb double mutant (Fig. 3). Mitochondrial FUM was also higher in the trxo1 mutant. Although MDH activity was not significantly affected by the addition of the recombinant TRXs, FUM showed a striking response to each protein, especially to TRXh2. These factors combined to result in an unusual higher activity of mitochondrial FUM than mitochondrial MDH. However, importantly, these activities are very similar in the absence of the recombinant TRXs. Activity of the enzyme was strongly reduced in all genotypes following the addition of TRXo1 recombinant protein, but, surprisingly, its activity was greatly increased following the addition of TRXh2 (Fig. 3). To our knowledge, this result provides the first example of an enzyme that is regulated in opposing directions by two types of TRX: in this case, activation by TRXh2 and inhibition by TRXo1. This finding is potentially significant from the standpoint of control of carbon flux within the mitochondrion because there may be separate pools of TRXo1 and TRXh2. Further, in view of its dual localization (cytosol and mitochondria), TRXh2 could also function as a regulatory link between the mitochondrion and the cytosol by a yet to be defined interorganellar network. These possibilities will be the subject of future investigations.
Fig. 3.
Enzyme activities and protein-based complementation in WT, trxo1, and ntra ntrb mutants. CS, SCoAL, MDH, FUM, and SDH activities were measured in mitochondrial extracts, whereas ACL was measured in leaf extracts of WT, trxo1, and ntra ntrb mutants. These extracts were also used to perform protein-based complementation assays. The extracts were untreated (Control) or treated with TRXo1 (3 μg; 180 nM) or TRXh2 (3 μg; 210 nM) both reduced by NTRB (7.5 μg; 100 nM) and NADPH (100 μM). Data presented are mean ± SEM (n = 5). A number symbol indicates values significantly different from WT by the Student’s t test at 5% (#P < 0.05), and an asterisk indicates values significantly different from the control in the same genotype by the Student’s t test at 5% (*P < 0.05).
Similar to FUM, the activity of SDH increased in both trxo1 and ntra ntrb mutants, and the addition of TRXo1 recombinant protein decreased SDH activity in all genotypes (Fig. 3). However, in contrast to FUM, the activity of SDH was reduced in all genotypes following the addition of TRXh2 recombinant protein (Fig. 3). Additionally, very strong changes were observed in the activity of ACL. As noted above, both mutants were characterized as containing lower activities of this enzyme (Fig. 1). Furthermore, addition of the TRXo1 and TRXh2 recombinant proteins doubled or even tripled the activity of the enzyme in all genotypes (Fig. 3). The situation for CS is less clear. Despite strong increases in activity in both mutants (Fig. 1), we observed no change in protein abundance (Fig. S3) and no effect on activity following the addition of recombinant proteins to preparations of these genotypes. However, the addition of TRXo1, but not TRXh2, significantly elevated the activity in mitochondrial extracts of WT (Fig. 3). Despite the differences shown in the activities of FUM and SDH, few changes were observed in the other enzymes of the TCA cycle following the addition of recombinant proteins (Fig. 3 and Fig. S4). The activities of SCoAL and NAD-dependent IDH were lower in mitochondrial extracts of the trxo1 mutant and ntra ntrb double mutant, respectively, whereas ACO activity was higher in mitochondrial preparations of both the trxo1 mutant and ntra ntrb double mutant (Fig. 3 and Fig. S4). No differences were observed in the activity of 2-oxoglutarate dehydrogenase in the mutants and following the addition of recombinant proteins (Fig. S4).
To recapitulate, the perturbation of the mitochondrial TRX system modifies the activity of enzymes of, or associated with, the TCA cycle and also leads to higher accumulation of TCA cycle (citrate and malate) and associated (pyruvate, Gln, and Glu) metabolites. Furthermore, protein-based confirmation of the measurements made of metabolite pool size and enzyme activities in extracts from mutant plants allowed us to conclude that at least two enzymes of the TCA cycle (FUM and SDH) are deactivated by the mitochondrial TRX system in vivo (Fig. 4). Although the addition of TRXh2 suggested that FUM can be activated in vitro, the absence of data in mutants lacking TRXh2 activity did not allow us to form conclusions regarding this phenomenon. Another enzyme, CS, appeared to be activated in vitro (Fig. 3) as previously shown by Schmidtmann et al. (24), but the increase in the activity in leaf extracts (Fig. 1) and mitochondrial extracts (Fig. 3) in both mutants suggests that this activation may not occur in vivo. A fourth enzyme, ACL of the cytosol, by contrast, appears to be activated by TRX both in vitro and in vivo (Fig. 3). Lastly, the observation that the reduction of SCoAL activity seen in trxo1 was not observed in the ntra ntrb double mutant may suggest that it is due to pleiotropic effects not related to redox regulation. Given that SCoAL has an exclusive mitochondrial location, we cannot currently exclude the possibility that the lack of an effect in ntra ntrb may be due to the complexities of compartmentation, because it is conceivable that modification of the cytosolic redox poise somehow counteracts redox changes in the mitochondria. However, further experimentation will be required to determine whether this interpretation is valid. Collectively, the results provide clear evidence for a posttranslational role of TRX in the modulation of FUM, SDH, and ACL in vivo (Fig. 4).
