Significance
The anti-HIV drug KP1212 was designed to intentionally increase the mutation rate of HIV, thereby causing viral population collapse. Its mutagenicity and thus antiviral activity was proposed to be the result of tautomerization. We used 2D IR spectroscopy to identify rapidly interconverting tautomers under physiological conditions. The traditionally rare enol–imino tautomer for nucleobases was found to be the major species for KP1212, providing a structural support for the tautomer hypothesis. We further found that KP1212 is significantly protonated at physiological pH with a pKa of 7. The protonated KP1212 was shown to be mutagenic, revealing a bimodal mutagenic property of KP1212. The results could prove instrumental in developing the next-generation antiviral treatments.
Keywords: time-resolved spectroscopy, tautomerism, KP1461, viral decay acceleration
Abstract
Antiviral drugs designed to accelerate viral mutation rates can drive a viral population to extinction in a process called lethal mutagenesis. One such molecule is 5,6-dihydro-5-aza-2′-deoxycytidine (KP1212), a selective mutagen that induces A-to-G and G-to-A mutations in the genome of replicating HIV. The mutagenic property of KP1212 was hypothesized to originate from its amino–imino tautomerism, which would explain its ability to base pair with either G or A. To test the multiple tautomer hypothesis, we used 2D IR spectroscopy, which offers subpicosecond time resolution and structural sensitivity to distinguish among rapidly interconverting tautomers. We identified several KP1212 tautomers and found that >60% of neutral KP1212 is present in the enol–imino form. The abundant proportion of this traditionally rare tautomer offers a compelling structure-based mechanism for pairing with adenine. Additionally, the pKa of KP1212 was measured to be 7.0, meaning a substantial population of KP1212 is protonated at physiological pH. Furthermore, the mutagenicity of KP1212 was found to increase dramatically at pH <7, suggesting a significant biological role for the protonated KP1212 molecules. Overall, our data reveal that the bimodal mutagenic properties of KP1212 result from its unique shape shifting ability that utilizes both tautomerization and protonation.
The high mutation rate of viruses enables rapid evolution in response to environmental pressures, but also results in substantial genetic risk. If the replication error rate exceeds the “error catastrophe limit,” no viable progeny can be sustained and the viral population collapses (1). Based on this concept, an antiviral therapeutic strategy known as lethal mutagenesis seeks to extinguish viruses by elevating their already high mutation rates above the error catastrophe limit, while minimizing toxicity or mutagenicity to the host organism (2–4). Viral ablation by lethal mutagenesis has been demonstrated to occur when viruses are replicated in the presence of mutagenic nucleoside analogs that can be incorporated into viral genomes (4). Examples include 5-hydroxy-2′-deoxycytidine (5-OH-dC) (4–6) and 5,6-dihydro-5-aza-2′-deoxycytidine (KP1212) (7–10) against HIV, ribavirin against Hepatitis C virus (11), and T-705 against influenza virus (12). Despite the promise of lethal mutagenesis as a universal antiviral strategy, its underlying molecular mechanism has been elusive.
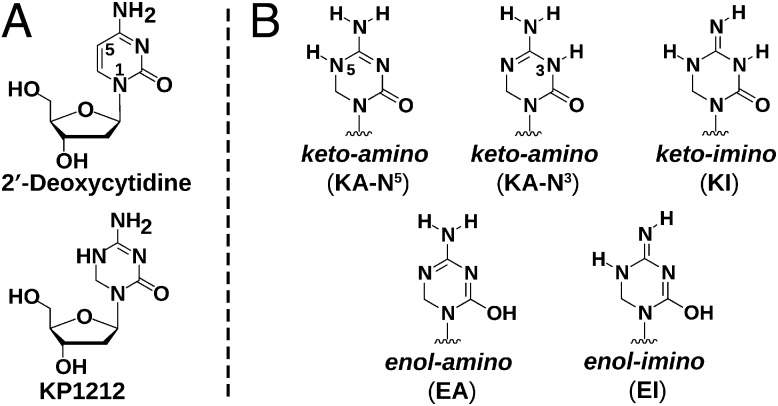
This study addresses the molecular mechanism of mutagenesis of KP1212 (Fig. 1A), a dC analog that has been shown to cause A-to-G and G-to-A mutations in biochemical and cell culture studies (7–9, 13) and a Phase IIa clinical trial (10). The leading proposal, the rare tautomer hypothesis, states that KP1212 exists in multiple tautomeric forms (Fig. 1B) and each tautomer displays distinct base-pairing preferences during replication, leading eventually to the observed mutations (8, 10). This explanation is supported by our recent study, which showed a 10% G-to-A mutation exclusively at the site opposite a single KP1212 base that was introduced site-specifically into a viral vector replicating in Escherichia coli (13). We also observed multiple KP1212 tautomers at −50 °C in dimethylformamide (DMF) (13); however, a detailed characterization of the tautomeric equilibrium under physiological conditions is lacking, hindering further testing of the tautomer hypothesis.
Fig. 1.
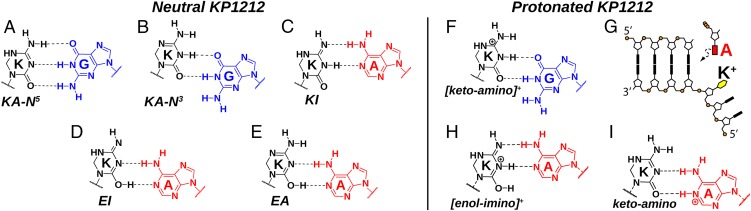
(A) Structure of 2′-deoxycytidine and KP1212 in the keto–amino form. (B) Structure of the base portion of five possible neutral KP1212 tautomers. Only cis-imino tautomers are shown, but the trans forms are also possible.
