Significance
DNA damage response is essential to preserve genomic stability. Here we report a previously unknown function for the baculovirus inhibitor of apoptosis protein repeat (BIR)-containing ubiquitin-conjugating enzyme (BRUCE) and ubiquitin-specific peptidase 8 (USP8) as activators of DNA damage response. They drive recruitment of the breast cancer susceptibility gene C terminus-repeat inhibitor of human telomerase reverse transcriptase expression (BRIT1) to DNA breaks by promoting BRIT1 deubiquitination. In contrast to the established regulation of repair foci formation by ubiquitination, our data demonstrate deubiquitination as a previously unrecognized critical step in promoting foci formation. Furthermore, we define a pathway by which BRUCE and USP8 activate BRIT1–switch/sucrose nonfermentable (SWI-SNF)–mediated chromatin relaxation to maximize cell responsiveness to DNA damage. Thus, BRUCE represents a novel component in safeguarding genomic stability and a promising therapeutic target in diseases of genomic instability such as cancer.
Keywords: inhibitor of apoptosis, DNA DSB repair, BRUCE, BRIT1
Abstract
The DNA damage response (DDR) is crucial for genomic integrity. BRIT1 (breast cancer susceptibility gene C terminus-repeat inhibitor of human telomerase repeat transcriptase expression), a tumor suppressor and early DDR factor, is recruited to DNA double-strand breaks (DSBs) by phosphorylated H2A histone family, member X (γ-H2AX), where it promotes chromatin relaxation by recruiting the switch/sucrose nonfermentable (SWI–SNF) chromatin remodeler to facilitate DDR. However, regulation of BRIT1 recruitment is not fully understood. The baculovirus IAP repeat (BIR)-containing ubiquitin-conjugating enzyme (BRUCE) is an inhibitor of apoptosis protein (IAP). Here, we report a non-IAP function of BRUCE in the regulation of the BRIT1–SWI–SNF DSB-response pathway and genomic stability. We demonstrate that BRIT1 is K63 ubiquitinated in unstimulated cells and that deubiquitination of BRIT1 is a prerequisite for its recruitment to DSB sites by γ-H2AX. We show mechanistically that BRUCE acts as a scaffold, bridging the ubiquitin-specific peptidase 8 (USP8) and BRIT1 in a complex to coordinate USP8-catalyzed deubiquitination of BRIT1. Loss of BRUCE or USP8 impairs BRIT1 deubiquitination, BRIT1 binding with γ-H2AX, the formation of BRIT1 DNA damage foci, and chromatin relaxation. Moreover, BRUCE-depleted cells display reduced homologous recombination repair, and BRUCE-mutant mice exhibit repair defects and genomic instability. These findings identify BRUCE and USP8 as two hitherto uncharacterized critical DDR regulators and uncover a deubiquitination regulation of BRIT1 assembly at damaged chromatin for efficient DDR and genomic stability.
BRUCE is a 528-kDa member of the inhibitor of apoptosis protein (IAP) family (1). It is a unique IAP that harbors a baculovirus IAP repeat (BIR) domain near its N terminus and a ubiquitin (Ub)-conjugating (UBC) domain near its C terminus (Fig. 1A) (1, 2). When overexpressed, BRUCE inhibits apoptosis by binding to cleaved caspases-3, -6, -7, and -9 (3–5). Recent data indicate that proteins containing a BIR domain also have many non-antiapoptotic functions in vivo (6–8). For BRUCE, one characterized non-IAP function is in the final stage of cytokinesis, abscission (8). During cytokinesis, BRUCE localizes to the midbody where it forms a platform to interact with mitotic regulators and components of the vesicle-targeting machinery to assist their delivery to the site of abscission (8). BRUCE may have other non-IAP functions yet to be identified, as suggested by the broad spectrum of phenotypes exhibited in BRUCE-mutant mice (3, 9, 10).
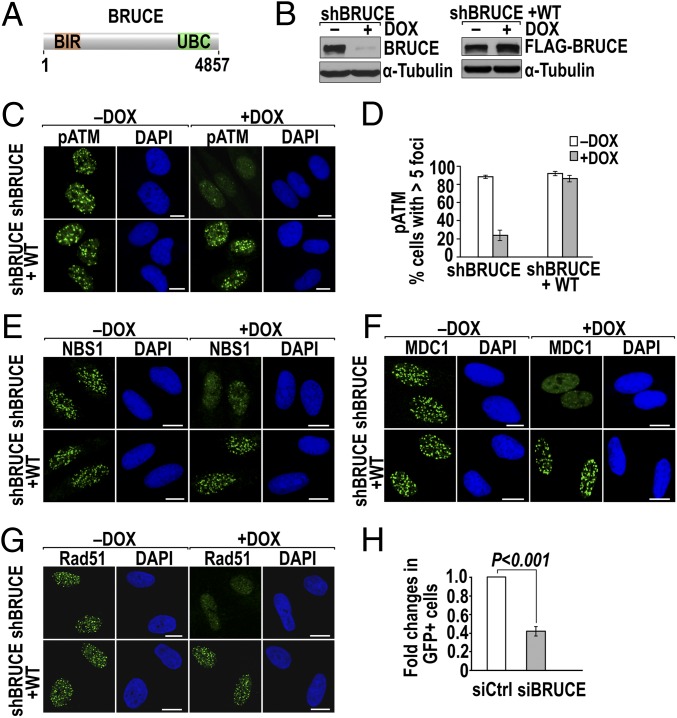
Fig. 1.
Depletion of BRUCE attenuates the formation of foci of DNA damage and impairs HR. (A) Schematic diagram of human BRUCE protein (4,857 aa) with BIR and UBC domains indicated. (B) shBRUCE-inducible U2OS cell line (shBRUCE) with endogenous BRUCE depleted after DOX treatment (Left) and an isogenic cell line with reconstituted expression of siBRUCE-resistant FLAG-tagged BRUCE (shBRUCE+WT) (Right). (C and D) BRUCE is required for pATM foci formation. (C) Immunostaining of pATM repair foci (green) in shBRUCE and shBRUCE+WT U2OS cells 1 h after IR exposure (5 Gy) with cell nuclei counterstained by DAPI (blue). (Scale bars, 10 µm.) (D) Quantification of pATM foci in C; error bars represent SD of two independent experiments. (E–G) Immunostaining of DNA damage foci of NBS1 (E), MDC1 (F), and Rad51 (G), following the experimental and analytic parameters for pATM foci shown in C and D. (H) HR assay. Fold change of GFP+ cells before and after BRUCE depletion; the %GFP+ cells treated with control siRNA (siCtrl) is set as 1. P < 0.001, Student’s two-tailed t test; error bars represent SEM from three independent experiments.
Unrepaired DNA double-strand breaks (DSBs) are highly toxic to cells. Mutation or depletion of DSB signaling and repair proteins often enhances cell sensitivity to DNA damage (11, 12). Previous studies have linked BRUCE levels with cell survival in response to the induction of DNA damage. Depletion of BRUCE sensitizes human cancer cell lines to several DSB-inducing agents, including ionizing radiation (IR), cisplatin, and etoposide. Conversely, elevated BRUCE protein levels increase cell resistance to these agents (2, 3, 9). Although BRUCE and DSB-response proteins alter cell viability similarly in the presence of DNA damage, the mechanisms by which BRUCE affects cell viability after DNA damage are unclear.
The DNA damage response (DDR) is a collective cellular-protective mechanism for detecting and repairing DNA lesions to maintain genomic integrity (13). DSB-response proteins usually accumulate at sites of damaged chromatin, forming cytologically distinct nuclear foci called “IR-induced foci” (IRIF) (14). Posttranslational modifications of DDR factors and histones flanking DSBs provide specificity and hierarchical recruitment of DDR factors by creating specific modular protein–protein interactions (15–17). Ubiquitination modification of DDR proteins plays essential roles in enabling their accumulation at damaged chromatin (17). Well-established examples include the monoubiquitination of Fanconi anemia complementation group D2 (FANCD2) as a prerequisite for its assembly at DNA damage sites (18, 19) and DSB-triggered ubiquitination of H2A and H2AX (H2A histone family, member X) at DSB-flanking regions as Ub-binding sites for breast cancer susceptibility gene 1 (BRCA1) complex A recruitment (20–26). The reverse process of ubiquitination, deubiquitination catalyzed by deubiquitinating enzymes (DUBs), also is important for DNA damage repair and cell-cycle checkpoints (16). In contrast to enabling the assembly of DDR factors at DSBs by Ub chains, it remains to be determined whether removal of Ub chains, i.e., deubiquitination, also is critical for enabling the assembly.