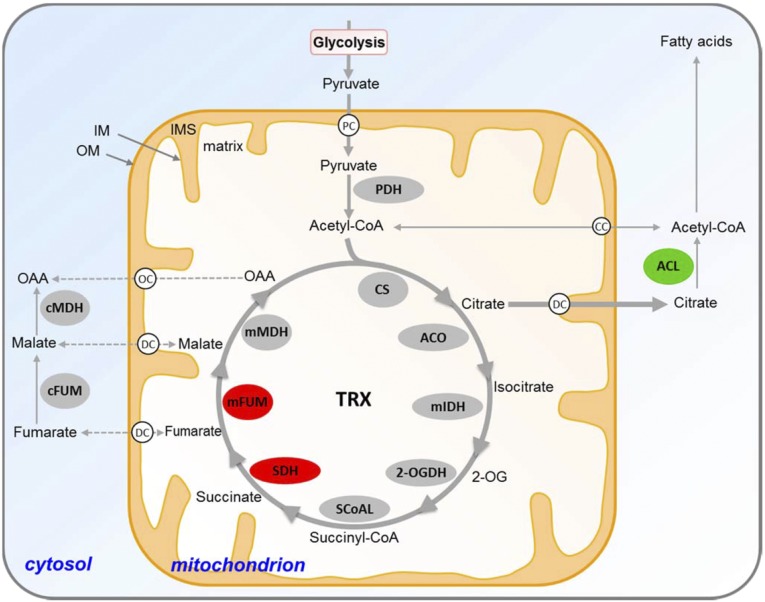
Fig. 4.
Schematic model of TRX regulation of the TCA cycle and associated with citrate metabolism enzymes. Enzymes presented in red and green are possible TRX targets to be deactivated and activated in vivo, respectively. SDH and mFUM are deactivated, whereas ACL is activated by TRX in vivo. We considered an enzyme to be the target of the TRX system in vivo only when the enzyme activity data in the mutants fit with the complementation assay using recombinant TRXs. Therefore, although our complementation data indicate that CS appears to be activated in vitro by TRXo1, the increase in the leaf enzyme activities in both mutants suggests that this activation may not occur in vivo. White spheres represent carriers or antiporters. CC, mitochondrial acetyl-CoA carrier; DC, dicarboxylate carrier; IM, inner mitochondrial membrane; IMS, intermembrane space; mFUM, mitochondrial fumarase; OAA, oxaloacetate; OC, putative oxaloacetate carrier; 2-OG, 2-oxoglutarate; 2-OGDH, 2-oxoglutarate dehydrogenase complex; OM, outer mitochondrial membrane; PC, pyruvate carrier; PDH, pyruvate dehydrogenase complex.