Although the rare tautomer hypothesis for spontaneous mutations was suggested long ago (14, 15), detecting minor tautomers of nucleobases under physiological conditions has proven difficult. High structural sensitivity and sufficiently high time resolution are required to distinguish tautomers that may exchange as fast as nanoseconds (16). Therefore, NMR—a method with a millisecond time resolution—cannot separate short-lived species and offers only an exchange-averaged characterization of the structure (17). By contrast, vibrational spectroscopy is more promising due to its structural sensitivity and picosecond time resolution (18, 19). For example, Raman spectroscopy of 5-OH-dC (also a G-to-A mutagen) was used to identify a <1% population of the anionic imino–keto tautomer at high pH, which was proposed to base pair with adenine (19). Nevertheless, even when using vibrational spectroscopy, tautomers remain difficult to separate due to peak overlap and uncertainty in peak assignments.
Ultrafast 2D IR spectroscopy can correlate the structural origin of different vibrational resonances in a congested IR vibrational spectrum, thereby offering unambiguous peak assignments and resolving structural isomers in a mixture (20). Our recent experiments on pyridone derivatives demonstrated that 2D IR can distinguish their lactam and lactim tautomers (21) and measure their tautomerization kinetics (16). Here, combining IR spectroscopy, 2D IR, and density functional theory (DFT) calculations, we identify the presence of multiple KP1212 tautomers in aqueous solution at physiological temperatures. Unexpectedly, we find that the dominant species is the enol–imino form. Enol tautomers can in principle base pair with adenine better than the canonical keto–amino form, supporting the hypothesis that tautomerism underlies the lethally mutagenic properties of KP1212. Furthermore we reveal the pKa of KP1212 as 7.0, indicating that protonated KP1212 is simultaneously present alongside the neutral tautomers at physiological pH. The consequences are significant as evidenced by the increasing percentage of adenine incorporated opposite KP1212 by a replicating polymerase at pH <7. The discovery of the mutagenicity of protonated KP1212 offers a strategy for fine-tuning the mutagenesis of nucleoside analogs by adjusting their pKas, which could enable the development of the next-generation lethal mutagens for combating a variety of viral diseases.
Results
Extracting the pKa of KP1212 from FTIR Spectra.
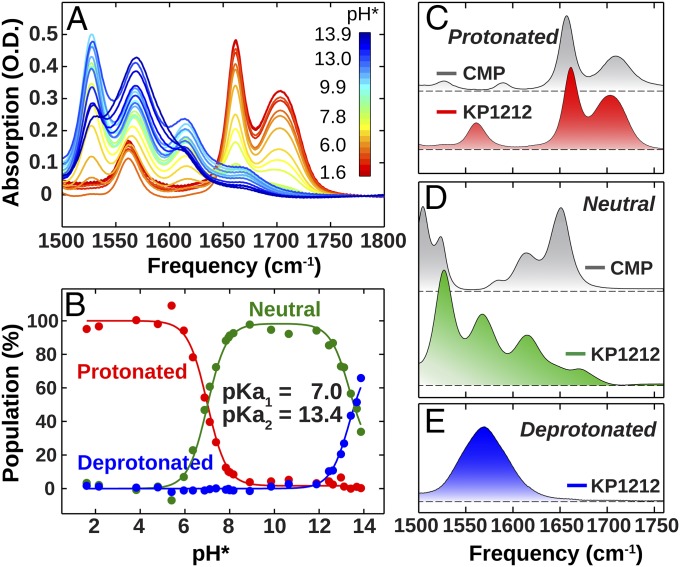
Because tautomerization is closely related to the molecule’s protonation state, we first characterized the pKa of KP1212 by measuring the FTIR spectra between pH* 1.6 and 13.9. The pH* notation refers to direct pH meter readings for deuterated water solutions (not corrected, see SI Appendix), because D2O was used to remove interference from the H2O bend absorption at 1,650 cm−1. The pH-dependent FTIR spectra of KP1212 (Fig. 2A) display intricate behavior in the in-plane base vibration region from 1,450 to 1,750 cm−1, which contains the carbonyl stretches (1,630–1,750 cm−1) and the combinations of C=C stretching, C=N stretching, and ND2 bending (1,450–1,650 cm−1). Even though KP1212 nucleoside lacks phosphate groups, previous studies have shown that phosphate groups do not alter the in-plane base vibrations (22).
Fig. 2.
(A) pH-dependent FTIR spectra of KP1212 from pH* = 1.6 (red) to 13.9 (blue) at 25 °C. (B) Populations of protonated, neutral, and deprotonated KP1212 as a function of pH* obtained from the first three components of SVD analysis. The solid lines show fits of the Henderson–Hasselbalch equation with pKa1 and pKa2 (see SI Appendix for details). (C–E) SVD reconstructed IR spectra representative for the protonated, neutral, and deprotonated KP1212 spectra. The gray curves in C and D are the experimental FTIR spectra of CMP at pH* 1.6 and 7.4, respectively.