BRIT1, also known as microcephalin (MCPH1), is an early DDR protein containing three BRCA1 C-terminal (BRCT) domains (27), which have conserved phosphor-peptide binding function (28). The C-terminal tandem BRCT2 and BRCT3 domains of BRIT1 mediate its recruitment to DSBs through binding to phosphorylated serine 139 (pSer139) of H2AX (29, 30). Once at a DSB, the BRIT1 N-terminal region interacts with and recruits the chromatin remodeler switch/sucrose nonfermentable (SWI–SNF), which in turn alters the nucleosome structure to relax DSB-flanking chromatin, facilitating the access of many repair factors to DSBs, including Nijmegen breakage syndrome (NBS1), mediator of DNA-damage checkpoint 1 (MDC1), BRCA1, 53BP1, and recombinase Rad51 (31). Inactivation of BRIT1 function results in a compact chromatin structure that impedes the recruitment of repair proteins to DNA lesions. As a result, DSB repair is compromised, and chromosomal aberrations accumulate (32). Studies of BRIT1-knockout mice reveal an essential role of BRIT1 in meiotic and mitotic recombination repair of DNA damage and in the preservation of genomic stability (33). Recently, it has been reported that BRIT1 also acts as a tumor suppressor (34). Depletion of BRIT1 promotes oncogenic transformation of normal mammary epithelial cells, and low levels of BRIT1 protein in multiple human cancers correlate with enhanced genomic instability and metastasis (34). Despite the critical role of BRIT1 in DDR and tumor suppression, it remains unclear how the recruitment of BRIT1 to DSBs by γ-H2AX is regulated.
Prompted by the close connection of BRUCE with cell sensitivity in the context of DNA damage, we investigated whether BRUCE is involved in DDR. Here, we report a non-IAP function of BRUCE in the regulation of DDR. We demonstrate that deubiquitination of BRIT1 is a prerequisite for BRIT1 recruitment to DSB sites by γ-H2AX. We show mechanistically that BRUCE acts as a scaffold, bridging the deubiquitinase Ub-specific peptidase 8 (USP8) and BRIT1 in a complex to coordinate USP8-catalyzed deubiquitination of BRIT1. In mice, BRUCE participates in the preservation of genomic stability by promoting DNA damage repair. These findings identify two previously uncharacterized DDR regulators, BRUCE and USP8, and uncover a deubiquitination regulation of BRIT1 assembly at damaged chromatin in the early steps of DSB response.
Results
BRUCE Depletion Impairs DNA DSB Repair.
To assess whether BRUCE is involved in DNA damage repair, we established a human osteosarcoma U2OS cell line expressing shBRUCE after doxycycline (DOX) induction (U2OS-shBRUCE) and an isogenic cell line expressing FLAG-tagged wild-type BRUCE resistant to the shBRUCE (U2OS-shBRUCE+WT) (Fig. 1B and Fig. S1A). After depletion of endogenous BRUCE by DOX treatment, a reduced incidence of IR-induced foci formed by phosphorylated ataxia telangiectasia mutated (ATM) (pATM), NBS1, and MDC1 was observed, whereas expression of exogenous BRUCE restored their foci formation (Fig. 1 C–F and Fig. S1 B and C). This phenomenon likely is independent of cell-cycle position because the reduction of IR-induced pATM foci occurred in both G2-phase (cyclin B1+) and G1/S-phase (cyclin B1−) cells (Fig. S1D). Together, these results suggest that BRUCE is required for the accumulation of critical DSB factors at DNA breaks.
Assembly of ATM, NBS1, and MDC1 at DNA breaks is essential for DSB repair. To examine directly whether BRUCE affects DSB repair, we determined if depletion of BRUCE impairs homologous recombination (HR), the major error-free DSB repair pathway in S- and G2-phase cells (35). Rad51 is a marker for HR repair and binds to ssDNA to aid the strand homology search (36–38). Foci formation by Rad51 was reduced significantly in BRUCE-depleted cells (Fig. 1G and Fig. S1E). Moreover, depletion of BRUCE by siRNA in the HR reporter cell line U2OS-DR-GFP (35, 39–41) resulted in a twofold reduction in the percentage of GFP+ cells (Fig. 1H and Fig. S1F), indicating a defect in HR repair. This degree of HR reduction is the same as that seen with the knockdown of the well-characterized DNA damage and repair proteins BRIT1 (31), CtIP (39, 42), and BACH1 (42). In contrast to HR, BRUCE depletion does not have a significant effect on nonhomologous end joining (NHEJ), an error-prone DSB repair pathway, analyzed in U2OS-EJ5-GFP reporter cells (Fig. S1 G and H). The observed defect in HR repair prompted us to assess whether BRUCE also is present in the cell nucleus, because previously it was found mainly in the cytoplasm (1). Subcellular fractionation analysis indeed identified a fraction of total BRUCE protein localized in the cell nucleus and a chromatin-enriched fraction in a variety of cell types, as exemplified in U2OS cells (Fig. S1 I and J). Together, these results identify BRUCE as a new player in DDR that acts at an early, upstream step of the repair pathway.
BRUCE, USP8, and BRIT1 Form a Protein Complex in the Cell Nucleus.
We speculated that BRUCE might interact with another protein component of DSB response. To test this notion, we used a U2OS cell line stably expressing FLAG-BRUCE to isolate BRUCE-interacting proteins by FLAG-affinity immunoprecipitation (IP). As previously reported, the DUB USP8 interacts and coimmunoprecipitates with BRUCE (8). Thus, USP8 was used as a positive control for our BRUCE IP (Fig. 2A). A selection of DDR proteins was examined by immunoblotting for their ability to coimmunoprecipitate with BRUCE in chromatin-containing whole-cell lysate. Using this approach, the early DDR protein BRIT1 was identified in BRUCE IP products (Fig. 2A). To confirm the interaction, a reciprocal IP of endogenous BRIT1 was performed, and endogenous BRUCE also was detected in the BRIT1 IP complex (Fig. 2B). Surprisingly, USP8 also was present in the BRIT1 IP complex (Fig. 2B), suggesting an association of USP8 with BRUCE-BRIT1. This suggestion was confirmed by the presence of both BRUCE and BRIT1 in the reciprocal IP products of the catalytic mutant USP8 C786A (Fig. 2C). Furthermore, this result indicates that inactivating USP8’s enzymatic function by introducing the C786A mutation did not block its interaction with BRUCE and BRIT1. Together, these coimmunoprecipitation results indicate that these three proteins interact as part of a large complex.
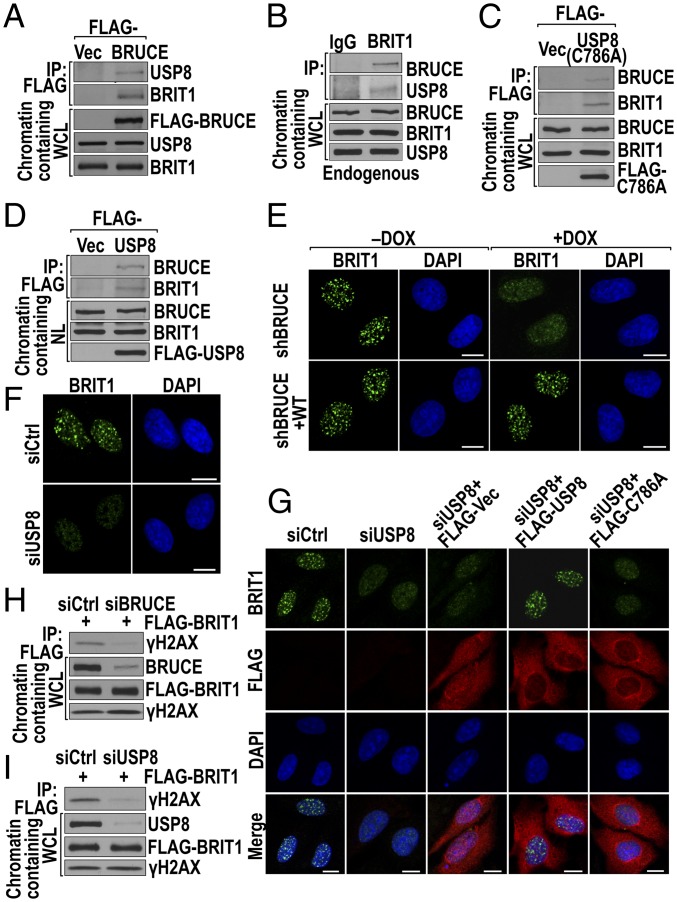
Fig. 2.