Discussion
Proteomic approaches have recently increased the number of putative TRX targets in photosynthetic organisms, broadening the role of the TRX system to encompass not just the classical photosynthetic targets of chloroplasts but also such diverse processes as starch biosynthesis in amyloplasts (39), chloroplast protein import (40), germination facilitation (41), and intercellular communication (38). In addition, several studies have begun to unravel the role of the TRX system with respect to both cytosolic and mitochondrial metabolism (13, 19, 26, 42). By use of affinity chromatography, over 100 potential mitochondrial targets for TRX have been identified in spinach leaf, Arabidopsis shoots, and potato tubers (13, 19). Moreover, 18 redox-active proteins have been identified in mitochondria isolated from Arabidopsis cell suspension cultures via 2D oxidant/reductant diagonal SDS/PAGE (43). Many of these proteins were previously reported to be TRX or GRX targets (13, 19, 44). The physiological significance of these changes in the proteins identified was, however, not determined, leaving the mechanism of mitochondrial redox regulation largely unexplored. Thus, despite the fact that it has long been established that light modifies both the NAD(P)H/NAD(P)+ ratio and the ubiquinone reduction level (9, 45), the significance of redox and the elusive mechanism for feedback inhibition of the TCA cycle (16, 46, 47) are attractive research topics in plant mitochondria. Previous studies have identified all eight core enzymes of the TCA cycle, as well as pyruvate dehydrogenase and NAD-malic enzyme, as potential targets of the TRX system in land plants (13, 19). The enzymes isocitrate lyase and acetyl-CoA synthetase, which have a possible role in acetate metabolism, appeared to be targets in Chlamydomonas reinhardtii (42). Moreover, the AOX, uncoupling protein (UCP), and several peptides associated with the mitochondrial electron transport chain, alongside Gly decarboxylase, Ser hydroxymethyltransferase, several amino transferases, and enzymes of branched chain amino acid metabolism, were also identified as putative targets (13, 19, 22). Notably, in nonplant systems, other enzymes associated with the mitochondrial electron transport chain (e.g., complex II, UCP) have been identified as being redox-regulated (48, 49). Here, we adopted a combination of metabolic and isotope tracer profiling approaches alongside mutant analysis and protein-based complementation assays to validate TRX targets of, and closely associated with, the TCA cycle in Arabidopsis.
Data currently presented on the metabolic characterization of the ntra ntrb and trxo1 mutants corroborate a role for the TRX system in regulating not only the TCA cycle but also photorespiration, amino acid, glucosinolate, and flavonol metabolism. Perhaps predictably, the ntra ntrb double mutant showed far greater change than trxo1, a feature ascribed to a certain degree of redundancy within the mitochondrially localized TRXs. It is important to note that overlapping functions have previously been identified between TRXs and glutaredoxins, suggesting that mitochondrial and cytosolic redox pools are highly and tightly regulated (50). The changes reported for organic acid metabolism in the mutants can be clearly related to the expected effects a deficiency in TRX would have on flux through the TCA cycle if it were inhibited by TRX in the clockwise direction between malate and succinate (which indeed appears to be the case) (Fig. 4). Similarly, changes in the levels or redistribution of isotope to Ala, Asp, and Ser are likely a result of a lack of regulation of TRX-linked Ala or Asp aminotransferases and the mitochondrial enzymes of the photorespiratory pathway, respectively. The close linkage between Ala and photorespiration with the TCA cycle activity has precedence in the literature (10, 51–54). Moreover, the change in the amount of Val is likely the result of a deregulation of the branched chain amino acid dissimilation pathway, which has recently been demonstrated to augment mitochondrial respiration in times of stress (55). Although we did not validate all of these changes at the enzyme level, we did so for members of the TCA cycle.
Enzymes of the TCA cycle are known to be highly susceptible to oxidative stress (52, 56), and CS was previously demonstrated to form intra- and intermolecular disulfide bridges under oxidizing conditions (43). Moreover, the activity of the cytosolic enzyme ACL, a central enzyme of the citrate shunt important in fatty acid and wax metabolism (57), was dramatically reduced in both mutants. These data were confirmed in experiments in which extracts from WT or mutants were incubated in the presence or absence of TRXo1 and TRXh2. Interestingly, four enzymes (CS, SDH, FUM, and ACL) could be confirmed as being regulated by TRX in vitro. Unexpectedly, and potentially significantly, the activity of cytosolic ACL was altered in the mutant deficient in a TRX specific to mitochondria, trxo1. Our results are not sufficiently comprehensive to enable us to provide a precise mechanism by which the observed changes generated in the trxo1 mutant could modulate the activity of a cytosolic enzyme. However, they are in keeping with data from a recent study suggesting posttranslational regulation of ACL (58).