We analyzed the pH-dependent spectra using singular value decomposition (SVD), which linearly decomposes these spectra into basis spectra that share common pH dependence (SI Appendix, Fig. S1). Fitting the population of these spectra revealed pKas at 7.0 and 13.4. We applied a matrix transformation to the first three SVD components to reconstruct the spectra and pH-dependent populations for protonated, neutral, and deprotonated KP1212 (Fig. 2 B–E). The pKa1 at 7.0 corresponds to protonation of KP1212, and is significantly higher than that of dC (pKa = 4.3 at N3 atom) (19). This value could be even higher as pKas of some nucleosides have been found to increase when converted to nucleotides (23). The presence of the 1,703-cm−1 mode suggests that protonated KP1212 is in the keto form because a similar high-frequency C=O stretch has been observed for protonated cytidine 5′-monophosphate (CMP) (Fig. 2C). The pKa2 at 13.4 reports on deprotonation; the formation of enolate ions is revealed by the complete loss of the C=O stretch. The neutral spectrum is more complicated as neutral KP1212 can interconvert among the five tautomers by keto–enol or amino–imino tautomerization (Fig. 1B). Also, cis- and trans-imine isomers may be present. Although the canonical form of dC is the keto–amino tautomer, which features a strong C=O peak at 1,651 cm−1 (Fig. 2D), the spectrum of neutral KP1212 shows only one peak above 1,650 cm−1, whose weak intensity compared with that of CMP indicates a strongly reduced keto population.
Identifying Distinct KP1212 Tautomers Using 2D IR Spectroscopy.
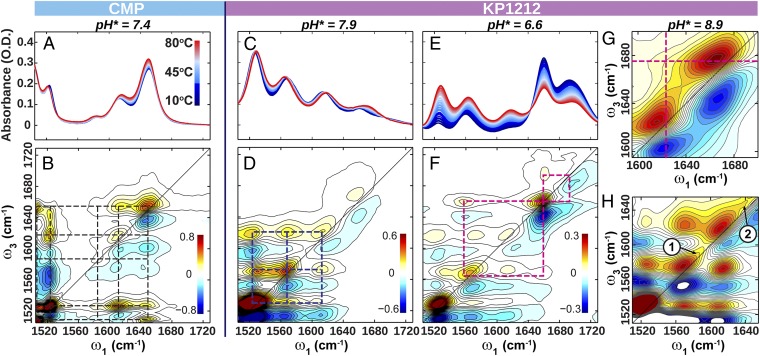
To assist in assigning and separating tautomers, we acquired 2D IR spectra of KP1212 at different pH* and temperatures (Fig. 3). Analogous to 2D NMR spectroscopy, 2D IR employs sequences of ultrafast IR pulses to excite molecular vibrations and detect the energy flow to other vibrations. The diagonal features in a 2D IR spectrum correspond to IR absorption peaks in the FTIR spectrum. Each peak consists of a doublet with a positive (red) peak at the FTIR frequency and a down-shifted negative (blue) peak from induced absorption. Off-diagonal cross-peaks correlate the excitation (ω1) and detection (ω3) frequencies and encode vibrational couplings. Previous studies of DNA bases and base analogs demonstrated that the in-plane stretching vibrations for these species are always strongly coupled, leading to sharp cross-peaks between all resonances for a given base (24). Therefore, the cross-peak patterns allow for separation of different tautomers (21).
Fig. 3.
Variable temperature FTIR spectra and 2D IR spectra with perpendicular polarization. (A and B) Spectra of CMP at pH* 7.4. (C and D) Spectra of KP1212 at pH* 7.9. (E and F) Spectra of KP1212 at pH* 6.6. (G and H) Zoom-in view of the 2D IR spectra of KP1212 at pH* 8.9 in the C=O region at 80 °C (G), and in the enol vibration region at 4 °C (H). Grids are drawn connecting the positive cross-peaks to emphasize connectivity.
The 2D IR spectrum of KP1212 at 37 °C and pH* 7.9 (Fig. 3D) shows a pronounced grid of cross-peaks between the three low-frequency modes at 1,529, 1,572, and 1,612 cm−1, indicating that these vibrations originate from the same molecular species. In contrast, the carbonyl peak at 1,662 cm−1 does not have cross-peaks to these three lower-frequency modes, suggesting that this vibration is due to a chemically distinct species. This is in sharp contrast with the 2D IR spectrum of CMP (Fig. 3B), in which cross-peaks between all of the diagonal peaks occur. Because of the characteristic carbonyl stretch frequency, we assign the 1,662-cm−1 mode of KP1212 to a keto C=O vibration, and the three lower-frequency modes to enol tautomers. Further evidence for this assignment comes from low pH* spectra. At pH* = 1, the 2D IR spectrum of KP1212 resembles that of CMP (SI Appendix, Fig. S2), indicating that both protonated bases are only in the keto–amino form. Because of the pKa1 value of 7.0, KP1212 exists as a mixture of protonated and neutral forms in the physiological pH range of 6–8. The IR spectra at pH* 6.6 (Fig. 3 E and F) show that compared with pH* 7.9, the protonated keto population is elevated as highlighted by the pink grids in Fig. 3F. The remaining neutral KP1212 population is distributed between the keto and enol tautomers, as shown by the presence of the multiple modes with frequencies <1,640 cm−1.