BRUCE–USP8–BRIT1 forms a protein complex, and BRUCE and USP8 are required for the formation of BRIT1 DNA damage foci. (A) BRUCE interacts with endogenous USP8 and BRIT1. BRUCE was immunoprecipitated from chromatin-containing whole-cell lysate (WCL) by FLAG M2-beads from a U2OS cell line with stable expression of pCI-Neo-FLAG (FLAG-Vec) or pCI-Neo-FLAG–tagged BRUCE. The IP products and whole-cell lysate were immunoblotted with the indicated antibodies. (B) Interaction of endogenous BRIT1, BRUCE, and USP8. BRIT1 immunoprecipitated from chromatin-containing WCL were immunoblotted for BRUCE and USP8; IgG served as a negative IP control. (C) C786A mutant USP8 interacts with endogenous BRUCE and BRIT1. USP8 was immunoprecipitated from U2OS cells expressing FLAG-USP8 C786A, and the IP products were immunoblotted for endogenous BRUCE and BRIT1. (D) The BRUCE–USP8–BRIT1 complex is present in chromatin-containing nuclear lysate (NL). USP8 was immunoprecipitated from U2OS cells expressing FLAG-USP8. The IP products were immunoblotted for endogenous BRUCE and BRIT1. (E) Depletion of BRUCE attenuates BRIT1 foci formation. U2OS cells depleted of BRUCE by DOX treatment were immunostained for BRIT1. (Scale bars, 10 µm.) (F) Depletion of USP8 attenuates BRIT1 foci formation. Immunofluorescent staining of BRIT1 in irradiated (5 Gy) U2OS cells with USP8 depleted by siRNA. (Scale bars, 10 µm.) (G) USP8 DUB activity is needed for BRIT1 foci formation. U2OS cells treated with USP8 siRNA were transfected with vectors expressing FLAG alone (FLAG-Vec), FLAG fused with USP8 (FLAG-USP8), or C786A (FLAG-C786A) (all siRNA resistant). Cells were immunostained for BRIT1 (green) and FLAG (red). (Scale bars, 10 µm.) (H and I) Depletion of BRUCE or USP8 disrupts the binding of BRIT1 to γ-H2AX. U2OS cells were depleted for BRUCE (H) or USP8 (I) followed by transfection with FLAG-BRIT1. After irradiation, BRIT1 was immunoprecipitated (anti-FLAG) in chromatin-containing WCL and immunoblotted for γ-H2AX.
Because BRIT1 is a nuclear protein, and a fraction of total BRUCE and USP8 proteins also are present in the cell nucleus (Fig. S1 I and J), we investigated specifically whether the three-protein complex was present in the cell nucleus. Analysis of immunoprecipitated FLAG-USP8 products from chromatin-containing nuclear extracts detected both BRUCE and BRIT1 (Fig. 2D). Moreover, although the cytosolic level of USP8 is higher than its level in the cell nucleus, the binding of USP8 with BRIT1 was detected only in the nuclear extracts (Fig. S2A), implying that the interaction reflects a nuclear-specific function. Together, these results indicate the presence of a preexisting BRUCE–USP8–BRIT1 complex in the cell nucleus under normal growth conditions.
BRIT1 Localization to DSBs Requires both BRUCE and USP8.
An early and critical cellular event following IR exposure is the recruitment of BRIT1 to DSB sites by binding to γ-H2AX, forming nuclear foci that colocalize with γ-H2AX foci (29, 30). Prompted by the presence of BRUCE and USP8 in a complex with BRIT1, we suspected that BRUCE and USP8 modulate BRIT1 function during DDR. We therefore examined whether BRUCE and USP8 are required for BRIT1 localization to DSBs. Depletion of BRUCE in U2OS cells significantly attenuated the number of BRIT1 foci formed by 1 h after IR exposure, and the foci were restored by ectopic expression of shRNA-resistant BRUCE (Fig. 2E and Fig. S2B). Similarly siRNA ablation of USP8 also significantly attenuated the formation of IR-induced BRIT1 foci, and the foci were restored by expressing siRNA-resistant USP8 (Fig. 2 F and G and Fig. S2C). IP studies confirmed that the impaired formation of BRIT1 foci was caused by impaired BRIT1 binding to γ-H2AX (Fig. 2 H and I). These results demonstrate that both BRUCE and USP8 are required for BRIT1 recruitment to DNA breaks. Moreover, the results of the DSB-detecting neutral-pH comet assay showed DSB repair defects in USP8-knockdown cells (Fig. S2D), supporting the idea that USP8 is involved in DNA damage repair.
The Integrity of the BRUCE–USP8–BRIT1 Complex Is Required for BRIT1 Recruitment to DSB Sites.
To understand the interaction of the three proteins, we mapped the BRUCE region that mediates the complex formation. Each of the FLAG-tagged BRUCE expression vectors (Fig. 3A), along with Myc-tagged BRIT1 or USP8, was transfected into the cells. Analysis of FLAG-BRUCE IP showed that the N-terminal region of BRUCE interacts with both BRIT1 (Fig. 3B) and USP8 (Fig. 3C). Notably, although the BIR domain is present in this region, it is dispensable for BRUCE and BRIT1 interaction, because deletion or mutation of the BIR domain did not affect the interaction of BRUCE with BRIT1 (Fig. S3). The dispensability of the BIR domain suggests that the DNA repair function of BRUCE is separate from its IAP function. We next made truncated BRIT1 constructs (Fig. 3D) and found that deletion of the BRCT3 domain (B10, Fig. 3D), which inactivates the function of the tandem BRCT2–BRCT3 domains (29, 30), abolished BRIT1 binding to BRUCE (Fig. 3E). Notably, the same deletion also abolishes BRIT1 binding to γ-H2AX (29, 30), suggesting that BRUCE binding may mask the binding of BRIT1 to γ-H2AX.
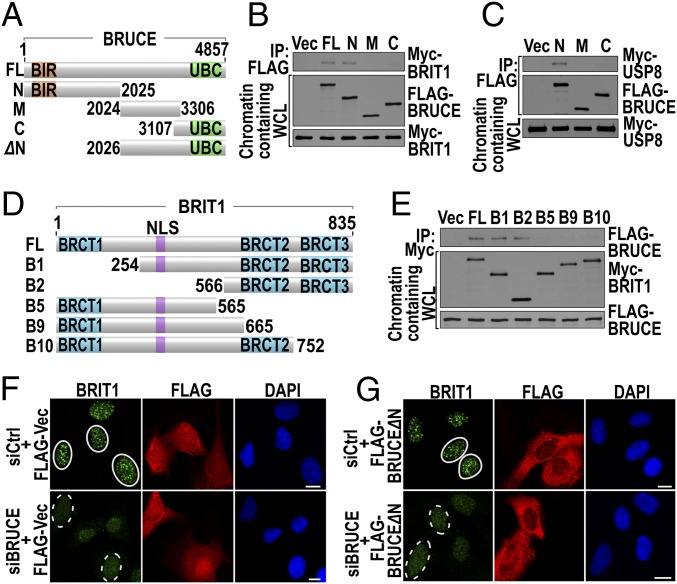
Fig. 3.
The BRUCE N-terminal region is needed for the formation of BRIT1 DNA damage foci. (A–C) The N-terminal region of BRUCE binds to BRIT1 and USP8. (A) Diagram of full-length and various truncated BRUCE constructs; N (1–2025); M (2024–3306); C (3107–4857); and ΔN (2026–4857). (B) U2OS cells were cotransfected with Myc-BRIT1 and various FLAG-BRUCE constructs as indicated and were immunoprecipitated for BRUCE and immunoblotted for BRIT1. (C) HEK293T cells were cotransfected with Myc-USP8 and various BRUCE fragments and were immunoprecipitated for BRUCE and immunoblotted for USP8. (D and E) Deletion of the BRIT1 C-terminal BRCT3 domain abolishes its binding to BRUCE. (D) Diagram of full-length and various truncated BRIT1 constructs. The BRCT domains and nuclear localization signal (NLS) are indicated. (E) U2OS cells stably expressing FLAG-BRUCE were transiently transfected with various Myc-BRIT1 constructs indicated in D. After 36 h, cell lysates were immunoprecipitated for BRIT1 and immunoblotted for BRUCE. (F and G) The BRUCE fragment BRUCE∆N depicted in A failed to support the formation of BRIT1 foci. U2OS cells depleted of BRUCE were transfected with pCI-Neo-FLAG alone (FLAG-Vec) (F) and pCI-Neo-FLAG-BRUCE∆N (FLAG-BRUCE∆N) (G) followed by immunofluorescent staining of BRTI1 (green) and FLAG (red). Foci formed (solid circles) and not formed (dashed circles) are shown. (Scale bars, 10 µm.)