Several metabolites (e.g., Gly, Thr, Ser, pyruvate) showed different accumulation in the trxo1 mutant. Additionally, the lack of convincing complementation of the increased activity of CS in the mutant preparations following the addition of recombinant proteins suggests, for this enzyme at least, that the change in activity may be rather an indirect effect. The explanation for this unclear situation would appear to reside in the complex effect of TRX on the dimerization of mitochondrial CS. Dimerization of this isoform is required for activation. A recent in vitro study suggests that TRX cleaves a disulfide bond between two subunits, enhancing activity by facilitating the regeneration of the active dimer from inactive dimers (24) similar to the effect observed here. The higher CS activity in the mutants is, however, likely due to the inability to cleave active dimer to form inactive monomers, thus suggesting that TRX is a direct negative regulator of mitochondrial CS in vivo. Nonetheless, the exact mechanism underlying this observation remains to be clarified. The lack of a direct effect of TRX on CS in the mutants in the current study is supported by previous work indicating that although DTT clearly affected activity in some plant species, it activated rather than repressed these activities (23). Intriguingly, however, DTT did not appear to have a significant effect on Arabidopsis CS, although it did in pumelo extracts. Although the reasons for the difference were not found in the previous study, it is tempting to suggest that this observation may be due to the presence of two CSs in Arabidopsis, with the possibility that the isoforms display differential properties (24).
This point notwithstanding, the presence of redox shuttles between the mitochondria and cytosol is sufficiently well established (8, 20, 46, 59) to enable one to visualize that a transported metabolite could transmit altered redox status of the mitochondria to the cytosol. Possibly, the increased concentration of citrate in both trxo1 and ntra ntrb mutants (Table 1) reflects a defect in the redox transport system. This possibility is consistent with the notion that changes in the NAD(P) reduction states, which are carried out by NAD(P)H dehydrogenases present in plant mitochondria (60) and are undetectable in whole tissues, may be sufficient in vivo to allow significant changes in redox potential, and hence signal amplification (61). It seems that the NTR/TRX system may use the NAD(P)H redox state present in the mitochondrial matrix to regulate the flux through the TCA cycle as previously suggested (62). However, how the NAD(P)H redox state of mitochondria and cytosol is regulated in vivo and how the redox state is transmitted between mitochondria and cytosol remain unclear.
That the TCA cycle is regulated in such a manner is highly attractive in light of data from other studies. First, although it has been subject to relatively little study, ACL has been shown to be of critical importance in Arabidopsis and was identified to be regulated by TRX as a member of the reverse TCA cycle in Chlorobium tepidum (63). Thus, ensuring that this enzyme is highly active at a time when ATP production is maximal is an effective strategy. Second, redox-based regulation of the cycle at reactions involved in citrate metabolism would allow a reduced flux through the mitochondrial TCA cycle without compromising reactions required to maintain a nitrate assimilation rate capable of ensuring optimal photosynthesis (64, 65). This strategy is consistent with recent experimental observations on the cycle (47, 66) and also with evidence that the AOX is regulated by redox [i.e., the enzyme is reduced by TRX, thereby allowing activation by pyruvate (22)]. Third, the reactions catalyzed by SDH and FUM include two of the four enzymes in which the majority of the control of flux through this pathway was revealed to reside following metabolic control analysis (16).
Finally, although yet to be validated, numerous other putative redox-regulated enzymes are involved in alternative respiratory pathways. This situation raises two possibilities: Either they are coordinately regulated with the TCA cycle to optimize cellular efficiency or, as has already been seen under conditions of stress (67), they are regulated in an opposite manner to sustain mitochondrial function under conditions in which classical respiration is inhibited. For either possibility, it will be of great interest to analyze the role of TRX in regulating these alternative pathways under both optimal and suboptimal growth conditions, as well as to define the role of TRX fully in the interorganellar coordination of energy metabolism. Similarly, a more detailed analysis of the role of TRX in orchestrating changes in secondary plant metabolism, a finding provided further support by an as yet undefined role of flavonols in redox regulation (68), will likely be highly informative.
Concluding Remarks
Our results have identified the NADP-linked TRX system as a regulator of the TCA cycle in mitochondria as well as root and plant growth. The cycle also links the decrease in biomass due to TRX inhibition to a metabolic compensating mechanism that adjusts primary metabolism to cope with prevailing growth conditions. Further, our metabolite analyses have uncovered several pathways directly or indirectly dependent on the substrates or products generated by enzymes under the control of the TRX system, including enzymes of, or associated with, the TCA cycle.