The temperature dependence of the KP1212 spectra (Fig. 3 C and E) also provides strong evidence for the existence of multiple tautomers. Unlike the FTIR of CMP (Fig. 3A), which shows marginal thermal changes, the KP1212 spectra display marked temperature dependence such as peak shift, intensity variation, and clear isosbestic points, especially at pH* 6.6. At pH* 7.9, the keto population increases with increasing temperature. A two-state model that considers only the broader categories of keto and enol species was used to analyze the spectra and obtain the thermodynamic parameters (SI Appendix, Fig. S3), which revealed that 66% of KP1212 species are enol tautomer at 37 °C. At pH* 6.6, KP1212 is substantially protonated in the keto form, whose population drops with temperature. Simultaneously, the neutral enol tautomer gains in population, reflecting that the temperature-dependent spectra largely report on protonation of KP1212. A two-state model was used to approximate the temperature dependence of pKa (SI Appendix, Fig. S5). A minimal 0.18-unit decrease in pKa from 25 °C to 37 °C was determined and is similar to reported values of canonical bases (25).
Peak Assignments Based on DFT Calculations.
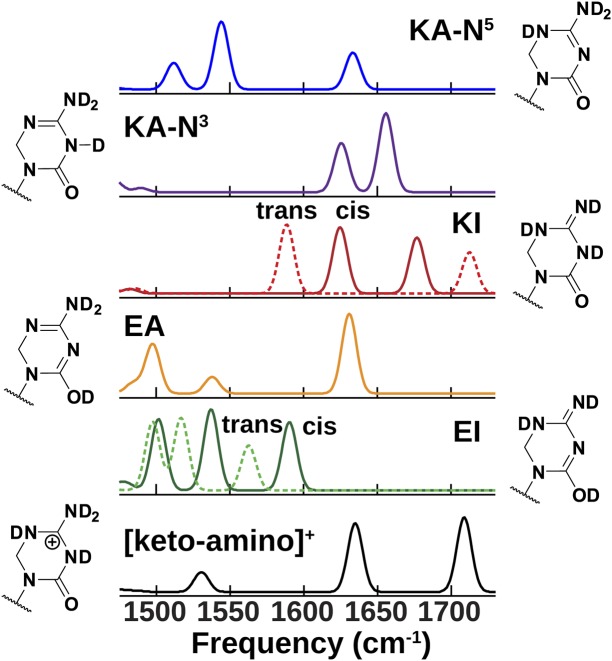
To assign vibrational modes to specific tautomers, we calculated the IR absorption spectra for the five tautomers shown in Fig. 1B using DFT calculations of harmonic vibrations [Becke, three-parameter, Lee–Yang–Parr (B3LYP) functional with 6-31G(d,p) basis set]. Similar to previous observations (21, 26), we found it necessary to include enough explicit water molecules in these calculations to hydrate solvation sites that contain or accept labile hydrogens (SI Appendix, Fig. S6). Fig. 4 displays the calculated IR absorption spectra with five explicit water molecules, for which we believe that vibrational frequencies within ±30 cm−1 can be used for assigning tautomers. For protonated KP1212, the calculated spectrum of the keto–amino form matches well with the experimental spectrum (Fig. 2C), in agreement with our tautomer assignment. A more detailed analysis of ionized KP1212 is presented in SI Appendix, Fig. S7.
Fig. 4.
DFT calculated IR spectra for KP1212 tautomers with five explicit water molecules. The frequencies have been scaled by 0.9614 and σ = 5-cm−1 Gaussian broadening has been applied. Spectra for the cis- and trans-imino isomers are plotted with solid and dashed lines, respectively. Calculations for other ionic species are shown in SI Appendix, Fig. S7.
For neutral KP1212, qualitative similarities between calculated and experimental spectra support the separation of spectra into keto and enol species. In particular, the triplet pattern of the cis-enol–imino (cis-EI) spectrum at 1,502, 1,537, and 1,591 cm−1 matches qualitatively with the peak positions in the 1,500–1,620-cm−1 region of the experimental spectra (blue grids in Fig. 3D). Although neither the cis- nor trans-isomer calculations predict the experimentally observed intensity pattern, we find that the calculated intensities are more easily influenced by solvent structural details than frequencies. Keto–amino-N3 (KA-N3) and keto–imino (KI) calculations both predict two transitions >1,600 cm−1. Keto–amino-N5 (KA-N5) is the only keto tautomer predicted to have a strong transition near 1,550 cm−1, which should result in a strong cross-peak from ∼1,550 cm−1 to the C=O stretch. Such a cross-peak is not observed in experimental spectra of the uncharged KP1212, meaning that if any KA-N5 is present, its population fraction is estimated to be <5%.
Assignment of minor tautomers requires careful analysis of 2D lineshape contours, which we performed at pH* 8.9 to eliminate contributions from the protonated KP1212. Detailed lineshape analysis and spectral simulations are presented in SI Appendix, Figs. S8–S12. We present zoom-ins of the keto and enol regions in Fig. 3 G and H. Fig. 3G shows that the C=O peak at 80 °C is about two times broader than that of CMP (Fig. 3B) and exceeds typical broadening due to solvent hydrogen bonding (27), implying the presence of two overlapped keto transitions from separate tautomers (21). Whereas changing solvation state can lead to frequency shift and peak broadening, the appearance of a new tautomer leads to a distinct cross-peak pattern (21). A cross-peak from the C=O stretch to a weak peak between 1,620 and 1,640 cm−1 (only observed at higher temperatures) is suggested by a ridge extending along ω1 at ω3 = 1,675 cm−1. This pair of peaks could be attributed to either KI or KA-N3. The C=O frequency of N3-methylcytidine, which adopts the keto–imino form, is blue-shifted to ∼1,670 cm−1 (19). Combining this observation with our DFT calculations, which predict a larger peak splitting for KI than for KA-N3, we conclude that the 1,675-cm−1 peak corresponds to the C=O stretch of the KI tautomer. As a result, the dominant 1,662-cm−1 peak at lower temperature corresponds to KA-N3.