To investigate the functional significance of the BRUCE–USP8–BRIT1 interaction for BRIT1 localization at DSBs, an expression construct of BRUCE∆N, deleted of the N-terminal amino acid residues 1–2025 (Fig. 3A, siBRUCE resistant), a segment that is required for the binding of BRUCE to BRIT1 and USP8, was transfected into cells in which endogenous BRUCE had been predepleted by siRNA. As in the control (Fig. 3F), BRUCE∆N failed to support the formation of BRIT1 foci (Fig. 3G), indicating that the integrity of the BRUCE–USP8–BRIT1 complex is required for BRIT1 targeting to DSBs.
BRIT1 Is Modified by K63 Polyubiquitination.
Depletion of BRUCE or USP8 did not decrease IR-induced γ-H2AX levels (Fig. 2 H and I), suggesting that the two proteins modulate cellular process other than γ-H2AX levels for BRIT1 foci formation. Because BRUCE and USP8 have ubiquitination and deubiquitination activity, respectively (4, 43, 44), we analyzed BRIT1 ubiquitination/deubiquitination. An ubiquitination assay was set up by coexpressing FLAG-BRIT1 and Myc-Ub in U2OS cells, followed by IP isolation of ubiquitinated proteins from the cell lysates with anti-Myc antibody. Immunoblotting for BRIT1 in the Ub IP products revealed multiple Ub-conjugated BRIT1 polypeptides with the typical ubiquitinated ladder pattern (Fig. 4A, bracket), indicating that BRIT1 is polyubiquitinated. Ub conjugation via K48 linkage generally targets the modified proteins for proteasomal degradation, whereas K63-linked conjugation often generates a role in signaling transduction (45). To ascertain the type of Ub linkage on BRIT1, the ubiquitination assay was repeated in cells expressing wild-type, K48R, or K63R Ub. Compared with wild-type Ub, BRIT1 ubiquitination was slightly reduced in cells expressing K48R Ub (Fig. 4B), possibly reflecting the basal turnover of BRIT1 by the proteasomal degradation pathway. In contrast to K48R, BRIT1 ubiquitinated products were nearly abolished by the expression of K63R Ub (Fig. 4B), demonstrating that a significant amount of conjugated BRIT1 is by K63 Ub. Subcellular fractionation further revealed that more of the K63 ubiquitinated BRIT1 was in the chromatin-enriched fraction than in the cytosol and nucleoplasm (Fig. 4C). Moreover, the endogenous BRIT1 also was polyubiquitinated in the chromatin-enriched fraction (Fig. 4D, bracket), eliminating the possibility that ubiquitination of ectopic BRIT1 was an overexpression artifact. Together, these results indicate that under normal growth conditions the K63 polyubiquitinated BRIT1 is present and is associated with chromatin. To eliminate the interference of K48-conjugated BRIT1, we used K48R Ub for most of our following studies, unless otherwise specified.
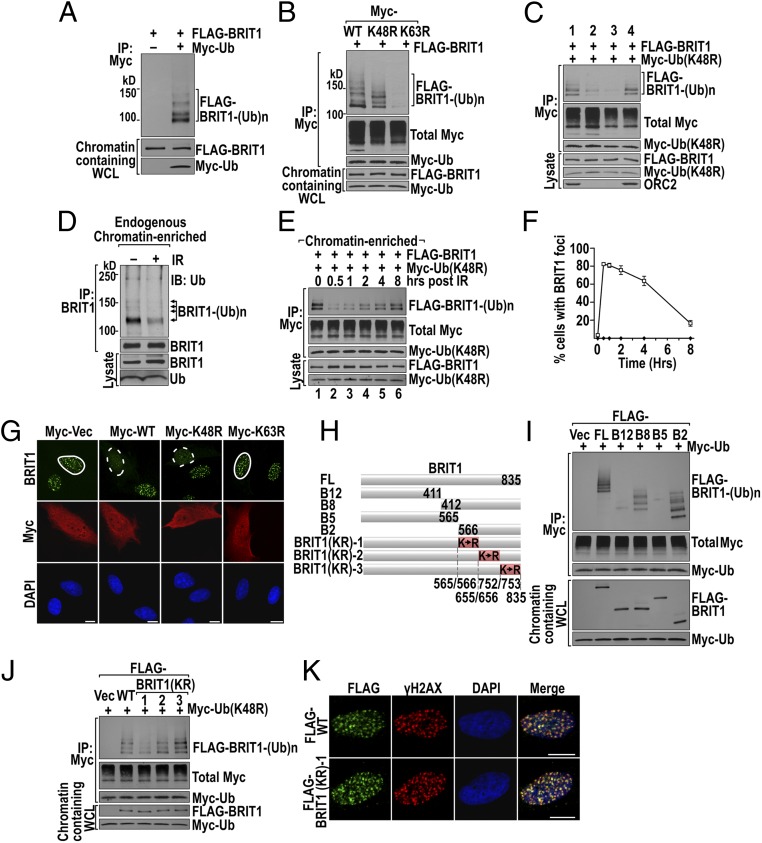
Fig. 4.
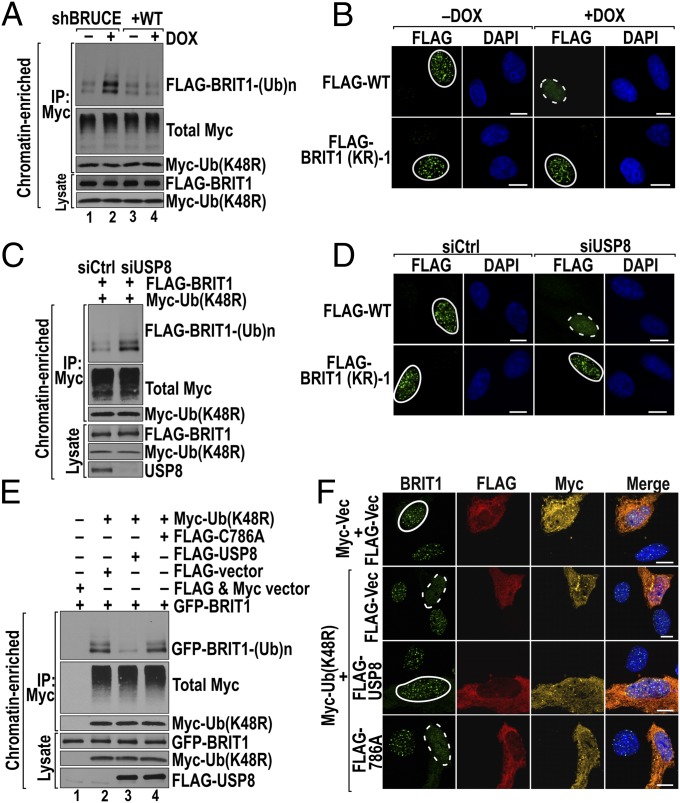
DSB-activated BRIT1 deubiquitination correlates with formation of BRIT1 foci induced by DNA damage. (A) BRIT1 is ubiquitinated in unstimulated U2OS cells. Cells cotransfected with FLAG-BRIT1 and Myc-Ub were subject to BRIT1 Ub assay (see SI Materials and Methods for details). IP of Ub was done in chromatin-containing whole-cell lysate (WCL) followed by immunoblotting for BRIT1. (B) BRIT1 ubiquitination is mainly K63-linked. The BRIT1 Ub assay was conducted as in A except that cells were transfected with K48R- and K63R-mutant Ub constructs. (C) BRIT1-ubiquitinated products are predominantly associated with the chromatin-enriched fraction. BRIT1 Ub assays were conducted in four subcellular fractions of chromatin containing WCL, cytosol, nucleoplasm, and chromatin (lanes 1–4, respectively). (D) Endogenous ubiquitinated BRIT1 in the chromatin fraction was reduced 1 h after IR (10 Gy) exposure. Immunoprecipitated endogenous BRIT1 products from U2OS cells were immunoblotted for Ub; ubiquitinated BRIT1 products are indicated by arrows. (E and F) BRIT1 deubiquitination correlates with its foci formation 8 h after IR (5 Gy) exposure. (E) Changes in the level of BRIT1 Ub products on chromatin. (F) Quantification of the time-course study of BRIT1 foci formation. (G) K63-Ub inhibits the formation of BRIT1 foci. U2OS cells transfected with constructs of Myc alone (Myc-Vec) and Myc-Ub (wild type, K48R, or K63R) were immunostained for BRIT1 (green) and Myc-Ub (red). Foci formed (solid circles) and inhibited (dashed circles) are shown. (Scale bars, 10 µm.) The quantified foci results are shown in Fig. S4A. (H) Diagram of truncated BRIT1 fragments and BRIT1 KR mutants with all lysine residues within each region (shaded in red) mutated to arginine. (I and J) Mapping of the ubiquitination region to the B2 fragment of BRIT1 (I) and further mapped to amino acid segment 566–655 (J) by BRIT1 Ub assay. (K) BRIT1 (KR)-1 forms IR-induced foci colocalizing with γ-H2AX foci. U2OS cells were irradiated and immunofluorescent stained for FLAG (green) and γ-H2AX (red). (Scale bars, 10 µm.)