The results of this study highlight the importance of the TRX system in mediating metabolic control of the TCA cycle. Evidence for this conclusion is several-fold. First, mutants of ntra ntrb and trxo1 both displayed enhanced levels of the cycle intermediates citrate and malate, and ntra ntrb additionally displayed enhanced levels of succinate. Second, both trxo1 and ntra ntrb mutants displayed elevated maximal catalytic activity of the mitochondrial isoforms of ACO, CS, SDH, and FUM, and markedly decreased activity of cytosolic ACL. Third, the redistribution of isotope in flux experiments, particularly with the ntra ntrb mutant, strongly suggested increased flux through the TCA cycle. Finally, assays of enzymes with either WT or mutant extracts revealed that changes in the levels of measured activities in the mutants for FUM, SDH, and ACL, at least, could be convincingly reverted, or altered in the case of WT plants, in a manner fully consistent with a regulatory role of the TRX system. Taken together, these data identify TRX as a master regulator of both the mitochondrial TCA cycle itself and the associated citrate shunt pathway by which intermediates of the cycle are exported to the cytosol to support biosynthesis. The results also implicate Trx in the regulation of mitochondrial electron transport indirectly through FUM, which is linked to the build-up of electrons from NADH, and directly via SDH and the attendant entry of electrons into the electron transport chain at complex II.
A remaining question concerns the mechanism by which the redox status of mitochondria is conveyed to the cytosol. This question assumes a paramount position in efforts to achieve a more complete understanding of the mitochondrial TRX system and its function within and, indirectly, beyond the organelle. One final point is that although the present results clearly demonstrate the importance of TRX in controlling the TCA cycle in Arabidopsis, the cross-kingdom conservation of Cys residues suggests that redox may be a broadly operative mechanism for regulating the cycle in other plants and in other types of organisms as well.
Materials and Methods
Plant Material.
All A. thaliana L. plants used in this study were of the Columbia (Col-0) ecotype. The ntra ntrb double-KO mutant was previously described (26), whereas the T-DNA insertion mutant in the trxo1 gene (At2g35010) from the Salk collection (SALK 042792) (27) was characterized in this study. Arabidopsis seeds were handled as described previously (26). Fully expanded rosette leaves of 4-wk-old plants were harvested for subsequent analysis.
Isolation of Mitochondria.
Mitochondria were isolated from the aerial part of 7-wk-old Arabidopsis plants grown under short-day photoperiod conditions as described previously (69). A brief description of the isolation protocol is given in SI Materials and Methods. The purity of the mitochondria was assessed by determining the activity of marker enzymes as well as by measuring the chlorophyll content (Table S5).
Enzyme Assays.
Enzymes assays were performed as described in SI Materials and Methods.
In Vitro Protein-Based Complementation and TRX Activity Assays.
For in vitro protein-based complementation assays, 3 μg of TRXo1 (180 nM) and 3 μg of TRXh2 (210 nM) were first incubated with or without 7.5 μg (100 nM) of NTRB and 100 μM NADPH at room temperature for 10 min. The mixture was incubated with leaf or mitochondrial protein extracts, and enzyme activity was measured as described above. To confirm that NTRB reduces TRXo1 and TRXh2 in vitro, a TRX activity assay was performed in the presence of 5,5′-Dithiobis(2-nitrobenzoic acid) (DTNB). For this purpose, 1 μg (60–70 nM) of TRX was reduced by 5 μg (67 nM) of NTRB and 100 μM NADPH. TRX activity was determined by following DTNB reduction at 412 nm (Fig. S5).
Metabolite Assays.
Metabolite assays were performed as described in SI Materials and Methods. The metabolite reporting guidelines and the overview of the metabolite reporting list are shown in Datasets S2 and S3, respectively.
Analysis of [U-13C]-Glc–, [U-13C]-Malate–, and [U-13C]-Pyruvate–Labeled Samples.
Arabidopsis leaves of similar size but from different genotypes were fed via the petiole with 15 mM [U-13C]-Glc, [U-13C]-malate, or [U-13C]-pyruvate (from Cambridge Isotope Laboratories) by incubation in buffered solution [10 mM MES-KOH (pH 6.5)] for 4 h. At the end of the incubation, leaves were snap-frozen in liquid nitrogen. They were subsequently extracted in 100% methanol, the fractional enrichment of metabolite pools was determined as described previously (35, 37), and label redistribution was expressed according to the method of Studart-Guimarães et al. (70).
Statistical Analysis.
Data were statistically examined using the Student’s t test (P < 0.05). The term “significant” is used in the text only when the change in question has been confirmed to be significant (P < 0.05) with this test. All of the statistical analyses were performed using the algorithm embedded into Microsoft Excel.