Whereas EI emerges from the data as the dominant species, a closer look at the enol region of the 2D IR spectrum shows the presence of multiple species. In Fig. 3D, the position of negative and positive peaks does not align on the same vertical excitation frequency (e.g., ω1 ∼ 1,530 cm−1), indicating multiple species in the sample. Because the imino tautomers can be either in the trans- or cis form, and the variations in frequency of their vibrational transitions are on the order of the vibrational linewidths, the observed broad peaks may be explained by a superposition of these isomers (e.g., feature 1 in Fig. 3H). Similarly, the enol–amino (EA) tautomer may be present in a small proportion. Calculated EA spectrum also has a three-peak pattern like EI, but with a blue-shifted ring mode at 1,631 cm−1. The 2D IR spectrum at 4 °C and pH* 8.9 shows an additional weak diagonal peak at 1,638 cm−1 (feature 2 in Fig. 3H). Moreover, ω1 frequency slice (SI Appendix, Fig. S10) suggests a cross-peak (at 1,638 cm−1, 1,529 cm−1)—a peak pattern that resembles the calculated EA spectrum. The populations of the minor tautomers such as KI and EA are estimated to be ∼5% based on spectral simulation shown in SI Appendix.
The combination of the experimental constraints from 2D IR spectra and the DFT calculations provides consistent evidence that under physiological conditions, KP1212 exists as an ensemble of varying structures including ionic and tautomeric species. Protonated KP1212 is primarily in the keto–amino form and neutral KP1212 adopts multiple tautomers with the enol–imino tautomer being the dominant species.
pH-Dependent Mutagenicity and DNA Stability.
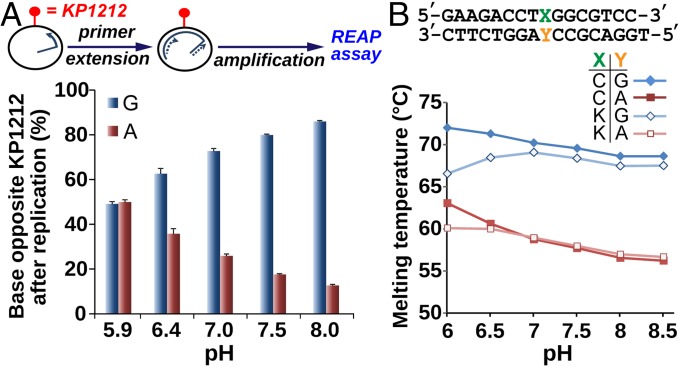
Because the pKa of KP1212 is unusually high at 7.0, a substantial proportion of KP1212 is protonated at physiological pH. This suggests that both neutral and cationic forms of KP1212 should be considered when examining its biological consequences. To investigate, we first measured the in vivo mutagenicity of KP1212 as a function of pH from pH 6.0 (predominantly protonated) to pH 8.0 (predominantly neutral). A primer extension reaction was carried out using the Klenow exo- polymerase on an M13 ssDNA genomic template containing a single KP1212 base at a defined site. The newly synthesized strand was specifically PCR amplified and then the base placed across the KP1212 site was quantified using the previously published restriction endonuclease and postlabeling (REAP) assay (28). KP1212 paired ∼10% of the time with A and 90% of the time with G around pH 8 (Fig. 5A). As the pH dropped, the type of mutation remained the same, but the percentage of inserted A increased strikingly reaching ∼50% at pH 5.9. This result underscores the complexity of the KP1212 mutagenesis mechanism and, more specifically, it spotlights the importance of the protonated version of the nucleoside.
Fig. 5.
(A) Percentage of base placed opposite KP1212 at different pHs during an in vitro primer extension reaction. A long primer containing a unique, noncomplementary overhang is annealed to a circular, single-stranded M13 genomic template containing single KP1212 base (the red lollipop) at a specific site, and extended by the Klenow fragment exo- polymerase. The newly synthesized strand is then PCR amplified, and analyzed using the REAP assay. Each data point is an average of three independent measurements, with 1 SD error bars. (B) The melting temperature of DNA duplexes in phosphate buffer solutions at pH 6.0–8.5. Base X is either C (closed symbols) or KP1212 (open symbols). Base Y is either G (blue curves) or A (red curves). Each data point is an average of three independent measurements, with less than 0.5 °C SD (not shown for clarity).
Additionally, we examined the stability of KP1212 pairing with G and A at different pHs, as evidenced by the melting temperatures (Tm) of 16mer oligonucleotides containing KP1212 (or dC) at the ninth position. The complementary strands were designed with either G or A opposite the ninth base. Fig. 5B displays the Tm of the four duplexes from pH 6–8.5. The Tm of the C•G duplex control was relatively unaffected by pH. The C•A duplex, featuring a mismatched pair in the middle of the duplex, had a considerably lower Tm by ∼10 °C at high pH. However, as the pH decreased below 7, the Tm of the C•A duplex increased, suggesting the formation of a stabilizing interaction, for example the wobble base pair proposed between C and protonated A (25). The K•G duplex (where “K” refers to KP1212) exhibited stability comparable with the C•G duplex at pH ≥7.0 but was destabilized at lower pH. The K•A duplex had a Tm trend similar to the C•A duplex with a smaller increase in Tm approaching pH 6.0. The progressive stabilization of the K•A duplex and destabilization of the K•G duplex as pH decreased from 8.0 to 6.0 is consistent with the increase in G-to-A mutations at lower pH.