DSB-Induced Deubiquitination of BRIT1 Enables Its Accumulation at DSBs.
The ubiquitination modification of DDR proteins plays essential roles in promoting their accumulation at damaged chromatin (17). To understand the role of BRIT1 ubiquitination, we first examined whether exposure to IR up-regulates the levels of ubiquitinated endogenous BRIT1. Contrary to our expectation, IR resulted in a reduction of BRIT1ubiquitination (Fig. 4D, right lane). We next examined how ubiquitination/deubiquitination affects BRIT1 localization to DSBs. As with endogenous BRIT1, IR also triggered a significantly reduced ubiquitination of exogenously expressed FLAG-BRIT1 at the 0.5-h time point (Fig. 4E, Upper, lane 2). This reduction correlated with a concomitant increase in the formation of BRIT1 foci at the same time point (Fig. 4F). Over the next 8 h, the levels of ubiquitinated BRIT1 increased gradually (Fig. 4E, Upper, lanes 3–6), whereas BRIT1 foci underwent a corresponding decrease (Fig. 4F). These results suggest that the removal of K63 Ub conjugates is required for BRIT1 recruitment to DSBs. This suggestion is confirmed by the results demonstrating that enhancing BRIT1 ubiquitination by ectopic expression of wild-type or K48R Ub suppresses BRIT1 foci formation, and, conversely, that preventing K63 Ub modification of BRIT1 by the expression of K63R permits the formation of BRIT1 foci (Fig. 4G and Fig. S4A).
BRIT1 Is K63-Ubiquitinated Primarily in the Region of Amino Acids 566–655.
To identify the ubiquitinated region, constructs encoding the N-terminal (B5, B12) and C-terminal (B2, B8) fragments of BRIT1 (Fig. 4H) were individually cotransfected with an Ub expression vector into U2OS cells, and their ubiquitination status was determined. Ubiquitination occurred primarily on the overlapping B2 and B8 fragments (Fig. 4I). Therefore the B2 region was divided further into three contiguous subsegments (Fig. 4H, shaded in red), and all lysine (K) residues in each subsegment were mutated to arginine (R) to block ubiquitination, generating the constructs BRIT1 (KR)-1, -2, and -3 (Fig. 4H and Fig. S4B). Ubiquitination analysis indicated that only BRIT1 (KR)-1 (amino acids 566–655) exhibited decreased Ub conjugation (Fig. 4J), and therefore the segment consisting of amino acids 566–655 is the primary region for BRIT1 ubiquitination and deubiquitination. Importantly, IR-induced BRIT1 (KR)-1 foci were colocalized with γ-H2AX foci (Fig. 4K and Fig. S4C), demonstrating the correct localization of BRIT1 (KR)-1 to the sites of DNA damage.
BRUCE and USP8 Are Required for BRIT1 Deubiquitination and Targeting to DSBs.
Our results showed that BRUCE, USP8, and deubiquitination are essential for the formation of BRIT1 foci. We next investigated whether BRUCE and USP8 are required for IR-induced BRIT1 deubiquitination. Depletion of BRUCE or USP8 resulted in elevated steady-state levels of K63-ubiquitinated BRIT1 in the chromatin-enriched fraction (Fig. 5 A and C, respectively). If BRUCE- and USP8-mediated BRIT1 deubiquitination is the mechanism allowing BRIT1 recruitment to DSBs, then recruitment to DSBs of the BRIT1 (KR)-1 variant, which mimics deubiquitinated BRIT1, should not require BRUCE or USP8. Indeed, BRIT1 (KR)-1 readily formed nuclear foci in irradiated U2OS cells depleted of BRUCE or USP8 (Fig. 5 B and D and Fig. S4C). These results demonstrate that BRUCE and USP8 do not directly recruit BRIT1 to DSBs but rather promote BRIT1 deubiquitination for its subsequent recruitment to DSBs.
Fig. 5.
BRUCE and USP8 both promote deubiquitination and formation of BRIT1 foci. (A) Depletion of BRUCE increases BRIT1 ubiquitination. U2OS cells depleted of BRUCE were transfected with FLAG-BRIT1 and Myc-Ub (K48R). BRIT1 ubiquitination in the chromatin-enriched fraction was assayed. (B) BRIT1 (KR)-1 forms DNA damage foci in BRUCE-depleted cells. U2OS cells depleted of BRUCE were transfected with wild-type BRIT1 or BRIT1 (KR)-1 and were immunostained for BRIT1 foci (anti-FLAG) 1 h after IR (5 Gy) exposure. Foci formed (solid circles) and abolished (dashed circle) are shown. (Scale bars, 10 µm.) (C) Depletion of USP8 increases BRIT1 ubiquitination. U2OS cells depleted of USP8 were assayed for BRIT1 ubiquitination following the method in A. (D) BRIT1 (KR)-1 formed DNA damage foci in USP8-depleted U2OS cells following the method in B. (E) The DUB activity of USP8 mediates BRIT1 deubiquitination. Ub conjugates were isolated from the chromatin fraction of U2OS cells transfected with various constructs as indicated above the blots and were immunoblotted for the proteins indicated at the right. (F) USP8 DUB activity is required for the recovery of BRIT1 foci preinhibited by Ub overexpression. U2OS cells cotransfected with the constructs indicated at the left were irradiated and stained for BRTI1 (green), FLAG (red), and Myc (yellow). Foci formed (solid circles) and abolished (dashed circles) are shown (Scale bars, 10 µm.)
The USP8 catalytic mutant C786A is dominant negative and inhibits endogenous USP8 activity (46). The C786A mutant still binds to BRIT1, and the binding is slightly stronger than that of wild-type USP8 (Fig. S5A). Next, we assessed whether USP8 DUB activity is required. K63-ubiquitinated BRIT1 products were generated by cotransfection of U2OS cells with BRIT1 and Ub constructs (Fig. 5E, lane 2). As expected, the ubiquitination of BRIT1 was inhibited (Fig. 5E, lane 3), and the suppressed formation of BRIT1 foci by Ub overexpression was desuppressed by overexpression of wild-type USP8 (Fig. 5F and Fig. S5B). However, overexpression of the catalytic mutant C786A USP8 failed to promote BRIT1 deubiquitination (Fig. 5E, lane 4) and foci formation (Fig. 5F and Fig. S5B). Collectively, these data indicate that USP8 DUB activity is required for BRIT1 deubiquitination and recruitment to DSBs by γ-H2AX.
BRUCE Acts as a Scaffold Platform for DSB-Induced USP8 Deubiquitination of BRIT1.
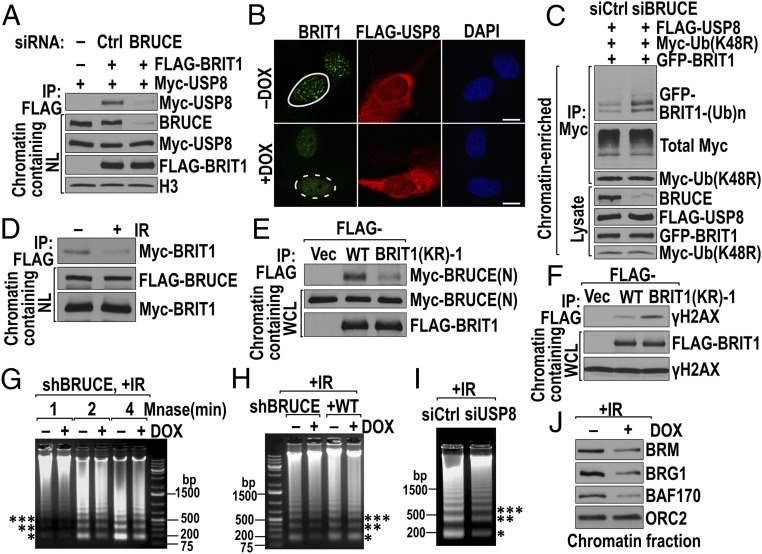
Large proteins can act as scaffolds, bringing together multiple members of a signaling pathway to coordinate their functions (47). We next investigated whether BRUCE is required for USP8-mediated BRIT1 deubiquitination and localization to DSBs. Because the integrity of the three-protein complex is required for targeting BRIT1 to DSBs (Fig. 3G), we investigated whether BRUCE acts as a scaffold. Depletion of BRUCE by siRNA disrupted the interaction of USP8 and BRIT1 in the cell nucleus (Fig. 6A), indicating that BRUCE is critical to retain USP8 and BRIT1 in the complex. In support of this observation, overexpression of USP8 in BRUCE-depleted cells could not enable BRIT1 foci formation (Fig. 6B) because of failed BRIT1 deubiquitination (Fig. 6C). These data establish that BRUCE acts as a scaffold tethering USP8 and BRIT1 into the complex, and this tethering is essential for DSB-induced BRIT1 deubiquitination by USP8.