Supplementary Material
Acknowledgments
This work was supported by funding from the Max Planck Society (W.L.A. and A.R.F.), the National Council for Scientific and Technological Development CNPq-Brazil (CNPq) (Grant 483525/2012-0 to W.L.A.), and the Agence Nationale de la Recherche (Grant ANR-12-BSV6-0011 to J.-P.R.). D.M.D. was the recipient of a scholarship from Foundation for Research Assistance of the Minas Gerais State, Brazil and the CNPq. B.B.B. acknowledges support from an Alexander von Humboldt Research Award that catalyzed the launching of this project at the Max-Planck-Institut für Molekulare Pflanzenphysiologie in Golm.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 3180.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424840112/-/DCSupplemental.
References
- 1.Krebs HA, Johnson WA. Metabolism of ketonic acids in animal tissues. Biochem J. 1937;31(4):645–660. doi: 10.1042/bj0310645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang S, Bryant DA. The tricarboxylic acid cycle in cyanobacteria. Science. 2011;334(6062):1551–1553. doi: 10.1126/science.1210858. [DOI] [PubMed] [Google Scholar]
- 3.Beevers H. Respiratory Metabolism in Plants. Harper and Row; New York: 1961. [Google Scholar]
- 4.Nunes-Nesi A, Araújo WL, Obata T, Fernie AR. Regulation of the mitochondrial tricarboxylic acid cycle. Curr Opin Plant Biol. 2013;16(3):335–343. doi: 10.1016/j.pbi.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Millar AH, Whelan J, Soole KL, Day DA. Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol. 2011;62(1):79–104. doi: 10.1146/annurev-arplant-042110-103857. [DOI] [PubMed] [Google Scholar]
- 6.Sweetlove LJ, Beard KFM, Nunes-Nesi A, Fernie AR, Ratcliffe RG. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010;15(8):462–470. doi: 10.1016/j.tplants.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Raghavendra AS, Padmasree K. Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 2003;8(11):546–553. doi: 10.1016/j.tplants.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Scheibe R, Dietz K-J. Reduction-oxidation network for flexible adjustment of cellular metabolism in photoautotrophic cells. Plant Cell Environ. 2012;35(2):202–216. doi: 10.1111/j.1365-3040.2011.02319.x. [DOI] [PubMed] [Google Scholar]
- 9.Scheibe R, Backhausen JE, Emmerlich V, Holtgrefe S. Strategies to maintain redox homeostasis during photosynthesis under changing conditions. J Exp Bot. 2005;56(416):1481–1489. doi: 10.1093/jxb/eri181. [DOI] [PubMed] [Google Scholar]
- 10.Bauwe H, Hagemann M, Fernie AR. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010;15(6):330–336. doi: 10.1016/j.tplants.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Leister D, Wang X, Haberer G, Mayer KFX, Kleine T. Intracompartmental and intercompartmental transcriptional networks coordinate the expression of genes for organellar functions. Plant Physiol. 2011;157(1):386–404. doi: 10.1104/pp.111.177691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pick TR, et al. PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc Natl Acad Sci USA. 2013;110(8):3185–3190. doi: 10.1073/pnas.1215142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balmer Y, et al. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA. 2004;101(8):2642–2647. doi: 10.1073/pnas.0308583101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaNoue KF, Bryla J, Williamson JR. Feedback interactions in the control of citric acid cycle activity in rat heart mitochondria. J Biol Chem. 1972;247(3):667–679. [PubMed] [Google Scholar]
- 15.Ohné M. Regulation of the dicarboxylic acid part of the citric acid cycle in Bacillus subtilis. J Bacteriol. 1975;122(1):224–234. doi: 10.1128/jb.122.1.224-234.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araújo WL, Nunes-Nesi A, Nikoloski Z, Sweetlove LJ, Fernie AR. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 2012;35(1):1–21. doi: 10.1111/j.1365-3040.2011.02332.x. [DOI] [PubMed] [Google Scholar]
- 17.Banze M, Follmann H. Organelle-specific NADPH thioredoxin reductase in plant mitochondria. J Plant Physiol. 2000;156(1):126–129. [Google Scholar]
- 18.Laloi C, et al. Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci USA. 2001;98(24):14144–14149. doi: 10.1073/pnas.241340898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida K, Noguchi K, Motohashi K, Hisabori T. Systematic exploration of thioredoxin target proteins in plant mitochondria. Plant Cell Physiol. 2013;54(6):875–892. doi: 10.1093/pcp/pct037. [DOI] [PubMed] [Google Scholar]
- 20.Kolbe A, et al. Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiol. 2006;141(2):412–422. doi: 10.1104/pp.106.081208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcus F, et al. Plant thioredoxin h: An animal-like thioredoxin occurring in multiple cell compartments. Arch Biochem Biophys. 1991;287(1):195–198. doi: 10.1016/0003-9861(91)90406-9. [DOI] [PubMed] [Google Scholar]