Discussion
Mutagenicity of Neutral KP1212.
Although minor tautomers of nucleobases have been proposed to form mismatched base pairs and cause spontaneous mutations (14, 15, 29), their low abundance and fast exchange rates make it challenging to obtain structural evidence for their role in mismatch events. The present work demonstrates that 2D IR spectroscopy is effective at probing the population distribution and thermodynamics of nucleic acid tautomers. We provide evidence of the existence of multiple tautomers for KP1212 and show that the traditionally rare enol–imino tautomer is the major species under physiological conditions. We also report that pH-dependent mutagenesis and the Tm of DNA oligonucleotides containing KP1212 are both correlated with the degree of KP1212 protonation.
A number of possibilities exist for the mechanism by which KP1212 induces G-to-A mutations. Among the various factors that influence base pair stability, notably hydrogen bond formation, base pair geometry, and base stacking, we first consider how neutral KP1212 may doubly or triply H-bond with canonical DNA purine bases. Although KP1212 was believed to be predominantly in the KA-N5 tautomeric form and pair with G in a Watson–Crick (WC) geometry (Fig. 6A), our results reveal the presence of multiple tautomers which could potentially pair with G or mispair with A. The keto–amino tautomer KA-N3 can pair with G in a wobble arrangement by shifting H-bond registry by 1 (Fig. 6B). More importantly, under physiological conditions the dominant enol–imino tautomer can potentially base pair with A in the wobble position (Fig. 6D). Although in low abundance, the other observed enol tautomer EA could pair with A in the same configuration (Fig. 6E). Each of these K•A pairs offers an explanation for the role of tautomerism in KP1212's promiscuous base-pairing ability and mutagenicity.
Fig. 6.
Proposed models to explain the G-to-A mutation induced by neutral (A–E) or protonated KP1212 (F–I). These schemes are examples illustrating that KP1212 tautomers can pair with either G or A, but other base pair geometries are also possible (SI Appendix, Figs. S13 and S14). (G) Protonated KP1212 flips out to create an abasic site-like intermediate, and the polymerase follows the A rule to insert A opposite KP1212.
The keto–imino tautomer was proposed to explain the mutagenicity of KP1212 as shown in Fig. 6C (7, 9). However, the KI population was found to be small and only detectable at elevated temperatures, making it less likely to contribute to KP1212s mutagenicity compared with the abundant EI tautomer. Although the proposed schemes for the enol tautomers (Fig. 6 D and E) are not WC base pairs, the wobble configuration is also a common structural motif and exhibits comparable stability (30). For instance, crystal structures of a wobble G•T mismatch within a polymerase have been reported (31). However, the low Tm of the K•A duplex (Fig. 5B) suggests that the K•A base pair may only be stabilized at the active site of the polymerase.
Tautomeric Distribution and Mutagenicity.
Whereas the high population of EI—a presumed base-pairing partner for A—clearly indicates KP1212's elevated mutation propensity, these solution measurements alone are not a basis for quantitatively predicting the G-to-A mutation rate when KP1212 is present in the template strand. We previously used the tautomer distribution obtained in DMF and a limited set of base-pairing schemes to explain the final incorporation rate of A (13); however, given the altered tautomeric equilibrium shown here in aqueous solution, additional factors should be considered. First, if the tautomer exchange rate is slower than the DNA synthesis steps, the mutagenicity should be governed by the tautomeric equilibrium; alternatively, if the exchange rate is faster, the less-populated KP1212 tautomers could be transiently stabilized during nucleotide incorporation. Similar to our previous work on nucleobase analogs (16), temperature-jump experiments revealed ∼20-ns relaxation rates (SI Appendix, Fig. S15). The fast time scale suggests that the latter case should apply to KP1212 and hence the tautomeric distribution is not a direct measure of mutation rates. Another consideration is that the spectroscopic work was performed on the free nucleoside whereas the mutation rates reflect the properties of KP1212 in DNA oligonucleotides. However, ongoing investigation has revealed similar tautomeric distributions, including the enol dominance, for KP1212 in free solution and in DNA oligomers (SI Appendix, Fig. S16).
The ability of KP1212 to select different base-pairing partners during replication is exemplified in Fig. 6. Yet, knowing the fast interconversion of tautomers, we now have to consider a wider range of base-pairing options, which may involve cis-/trans-isomerization of the imine bond, anti–syn rotamerism of the base, or rotation of the enol hydroxyl group, as shown in SI Appendix, Fig. S13. Overall, the canonical keto–amino forms are proposed to pair exclusively with G, whereas the KI, EA, and EI tautomers may pair with either A or G. Finally, base incorporation in a DNA polymerase involves a change in solvation patterns and electrostatics which could alter the tautomeric distribution. To this effect, IR spectra of KP1212 recorded in the aprotic solvent dimethyl sulfoxide showed that the keto population increased while still preserving some enol tautomers (SI Appendix, Fig. S17). Even though determining the mutation rate directly from the tautomeric distribution remains a challenging task, the present study shows that the high fraction of enol species in solution and in a KP1212 oligomer correlates with the high G-to-A mutation rate, therefore providing compelling structural evidence for the tautomer hypothesis.
Mutagenicity of Protonated KP1212.