Fig. 6.
The integrity of the BRUCE–USP8–BRIT1 complex is essential for BRIT1 function. (A) Depletion of BRUCE abolishes interaction between USP8 and BRIT1. HEK293T cells depleted of BRUCE by siRNA were cotransfected with USP8 and BRIT1 constructs. BRIT1 IP (anti-FLAG) products from chromatin-containing nuclear lysate (NL) were immunoblotted for USP8 (anti-Myc). (B) Depletion of BRUCE abolishes the formation of BRIT1 foci in cells overexpressing USP8. U2OS cells treated with DOX or left untreated were transfected with FLAG-USP8 and were immunostained for endogenous BRIT1 (green) and USP8 (anti-FLAG, red) 1 h after IR (5 Gy) exposure. Foci formed (solid circle) or abolished (dashed circle) are shown. (Scale bars, 10 µm.) (C) Depletion of BRUCE abolishes USP8-catalyzed BRIT1 deubiquitination. U2OS cells treated with siRNA were transfected with the constructs indicated above the blot and were analyzed for BRIT1 ubiquitination in the chromatin-enriched fraction. (D) IR reduces the interaction of BRIT1 and BRUCE. The U2OS FLAG-BRUCE stable cell line was transfected with Myc-BRIT1 and then was irradiated (10 Gy). The amount of BRIT1 bound to BRUCE was examined in the BRUCE IP products. (E) BRIT1 (KR)-1 binds more weakly than wild-type BRIT1 to BRUCE. U2OS cells were cotransfected with the indicated BRIT1 and BRUCE-N (Fig. 3A) constructs. The amount of BRUCE that bound to wild-type or (KR)-1 mutant BRIT1 was analyzed in BRIT1 IP products. (F) BRIT1 (KR)-1 binds more strongly than wild-type BRIT1 to γ-H2AX. U2OS cells transfected with BRIT1 constructs were irradiated (10 Gy). The amount of γ-H2AX that bound to wild-type and (KR)-1 mutant BRIT1 was analyzed in BRIT1 IP products. (G–I) Depletion of BRUCE or USP8 impairs chromatin relaxation. Irradiated U2OS cells were digested with MNase for 1–4 min after BRUCE knockdown (G), for 2 min with cells reconstituted of BRUCE (H), and for 3 min after USP8 knockdown (I). Chromatin relaxation was monitored by the release of nucleosomes with mono(*), di-(**), and trinucleosomes (***) indicated. (J) Depletion of BRUCE reduces chromatin association of SWI–SNF subunits BRM, BRG1, and BAF170. ORC2 served as control.
Deubiquitinated BRIT1 Is Released from the BRUCE Complex and Subsequently Is Recruited to DSBs by γ-H2AX.
Ubiquitination/deubiquitination could alter protein interaction and subcellular localization. Because BRIT1 is deubiquitinated by IR exposure, we examined whether deubiquitination alters BRIT1’s interaction with its binding partners by comparing the amount of BRIT1 associated with BRUCE before and after IR. As shown in Fig. 6D, the amount of BRIT1 that interacted with BRUCE was reduced significantly after IR exposure, suggesting that BRIT1 is released from BRUCE. Because the level of deubiquitinated BRTI1 correlates with the amount of BRIT1 foci formation (Fig. 4 E and F), we postulated that the released, deubiquitinated BRIT1 subsequently will translocate to the damaged chromatin, forming nuclear foci. In support of this notion, the deubiquitinated BRIT1 (KR)-1 exhibited weaker binding with BRUCE than wild-type BRIT1 (Fig. 6E). Meanwhile, larger amounts of deubiquitinated BRIT1 (KR)-1 than wild-type BRIT1 bound to γ-H2AX in irradiated cells (Fig. 6F). Together, these data suggest that deubiquitination of BRIT1 occurs on the BRUCE–USP8 complex before BRIT1 is recruited to damaged chromatin by γ-H2AX.
The BRUCE–USP8–BRIT1 Complex Facilitates Chromatin Relaxation in the DSB Response.
One major function of BRIT1 is to recruit SWI–SNF to DSB-flanking chromatin, relaxing the chromatin structure to allow access by downstream DNA damage signaling and repair proteins (31). Depletion of BRUCE and USP8, which prevents BRIT1 localization to DSBs, is anticipated to have an effect similar to BRIT1 depletion by rendering chromatin to a more compact configuration in response to IR exposure. Indeed, depletion of BRUCE resulted in decreased nucleosome release from irradiated cell chromatin after micrococcal nuclease (MNase) digestion (Fig. 6G and Fig. S6A). This effect is specific to BRUCE, because it could be reversed by BRUCE restoration (Fig. 6H). Similarly, USP8 depletion also resulted in chromatin compaction (Fig. 6I and Fig. S6B). Supporting the results of MNase digestion, the amount of chromatin-associated SWI–SNF was reduced in BRUCE-depleted cells as exemplified by its essential ATPase subunits Brahma (BRM) and Brahma-related gene 1 (BRG1) and one key catalytic core subunit, BRG1-associated factor 170 (BAF170) (Fig. 6J). Furthermore, BRUCE depletion impaired the formation of pATM, NBS1, MDC1, and Rad51 foci (Fig. 1), all of which require the decondensation of chromatin structure mediated by the BRIT1–SWI–SNF pathway (31). Moreover, the chromatin-relaxing agents sodium butyrate (NaBu) and trichostatin A (TSA) restored pATM, NBS1, MDC1, and Rad51 foci (Fig. S6 C–F), but not BRIT1 foci (Fig. S6G), placing the chromatin-relaxation step downstream of BRIT1 localization to DSBs.
BRUCE-Mutant Mouse Embryonic Fibroblasts Exhibit Defective DSB Repair and Genomic Instability.
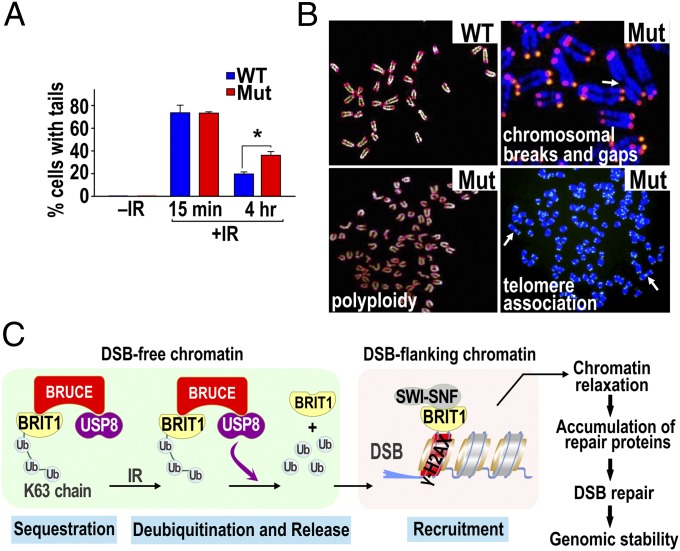
To elucidate further the significance of BRUCE function in DDR, we examined DNA repair activity in mouse embryonic fibroblasts (MEFs) isolated from BRUCE-mutant mice (9) by neutral-pH comet assay. As shown in Fig. 7A and Fig. S7A, BRUCE-mutant MEFs had significantly impaired DSB repair by 4 h after IR exposure compared with wild-type cells. DNA DSBs are highly toxic and, if unrepaired, can introduce genomic instability, a hallmark of cancer. We therefore performed telomere-FISH to assess chromosomal integrity and found a significant increase in various manifestations of genomic instability, including chromosomal breaks and gaps, telomere associations, and aneupolyploidy, in BRUCE-mutant MEFs (Fig. 7B and Fig. S7B), again demonstrating that BRUCE regulates DSB repair and preserves genomic stability.