- 22.Gelhaye E, et al. A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci USA. 2004;101(40):14545–14550. doi: 10.1073/pnas.0405282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens FJ, Li AD, Lateef SS, Anderson LE. Identification of potential inter-domain disulfides in three higher plant mitochondrial citrate synthases: Paradoxical differences in redox-sensitivity as compared with the animal enzyme. Photosynth Res. 1997;54(3):185–197. [Google Scholar]
- 24.Schmidtmann E, et al. Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol Plant. 2014;7(1):156–169. doi: 10.1093/mp/sst144. [DOI] [PubMed] [Google Scholar]
- 25.Møller IM. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- 26.Reichheld J-P, et al. Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell. 2007;19(6):1851–1865. doi: 10.1105/tpc.107.050849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 28.van der Merwe MJ, et al. Tricarboxylic acid cycle activity regulates tomato root growth via effects on secondary cell wall production. Plant Physiol. 2010;153(2):611–621. doi: 10.1104/pp.109.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angelovici R, Fait A, Fernie AR, Galili G. A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol. 2011;189(1):148–159. doi: 10.1111/j.1469-8137.2010.03478.x. [DOI] [PubMed] [Google Scholar]
- 30.Cejudo FJ, Meyer AJ, Reichheld J-P, Rouhier N, Traverso JA. Thiol-based redox homeostasis and signaling. Front Plant Sci. 2014;5:266. doi: 10.3389/fpls.2014.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1(1):387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 32.Tohge T, Fernie AR. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat Protoc. 2010;5(6):1210–1227. doi: 10.1038/nprot.2010.82. [DOI] [PubMed] [Google Scholar]
- 33.Bashandy T, Taconnat L, Renou J-P, Meyer Y, Reichheld J-P. Accumulation of flavonoids in an ntra ntrb mutant leads to tolerance to UV-C. Mol Plant. 2009;2(2):249–258. doi: 10.1093/mp/ssn065. [DOI] [PubMed] [Google Scholar]
- 34.Szecowka M, et al. Metabolic fluxes in an illuminated Arabidopsis rosette. Plant Cell. 2013;25(2):694–714. doi: 10.1105/tpc.112.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tieman D, et al. Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci USA. 2006;103(21):8287–8292. doi: 10.1073/pnas.0602469103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timm S, et al. A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell. 2008;20(10):2848–2859. doi: 10.1105/tpc.108.062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roessner-Tunali U, et al. Kinetics of labelling of organic and amino acids in potato tubers by gas chromatography-mass spectrometry following incubation in (13)C labelled isotopes. Plant J. 2004;39(4):668–679. doi: 10.1111/j.1365-313X.2004.02157.x. [DOI] [PubMed] [Google Scholar]
- 38.Meng L, Wong JH, Feldman LJ, Lemaux PG, Buchanan BB. A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc Natl Acad Sci USA. 2010;107(8):3900–3905. doi: 10.1073/pnas.0913759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P. NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA. 2009;106(24):9908–9913. doi: 10.1073/pnas.0903559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balsera M, Soll J, Buchanan BB. Redox extends its regulatory reach to chloroplast protein import. Trends Plant Sci. 2010;15(9):515–521. doi: 10.1016/j.tplants.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Wong JH, et al. Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Natl Acad Sci USA. 2002;99(25):16325–16330. doi: 10.1073/pnas.212641999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemaire SD, et al. New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2004;101(19):7475–7480. doi: 10.1073/pnas.0402221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winger AM, Taylor NL, Heazlewood JL, Day DA, Millar AH. Identification of intra- and intermolecular disulphide bonding in the plant mitochondrial proteome by diagonal gel electrophoresis. Proteomics. 2007;7(22):4158–4170. doi: 10.1002/pmic.200700209. [DOI] [PubMed] [Google Scholar]
- 44.Rouhier N, et al. Identification of plant glutaredoxin targets. Antioxid Redox Signal. 2005;7(7-8):919–929. doi: 10.1089/ars.2005.7.919. [DOI] [PubMed] [Google Scholar]
- 45.Igamberdiev AU, Gardeström P. Regulation of NAD- and NADP-dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim Biophys Acta. 2003;1606(1-3):117–125. doi: 10.1016/s0005-2728(03)00106-3. [DOI] [PubMed] [Google Scholar]