The evolutionary optimal pKa values of canonical nucleobases are thought to be far from 7 to prevent the base pair ambiguity that may come from ionization (32). In contrast, KP1212, having a pKa near neutrality, is significantly protonated at physiological pH. The enhanced mutagenicity of protonated KP1212 raises additional mechanistic questions. Our results show that positively charged KP1212 adopts a protonated keto–amino form, which can pair with G in a wobble configuration (Fig. 6F) but cannot form more than one H bond with A.
As the mutagenicity of protonated KP1212 is insufficiently explained by its predominant tautomeric form, we propose three other models by which the polymerase replicates across the lesion. One possibility involves the “A rule,” where a polymerase replicating across a bulky damaged base or an abasic site is more likely to place an adenine in the opposing strand (33). The charged state of KP1212 may promote its ability to flip out of the helix stack, presenting the polymerase an intermediate structure reminiscent of an abasic site (Fig. 6G). The absence of a K+•A base pair, however, is not consistent with the Tm measurements, which show increased stability for the K•A duplex from pH 7–6. The second possibility is that the protonated KP1212 has a direct stabilizing interaction with A. Even though the predominant keto–amino tautomer can only pair with G, other isomers such as protonated enol tautomers could form two H bonds with A, either in WC or wobble configurations as illustrated in Fig. 6H and SI Appendix, Fig. S14. This interaction is similar to previously observed C•A pairs involving the imino tautomer of C (29). Whereas these protonated enol tautomers do not exist in detectable amounts in free solution, they could be transiently stabilized in the polymerase active site to allow pairing with A. However, we do not have any direct evidence of such a possibility. The last mechanism presumes the ability of KP1212 to function as a general acid and protonate the incoming adenine at N1 (Fig. 6I). Although the pKa of adenine is relatively low (3.8), the protonated adenine forms a wobble pair with the neutral keto–amino KP1212, and this stabilizing interaction may shift the acid–base equilibrium. Such an interaction has been observed for C•A+ pairs (25). Further spectroscopic and biochemical work is needed to investigate these possibilities.
In summary, our studies show that both tautomerism and protonation are essential for KP1212's base-pairing promiscuity and therefore high mutagenicity. At pH values where KP1212 is uncharged, the observed G-to-A mutation patterns from in vitro, in vivo, and clinical studies fit logically with the tautomerism model. Our experiments establish the presence of various KP1212 tautomers with the enol–imino isomer being the major species, which can potentially base pair with A. Protonated KP1212 was found to exist primarily in the keto–amino form and exhibit even greater mutagenicity. The mode of action of KP1212 parallels that of the C•A mismatch where a minor imino tautomer of C is involved at high pH and a protonated C•A+ pair is formed at low pH. The difference is that KP1212's unusually high pKa and unique property to adopt various tautomers leads to much higher mutation rates. The findings advance the notion that the tautomeric equilibria and pKa of nucleobases can be engineered with therapeutically useful chemical modifications. Finally, this work forms the basis for future experiments that will focus on directly observing K•A base pairing to elucidate the mechanism of mutagenesis.
Methods
Spectroscopy.
KP1212 and CMP were purchased from Berry & Associates, Inc. and Sigma-Aldrich, respectively. The samples were used without further purification, and were dissolved in potassium phosphate buffer in D2O at 20 mg/mL; the pH was adjusted with DCl and NaOD. Two-dimensional IR experiments were performed following the methods described in SI Appendix.
In Vitro Mutagenesis and Melting Point Analysis.
The M13 single-stranded genomes, containing one KP1212 base at a specific site, were constructed and purified as previously reported (13). The details of the primer extension reaction are described in SI Appendix and the REAP assay is described in refs. 13, 28. The 16mer oligonucleotide 5′-GAAGACCTXGGCGTCC-3′, where X is KP1212, was synthesized and purified as described previously (13). All other oligonucleotides were from Integrated DNA Technologies. Thermal denaturation of DNA duplexes in the presence of a saturating fluorescent dye was carried out on a Roche LightCycler480 machine using the procedure described in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grants CHE-1212557 and CHE-1414486, the Massachusetts Institute of Technology (MIT) Center for Environmental Health Sciences [National Institutes of Health (NIH) Center Grant P30-ES002109], and the MIT Laser Biomedical Research Center (NIH Center Grant P41-EB015871). V.S. was supported by NIH Traineeship T32 ES007020. J.M.E. was supported by NIH Grants CA080024 and CA26731.
Footnotes
Conflict of interest statement: J.M.E. is a cofounder and advisor for a pharmaceutical company interested in developing mutagenic inhibitors of HIV.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415974112/-/DCSupplemental.