Fig. 7.
BRUCE is required for DNA repair and chromosomal integrity in mouse cells. (A) Comet assay results showing sustained amount of DNA DSBs in BRUCE mutant MEFs compared with wild type 4 h after exposure to IR (5 Gy); *P < 0.05; two-way ANOVA, posthoc test. (B) Telomere-FISH of MEFs showing chromosomal gaps and breaks (arrow), telomere association (arrows), and aneupolyploidy in mutant MEFs. Quantification results showed statistical significance (P < 0.05; χ2 analysis; Fig. S7B). (C) A sequestration and release model showing how the BRUCE–USP8–BRIT1 complex regulates DSB response. See text for details.
A Working Model.
Based on the data presented above, we propose a “sequestration and release” model (Fig. 7C): Ubiquitinated BRIT1, as part of the BRUCE–USP8–BRIT1 complex, is sequestered in a DSB-free chromatin region in unstimulated cells. In response to DSB induction, the scaffold BRUCE promotes USP8 deubiquitination of BRIT1. Once deubiquitinated, BRIT1 is released from the complex and subsequently is recruited to DSB-flanking chromatin by binding to γ-H2AX. Consequently, BRUCE and USP8 link to the BRIT1–SWI–SNF pathway to open chromatin structures for timely DNA repair and preservation of genomic stability. Thus, BRUCE and USP8 represent novel players in safeguard of genomic stability.
Discussion
An increasing number of reports have demonstrated important non-antiapoptotic functions for IAP proteins. However, to our knowledge, none has yet identified any IAP protein that functions in the regulation of early signaling events of DSB response. Moreover, in contrast to ubiquitination modification, deubiquitination in enabling assembly of DDR factors at damaged chromatin is largely unknown. In this study, we identify a new non-IAP function for BRUCE in DSB response and a new deubiquitination regulation of BRIT1 by USP8 for enabling BRIT1 foci formation. Our results indicate the presence of a nuclear BRUCE–USP8–BRIT1 DDR complex under normal growth conditions. BRUCE acts as a scaffold, localizes USP8 and BRIT1 onto the complex, and coordinates DSB-activated USP8 deubiquitination of BRIT1 for subsequent recruitment of BRIT1 to DSBs by γ-H2AX. Our data support a sequestration and release model illustrated in Fig. 7C. The preexisting BRUCE–USP8–BRIT1 complex localized to the DSB-free chromatin region sequesters BRIT1 on the BRUCE platform. Such localization prevents inadvertent DSB response and brings the deubiquitinase USP8 and the substrate BRIT1 in close proximity to ensure that rapid and efficient BRIT1 deubiquitination occurs in the face of DNA damage. Once deubiquitinated, BRIT1 is released from the complex and is recruited by γ-H2AX to the subcellular compartment of DSB-flanking chromatin, resulting in the previously known recruitment of the SWI–SNF complex for chromatin relaxation. Thus, BRUCE and USP8 are new regulators of DSB responses to facilitate BRIT1 deubiquitination and recruitment to DSB-flanking chromatin for subsequent timely DNA repair and preservation of chromatin stability.
Deubiquitination Regulates BRIT1 Function in DSB Response.
Modification by Ub is a well-established mechanism by which DDR factors accumulate at DSBs. One classic example is the monoubiquitination of the Fanconi anemia factor FANCD2 for its localization to sites of DNA damage during the repair of DNA interstrand cross-links (18, 19). Another well-established ubiquitination cascade is the MDC1-dependent K63 polyubiquitination of histones H2A and H2AX at damaged chromatin, creating Ub-binding sites for RAP80 and BRAC1 accumulation at DNA breaks (20–26, 36). Another is the monoubiquitination of histone H2B by the heterodimeric E3 ligases RNF20 and RNF40 and their functional partner WAC; together they facilitate chromatin relaxation for DNA repair, linking gene expression with DDR (48–51).
Deubiquitination is equally important for DDR (16) but is better known for its role in inducing dissociation of factors from DNA breaks and for allowing the next round of ubiquitination to occur, as in the deubiquitination of FANCD2 by USP1 (52). In this study we investigated whether deubiquitination enables the accumulation of DSB-response factors at damaged chromatin. We show evidence that the fate of BRIT1—being sequestered on the BRUCE complex or unleashed to anchor at DSBs—is determined by Ub modification and deubiquitination, respectively. One question that remains is why deubiquitination is needed, because deubiquitinated BRIT1 already is present in unstimulated cells. Because not all BRIT1 is associated with BRUCE and USP8 in our IP analysis, we propose that not all deubiquitinated BRIT1 is functionally the same, because of posttranslational modification, subnuclear compartment localization, and interacting partners. Indeed, BRIT1 has many functions. Mutation of BRIT1 results in defective G2/M checkpoint arrest and is implicated in primary autosomal recessive microcephaly and in the premature chromosome condensation syndrome (53). Thus, deubiquitinated BRIT1 could function in these different cellular processes.
BRUCE and USP8 Promote BRIT1 Deubiquitination at DSB-Free Subnuclear Compartments.
One hallmark of most DDR proteins is the formation of foci of DNA damage. However, not all DNA damage and repair factors form foci. Chk1 and Chk2, two effector protein kinases in DDR, are activated at DNA breaks but do not form IR-induced nuclear foci. They dissociate promptly and distribute to the entire cell nucleus to reach their targets in undamaged, DSB-free subnuclear compartments (54, 55). Similarly, not all DDR regulators accumulate at DSBs and therefore are not expected to form foci of DNA damage. For instance, the regulators Polo-like kinase 1 (Plk1) and casein kinase 2 (CK2) facilitate Rad51 recruitment to DNA breaks by phosphorylation of Rad51. However, Plk1 and CK2 do not localize to the DSB site (56). Our data indicate that BRUCE and USP8 work in a similar manner, not forming discernible IRIF, as examined by multiple antibody staining of endogenous BRUCE and USP8 and by anti-FLAG antibody staining of ectopically expressed FLAG-tagged BRUCE or USP8. These observations suggest that upon IR exposure, BRIT1 deubiquitination by the BRUCE–USP8–BRIT1 complex occurs in DSB-free subnuclear compartments. Considering the complexity of chromatin structures and the large number of DDR regulators with distinct functions, it is conceivable that the regulatory events occurring at DSB-free subnuclear compartments are as important as those occurring at DSB-flanking chromatin. Elucidation of the mechanism by which regulators distal to the DSB work will provide novel insight into the complicated DSB response. As for USP8, how it is activated by IR remains an open question. Here we propose several mechanisms. IR could change the posttranslational modification of USP8, for instance, its ubiquitination and phosphorylation, because these modifications play critical roles in the activation of many DDR proteins. In addition, IR could trigger the removal of inhibitory protein(s) from the complex to allow USP8 activation. Finally, IR could activate USP8 by changing its conformation. Certainly, future work will be directed toward identifying the mechanism.
How Deubiquitination May Regulate BRIT1 Anchorage at Sites of DNA Damage.
The C-terminal BRCT2–BRCT3 domains of BRIT1 interact with γ-H2AX. Our data suggest that the ubiquitination status of BRIT1 regulates the binding selectivity of its C-terminal BRCT domains. The identified ubiquitination and deubiquitination within the region of amino acids 566–655, localized upstream and adjacent to BRCT2, could serve as a switch to regulate the conformation of BRIT1 to favor the interaction of its BRCT domains with one protein partner over the other. Specifically, in the absence of DSBs, the conformation of BRIT1 resulting from modification by K63 polyubiquitination favors the interaction of its BRCT domains with BRUCE. In the presence of DSB, removal of the Ub chain switches the conformation toward one optimal for interaction with pS139 of H2AX. Another, and not mutually exclusive, possibility is that, although optimal for binding to BRUCE, the Ub chains conjugated to BRIT1 may be a structural hindrance for BRIT1 interaction with pS139 of H2AX; deubiquitination would enable the interaction. Future structural studies comparing the differences between ubiquitinated and deubiquitinated BRIT1 will provide the molecular basis. Nonetheless, our data demonstrate that the assembly of DSB factors at DSB-flanking chromatin can be regulated by deubiquitination in addition to ubiquitination.
BRUCE Is a Nuclear Scaffold and Acts as an Assembly Platform for BRIT1 Deubiquitination.