- 46.Noctor G, De Paepe R, Foyer CH. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007;12(3):125–134. doi: 10.1016/j.tplants.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Tcherkez G, Boex-Fontvieille E, Mahé A, Hodges M. Respiratory carbon fluxes in leaves. Curr Opin Plant Biol. 2012;15(3):308–314. doi: 10.1016/j.pbi.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Mailloux RJ, et al. Glutaredoxin-2 is required to control proton leak through uncoupling protein-3. J Biol Chem. 2013;288(12):8365–8379. doi: 10.1074/jbc.M112.442905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y-R, Chen C-L, Pfeiffer DR, Zweier JL. Mitochondrial complex II in the post-ischemic heart: Oxidative injury and the role of protein S-glutathionylation. J Biol Chem. 2007;282(45):32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 50.Reichheld J-P, Meyer E, Khafif M, Bonnard G, Meyer Y. AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett. 2005;579(2):337–342. doi: 10.1016/j.febslet.2004.11.094. [DOI] [PubMed] [Google Scholar]
- 51.Rocha M, et al. Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol. 2010;152(3):1501–1513. doi: 10.1104/pp.109.150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obata T, et al. Alteration of mitochondrial protein complexes in relation to metabolic regulation under short-term oxidative stress in Arabidopsis seedlings. Phytochemistry. 2011;72(10):1081–1091. doi: 10.1016/j.phytochem.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Timm S, et al. High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS ONE. 2012;7(8):e42809. doi: 10.1371/journal.pone.0042809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niessen M, et al. Two alanine aminotranferases link mitochondrial glycolate oxidation to the major photorespiratory pathway in Arabidopsis and rice. J Exp Bot. 2012;63(7):2705–2716. doi: 10.1093/jxb/err453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Araújo WL, et al. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell. 2010;22(5):1549–1563. doi: 10.1105/tpc.110.075630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verniquet F, Gaillard J, Neuburger M, Douce R. Rapid inactivation of plant aconitase by hydrogen peroxide. Biochem J. 1991;276(Pt 3):643–648. doi: 10.1042/bj2760643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fatland BL, Nikolau BJ, Wurtele ES. Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. Plant Cell. 2005;17(1):182–203. doi: 10.1105/tpc.104.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xing S, et al. ATP citrate lyase activity is post-translationally regulated by sink strength and impacts the wax, cutin and rubber biosynthetic pathways. Plant J. 2014;79(2):270–284. doi: 10.1111/tpj.12559. [DOI] [PubMed] [Google Scholar]
- 59.Foyer CH, Noctor G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011;155(1):2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rasmusson AG, Geisler DA, Møller IM. The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion. 2008;8(1):47–60. doi: 10.1016/j.mito.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer CH. Glutathione. Arabidopsis Book. 2011;9:e0142. doi: 10.1199/tab.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Møller IM, Rasmusson AG. The role of NADP in the mitochondrial matrix. Trends Plant Sci. 1998;3(1):21–27. [Google Scholar]
- 63.Hosoya-Matsuda N, Inoue K, Hisabori T. Roles of thioredoxins in the obligate anaerobic green sulfur photosynthetic bacterium Chlorobaculum tepidum. Mol Plant. 2009;2(2):336–343. doi: 10.1093/mp/ssn077. [DOI] [PubMed] [Google Scholar]
- 64.Nunes-Nesi A, Fernie AR, Stitt M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant. 2010;3(6):973–996. doi: 10.1093/mp/ssq049. [DOI] [PubMed] [Google Scholar]
- 65.Lawlor DW. Carbon and nitrogen assimilation in relation to yield: Mechanisms are the key to understanding production systems. J Exp Bot. 2002;53(370):773–787. [PubMed] [Google Scholar]
- 66.Tcherkez G, et al. Respiratory metabolism of illuminated leaves depends on CO2 and O2 conditions. Proc Natl Acad Sci USA. 2008;105(2):797–802. doi: 10.1073/pnas.0708947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. Protein degradation—An alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011;16(9):489–498. doi: 10.1016/j.tplants.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 68.Nakabayashi R, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77(3):367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meyer EH, et al. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 2009;151(2):603–619. doi: 10.1104/pp.109.141770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Studart-Guimarães C, et al. Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the γ-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol. 2007;145(3):626–639. doi: 10.1104/pp.107.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.