References
- 1.Eigen M. Error catastrophe and antiviral strategy. Proc Natl Acad Sci USA. 2002;99(21):13374–13376. doi: 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perales C, Martín V, Domingo E. Lethal mutagenesis of viruses. Curr Opin Virol. 2011;1(5):419–422. doi: 10.1016/j.coviro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Bonnac LF, Mansky LM, Patterson SE. Structure-activity relationships and design of viral mutagens and application to lethal mutagenesis. J Med Chem. 2013;56(23):9403–9414. doi: 10.1021/jm400653j. [DOI] [PubMed] [Google Scholar]
- 4.Loeb LA, et al. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci USA. 1999;96(4):1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feig DI, Sowers LC, Loeb LA. Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl Acad Sci USA. 1994;91(14):6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C —> T transitions in vivo. Proc Natl Acad Sci USA. 1998;95(7):3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daifuku R. Stealth nucleosides: mode of action and potential use in the treatment of viral diseases. BioDrugs. 2003;17(3):169–177. doi: 10.2165/00063030-200317030-00003. [DOI] [PubMed] [Google Scholar]
- 8.Harris KS, Brabant W, Styrchak S, Gall A, Daifuku R. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res. 2005;67(1):1–9. doi: 10.1016/j.antiviral.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Murakami E, Basavapathruni A, Bradley WD, Anderson KS. Mechanism of action of a novel viral mutagenic covert nucleotide: Molecular interactions with HIV-1 reverse transcriptase and host cell DNA polymerases. Antiviral Res. 2005;67(1):10–17. doi: 10.1016/j.antiviral.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Mullins JI, et al. Mutation of HIV-1 genomes in a clinical population treated with the mutagenic nucleoside KP1461. PLoS ONE. 2011;6(1):e15135. doi: 10.1371/journal.pone.0015135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crotty S, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6(12):1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 12.Baranovich T, et al. T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol. 2013;87(7):3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, et al. Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2′-deoxycytidine. Proc Natl Acad Sci USA. 2014;111(32):E3252–E3259. doi: 10.1073/pnas.1405635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171(4361):964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 15.Topal MD, Fresco JR. Complementary base pairing and the origin of substitution mutations. Nature. 1976;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 16.Peng CS, Baiz CR, Tokmakoff A. Direct observation of ground-state lactam-lactim tautomerization using temperature-jump transient 2D IR spectroscopy. Proc Natl Acad Sci USA. 2013;110(23):9243–9248. doi: 10.1073/pnas.1303235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhne RO, Schaffhauser T, Wokaun A, Ernst RR. Study of transient chemical reactions by NMR. Fast stopped-flow Fourier transform experiments. J Magn Reson. 1979;35(1):39–67. [Google Scholar]
- 18.Miles HT. Tautomeric forms in a polynucleotide helix and their bearing on the structure of DNA. Proc Natl Acad Sci USA. 1961;47:791–802. doi: 10.1073/pnas.47.6.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suen W, Spiro TG, Sowers LC, Fresco JR. Identification by UV resonance Raman spectroscopy of an imino tautomer of 5-hydroxy-2′-deoxycytidine, a powerful base analog transition mutagen with a much higher unfavored tautomer frequency than that of the natural residue 2′-deoxycytidine. Proc Natl Acad Sci USA. 1999;96(8):4500–4505. doi: 10.1073/pnas.96.8.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messmer AT, Lippert KM, Schreiner PR, Bredenbeck J. Structure analysis of substrate catalyst complexes in mixtures with ultrafast two-dimensional infrared spectroscopy. Phys Chem Chem Phys. 2013;15(5):1509–1517. doi: 10.1039/c2cp42863f. [DOI] [PubMed] [Google Scholar]
- 21.Peng CS, Tokmakoff A. Identification of lactam–lactim tautomers of aromatic heterocycles in aqueous solution using 2D IR spectroscopy. J Phys Chem Lett. 2012;3(22):3302–3306. doi: 10.1021/jz301706a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banyay M, Sarkar M, Gräslund A. A library of IR bands of nucleic acids in solution. Biophys Chem. 2003;104(2):477–488. doi: 10.1016/s0301-4622(03)00035-8. [DOI] [PubMed] [Google Scholar]
- 23.La Francois CJ, Jang YH, Cagin T, Goddard WA, 3rd, Sowers LC. Conformation and proton configuration of pyrimidine deoxynucleoside oxidation damage products in water. Chem Res Toxicol. 2000;13(6):462–470. doi: 10.1021/tx990209u. [DOI] [PubMed] [Google Scholar]
- 24.Peng CS, Jones KC, Tokmakoff A. Anharmonic vibrational modes of nucleic acid bases revealed by 2D IR spectroscopy. J Am Chem Soc. 2011;133(39):15650–15660. doi: 10.1021/ja205636h. [DOI] [PubMed] [Google Scholar]
- 25.Siegfried NA, O’Hare B, Bevilacqua PC. Driving forces for nucleic acid pK(a) shifting in an A(+).C wobble: Effects of helix position, temperature, and ionic strength. Biochemistry. 2010;49(15):3225–3236. doi: 10.1021/bi901920g. [DOI] [PubMed] [Google Scholar]
- 26.Lee C, Cho M. Vibrational dynamics of DNA. II. Deuterium exchange effects and simulated IR absorption spectra. J Chem Phys. 2006;125(11):114509. doi: 10.1063/1.2213258. [DOI] [PubMed] [Google Scholar]
- 27.Ham S, Kim J-H, Lee H, Cho M. Correlation between electronic and molecular structure distortions and vibrational properties. II. Amide I modes of NMA–nD2O complexes. J Chem Phys. 2003;118(8):3491–3498. [Google Scholar]
- 28.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci USA. 2011;108(43):17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varani G, McClain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1(1):18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116(6):803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 32.Sowers LC, Shaw BR, Veigl ML, Sedwick WD. DNA base modification: Ionized base pairs and mutagenesis. Mutat Res. 1987;177(2):201–218. doi: 10.1016/0027-5107(87)90003-0. [DOI] [PubMed] [Google Scholar]
- 33.Obeid S, et al. Replication through an abasic DNA lesion: Structural basis for adenine selectivity. EMBO J. 2010;29(10):1738–1747. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.