Large proteins can act as scaffolds to tether other proteins to coordinate their functions. The large mass of the BRUCE protein (528 kDa, 4,857 aa), which is even larger after self-dimerization (4), makes it eligible to be a scaffold in DDR. Consistent with its scaffolding role in cytokinesis, our data suggest that BRUCE also acts as a scaffold in DDR by tethering USP8 and BRIT1 on the complex. However, the BIR domain does not participate in the scaffolding function despite its presence within the N-terminal–interacting region. In addition to the BIR, we identified putative WD40/YVTN repeats and coiled-coil domains in the N-terminal–binding region by bioinformatic analyses using InterPro at The European Bioinformatics Institute. Although experimental validation is needed, these repeats and domains could be involved in the interaction of BRUCE with USP8 and BRIT1. Our study extends the scaffold feature of BRUCE from cytokinesis to a distinctly different cellular process of DNA repair. Therefore, scaffolding may be a common feature of BRUCE that is responsible for other, yet to be characterized cellular functions indicated by the diverse phenotypes of BRUCE-mutant mice (3, 9, 10).
The DDR Function of BRUCE Is Separate from Its Roles in Antiapoptosis and Procytokinesis.
Two of the hallmarks of apoptotic cells are DNA condensation and caspase activation. In our experimental conditions IR exposure (5–10 Gy) was followed by a short period of continued culture up to 1 h. Such conditions are insufficient to activate apoptosis, as shown by the normal nuclear morphology, homogenous nuclear staining by DAPI, and the absence of active caspases including caspases-7. Moreover, the BIR domain of BRUCE, although essential for suppression of apoptosis, is not required for the formation of the BRUCE–USP8–BRIT1 complex or for USP8 deubiquitination of BRIT1 (Fig. S3). The function of BRUCE in DDR also appears to be separated from its role in cytokinesis because BRUCE- or USP8-depleted cells with impaired DDR are still single nucleated rather than di- or multinucleated, a hallmark of aborted cytokinesis.
Concluding Remarks.
For the first time, to our knowledge, this study identifies a critical role for BRUCE and USP8 in DDR and a novel regulation of BRIT1 by deubiquitination. This work lays the foundation for further study of BRUCE, USP8, and BRIT1 in DDR. Defective HR is tightly associated with genomic instability that accelerates cancer by promoting mutations. The importance of BRUCE and USP8 and of BRIT1 deubiquitination in DDR and genomic stability provides new insight into the pathogenesis of diseases resulting from DNA repair defects and genomic instability.
Materials and Methods
Human U2OS and HEK 293T cell lines purchased from ATCC were transfected with plasmid and siRNA by Lipofectamine 2000 and Lipofectamine RNAiMAX (Invitrogen), respectively. For immunofluorescent staining of foci of DNA damage, cells were irradiated with a Faxitron X-ray system (RX-650). After fixation and permeabilization, cells were incubated with primary antibodies followed with fluor-conjugated secondary antibodies and were examined under a Zeiss LSM 710 confocal microscope. HR and NHEJ were assayed in U2OS-DR-GFP and U2OS-EJ5-GFP cell lines, respectively. Detailed methods and statistical data analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Jeremy Stark at City of Hope for DR-GFP and EJ5-GFP U2OS cells and James (Zhijian) Chen at the University of Texas Southwestern Medical Center for ubiquitin expression constructs. We also thank our colleagues Drs. Sohaib Khan and David Plas for critical comments on the manuscript, Rachid Drissi for the micrococcal nuclease digestion protocol, Glenn Doerman for making the figures, Belinda Peace for English editing, G. Sharma in the T.K.P. laboratory (Washington University) for technical help, members of the C.D. laboratory for discussion, and Xingyuan Yang for assistance with BRUCE and USP8 expression analysis (not included in this article). This work was supported by the National Institutes of Health Grants CA158323 (to C.D.), CA129537, CA154320, and GM109768 (to T.R.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418335112/-/DCSupplemental.
References
- 1.Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S. A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol. 1998;141(6):1415–1422. doi: 10.1083/jcb.141.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z, et al. A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun. 1999;264(3):847–854. doi: 10.1006/bbrc.1999.1585. [DOI] [PubMed] [Google Scholar]
- 3.Hao Y, et al. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6(9):849–860. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- 4.Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14(6):801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Qiu XB, Markant SL, Yuan J, Goldberg AL. Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J. 2004;23(4):800–810. doi: 10.1038/sj.emboj.7600075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubrez-Daloz L, Dupoux A, Cartier J. IAPs: More than just inhibitors of apoptosis proteins. Cell Cycle. 2008;7(8):1036–1046. doi: 10.4161/cc.7.8.5783. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasula SM, Ashwell JD. IAPs: What’s in a name? Mol Cell. 2008;30(2):123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132(5):832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Ren J, et al. The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc Natl Acad Sci USA. 2005;102(3):565–570. doi: 10.1073/pnas.0408744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lotz K, Pyrowolakis G, Jentsch S. BRUCE, a giant E2/E3 ubiquitin ligase and inhibitor of apoptosis protein of the trans-Golgi network, is required for normal placenta development and mouse survival. Mol Cell Biol. 2004;24(21):9339–9350. doi: 10.1128/MCB.24.21.9339-9350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosoya N, Miyagawa K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014;105(4):370–388. doi: 10.1111/cas.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powell SN, Bindra RS. Targeting the DNA damage response for cancer therapy. DNA Repair (Amst) 2009;8(9):1153–1165. doi: 10.1016/j.dnarep.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011;25(5):409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152(6):1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacq X, Kemp M, Martin NM, Jackson SP. Deubiquitylating enzymes and DNA damage response pathways. Cell Biochem Biophys. 2013;67(1):25–43. doi: 10.1007/s12013-013-9635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49(5):795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Andreassen PR, D’Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24(13):5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7(2):249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 20.Bekker-Jensen S, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol. 12(1):80–86; sup pp 81-12. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 21.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 22.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131(5):901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318(5856):1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131(5):887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 25.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136(3):420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, et al. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol Cell Biol. 2009;29(3):849–860. doi: 10.1128/MCB.01302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin SY, Rai R, Li K, Xu ZX, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci USA. 2005;102(42):15105–15109. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung CC, Glover JN. BRCT domains: Easy as one, two, three. Cell Cycle. 2011;10(15):2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood JL, Singh N, Mer G, Chen J. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem. 2007;282(48):35416–35423. doi: 10.1074/jbc.M705245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffers LJ, Coull BJ, Stack SJ, Morrison CG. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene. 2008;27(1):139–144. doi: 10.1038/sj.onc.1210595. [DOI] [PubMed] [Google Scholar]
- 31.Peng G, et al. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol. 2009;11(7):865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rai R, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10(2):145–157. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Y, et al. BRIT1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice. PLoS Genet. 2010;6(1):e1000826. doi: 10.1371/journal.pgen.1000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, et al. BRIT1 regulates p53 stability and functions as a tumor suppressor in breast cancer. Carcinogenesis. 2013;34(10):2271–2280. doi: 10.1093/carcin/bgt190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5(11):a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467(7316):678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17(10):1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorslund T, et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17(10):1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4(6):e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12(6):224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 41.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, et al. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011;25(7):685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niendorf S, et al. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27(13):5029–5039. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Row PE, Prior IA, McCullough J, Clague MJ, Urbé S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281(18):12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- 45.Pickart CM, Fushman D. Polyubiquitin chains: Polymeric protein signals. Curr Opin Chem Biol. 2004;8(6):610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Naviglio S, et al. UBPY: A growth-regulated human ubiquitin isopeptidase. EMBO J. 1998;17(12):3241–3250. doi: 10.1093/emboj/17.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Good MC, Zalatan JG, Lim WA. Scaffold proteins: Hubs for controlling the flow of cellular information. Science. 2011;332(6030):680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiloh Y, Shema E, Moyal L, Oren M. RNF20-RNF40: A ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 2011;585(18):2795–2802. doi: 10.1016/j.febslet.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura K, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41(5):515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 50.Moyal L, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41(5):529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell. 2011;41(4):384–397. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JM, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16(2):314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venkatesh T, Suresh PS. Emerging roles of MCPH1: Expedition from primary microcephaly to cancer. Eur J Cell Biol. 2014;93(3):98–105. doi: 10.1016/j.ejcb.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5(3):255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 55.Smits VA, Reaper PM, Jackson SP. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol. 2006;16(2):150–159. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 56.Yata K, et al. Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol Cell. 2012;45(3):371–383. doi: 10.1016/j.molcel.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.